Abstract
HIV-1 particle assembly is driven by the structural protein Gag. Gag binds to and multimerizes on the inner leaflet of the plasma membrane, eventually resulting in formation of spherical particles. During virus spread among T cells, Gag accumulates to the plasma membrane domain that, together with target cell membrane, forms a cell junction known as the virological synapse. While Gag association with plasma membrane microdomains has been implicated in virus assembly and cell-to-cell transmission, recent studies suggest that, rather than merely accumulating to pre-existing microdomains, Gag plays an active role in reorganizing the microdomains via its multimerization activity. In this paper, we will discuss this emerging view of Gag microdomain interactions. Relationships between Gag multimerization and microdomain association will be further discussed in the context of Gag localization to T-cell uropods and virological synapses.
1. Introduction
Microdomain-based compartmentalization of the plasma membrane is implicated in many aspects of the HIV-1 life cycle. In particular, during events in the late phase of the HIV-1 life cycle such as assembly and cell-to-cell transmission, these microdomains have been thought to serve as preformed platforms that facilitate concentration of viral components (e.g., Gag and Env) or delivery of these proteins to specific locations in cells. However, recent studies suggest that Gag is not a simple passenger of microdomains but rather plays an active role in reorganizing microdomains via its membrane-binding and multimerization activities. In this paper, we focus on recent findings on this active role played by Gag during microdomain association. In light of this new view, we will also discuss the implications of plasma membrane microdomains and large-scale domains in cell-to-cell transmission. Microdomains are also thought to affect virion infectivity, attachment of virions to target cells, and virus-cell fusion, in which they modulate distributions and/or activities of Env, Nef, and virus receptors. For these topics, interested readers are referred to more comprehensive papers published in recent years [1–5].
2. HIV-1 Assembly at the Plasma Membrane
The viral structural polyprotein Gag is necessary and sufficient for the assembly of virus-like particles. HIV-1 Gag is synthesized as a 55 kDa polyprotein composed of 4 major structural domains (and 2 spacer polypeptides), as defined by cleavage by the viral protease: matrix (MA), capsid (CA), nucleocapsid (NC), and p6. However, proteolytic cleavage occurs largely after virion assembly and release; thus, its constituents must work together in the context of the full-length Gag polyprotein to drive particle assembly. After its synthesis in the cytosol, Gag traffics to the site of assembly, binds cellular membranes, multimerizes, buds through the membrane, and recruits host factors that mediate membrane scission, releasing an immature particle [6, 7]. It is increasingly apparent that many of these steps occur in a coordinated, interdependent fashion. Among them, Gag membrane binding and multimerization are implicated in association of virus assembly with membrane microdomains.
Gag membrane binding is mediated by its N-terminal MA domain, containing bipartite membrane binding motifs. The MA domain is cotranslationally myristoylated and contains a highly basic region (HBR) that binds the plasma-membrane-specific acidic phospholipid phosphatidylinositol-(4,5)-bisphosphate [PI(4,5)P2] [8–16] (reviewed in [17]). It has been suggested that exposure of the myristoyl moiety is regulated through a mechanism known as the myristoyl switch [18, 19]. Indeed, NMR studies demonstrated that prior to membrane binding, the myristoyl moiety is sequestered in a hydrophobic cavity of the MA domain. Upon Gag multimerization or PI(4,5)P2 binding, the myristoyl chain is exposed to promote membrane binding [14, 20, 21]. As for MA HBR, RNA appears to competitively regulate its binding to acidic membrane lipids. Studies using in vitro assays collectively support a model in which RNA bound to HBR prevents MA from binding to prevalent acidic lipids like phosphatidylserine, but allows MA binding to PI(4,5)P2, thereby enhancing the specificity of Gag binding to PI(4,5)P2-containing membranes, that is, the plasma membrane [10, 22–25].
Two major functional regions that contribute to Gag multimerization are the C-terminal region of the CA domain (CA-CTD) and NC. CA-CTD forms an interface that mediates Gag homodimerization [26–29]. The NC domain is thought to contribute to Gag multimerization via its ability to bind RNA [30–34]. Notably, heterologous leucine zipper dimerization motifs can substitute for NC in Gag multimerization and particle assembly [35–39]. These findings suggest a model in which RNA binding to NC serves a structural role, either as a scaffold or a trigger for CA dimerization. In addition to CA and NC, the Spacer Peptide 1 (SP1) between CA and NC plays an important role in regulating the multimerization process [40].
Higher-order Gag multimerization induces outward curvature of the plasma membrane area where the Gag multimer is bound. This step is likely driven by the inherent curvature of the Gag hexameric lattice, formation of which relies on CA [41]. Consistent with this, a number of CA mutations lead to a budding arrest phenotype, characterized by many electron-dense patches underneath the plasma membrane [29, 42]. Release of nascent particles is driven by the cellular ESCRT (endosomal sorting complexes required for transport) that is recruited to assembling virions through interactions with the NC and p6 domains [43].
HIV-1 has been observed to assemble at the plasma membrane in T cells and some laboratory cell lines such as HeLa cells (see [44] for a review). Assembly in macrophages was originally thought to occur at late endosomes/multivesicular bodies (LE/MVB), based on the apparently intracellular location of assembling Gag and the presence of LE/MVB markers, such as the tetraspanin protein CD63 and ESCRT [45, 46]. However, the sites of assembly in macrophages were found to be actually deep invaginations of the plasma membrane, now known as virus-containing compartments (VCC) (although the architecture of the VCC, in particular whether VCCs are all connected to the plasma membrane, is still under intense investigation) [47–53]. Moreover, markers like CD63 strongly colocalize with Gag, even at assembly sites that are unambiguously on the plasma membrane (e.g., [54, 55]). Therefore, the currently accepted idea is that for most cell types including macrophages, the primary site of HIV-1 assembly is the plasma membrane or its specific domains [56, 57].
3. Plasma Membrane Microdomains Associated with HIV-1 Assembly
The plasma membrane consists of diverse microdomains. This partitioning of membrane components is regulated by lipid-lipid, protein-protein, and protein-lipid interactions and compartmentalizes cellular processes [58]. As with many diverse enveloped viruses, HIV-1 was initially proposed to assemble at lipid rafts, based on sensitivity to cellular cholesterol depletion and cofractionation of viral components with detergent-resistant membranes (DRM). Subsequently, HIV-1 assembly was also proposed to occur at tetraspanin-enriched microdomains based on microscopy.
3.1. Lipid Rafts
Spontaneous partitioning of lipids into an ordered phase and a disordered phase has been observed in chemically defined model membranes and model membranes reconstituted from cellular membrane components [58]. The ordered phase is enriched in cholesterol and saturated lipids, and the disordered phase is enriched in unsaturated lipids. This biophysical phenomenon of lipid phase separation in model membranes has been hypothesized to underlie the phenomenon of lipid rafts in cells. In contrast to model membranes, however, cellular membranes contain a greater diversity of lipids and proteins. The partitioning of these molecules is governed by a much greater complexity of lipid-lipid, protein-lipid, and protein-protein interactions. Thus, the current consensus is that lipid rafts are highly dynamic, submicroscopic membrane domains enriched in sterols and sphingolipids, which can be stabilized to form larger platforms through protein-protein and protein-lipid interactions [58].
To assess the involvement of lipid rafts in HIV-1 assembly processes, biochemical assays that measure either resistance to nonionic detergents or sensitivity to cellular cholesterol depletion have been widely used. Results from these assays generally support lipid raft association of the HIV-1 assembly process [59–71]. Both cholesterol depletion and substitution of the Gag myristoyl moiety with an unsaturated acyl analogue inhibit virus particle production, suggesting a functional role for association between HIV-1 Gag and lipid rafts during virus assembly [66, 68, 69]. While biochemical methods used in these studies require cautious interpretations of data due to their inherent limitations [72–76], studies using different approaches, such as microscopy and virion content analyses described below, also generally support raft association with the HIV-1 assembly process.
Because of the dynamic and submicroscopic nature of lipid rafts, cross-linking of cell-surface proteins, which stabilizes the microdomains they associate with, is often used to observe protein partitioning into microdomains by standard fluorescence microscopy. When two microdomain markers are independently clustered using specific antibodies or toxins, these markers can colocalize within the same patch, or “copatch”, indicating propensity of these markers to partition into the same microdomains [77–80]. Consistent with biochemical analysis described above, Gag puncta that represent assembled particles or multimerizing Gag are observed to colocalize or copatch with raft markers, such as the glycosphingolipid GM1 and GPI-anchored proteins [42, 64, 67, 81–83]. However, a recent super-resolution microscopy study showed that GM1 does not colocalize with Gag clusters, at least in the particular cell type used [84]. Therefore, while GM1 may have a propensity to associate with lipid rafts, codistribution of this lipid with other raft components may occur only when raft partitioning is stabilized by crosslinking. These new super-resolution microscopy technologies will likely allow us to define the native distribution of each raft component associated with HIV-1 assembly sites.
Finally, analyses of cellular molecules incorporated into HIV-1 particles also support lipid raft involvement during the HIV-1 assembly process. Biochemical, proteomics, and lipidomics studies have shown that the HIV-1 envelope is enriched in many of lipids and proteins that are also components of lipid rafts [85–91]. Of note, the cholesterol content of virions may be upregulated via activities of viral proteins such as HIV-1 Nef [92–94] and MLV glyco-Gag [95]. Importantly, by measuring spectral shift of the lipophilic fluorescent dye laurdan, which is sensitive to ordered packing of its surrounding lipids [96], HIV-1 envelope was shown to contain liquid-ordered domains [97].
3.2. Tetraspanin-Enriched Microdomains
Tetraspanin-enriched microdomains (TEMs) are plasma membrane microdomains organized by the homo- and heterooligomerization of tetraspanins, a family of homologous proteins with four transmembrane domains. Proteomics studies have identified a wide variety of proteins associated with TEMs. Most notably, tetraspanins interact with cell-adhesion molecules, integrins, and cell-signaling proteins, suggesting that TEMs serve as a platform to spatially organize cell-cell and cell-extracellular matrix adhesion and signaling [4, 98, 99]. Tetraspanins CD63 and CD81 have been shown to associate with phosphatidylinositol 4-kinase, a critical enzyme in creating a precursor for PI(4,5)P2 [100]. Importantly, different tetraspanins appear to be at least partially redundant in the cell functions measured in some of these studies.
The first evidence of association between tetraspanin proteins and HIV-1 assembly were early studies that found the tetraspanin protein CD63 enriched in the envelopes of HIV-1 particles. This was taken as evidence that Gag traffics through, or assembles at, an endosomal compartment, such as the LE/MVB. However, it was later shown that Gag associates with CD63 and other tetraspanin proteins at discrete microdomains on the plasma membrane [54, 55].
Tetraspanins, including CD9, CD63, and CD81, are incorporated into virus particles [45, 55, 88, 101–107], coimmunoprecipitate with Gag-laden cellular membranes [108], and strongly colocalize/copatch with Gag by immunofluorescence microscopy assays (e.g., [54, 55, 108]). As for functions, a variety of studies have suggested roles for tetraspanins and TEMs in different phases of the HIV-1 replication cycle such as virus entry (see [4] for a review). However, the role of tetraspanins and TEMs in Gag assembly remains currently unclear. The gross effects of perturbing tetraspanins by siRNA knockdown or overexpression are so far contradictory: some studies report perturbation reduces particle production [108, 109], while others report no effect [107, 110, 111]. In contrast, it is well accepted that tetraspanins incorporated into virus particles have an inhibitory effect on subsequent virus entry [107, 108, 110].
3.3. Gag Determinants for Interactions with Microdomains
While association of Gag with microdomains has been well documented, how this association occurs is only beginning to be elucidated. As saturated acyl chains mediate raft association of many cytoplasmic proteins, it is straightforward to imagine that the N-terminal myristoyl moiety of Gag plays a role. Consistent with this notion, incorporation of an unsaturated myristate analogue in the place of myristate impairs Gag recovery into DRM fractions [66]. Interestingly, an NMR study of MA bound to a soluble PI(4,5)P2 (with short acyl chains, allowing it to remain in aqueous solution) showed that, while PI(4,5)P2 binding induces myristoyl exposure, a hydrophobic cleft of the MA domain sequesters the typically unsaturated sn2 acyl chain of PI(4,5)P2—effectively exchanging an unsaturated acyl chain from PI(4,5)P2 for the saturated myristoyl chain of Gag [14]. This sequestration of the unsaturated sn2 acyl chain of PI(4,5)P2 has been hypothesized to facilitate Gag association with lipid rafts [14]. It remains to be seen if this acyl chain exchange occurs in the more authentic case of Gag binding a lipid bilayer, as opposed to interaction between isolated MA domains and water-soluble lipids.
HIV-1 Gag multimerization has also been observed to enhance microdomain association. Biochemical studies showed that the presence of NC and other Gag regions necessary for multimerization affect the steady-state association of Gag with DRM [61, 65]. The presence of NC is also required for colocalization of Gag with markers for microdomains termed endosome-like domains (ELD), which appear to be a subset of TEMs [54, 112]. ELD association of Gag and other multimeric proteins was reported to be independent of a membrane-binding interface; a variety of plasma membrane targeting motifs were observed to mediate ELD association of a normally-cytosolic oligomeric protein, TyA [113]. Altogether, these results are consistent with a notion that Gag multimerization plays a key role in stable association with specific microdomains at the plasma membrane.
In the context of assembly of many enveloped viruses, membrane microdomains are often regarded as preexisting platforms that accumulate viral structural components, thereby facilitating virus assembly. However, as alluded to earlier, protein-protein interactions are thought to stabilize or recruit microdomains [114, 115]. Therefore, Gag multimerization was postulated to modulate structure and/or size of Gag-associated microdomains [11, 65, 68, 86]. Consistent with this protein-centric view of microdomains, recent studies suggest that HIV-1 Gag is not just passively accumulated in microdomains but rather actively stabilize, recruit, or reorganize microdomains at the plasma membrane through its multimerization. Fluorescence recovery after photobleaching and single-molecule tracking analyses showed that Gag multimers trap the tetraspanin CD9 and, to a lesser extent, the raft markers GM1 and CD55 and clusters these microdomain components in a Gag-multimerization-dependent manner [82]. Furthermore, copatching and fluorescence resonance energy transfer analyses showed that HIV-1 coalesces TEMs and lipid rafts [42], two microdomains that are otherwise distinct and do not colocalize in cells that do not express Gag [98, 116–120]. Interestingly, correlative fluorescence and scanning electron microscopy showed that copatching between raft and TEM markers does not occur at assembly sites of a Gag mutant that forms multimeric Gag patches but fails to form spherical particles [42]. Therefore, raft-TEM coalescence appears to depend on membrane curvature induced by Gag multimerization. Altogether, Gag is likely to direct the formation of its own microdomains by recruiting and coalescing membrane proteins and microdomains, in a manner dependent on the process of virus assembly.
What determines microdomain recruitment to Gag multimers? Since MA functions as the interface of Gag with lipid bilayer, it is conceivable that MA or MA-interacting molecules drive recruitment of lipid raft and TEM markers. For example, the combination of the N-terminal myristoyl moiety and a saturated acyl chain of PI(4,5)P2, which is postulated to direct Gag to lipid rafts [14], may also direct small lipid rafts to Gag assembly sites. This is also consistent with the enrichment of specific lipids to the viral envelope, relative to the plasma membrane [86, 87]. However, copatching studies suggest that coalescence of lipid rafts and TEMs at assembly sites occur even when MA was replaced with a triple acylation motif or a heterologous lipid-binding domain [42]. Therefore, the MA sequence per se is not essential for reorganization of lipid rafts and TEMs.
As described below, Gag multimerization is also important for Gag localization to larger membrane domains.
4. Large-Scale Membrane Domains Implicated in HIV-1 Spread
In addition to the microdomains described above, larger plasma membrane domains are implicated in HIV-1 spread. One of such domains is the VCC [47–53], which may serve as a virus reservoir that can transfer viruses upon contact with T cells [121–123]. A similar surface-accessible intracellular compartment in dendritic cells also promotes transmission of captured viruses to T cells via cell contacts during trans-infection [124–128]. In this section, however, we focus on membrane domains implicated in T-cell-to-T-cell virus transmission and their relationships with microdomains.
4.1. Virological Synapses
HIV-1 virions released from infected cells may travel in the extracellular space until they come in contact with a target cell by chance (random three-dimensional diffusion and fluid flow). However, this cell-free infection route is much less efficient than cell-to-cell transmission, in which an infected cell physically contacts a target cell and directly transfers the virus. In contrast to cell-free transmission, cell-to-cell transmission of HIV-1 is 10- to several-thousand fold more efficient in cultured T cells [129–132] and is believed to be the major form of transmission for HIV-1 in vivo. In addition to HIV-1, direct cell-to-cell transfer is likely to be important for efficient spreading of several other retroviruses such as human T-lymphotropic virus-1 (HTLV-1) [133–136] and murine leukemia virus [137–139] as well as other pathogens (reviewed in [140, 141]). Moreover, a recent study suggested that cell-to-cell transmission enhances resistance of HIV-1 to antiretroviral drugs and therefore potentially constitutes a mechanism by which HIV-1 maintains an active reservoir in infected individuals undergoing combination drug therapy [142].
Cell-to-cell transmission occurs through several distinct plasma membrane structures. These structures include cytonemes [137–139], membrane nanotubes [143], and virological synapses (VSs) [124, 127, 134, 144–146]. Because involvement of membrane microdomains in the first two structures has yet to be described, in this paper we focus on VSs. VSs formed between HIV-1 infected and uninfected T cells are contact structures enriched in Gag, Env, and viral receptors. Stable VS formation between two T cells is primarily mediated by the Env-CD4 interaction [129, 145–148] unlike VS formed by monocyte-derived macrophages [121]. Consistent with this, antibodies that block the Env-CD4 interaction blocks VS formation and cell-to-cell virus transfer [129, 130, 145, 147, 149] (although neutralization by patient-derived antibodies is ineffective perhaps due to the delayed virion maturation during transfer at the VS [129, 144, 150]). The VS is also enriched in adhesion molecules such as LFA-1, although the significance of these adhesion molecules in VS formation and virus transfer/transmission varies depending on the experimental systems [146–148, 151].
VSs were first described for HTLV-1 [134]. Early studies including this HTLV-1 study and subsequent studies on HIV-1 VS have pointed to the importance of cytoskeleton in VS stability and formation [134, 145, 152–154]. Recent studies further suggest that polarization of HIV-1 Env is dependent on the microtubules and microtubule-dependent trafficking of secretory lysosomes that bear Env [155]. Consistent with this finding, Zap70, which regulates cell polarization in the immunological synapses by controlling localization of the microtubule organizing center (MTOC) [156], facilitates formation of VSs and cell-to-cell transmission [157]. The actin cytoskeleton is also important for VS formation, as evident from the impact of actin depolymerizing agents and a myosin light chain kinase inhibitor on VS formation, cell-to-cell virus transfer and transmission [145, 152, 158].
In addition to cytoskeleton, lipid rafts and TEMs are implicated in VS formation as well. Markers for both microdomains accumulate to VS [110, 146, 159, 160]. Consistent with a role for lipid rafts in VS formation, cholesterol depletion was observed to diminish formation of VS, as defined by the accumulation of CD4 (on the target cell) and HIV antigens (in the donor cell) at the cell-cell interface [159]. However, whether this impact was due to disruption of lipid rafts or inhibition of other cholesterol-dependent processes is unknown. If the former is the case, what particular role lipid rafts play in VS formation also remains to be determined.
As for the role of TEMs, multiple and potentially opposing roles played by tetraspanins (for a review, see Thali [4]) make it difficult to assess the contribution of TEMs to VS formation. Anti-tetraspanin antibodies were observed to reduce VS formation albeit modestly [160]. Consistent with the inhibitory effect of tetraspanins on infectivity of virions [107, 108, 110], tetraspanins also prevent Env-mediated cell-cell fusion [161], inhibition of which was suggested to help preserve productive VSs [110]. Moreover, the presence of CD81, but not other tetraspanins, was shown to facilitate polarized localization of Gag [108]. On the other hand, CD81 was observed to decrease cell-to-cell virus transmission, perhaps via inhibition of virion infectivity [110]. Therefore, it remains to be determined whether and in what context TEMs or tetraspanins play a positive or negative role in cell-to-cell transmission of HIV-1 via VSs.
4.2. Uropods
A majority of T cells in lymph nodes where cell-to-cell transmission likely occurs frequently are highly motile and adopt a polarized morphology [162–166]. The front end of a polarized T cell is called the leading edge, and the protrusion at the rear is called a uropod [167–169]. Functionally, uropods seem to promote T-cell migration by facilitating deadhesion of integrins such as LFA-1 that mediates substrate adhesion at the leading edge [169]. During T-cell migration, uropods also mediate contact with other T cells [170] and recruit bystander T cells to sites of inflammation [171]. Interestingly, Gag accumulates to the plasma membrane area constituting the uropod surface in polarized T cells [67, 129, 158] (Figure 1(a)). Moreover, upon contact with uninfected T cells, this plasma membrane domain participates in the VS, as supported by the observation that Gag and uropod markers on the infected cell and CD4 on the uninfected cell accumulate at the site of cell-cell contact [158]. These findings suggest a model in which the uropod surface of polarized HIV-1-infected T cells serves as a preformed platform that participates in VS formation.
Figure 1.
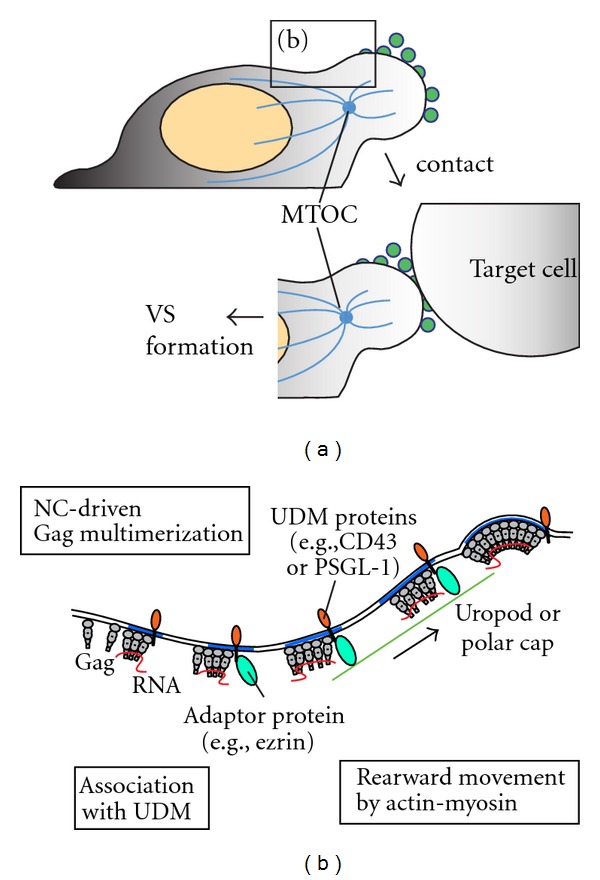
(a) Gag accumulates at the uropod surface. While it remains to be determined whether the first contact between virus-producing and target cells occurs right at the uropod or elsewhere during VS formation, virus-laden uropods do participate in VS formation as determined by concentration of uropod markers at the VS. (b) A working model for a mechanism by which Gag multimers associate with rearward actin flow that directs Gag to the uropod. NC-dependent Gag multimerization underneath the plasma membrane promotes association between Gag multimer and UDM. Of note, in contrast to lipid raft and TEM markers, UDM proteins appear to accumulate at assembly sites of wild-type Gag as well as those of a Gag mutant that multimerizes but fails to bud (GNL, unpublished data).
Which part of an HIV-1-infected cell mediates the initial contact with a target cell remains to be determined. It is possible that uropods, where the virus is concentrated, establish the initial contacts, and these contacts eventually develop into VSs without large-scale shift in cell polarity (as shown in Figure 1(a)). Consistent with this possibility, uropods are enriched in various adhesion molecules that help promote contact with other cells. However, it is also possible that initial contacts may be established at other regions of cells such as the leading edge. Under this scenario, after initial contact, viral proteins and VS components that are preaccumulated at the uropod would subsequently move laterally to the cell-cell junction. In support of this latter possibility, patches containing HIV-1 Gag have been observed to move laterally over the cell surface to the VS [144, 146]. Regardless of the pathways taken by Gag to the uropod and cell junctions, this preaccumulation of Gag at the uropod may constitute an important early step in VS formation.
The molecular mechanisms of Gag localization to the uropod also remain to be determined. Notably, Gag accumulation to uropods requires higher-order multimerization driven by NC [158, 172], while the dimerization function of CA-CTD is neither sufficient nor necessary [158]. In this regard, it is important to note that crosslinking of cell surface proteins with antibodies induces polarized localization of these proteins in leukocytes and other cell types [173]. Such “capping” has also been observed for lipids cross-linked by pentavalent cholera toxin [174, 175]. During T-cell polarization, similar polar cap formation occurs spontaneously at the cell surface from which a uropod originates [169, 176]. These capping phenomena depend on myosin II-driven rearward actin flow [177–181]. Thus, in a manner similar to capping, higher-order Gag multimerization might trigger Gag association with the actin flow, which in turn drives accumulation of Gag in uropods. In support of this model (Figure 1(b)), a myosin light chain kinase inhibitor ML7, which inhibits myosin II, dispersed Gag all over the cell surface [145, 158].
The nature of the link between Gag multimers and retrograde actin flow is currently unknown. While NC has been implicated in interaction with actin [182, 183], this does not account for uropod localization of Gag-LZ in which NC was replaced with a heterologous leucine zipper [158, 172]. As Gag multimerization recruits and stabilizes lipid rafts and TEMs at assembly sites (discussed earlier), it is conceivable that reorganization of these microdomains, as well as cellular proteins associated with these microdomains, is involved in polarized localization of Gag multimers to uropods and subsequently to the VS. In support of this hypothesis, markers for both microdomains are found to accumulate at uropods [169] and VSs [110, 146, 159, 160]. Indeed, in HIV-1-expressing T cells, both a raft marker CD59 and a tetraspanin CD81 copolarize with Gag to uropods. However, using a T-cell line that polarizes spontaneously, we observed that Gag copatches with CD59 only to a very low extent prior to cell polarization (GNL unpublished data). Even within uropods, copatching between Gag and CD59 was poor (GNL unpublished data). Copatching between Gag and CD81 was shown to be higher but still at a modest level [158]. In contrast, uropod markers PSGL-1, CD43, and CD44 strongly copatch with Gag both before and after polarization [158] (GNL unpublished data). Therefore, at least in these T cells, Gag appears to associate predominantly with a specific microdomain enriched in uropod-directed proteins (uropod-directed microdomain or UDM), which is likely to be distinct from CD59-positive lipid rafts.
One can postulate that in T cells Gag multimerization induces recruitment of UDMs more efficiently than that of lipid rafts or perhaps TEMs. UDM association may in turn promote association between Gag multimers and actin flow and thereby facilitate Gag localization to the uropod (Figure 1(b)). In support of this possibility, PSGL-1 comigrates with Gag toward the uropod as T cells polarize [158]. This possibility is further supported by the observation that actin-binding proteins such as ezrin and moesin, which are found in HIV-1 virions [90], bind cytoplasmic domains of several uropod-specific transmembrane proteins and promote localization of these proteins to uropods [169]. Alternatively, it is possible that while PSGL-1 and other UDM proteins are recruited to Gag multimers, Gag might not require UDM association for localization to the uropod. In such case, recruited UDM proteins may serve other functions in cell-to-cell transmission. Elucidating the mechanism by which Gag multimers associate with UDMs will likely allow us to determine the potential role of this association in Gag localization and cell-to-cell HIV-1 transmission.
5. Future Perspectives
The plasma membrane microdomains that constitute virus assembly sites have been frequently depicted as stable preformed platforms. However, a more nuanced view of plasma membrane compartmentalization is that they exist along a continuum of size and stability. On one end, large domains such as the Gag-laden uropod surface may serve as a preformed stable structure poised to form cell-cell junctions or VSs. On the other extreme, microdomains are submicroscopic, dynamic, and unstable unless protein-protein and protein-lipid interactions drive their stabilization. At least for HIV-1, it is increasingly clear that Gag multimerization and/or membrane curvature reorganizes plasma membrane microdomains at assembly sites. With this new view, a number of new questions arise: What are the characteristics specific to virus-reorganized microdomains compared to those of original individual microdomains? What is the nature of association (or lack thereof) between monomeric Gag and microdomains? Do other enveloped viruses alter microdomain organization at their assembly sites, and if so, what are the differences in composition and function among these virus-reorganized microdomains?
Cellular proteins and lipids that specifically associate with membrane microdomains of virus assembly sites affect HIV-1 particle production and infectivity, either positively (e.g., cholesterol, see [5] for a review; sphingolipids [184]) or negatively (e.g., tetraspanins; see above). Incorporation of viral proteins such as Env into virus particles may also be modulated by microdomains [1, 2]. To fully understand incorporation of these molecules into virus particles, it is crucial to elucidate the mechanisms by which Gag multimerization reorganizes microdomains. Although even Gag derivatives with heterologous membrane-binding domains can induce coalescence of lipid rafts and TEMs, membrane-binding domains of Gag may still modulate compositions of reorganized microdomains via molecular interactions. In this regard, it is interesting to note that a highly basic protein can induce formation of a microdomain enriched in acidic lipids, which in turn attract other basic proteins [185, 186]. As Gag and other viral structural proteins contain highly basic regions, it is conceivable that multimerization of these viral proteins may induce acidic lipid clustering and thereby trigger association of basic-region-containing proteins to assembly sites. Whether such indirect mechanism, in addition to direct protein-protein interactions, modulates microdomain compositions will potentially be of functional significance.
Less well-characterized functions for reorganized microdomains include contribution to polarized localization and cell-to-cell transmission. On this front, future studies need to be directed to understanding (1) the relationships among UDMs, lipid rafts, and TEMs, (2) the mechanism by which Gag multimerization facilitates association of Gag with the retrograde actin flow, and (3) the role for UDM proteins in polarized localization and VS functions. These studies will help us further understand molecular mechanisms that facilitate VS formation and cell-to-cell transmission.
Authors' Contribution
I. B. Hogue and G. N. Llewellyn have contributed to this work equally.
Acknowledgments
The authors thank members of the Ono laboratory for helpful discussions. Studies in their laboratory related to the topics in this paper were supported by National Institute of Allergy and Infectious Diseases (R01 AI071727, R56 AI 089282, R21 AI095022), American Heart Association (0850133Z), and amfAR (107449-45-RGHF). Ian B. Hogue and G. N. Llewellyn were supported by NIH training Grants T32 GM007544 (the University of Michigan Genetics Training Program) and T32 GM007315 (the University of Michigan Cellular and Molecular Biology Training Program), respectively. The present addresses of the first and second authors are Department of Molecular Biology, Princeton University and Department of Molecular Microbiology and Immunology, University of Southern California, respectively.
References
- 1.Checkley MA, Luttge BG, Freed EO. HIV-1 envelope glycoprotein biosynthesis, trafficking, and incorporation. Journal of Molecular Biology. 2011;410(4):582–608. doi: 10.1016/j.jmb.2011.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johnson MC. Mechanisms for env glycoprotein acquisition by retroviruses. AIDS Research and Human Retroviruses. 2011;27(3):239–247. doi: 10.1089/aid.2010.0350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ono A. Relationships between plasma membrane microdomains and HIV-1 assembly. Biology of the Cell. 2010;102(6):335–350. doi: 10.1042/BC20090165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thali M. The roles of tetraspanins in HIV-1 replication. Current Topics in Microbiology and Immunology. 2009;339(1):85–102. doi: 10.1007/978-3-642-02175-6_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Waheed AA, Freed EO. Lipids and membrane microdomains in HIV-1 replication. Virus Research. 2009;143(2):162–176. doi: 10.1016/j.virusres.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adamson CS, Freed EO. Human immunodeficiency virus type 1 assembly, release, and maturation. Advances in Pharmacology. 2007;55:347–387. doi: 10.1016/S1054-3589(07)55010-6. [DOI] [PubMed] [Google Scholar]
- 7.Balasubramaniam EO, Freed M. New Insights into HIV Assembly and Trafficking. Physiology. 2011;26(4):236–251. doi: 10.1152/physiol.00051.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bryant M, Ratner L. Myristoylation-dependent replication and assembly of human immunodeficiency virus 1. Proceedings of the National Academy of Sciences of the United States of America. 1990;87(2):523–527. doi: 10.1073/pnas.87.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chukkapalli V, Hogue IB, Boyko V, Hu WS, Ono A. Interaction between the human immunodeficiency virus type 1 Gag matrix domain and phosphatidylinositol-(4,5)-bisphospnate is essential for efficient Gag membrane binding. Journal of Virology. 2008;82(5):2405–2417. doi: 10.1128/JVI.01614-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chukkapalli V, Oh SJ, Ono A. Opposing mechanisms involving RNA and lipids regulate HIV-1 Gag membrane binding through the highly basic region of the matrix domain. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(4):1600–1605. doi: 10.1073/pnas.0908661107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dalton AK, Ako-Adjei D, Murray PS, Murray D, Vogt VM. Electrostatic interactions drive membrane association of the human immunodeficiency virus type 1 Gag MA domain. Journal of Virology. 2007;81(12):6434–6445. doi: 10.1128/JVI.02757-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gottlinger HG, Sodroski JG, Haseltine WA. Role of capsid precursor processing and myristoylation in morphogenesis and infectivity of human immunodeficiency virus type 1. Proceedings of the National Academy of Sciences of the United States of America. 1989;86(15):5781–5785. doi: 10.1073/pnas.86.15.5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hill CP, Worthylake D, Bancroft DP, Christensen AM, Sundquist WI. Crystal structures of the trimeric human immunodeficiency virus type 1 matrix protein: Implications for membrane association and assembly. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(7):3099–3104. doi: 10.1073/pnas.93.7.3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saad JS, Miller J, Tai J, Kim A, Ghanam RH, Summers MF. Structural basis for targeting HIV-1 Gag proteins to the plasma membrane for virus assembly. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(30):11364–11369. doi: 10.1073/pnas.0602818103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shkriabai N, Datta SAK, Zhao Z, Hess S, Rein A, Kvaratskhelia M. Interactions of HIV-1 Gag with assembly cofactors. Biochemistry. 2006;45(13):4077–4083. doi: 10.1021/bi052308e. [DOI] [PubMed] [Google Scholar]
- 16.Zhou W, Parent LJ, Wills JW, Resh MD. Identification of a membrane-binding domain within the amino-terminal region of human immunodeficiency virus type 1 Gag protein which interacts with acidic phospholipids. Journal of Virology. 1994;68(4):2556–2569. doi: 10.1128/jvi.68.4.2556-2569.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chukkapalli V, Ono A. Molecular determinants that regulate plasma membrane association of HIV-1 Gag. Journal of Molecular Biology. 2011;410(4):512–524. doi: 10.1016/j.jmb.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spearman P, Horton R, Ratner L, Kuli-Zade I. Membrane binding of human immunodeficiency virus type 1 matrix protein in vivo supports a conformational myristyl switch mechanism. Journal of Virology. 1997;71(9):6582–6592. doi: 10.1128/jvi.71.9.6582-6592.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou W, Resh MD. Differential membrane binding of the human immunodeficiency virus type 1 matrix protein. Journal of Virology. 1996;70(12):8540–8548. doi: 10.1128/jvi.70.12.8540-8548.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saad JS, Loeliger E, Luncsford P, et al. Point mutations in the HIV-1 matrix protein turn off the myristyl switch. Journal of Molecular Biology. 2007;366(2):574–585. doi: 10.1016/j.jmb.2006.11.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang C, Loeliger E, Luncsford P, Kinde I, Beckett D, Summers MF. Entropic switch regulates myristate exposure in the HIV-1 matrix protein. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(2):517–522. doi: 10.1073/pnas.0305665101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alfadhli A, McNett H, Tsagli S, Bachinger HP, Peyton DH, Barklis E. HIV-1 matrix protein binding to RNA. Journal of Molecular Biology. 2011;410(4):653–666. doi: 10.1016/j.jmb.2011.04.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alfadhli A, Still A, Barklis E. Analysis of human immunodeficiency virus type 1 matrix binding to membranes and nucleic acids. Journal of Virology. 2009;83(23):12196–12203. doi: 10.1128/JVI.01197-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Datta SAK, Heinrich F, Raghunandan S, et al. HIV-1 Gag extension: conformational changes require simultaneous interaction with membrane and nucleic acid. Journal of Molecular Biology. 2011;406(2):205–214. doi: 10.1016/j.jmb.2010.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones CP, Datta SAK, Rein A, Rouzina I, Musier-Forsyth K. Matrix domain modulates HIV-1 Gag’s nucleic acid chaperone activity via inositol phosphate binding. Journal of Virology. 2011;85(4):1594–1603. doi: 10.1128/JVI.01809-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Datta SAK, Zhao Z, Clark PK, et al. Interactions between HIV-1 Gag molecules in solution: an inositol phosphate-mediated switch. Journal of Molecular Biology. 2007;365(3):799–811. doi: 10.1016/j.jmb.2006.10.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gamble TR, Yoo S, Vajdos FF, et al. Structure of the carboxyl-terminal dimerization domain of the HIV-1 capsid protein. Science. 1997;278(5339):849–853. doi: 10.1126/science.278.5339.849. [DOI] [PubMed] [Google Scholar]
- 28.Hogue IB, Hoppe A, Ono A. Quantitative fluorescence resonance energy transfer microscopy analysis of the human immunodeficiency virus type 1 Gag-Gag interaction: Relative contributions of the CA and NC domains and membrane binding. Journal of Virology. 2009;83(14):7322–7336. doi: 10.1128/JVI.02545-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Von Schwedler UK, Stray KM, Garrus JE, Sundquist WI. Functional surfaces of the human immunodeficiency virus type 1 capsid protein. Journal of Virology. 2003;77(9):5439–5450. doi: 10.1128/JVI.77.9.5439-5450.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Campbell S, Rein A. In vitro assembly properties of human immunodeficiency virus type 1 Gag protein lacking the p6 domain. Journal of Virology. 1999;73(3):2270–2279. doi: 10.1128/jvi.73.3.2270-2279.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Campbell S, Vogt VM. Self-assembly in vitro of purified CA-NC proteins from Rous sarcoma virus and human immunodeficiency virus type 1. Journal of Virology. 1995;69(10):6487–6497. doi: 10.1128/jvi.69.10.6487-6497.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cimarelli A, Sandin S, Höglund S, Luban J. Basic residues in human immunodeficiency virus type 1 nucleocapsid promote virion assembly via interaction with RNA. Journal of Virology. 2000;74(7):3046–3057. doi: 10.1128/jvi.74.7.3046-3057.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khorchid A, Halwani R, Wainberg MA, Kleiman L. Role of RNA in facilitating Gag/Gag-Pol interaction. Journal of Virology. 2002;76(8):4131–4137. doi: 10.1128/JVI.76.8.4131-4137.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muriaux D, Mirro J, Harvin D, Rein A. RNA is a structural element in retrovirus particles. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(9):5246–5251. doi: 10.1073/pnas.091000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Accola MA, Strack B, Göttlinger HG. Efficient particle production by minimal Gag constructs which retain the carboxy-terminal domain of human immunodeficiency virus type 1 capsid-p2 and a late assembly domain. Journal of Virology. 2000;74(12):5395–5402. doi: 10.1128/jvi.74.12.5395-5402.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Crist RM, Datta SAK, Stephen AG, et al. Assembly properties of human immunodeficiency virus type 1 gag-leucine zipper chimeras: Implications for retrovirus assembly. Journal of Virology. 2009;83(5):2216–2225. doi: 10.1128/JVI.02031-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johnson MC, Scobie HM, Ma YM, Vogt VM. Nucleic acid-independent retrovirus assembly can be driven by dimerization. Journal of Virology. 2002;76(22):11177–11185. doi: 10.1128/JVI.76.22.11177-11185.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klein KC, Reed JC, Tanaka M, Nguyen VT, Giri S, Lingappa JR. HIV Gag-leucine zipper chimeras form ABCE1-containing intermediates and RNase-resistant immature capsids similar to those formed by wild-type HIV-1 Gag. Journal of Virology. 2011;85(14):7419–7435. doi: 10.1128/JVI.00288-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Y, Qian H, Love Z, Barklis E. Analysis of the assembly function of the human immunodeficiency virus type 1 gag protein nucleocapsid domain. Journal of Virology. 1998;72(3):1782–1789. doi: 10.1128/jvi.72.3.1782-1789.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Datta SAK, Temeselew LG, Crist RM, et al. On the role of the SP1 domain in HIV-1 particle assembly: a molecular switch? Journal of Virology. 2011;85(9):4111–4121. doi: 10.1128/JVI.00006-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Briggs JAG, Kräusslich H-G. The molecular architecture of HIV. Journal of Molecular Biology. 2011;410(4):491–500. doi: 10.1016/j.jmb.2011.04.021. [DOI] [PubMed] [Google Scholar]
- 42.Hogue IB, Grover JR, Soheilian F, Nagashima K, Ono A. Gag induces the coalescence of clustered lipid rafts and tetraspanin-enriched microdomains at HIV-1 assembly sites on the plasma membrane. Journal of Virology. 2011;85(19):9749–9766. doi: 10.1128/JVI.00743-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weiss ER, Gottlinger H. The role of cellular factors in promoting HIV budding. Journal of Molecular Biology. 2011;410(4):525–533. doi: 10.1016/j.jmb.2011.04.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ono A. HIV-1 assembly at the plasma membrane: Gag trafficking and localization. Future Virology. 2009;4(3):241–257. doi: 10.2217/fvl.09.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pelchen-Matthews A, Kramer B, Marsh M. Infectious HIV-1 assembles in late endosomes in primary macrophages. Journal of Cell Biology. 2003;162(3):443–455. doi: 10.1083/jcb.200304008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Raposo G, Moore M, Innes D, et al. Human macrophages accumulate HIV-1 particles in MHC II compartments. Traffic. 2002;3(10):718–729. doi: 10.1034/j.1600-0854.2002.31004.x. [DOI] [PubMed] [Google Scholar]
- 47.Bennett AE, Narayan K, Shi D, et al. Ion-abrasion scanning electron microscopy reveals surface-connected tubular conduits in HIV-infected macrophages. PLoS Pathogens. 2009;5(9) doi: 10.1371/journal.ppat.1000591.e1000591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chu H, Wang J-J, Qi M, et al. The intracellular virus-containing compartments in primary human macrophages are largely inaccessible to antibodies and small molecules. PLoS ONE. 2012;7(5) doi: 10.1371/journal.pone.0035297.e35297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Deneka M, Pelchen-Matthews A, Byland R, Ruiz-Mateos E, Marsh M. In macrophages, HIV-1 assembles into an intracellular plasma membrane domain containing the tetraspanins CD81, CD9, and CD53. Journal of Cell Biology. 2007;177(2):329–341. doi: 10.1083/jcb.200609050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jouve M, Sol-Foulon N, Watson S, Schwartz O, Benaroch P. HIV-1 buds and accumulates in “Nonacidic” endosomes of macrophages. Cell Host and Microbe. 2007;2(2):85–95. doi: 10.1016/j.chom.2007.06.011. [DOI] [PubMed] [Google Scholar]
- 51.Koppensteiner H, Banning C, Schneider C, Hohenberg H, Schindler M. Macrophage internal HIV-1 is protected from neutralizing antibodies. Journal of Virology. 2012;86(5):2826–2836. doi: 10.1128/JVI.05915-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Welsch S, Groot F, Kräusslich HG, Keppler OT, Sattentau QJ. Architecture and regulation of the HIV-1 assembly and holding compartment in macrophages. Journal of Virology. 2011;85(15):7922–7927. doi: 10.1128/JVI.00834-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Welsch S, Keppler OT, Habermann A, Allespach I, Krijnse-Locker J, Kräusslich HG. HIV-1 buds predominantly at the plasma membrane of primary human macrophages. PLoS Pathogens. 2007;3(3) doi: 10.1371/journal.ppat.0030036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Booth AM, Fang Y, Fallon JK, et al. Exosomes and HIV Gag bud from endosome-like domains of the T cell plasma membrane. Journal of Cell Biology. 2006;172(6):923–935. doi: 10.1083/jcb.200508014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nydegger S, Khurana S, Krementsov DN, Foti M, Thali M. Mapping of tetraspanin-enriched microdomains that can function as gateways for HIV-1. Journal of Cell Biology. 2006;173(5):795–807. doi: 10.1083/jcb.200508165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jouvenet N, Neil SJ, Bess C, et al. Plasma membrane is the site of productive HIV-1 particle assembly. PLoS biology. 2006;4(12) doi: 10.1371/journal.pbio.0040435.e435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Finzi A, Orthwein A, Mercier J, Cohen EA. Productive human immunodeficiency virus type 1 assembly takes place at the plasma membrane. Journal of Virology. 2007;81(14):7476–7490. doi: 10.1128/JVI.00308-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lingwood D, Simons K. Lipid rafts as a membrane-organizing principle. Science. 2010;327(5961):46–50. doi: 10.1126/science.1174621. [DOI] [PubMed] [Google Scholar]
- 59.Bhattacharya J, Repik A, Clapham PR. Gag regulates association of human immunodeficiency virus type 1 envelope with detergent-resistant membranes. Journal of Virology. 2006;80(11):5292–5300. doi: 10.1128/JVI.01469-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ding L, Derdowski A, Wang JJ, Spearman P. Independent segregation of human immunodeficiency virus type 1 Gag protein complexes and lipid rafts. Journal of Virology. 2003;77(3):1916–1926. doi: 10.1128/JVI.77.3.1916-1926.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dou J, Wang JJ, Chen X, Li H, Ding L, Spearman P. Characterization of a myristoylated, monomeric HIV Gag protein. Virology. 2009;387(2):341–352. doi: 10.1016/j.virol.2009.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gomez CY, Hope TJ. Mobility of human immunodeficiency virus type 1 Pr55Gag in living cells. Journal of Virology. 2006;80(17):8796–8806. doi: 10.1128/JVI.02159-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Halwani R, Khorchid A, Cen S, Kleiman L. Rapid localization of Gag/GagPol complexes to detergent-resistant membrane during the assembly of human immunodeficiency virus type 1. Journal of Virology. 2003;77(7):3973–3984. doi: 10.1128/JVI.77.7.3973-3984.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Holm K, Weclewicz K, Hewson R, Suomalainen M. Human immunodeficiency virus type 1 assembly and lipid rafts: Pr55gag associates with membrane domains that are largely resistant to Brij98 but sensitive to triton X-100. Journal of Virology. 2003;77(8):4805–4817. doi: 10.1128/JVI.77.8.4805-4817.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lindwasser OW, Resh MD. Multimerization of human immunodeficiency virus type 1 Gag promotes its localization to barges, raft-like membrane microdomains. Journal of Virology. 2001;75(17):7913–7924. doi: 10.1128/JVI.75.17.7913-7924.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lindwasser OW, Resh MD. Myristoylation as a target for inhibiting HIV assembly: unsaturated fatty acids block viral budding. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(20):13037–13042. doi: 10.1073/pnas.212409999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nguyen DH, Hildreth JEK. Evidence for budding of human immunodeficiency virus type 1 selectively from glycolipid-enriched membrane lipid rafts. Journal of Virology. 2000;74(7):3264–3272. doi: 10.1128/jvi.74.7.3264-3272.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ono A, Freed EO. Plasma membrane rafts play a critical role in HIV-1 assembly and release. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(24):13925–13930. doi: 10.1073/pnas.241320298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ono A, Waheed AA, Freed EO. Depletion of cellular cholesterol inhibits membrane binding and higher-order multimerization of human immunodeficiency virus type 1 Gag. Virology. 2007;360(1):27–35. doi: 10.1016/j.virol.2006.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ono A, Waheed AA, Joshi A, Freed EO. Association of human immunodeficiency virus type 1 Gag with membrane does not require highly basic sequences in the nucleocapsid: use of a novel Gag multimerization assay. Journal of Virology. 2005;79(22):14131–14140. doi: 10.1128/JVI.79.22.14131-14140.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pickl WF, Pimentel-Muiñios FX, Seed B. Lipid rafts and pseudotyping. Journal of Virology. 2001;75(15):7175–7183. doi: 10.1128/JVI.75.15.7175-7183.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hancock JF. Lipid rafts: contentious only from simplistic standpoints. Nature Reviews Molecular Cell Biology. 2006;7(6):456–462. doi: 10.1038/nrm1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Heerklotz H. Triton promotes domain formation in lipid raft mixtures. Biophysical Journal. 2002;83(5):2693–2701. doi: 10.1016/S0006-3495(02)75278-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kwik J, Boyle S, Fooksman D, Margolis L, Sheetz MP, Edidin M. Membrane cholesterol, lateral mobility, and the phosphatidylinositol 4,5-bisphosphate-dependent organization of cell actin. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(2):13964–13969. doi: 10.1073/pnas.2336102100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lichtenberg D, Goñi FM, Heerklotz H. Detergent-resistant membranes should not be identified with membrane rafts. Trends in Biochemical Sciences. 2005;30(8):430–436. doi: 10.1016/j.tibs.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 76.Munro S. Lipid rafts: elusive or illusive? Cell. 2003;115(4):377–388. doi: 10.1016/s0092-8674(03)00882-1. [DOI] [PubMed] [Google Scholar]
- 77.Gri G, Molon B, Manes S, Pozzan T, Viola A. The inner side of T cell lipid rafts. Immunology Letters. 2004;94(3):247–252. doi: 10.1016/j.imlet.2004.05.012. [DOI] [PubMed] [Google Scholar]
- 78.Harder T, Scheiffele P, Verkade P, Simons K. Lipid domain structure of the plasma membrane revealed by patching of membrane components. Journal of Cell Biology. 1998;141(4):929–942. doi: 10.1083/jcb.141.4.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Janes PW, Ley SC, Magee AI. Aggregation of lipid rafts accompanies signaling via the T cell antigen receptor. Journal of Cell Biology. 1999;147(2):447–461. doi: 10.1083/jcb.147.2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shvartsman DE, Kotler M, Tall RD, Roth MG, Henis YI. Differently anchored influenza hemagglutinin mutants display distinct interaction dynamics with mutual rafts. Journal of Cell Biology. 2003;163(4):879–888. doi: 10.1083/jcb.200308142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Harrist AV, Ryzhova EV, Harvey T, González-Scarano F. Anx2 interacts with HIV-1 Gag at phosphatidylinositol (4,5) bisphosphate-containing lipid rafts and increases viral production in 293T cells. PLoS ONE. 2009;4(3) doi: 10.1371/journal.pone.0005020.e5020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Krementsov DN, Rassam P, Margeat E, et al. HIV-1 assembly differentially alters dynamics and partitioning of tetraspanins and raft components. Traffic. 2010;11(11):1401–1414. doi: 10.1111/j.1600-0854.2010.01111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ono EO, Freed A. The role of lipid rafts in virus replication. New York, NY, USA: Elsevier; 2005. [DOI] [PubMed] [Google Scholar]
- 84.Lehmann M, Rocha S, Mangeat B, et al. Quantitative multicolor super-resolution microscopy reveals tetherin HIV-1 interaction. PLoS Pathogens. 2011;7(12) doi: 10.1371/journal.ppat.1002456.e1002456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Aloia RC, Tian H, Jensen FC. Lipid composition and fluidity of the human immunodeficiency virus envelope and host cell plasma membranes. Proceedings of the National Academy of Sciences of the United States of America. 1993;90(11):5181–5185. doi: 10.1073/pnas.90.11.5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Brügger B, Glass B, Haberkant P, Leibrecht I, Wieland FT, Kräusslich HG. The HIV lipidome: a raft with an unusual composition. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(8):2641–2646. doi: 10.1073/pnas.0511136103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chan R, Uchil PD, Jin J, et al. Retroviruses human immunodeficiency virus and murine leukemia virus are enriched in phosphoinositides. Journal of Virology. 2008;82(22):11228–11238. doi: 10.1128/JVI.00981-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chertova E, Chertov O, Coren LV, et al. Proteomic and biochemical analysis of purified human immunodeficiency virus type 1 produced from infected monocyte-derived macrophages. Journal of Virology. 2006;80(18):9039–9052. doi: 10.1128/JVI.01013-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Graham DRM, Chertova E, Hilburn JM, Arthur LO, Hildreth JEK. Cholesterol depletion of human immunodeficiency virus type 1 and simian immunodeficiency virus with β-cyclodextrin inactivates and permeabilizes the virions: Evidence for virion-associated lipid rafts. Journal of Virology. 2003;77(15):8237–8248. doi: 10.1128/JVI.77.15.8237-8248.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ott DE. Cellular proteins detected in HIV-1. Reviews in Medical Virology. 2008;18(3):159–175. doi: 10.1002/rmv.570. [DOI] [PubMed] [Google Scholar]
- 91.Saifuddin M, Parker CJ, Peeples ME, et al. Role of virion-associated glycosylphosphatidylinositol-linked proteins CD55 and CD59 in complement resistance of cell line-derived and primary isolates of HIV-1. Journal of Experimental Medicine. 1995;182(2):501–509. doi: 10.1084/jem.182.2.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mujawar Z, Rose H, Morrow MP, et al. Human immunodeficiency virus impairs reverse cholesterol transport from macrophages. PLoS biology. 2006;4(11) doi: 10.1371/journal.pbio.0040365.e365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zheng YH, Plemenitas A, Fielding CJ, Peterlin BM. Nef increases the synthesis of and transports cholesterol to lipid rafts and HIV-1 progeny virions. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(14):8460–8465. doi: 10.1073/pnas.1437453100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zheng YH, Plemenitas A, Linnemann T, Fackler OT, Peterlin BM. Nef increases infectivity of HIV via lipid rafts. Current Biology. 2001;11(11):875–879. doi: 10.1016/s0960-9822(01)00237-8. [DOI] [PubMed] [Google Scholar]
- 95.Nitta T, Kuznetsov Y, McPherson A, Fan H. Murine leukemia virus glycosylated Gag (gPr80gag) facilitates interferon-sensitive virus release through lipid rafts. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(3):1190–1195. doi: 10.1073/pnas.0908660107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Dietrich C, Bagatolli LA, Volovyk ZN, et al. Lipid rafts reconstituted in model membranes. Biophysical Journal. 2001;80(3):1417–1428. doi: 10.1016/S0006-3495(01)76114-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lorizate M, Brügger B, Akiyama H, et al. Probing HIV-1 membrane liquid order by Laurdan staining reveals producer cell-dependent differences. Journal of Biological Chemistry. 2009;284(33):22238–22247. doi: 10.1074/jbc.M109.029256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Charrin S, Le Naour F, Silvie O, Milhiet PE, Boucheix C, Rubinstein E. Lateral organization of membrane proteins: tetraspanins spin their web. Biochemical Journal. 2009;420(2):133–154. doi: 10.1042/BJ20082422. [DOI] [PubMed] [Google Scholar]
- 99.Yáñez-Mó M, Barreiro O, Gordon-Alonso M, Sala-Valdés M, Sánchez-Madrid F. Tetraspanin-enriched microdomains: a functional unit in cell plasma membranes. Trends in Cell Biology. 2009;19(9):434–446. doi: 10.1016/j.tcb.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 100.Berditchevski F, Tolias KF, Wong K, Carpenter CL, Hemler ME. A novel link between integrins, transmembrane-4 superfamily proteins (CD63 and CD81), and phosphatidylinositol 4-kinase. Journal of Biological Chemistry. 1997;272(5):2595–2598. doi: 10.1074/jbc.272.5.2595. [DOI] [PubMed] [Google Scholar]
- 101.Gluschankof P, Mondor I, Gelderblom HR, Sattentau QJ. Cell membrane vesicles are a major contaminant of gradient-enriched human immunodeficiency virus type-1 preparations. Virology. 1997;230(1):125–133. doi: 10.1006/viro.1997.8453. [DOI] [PubMed] [Google Scholar]
- 102.Khurana S, Krementsov DN, De Parseval A, Elder JH, Foti M, Thali M. Human immunodeficiency virus type 1 and influenza virus exit via different membrane microdomains. Journal of Virology. 2007;81(22):12630–12640. doi: 10.1128/JVI.01255-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Meerloo T, Parmentier HK, Osterhaus ADME, Goudsmit J, Schuurman HJ. Modulation of cell surface molecules during HIV-1 infection of H9 cells. An immunoelectron microscopic study. AIDS. 1992;6(10):1105–1116. doi: 10.1097/00002030-199210000-00007. [DOI] [PubMed] [Google Scholar]
- 104.Meerloo T, Sheikh MA, Bloem AC, et al. Host cell membrane proteins on human immunodeficiency virus type 1 after in vitro infection of H9 cells and blood mononuclear cells. An immuno-electron microscopic study. Journal of General Virology. 1993;74(1):129–135. doi: 10.1099/0022-1317-74-1-129. [DOI] [PubMed] [Google Scholar]
- 105.Nguyen DG, Booth A, Gould SJ, Hildreth JEK. Evidence that HIV budding in primary macrophages occurs through the exosome release pathway. Journal of Biological Chemistry. 2003;278(52):52347–52354. doi: 10.1074/jbc.M309009200. [DOI] [PubMed] [Google Scholar]
- 106.Orentas RJ, Hildreth JEK. Association of host cell surface adhesion receptors and other membrane proteins with HIV and SIV. AIDS Research and Human Retroviruses. 1993;9(11):1157–1165. doi: 10.1089/aid.1993.9.1157. [DOI] [PubMed] [Google Scholar]
- 107.Sato K, Aoki J, Misawa N, et al. Modulation of human immunodeficiency virus type 1 infectivity through incorporation of tetraspanin proteins. Journal of Virology. 2008;82(2):1021–1033. doi: 10.1128/JVI.01044-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Grigorov B, Attuil-Audenis V, Perugi F, et al. A role for CD81 on the late steps of HIV-1 replication in a chronically infected T cell line. Retrovirology. 2009;6:p. 28. doi: 10.1186/1742-4690-6-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chen H, Dziuba N, Friedrich B, et al. A critical role for CD63 in HIV replication and infection of macrophages and cell lines. Virology. 2008;379(2):191–196. doi: 10.1016/j.virol.2008.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Krementsov DN, Weng J, Lambelé M, Roy NH, Thali M. Tetraspanins regulate cell-to-cell transmission of HIV-1. Retrovirology. 2009;6:p. 64. doi: 10.1186/1742-4690-6-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ruiz-Mateos E, Pelchen-Matthews A, Deneka M, Marsh M. CD63 is not required for production of infectious human immunodeficiency virus type 1 in human macrophages. Journal of Virology. 2008;82(10):4751–4761. doi: 10.1128/JVI.02320-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Fang Y, Wu N, Gan X, Yan W, Morrell JC, Gould SJ. Higher-order oligomerization targets plasma membrane proteins and HIV gag to exosomes. PLoS biology. 2007;5(6) doi: 10.1371/journal.pbio.0050158.e158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Shen B, Wu N, Yang M, Gould SJ. Protein targeting to exosomes/microvesicles by plasma membrane anchors. Journal of Biological Chemistry. 2011;286(16):14383–14395. doi: 10.1074/jbc.M110.208660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Langhorst MF, Reuter A, Stuermer CAO. Scaffolding microdomains and beyond: the function of reggie/flotillin proteins. Cellular and Molecular Life Sciences. 2005;62(19-20):2228–2240. doi: 10.1007/s00018-005-5166-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Parton RG, Simons K. The multiple faces of caveolae. Nature Reviews Molecular Cell Biology. 2007;8(3):185–194. doi: 10.1038/nrm2122. [DOI] [PubMed] [Google Scholar]
- 116.Barreiro O, Zamai M, Yáñez-Mó M, et al. Endothelial adhesion receptors are recruited to adherent leukocytes by inclusion in preformed tetraspanin nanoplatforms. Journal of Cell Biology. 2008;183(3):527–542. doi: 10.1083/jcb.200805076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Charrin S, Manié S, Oualid M, Billard M, Boucheix C, Rubinstein E. Differential stability of tetraspanin/tetraspanin interactions: role of palmitoylation. FEBS Letters. 2002;516(1–3):139–144. doi: 10.1016/s0014-5793(02)02522-x. [DOI] [PubMed] [Google Scholar]
- 118.Claas C, Stipp CS, Hemler ME. Evaluation of prototype transmembrane 4 superfamily protein complexes and their relation to lipid rafts. Journal of Biological Chemistry. 2001;276(11):7974–7984. doi: 10.1074/jbc.M008650200. [DOI] [PubMed] [Google Scholar]
- 119.Espenel C, Margeat E, Dosset P, et al. Single-molecule analysis of CD9 dynamics and partitioning reveals multiple modes of interaction in the tetraspanin web. Journal of Cell Biology. 2008;182(4):765–776. doi: 10.1083/jcb.200803010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Le Naour F, André M, Boucheix C, Rubinstein E. Membrane microdomains and proteomics: lessons from tetraspanin microdomains and comparison with lipid rafts. Proteomics. 2006;6(24):6447–6454. doi: 10.1002/pmic.200600282. [DOI] [PubMed] [Google Scholar]
- 121.Gousset K, Ablan SD, Coren LV, et al. Real-time visualization of HIV-1 GAG trafficking in infected macrophages. PLoS Pathogens. 2008;4(3) doi: 10.1371/journal.ppat.1000015.e1000015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Groot F, Welsch S, Sattentau QJ. Efficient HIV-1 transmission from macrophages to T cells across transient virological synapses. Blood. 2008;111(9):4660–4663. doi: 10.1182/blood-2007-12-130070. [DOI] [PubMed] [Google Scholar]
- 123.Sharova N, Swingler C, Sharkey M, Stevenson M. Macrophages archive HIV-1 virions for dissemination in trans. EMBO Journal. 2005;24(13):2481–2489. doi: 10.1038/sj.emboj.7600707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Arrighi JF, Pion M, Garcia E, et al. DC-SIGN-mediated infectious synapse formation enhances X4 HIV-1 transmission from dendritic cells to T cells. Journal of Experimental Medicine. 2004;200(10):1279–1288. doi: 10.1084/jem.20041356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Felts RL, Narayan K, Estes JD, et al. 3D visualization of HIV transfer at the virological synapse between dendritic cells and T cells. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(30):13336–13341. doi: 10.1073/pnas.1003040107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Garcia E, Pion M, Pelchen-Matthews A, et al. HIV-1 trafficking to the dendritic cell-T-cell infectious synapse uses a pathway of tetraspanin sorting to the immunological synapse. Traffic. 2005;6(6):488–501. doi: 10.1111/j.1600-0854.2005.00293.x. [DOI] [PubMed] [Google Scholar]
- 127.McDonald D, Wu L, Bohks SM, KewalRamani VN, Unutmaz D, Hope TJ. Recruitment of HIV and its receptors to dendritic cell-T cell junctions. Science. 2003;300(5623):1295–1297. doi: 10.1126/science.1084238. [DOI] [PubMed] [Google Scholar]
- 128.Yu HJ, Reuter MA, McDonald D. HIV traffics through a specialized, surface-accessible intracellular compartment during trans-infection of T cells by mature dendritic cells. PLoS pathogens. 2008;4(8):p. e1000134. doi: 10.1371/journal.ppat.1000134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Chen P, Hübner W, Spinelli MA, Chen BK. Predominant mode of human immunodeficiency virus transfer between T cells is mediated by sustained Env-dependent neutralization-resistant virological synapses. Journal of Virology. 2007;81(22):12582–12595. doi: 10.1128/JVI.00381-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Martin N, Welsch S, Jolly C, Briggs JAG, Vaux D, Sattentau QJ. Virological synapse-mediated spread of human immunodeficiency virus type 1 between T cells is sensitive to entry inhibition. Journal of Virology. 2010;84(7):3516–3527. doi: 10.1128/JVI.02651-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Bu G, Morton PA, Schwartz AL. Receptor-mediated endocytosis of plasminogen activators. Advances in Molecular and Cell Biology. 1994;8(C):87–131. [Google Scholar]
- 132.Sourisseau M, Sol-Foulon N, Porrot F, Blanchet F, Schwartz O. Inefficient human immunodeficiency virus replication in mobile lymphocytes. Journal of Virology. 2007;81(2):1000–1012. doi: 10.1128/JVI.01629-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Mazurov D, Ilinskaya A, Heidecker G, Lloyd P, Derse D. Quantitative comparison of HTLV-1 and HIV-1 cell-to-cell infection with new replication dependent vectors. PLoS Pathogens. 2010;6(2) doi: 10.1371/journal.ppat.1000788.e1000788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Igakura T, Stinchcombe JC, Goon PKC, et al. Spread of HTLV-I between lymphocytes by virus-induced polarization of the cytoskeleton. Science. 2003;299(5613):1713–1716. doi: 10.1126/science.1080115. [DOI] [PubMed] [Google Scholar]
- 135.Pais-Correia AM, Sachse M, Guadagnini S, et al. Biofilm-like extracellular viral assemblies mediate HTLV-1 cell-to-cell transmission at virological synapses. Nature Medicine. 2010;16(1):83–89. doi: 10.1038/nm.2065. [DOI] [PubMed] [Google Scholar]
- 136.Majorovits E, Nejmeddine M, Tanaka Y, Taylor GP, Fuller SD, Bangham CRM. Human T-Lymphotropic Virus-1 visualized at the virological synapse by electron tomography. PLoS ONE. 2008;3(5) doi: 10.1371/journal.pone.0002251.e2251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Jin J, Sherer NM, Heidecker G, Derse D, Mothes W. Assembly of the murine leukemia virus is directed towards sites of cell-cell contact. PLoS Biology. 2009;7(7) doi: 10.1371/journal.pbio.1000163.e1000163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Sherer NM, Lehmann MJ, Jimenez-Soto LF, Horensavitz C, Pypaert M, Mothes W. Retroviruses can establish filopodial bridges for efficient cell-to-cell transmission. Nature Cell Biology. 2007;9(3):310–315. doi: 10.1038/ncb1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Sherer NM, Mothes W. Cytonemes and tunneling nanotubules in cell-cell communication and viral pathogenesis. Trends in Cell Biology. 2008;18(9):414–420. doi: 10.1016/j.tcb.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Mothes W, Sherer NM, Jin J, Zhong P. Virus cell-to-cell transmission. Journal of Virology. 2010;84(17):8360–8368. doi: 10.1128/JVI.00443-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Sattentau Q. Avoiding the void: cell-to-cell spread of human viruses. Nature Reviews Microbiology. 2008;6(11):815–826. doi: 10.1038/nrmicro1972. [DOI] [PubMed] [Google Scholar]
- 142.Sigal A, Kim JT, Balazs AB, et al. Cell-to-cell spread of HIV permits ongoing replication despite antiretroviral therapy. Nature. 2011;477(7362):95–99. doi: 10.1038/nature10347. [DOI] [PubMed] [Google Scholar]
- 143.Sowinski S, Jolly C, Berninghausen O, et al. Membrane nanotubes physically connect T cells over long distances presenting a novel route for HIV-1 transmission. Nature Cell Biology. 2008;10(2):211–219. doi: 10.1038/ncb1682. [DOI] [PubMed] [Google Scholar]
- 144.Hübner W, McNerney GP, Chen P, et al. Quantitative 3D video microscopy of HIV transfer across T cell virological synapses. Science. 2009;323(5922):1743–1747. doi: 10.1126/science.1167525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Jolly C, Kashefi K, Hollinshead M, Sattentau QJ. HIV-1 Cell to cell transfer across an Env-induced, actin-dependent synapse. Journal of Experimental Medicine. 2004;199(2):283–293. doi: 10.1084/jem.20030648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Rudnicka D, Feldmann J, Porrot F, et al. Simultaneous cell-to-cell transmission of human immunodeficiency virus to multiple targets through polysynapses. Journal of Virology. 2009;83(12):6234–6246. doi: 10.1128/JVI.00282-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Puigdomènech I, Massanella M, Izquierdo-Useros N, et al. HIV transfer between CD4 T cells does not require LFA-1 binding to ICAM-1 and is governed by the interaction of HIV envelope glycoprotein with CD4. Retrovirology. 2008;5:p. 32. doi: 10.1186/1742-4690-5-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Vasiliver-Shamis G, Tuen M, Wu TW, et al. Human immunodeficiency virus type 1 envelope gp120 induces a stop signal and virological synapse formation in noninfected CD4+ T cells. Journal of Virology. 2008;82(19):9445–9457. doi: 10.1128/JVI.00835-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Massanella M, Puigdoménech I, Cabrera C, et al. Antigp41 antibodies fail to block early events of virological synapses but inhibit HIV spread between T cells. AIDS. 2009;23(2):183–188. doi: 10.1097/QAD.0b013e32831ef1a3. [DOI] [PubMed] [Google Scholar]
- 150.Dale BM, McNerney GP, Thompson DL, et al. Cell-to-cell transfer of HIV-1 via virological synapses leads to endosomal virion maturation that activates viral membrane fusion. Cell Host and Microbe. 2011;10(6):551–562. doi: 10.1016/j.chom.2011.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Jolly C, Mitar I, Sattentau QJ. Adhesion molecule interactions facilitate human immunodeficiency virus type 1-induced virological synapse formation between T cells. Journal of Virology. 2007;81(24):13916–13921. doi: 10.1128/JVI.01585-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Jolly C, Mitar I, Sattentau QJ. Requirement for an intact T-cell actin and tubulin cytoskeleton for efficient assembly and spread of human immunodeficiency virus type 1. Journal of Virology. 2007;81(11):5547–5560. doi: 10.1128/JVI.01469-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Lehmann M, Nikolic DS, Piguet V. How HIV-1 takes advantage of the cytoskeleton during replication and cell-to-cell transmission. Viruses. 2011;3(9):1757–1776. doi: 10.3390/v3091757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Nejmeddine M, Bangham CRM. The HTLV-1 virological synapse. Viruses. 2010;2(7):1427–1447. doi: 10.3390/v2071427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Jolly C, Welsch S, Michor S, Sattentau QJ. The regulated secretory pathway in cd4+ t cells contributes to human immunodeficiency virus type-1 cell-to-cell spread at the virological synapse. PLoS Pathogens. 2011;7(9) doi: 10.1371/journal.ppat.1002226.e1002226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Blanchard N, Di Bartolo V, Hivroz C. In the immune synapse, ZAP-70 controls T cell polarization and recruitment of signaling proteins but not formation of the synaptic pattern. Immunity. 2002;17(4):389–399. doi: 10.1016/s1074-7613(02)00421-1. [DOI] [PubMed] [Google Scholar]
- 157.Sol-Foulon N, Sourisseau M, Porrot F, et al. ZAP-70 kinase regulates HIV cell-to-cell spread and virological synapse formation. EMBO Journal. 2007;26(2):516–526. doi: 10.1038/sj.emboj.7601509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Llewellyn GN, Hogue IB, Grover JR, Ono A. Nucleocapsid promotes localization of HIV-1 gag to uropods that participate in virological synapses between T cells. PLoS pathogens. 2010;6(10):p. e1001167. doi: 10.1371/journal.ppat.1001167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Jolly C, Sattentau QJ. Human immunodeficiency virus type 1 virological synapse formation in T cells requires lipid raft integrity. Journal of Virology. 2005;79(18):12088–12094. doi: 10.1128/JVI.79.18.12088-12094.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Jolly C, Sattentau QJ. Human immunodeficiency virus type 1 assembly, budding, and cell-cell spread in T cells take place in tetraspanin-enriched plasma membrane domains. Journal of Virology. 2007;81(15):7873–7884. doi: 10.1128/JVI.01845-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Weng J, Krementsov DN, Khurana S, Roy NH, Thali M. Formation of syncytia is repressed by tetraspanins in human immunodeficiency virus type 1-producing cells. Journal of Virology. 2009;83(15):7467–7474. doi: 10.1128/JVI.00163-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Bajénoff M, Egen JG, Koo LY, et al. Stromal cell networks regulate lymphocyte entry, migration, and territoriality in lymph nodes. Immunity. 2006;25(6):989–1001. doi: 10.1016/j.immuni.2006.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Hugues S, Fetler L, Bonifaz L, Helft J, Amblard F, Amigorena S. Distinct T cell dynamics in lymph nodes during the induction of tolerance and immunity. Nature Immunology. 2004;5(12):1235–1242. doi: 10.1038/ni1134. [DOI] [PubMed] [Google Scholar]
- 164.Mempel TR, Henrickson SE, Von Andrian UH. T-cell priming by dendritic cells in lymph nodes occurs in three distinct phases. Nature. 2004;427(6970):154–159. doi: 10.1038/nature02238. [DOI] [PubMed] [Google Scholar]
- 165.Miller MJ, Wei SH, Cahalan MD, Parker I. Autonomous T cell trafficking examined in vivo with intravital two-photon microscopy. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(5):2604–2609. doi: 10.1073/pnas.2628040100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Miller MJ, Wei SH, Parker I, Cahalan MD. Two-photon imaging of lymphocyte motility and antigen response in intact lymph node. Science. 2002;296(5574):1869–1873. doi: 10.1126/science.1070051. [DOI] [PubMed] [Google Scholar]
- 167.Krummel MF, Macara I. Maintenance and modulation of T cell polarity. Nature Immunology. 2006;7(11):1143–1149. doi: 10.1038/ni1404. [DOI] [PubMed] [Google Scholar]
- 168.Sánchez-Madrid F, Del Pozo MA. Leukocyte polarization in cell migration and immune interactions. EMBO Journal. 1999;18(3):501–511. doi: 10.1093/emboj/18.3.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Sánchez-Madrid F, Serrador JM. Bringing up the rear: defining the roles of the uropod. Nature Reviews Molecular Cell Biology. 2009;10(5):353–359. doi: 10.1038/nrm2680. [DOI] [PubMed] [Google Scholar]
- 170.Mcfarland W, Heilman DH. Lymphocyte foot appendage: its role in lymphocyte function and in immunological reactions. Nature. 1965;205(4974):887–888. doi: 10.1038/205887a0. [DOI] [PubMed] [Google Scholar]
- 171.Del Pozo MA, Cabañas C, Montoya MC, Ager A, Sánchez-Mateos P, Sánchez-Madrid F. ICAMs redistributed by chemokines to cellular uropods as a mechanism for recruitment of T lymphocytes. Journal of Cell Biology. 1997;137(2):493–508. doi: 10.1083/jcb.137.2.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Shen B, Fang Y, Wu N, Gould SJ. Biogenesis of the posterior pole is mediated by the exosome/microvesicle protein-sorting pathway. Journal of Biological Chemistry. 2011;286(51):44162–44176. doi: 10.1074/jbc.M111.274803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Taylor RB, Duffus WP, Raff MC, de Petris S. Redistribution and pinocytosis of lymphocyte surface immunoglobulin molecules induced by anti-immunoglobulin antibody. Nature: New biology. 1971;233(42):225–229. doi: 10.1038/newbio233225a0. [DOI] [PubMed] [Google Scholar]
- 174.Kellie S, Patel B, Pierce EJ, Critchley DR. Capping of cholera toxin-ganglioside GM1 complexes on mouse lymphocytes is accompanied by co-capping of alpha-actinin. Journal of Cell Biology. 1983;97(2):447–454. doi: 10.1083/jcb.97.2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Révész T, Greaves M. Ligand-induced redistribution of lymphocyte membrane ganglioside GM1. Nature. 1975;257(5522):103–106. doi: 10.1038/257103a0. [DOI] [PubMed] [Google Scholar]
- 176.Lee JH, Katakai T, Hara T, Gonda H, Sugai M, Shimizu A. Roles of p-ERM and Rho-ROCK signalling in lymphocyte polarity and uropod formation. Journal of Cell Biology. 2004;167(2):327–337. doi: 10.1083/jcb.200403091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Bourguignon LYW, Singer SJ. Transmembrane interactions and the mechanism of capping of surface receptors by their specific ligands. Proceedings of the National Academy of Sciences of the United States of America. 1977;74(11):5031–5035. doi: 10.1073/pnas.74.11.5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Cai Y, Biais N, Giannone G, et al. Nonmuscle myosin IIA-dependent force inhibits cell spreading and drives F-actin flow. Biophysical Journal. 2006;91(10):3907–3920. doi: 10.1529/biophysj.106.084806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Holifield BF, Ishihara A, Jacobson K. Comparative behavior of membrane protein-antibody complexes on motile fibroblasts: Implications for a mechanism of capping. Journal of Cell Biology. 1990;111(6):2499–2512. doi: 10.1083/jcb.111.6.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Schreiner GF, Fujiwara K, Pollard TD, Unanue ER. Redistribution of myosin accompanying capping of surface Ig. Journal of Experimental Medicine. 1977;145(5):1393–1398. doi: 10.1084/jem.145.5.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Shih W, Yamada S. Myosin IIA dependent retrograde flow drives 3D cell migration. Biophysical Journal. 2010;98(8):L29–L31. doi: 10.1016/j.bpj.2010.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Liu B, Dai R, Tian C-J, Dawson L, Gorelick R, Yu X-F. Interaction of the human immunedeficiency virus type 1 nucleocapsid with actin. Journal of Virology. 1999;73(4):2901–2908. doi: 10.1128/jvi.73.4.2901-2908.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Wilk T, Gowen B, Fuller SD. Actin associates with the nucleocapsid domain of the human immunodeficiency virus Gag polyprotein. Journal of Virology. 1999;73(3):1931–1940. doi: 10.1128/jvi.73.3.1931-1940.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Hatch SC, Archer J, Gummuluru S. Glycosphingolipid composition of Human Immunodeficiency Virus type 1 (HIV-1) particles is a crucial determinant for dendritic cell-mediated HIV-1 trans-infection. Journal of Virology. 2009;83(8):3496–3506. doi: 10.1128/JVI.02249-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.McLaughlin S, Murray D. Plasma membrane phosphoinositide organization by protein electrostatics. Nature. 2005;438(7068):605–611. doi: 10.1038/nature04398. [DOI] [PubMed] [Google Scholar]
- 186.Van Den Bogaart G, Meyenberg K, Risselada HJ, et al. Membrane protein sequestering by ionic protein-lipid interactions. Nature. 2011;479(7374):552–555. doi: 10.1038/nature10545. [DOI] [PMC free article] [PubMed] [Google Scholar]


