Abstract
Aspergillus species are industrially and agriculturally important as fermentors and as producers of various secondary metabolites. Among them, fungal polyketides such as lovastatin and melanin are considered a gold mine for bioactive compounds. We used a phylogenomic approach to investigate the distribution of iterative polyketide synthases (PKS) in eight sequenced Aspergilli and classified over 250 fungal genes. Their genealogy by the conserved ketosynthase (KS) domain revealed three large groups of nonreducing PKS, one group inside bacterial PKS, and more than 9 small groups of reducing PKS. Polyphyly of nonribosomal peptide synthase (NRPS)-PKS genes raised questions regarding the recruitment of the elegant conjugation machinery. High rates of gene duplication and divergence were frequent. All data are accessible through our web database at http://metabolomics.jp/wiki/Category:PK.
Keywords: secondary metabolite, polyketide synthase, fungi, database, phylogeny
Background
Polyketide synthase (PKS) genes generate a class of structurally diverse products; they are found in all kingdoms except archaea, which do not synthesize fatty acids.1 While it is difficult to elucidate the biosynthetic mechanisms of each polyketide from only genomic sequences, the recognition of PKS genes is rather straightforward. They exhibit a pronounced domain architecture in common with fatty acyl synthase (FAS) genes, ie, three essential domains (ketosynthase (KS), acyltransferase (AT), and acyl carrier protein (ACP)), and optional tailoring domains such as ketoreductase (KR)-, dehydratase (DH)-, enoylreductase (ER)-, methyltransferase (ME)-, and thioesterase (TE) domains. Based on the domain architecture, research on PKS genes developed the type I, II and III paradigm.2,3 Type I, resembling yeast and animal FAS, is a large multi-domain enzyme that produces a variety of structures (details follow). Type II, resembling plant and bacterial FAS, is a single-module enzyme lacking the AT domain. In vivo, type II enzymes are thought to form multi-enzyme complexes to synthesize aromatic compounds. Type III, known as ‘chalcone synthase-like’, is mainly associated with plants and structurally and mechanistically distinct from type I and II; it does not contain the otherwise essential ACP domain. In this work we focus on type I iterative genes, mostly in fungi but also in other kingdoms.
Type I genes are at least 5 kbp in length and they harbor multiple catalytic domains. Their two classes, modular and iterative, are usually associated with bacteria and fungi, respectively. In bacteria, each domain or a module corresponds to one biosynthetic step and the arrangement of domains in the genome reflects the molecular structure of the final product; this is known as the ‘co-linearity rule’. The modules are not necessarily coded in a single gene; some are separated into multiple open reading frames. Such genes, called type I modular PKS, have been extensively investigated in bacteria.2
In fungi, on the other hand, the same domains are iteratively used in the course of chemical condensation. Such genes are designated type I iterative PKS (iPKS) genes. The control mechanisms that determine the number of iterations are not fully understood.4 Depending on the reduction degree of their final product, type I iterative genes are further classified into three categories, ie, non-reducing-, partially reducing-, and reducing PKS (NR-, PR-, and R-PKS, respectively). Fully reduced polyketides correspond to fatty acid derivatives; this also indicates the relationship between PKS and FAS.
The availability of fully-sequenced genomes provides a new avenue for the study of the evolutionary relationship and diversity of PKS genes in different organisms. Some fungi have type I modular genes that produce diketides and some bacteria possess type I iterative genes.5,6 The existence of nonribosomal peptide synthase (NRPS) genes further complicates the species-metabolite distribution. NRPS genes are megasynthases similar to PKS genes. When a PKS is adjacent to a NRPS, the final product may be a hybrid of polyketide and small peptides. A good example is the mycotoxin alpha-cyclopiazonic acid in Aspergillus species synthesized by the NRPS-PKS gene cpaA.7 While it seems difficult to evolutionarily re-invent the elegant mechanism(s) of PKS tethering, we found NRPS-PKS genes to be polyphyletic (see below).
The full genome sequences of eight Aspergillus species are a rich source of PKS information. These sequences contain hundreds of putative PKS genes but only a handful have been identified experimentally.8,9 The remarkable gene-expression strategy of Ahuja et al10 may increase the speed of identification, but its overall rate remains low. Filamentous fungi have been commercially exploited for centuries in Asian countries, especially Japan.11 Consequently, the prediction of their biosynthetic abilities and mechanisms presents a great challenge for computational biologists.
We created a web database to explore and understand the diversity of iPKS genes in Aspergilli in an effort to answer the following questions: (1) How many PKS genes are found in sequenced Aspergilli? (2) What are their domain structures and types of product? (3) How accurate are computational predictions and what are their limitations? We classified the domain structures of iPKS genes and performed phylogenomic analyses on eight Aspergilli (A. clavatus, -flavus, -fumigatus, -nidulans, -niger, -oryzae, -terreus, and Neosartorya fischeri). Our intent was to provide a data overview via a web-based resource so that users could easily check and reproduce the results of our analyses.
Methods
PKS genes and genome databases
We collected the amino acid sequences in the GenBank Database for eight Aspergillus species (A. clavatus NRRL1, -flavus NRRL3357, -fumigatus A1163/Af293, -nidulans FGSCA4, -niger CBS513.88, -oryzae RIB40, -terreus NIH2624, and Neosartorya fischeri NRRL181 also known as A. fischerianus). To obtain PKS genes in these species, we first availed ourselves of the CADRE database of University Hospital in Manchester, UK (release 3).12 For verification we referenced the AspGD database of the Broad Institute and Stanford University.13 We first collected all PKS sequences of more than 800 amino acid residues that feature KS and AT domains alignable to their consensus sequences. Then we collected candidate sequences of iPKS from over 100 species based on a BLAST search and literature sources (see http://metabolomics.jp/wiki/Category:PK/References).8,14 We also included bacterial iPKS sequences used in recent phylogenetic studies and added 10 FAS genes from higher and lower eukaryotes as an outgroup.4,10,15–21
Throughout our study we referenced several PKS-specific databases for domain information. For functionally identified PKS genes we used PKSDB and ITERDB (National Institute of Immunology, India), which contain information and references on 20 modularly- and 13 iteratively synthesized polyketides, respectively.22,23 The other information source was MapsiDB developed in Korea by SmallSoft Co., Ltd.24 This database provides genomic information on 45 modularly and 21 iteratively synthesized polyketides and is part of the MAPSI (Management and Analysis for Polyketide Synthase Type I) prediction system that is based on the hidden Markov model. We used this tool extensively in our domain predictions.
Assignment of catalytic domains and PKS types
Amino acid sequences were analyzed with MAPSI and the Conserved Domain Database (CDD) from the National Center for Biotechnology Information (NCBI).25 The use of multiple prediction tools is important because MAPSI recognizes neither the ME domain nor domains in NRPS genes. CDD, a general-purpose system not optimized for PKS, is based on a position-specific scoring matrix and yields many false positives. We manually removed unreasonable predictions such as domains unrelated to PKS. After determining the domain composition we assigned each gene to one of 7 PKS types (Table 1). Labels for NRPS-PKS and bacterial iPKS (bMSAS or bPR-PKS) were assigned only when such assignment was supported by the NCBI web resource (especially GenBank) or other literature sources. The remaining 4 types (6-MSAS, NR-PKS, PR-PKS, or R-PKS) were assigned based on literature information or the domain composition predicted by computational tools. Thus, we first assigned the 6-MSAS label to small genes with KS-AT-DH-KR-ACP domains in this order.15,16 Next, the NR-PKS label was assigned to genes without any DH, KR, and ER domains. We assigned the PR-PKS label to genes without the ER domain and the R-PKS label to the remaining genes. Whenever gene functions were experimentally identified we assigned labels according to the biosynthetic role of the genes. Fewer than 20% of all PKS genes, however, have been functionally identified by detailed experiments.
Table 1.
Labels for iterative PKS genes.
| Label | Explanation | Reasoning method |
|---|---|---|
| 6-MSAS | 6-methylsalicylic acid synthase | KS-AT-DH-KR-ACP in this order |
| NR-PKS | Non-reducing | No DH, KR, and ER domains |
| PR-PKS | Partially-reducing | No ER domain and not 6-MSAS type |
| R-PKS | Reducing | With DH, KR, or ER domain but not 6-MSAS type |
| NRPS-PKS | Hybrid synthase with nonribosomal peptide synthase (NRPS) | NCBI annotation and literature |
| bMSAS | Bacterial 6-MSAS | Experimental evidence from the literature |
| bPR-PKS | Bacterial PR-PKS | Experimental evidence from the literature |
| FAS | Fatty acid synthase | Outgroup |
Domain genealogy construction
According to our data curation policy, all genes must contain both KS and AT domains. Multiple alignments of amino acid sequences were performed by CLUSTALW and MUSCLE software embedded in the MEGA program package (Version 5.0) with manual corrections.26 We did not use sequences with incomplete domains. Phylogenetic inference was obtained with neighbor-joining (NJ)- and maximum- parsimony (MP) algorithms with a bootstrap test (1000 pseudo-replicates) in the MEGA package. Visualization was with the TreeDyn software package.27
The concise KS tree was created for a reduced set of domain sequences. After generating the complete tree with all KS domains we one-by-one chose reliable clades with a bootstrap value greater than 75 and fed them into the CD-HIT program to obtain representative sequences for each clade.28 Lastly, the chosen sequences were subjected to the phylogenetic algorithms to form a reduced tree.
Database construction
PKS genes from Aspergilli and related iPKS genes from other fungi and bacteria are registered in our wiki-based database. Each entry contains information on the (1) species, (2) gene product (if identified), (3) GenBank and UniProt identifiers, (4) domains predicted by computational tools, (5) domains based on literature and databases, (6) PKS type; additional information, if any, is also included. The website also provides a summary view that lists genes for each biological species, ie, UniRef grouping,29 PKS type, and domain patterns. The web interface was originally developed for different types of molecular biology data by one of the authors (M.A.).
Results and Discussion
Distribution of PKS-related genes
In total, we collected 400 type I iPKS-related genes from fungi and 71 from bacteria. To our knowledge, our collection is the largest publicly accessible, manually curated data resource focusing on iPKS in Aspergilli. Among fungi, 258 genes were from Aspergilli and Neosartorya. Hereafter, we use only epithets to refer to these species (eg, oryzae instead of A. oryzae). The distribution of gene length roughly followed the normal distribution; the average with standard deviation was 2331 aa ± 561 SD for fungi and 1945 aa ± 735 SD for bacteria. Although our list is not comprehensive in terms of taxonomy, fungi possessed longer iPKS genes than bacteria. Presumably, eubacteria elaborated modular PKS genes whose domains may span multiple open reading frames.
The statistics of the iPKS genes for the eight Aspergillus species (Table 2) are in good agreement with the literature.12–14,30–35 The total number of iPKS in the niger species, which is extensively used in industry, exceeds 40. Around 30 iPKS were found in flavus and its close relatives oryzae and terreus. Their number was fewer (around 15) in saprotrophic species, clavatus and its close relatives fumigatus and N. fischeri. A. nidulans belongs to a distant section (Nidulantes); it harbored around 30 genes.
Table 2.
The number of PKS genes in sequenced Aspergillus species.
| Species | CADRE | Aspergillus book (Ref. 14) | This study | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PKS | NRPS-PKS | Total | PKS | PKS-like NRPS | Total | 6-MSAS | NR-PKS | PR-PKS | R-PKS | PKS total | NRPS-PKS | Total | |
| clavatus | 16 | 3 | 19 | 16 | 1 | 17F | 2 | 3 | 3 | 9 | 17 | 4 | 21 |
| flavus | 25 | 2 | 27 | 35 | – | 35Y | 0 | 14 | 9 | 10 | 33 | 3 | 36 |
| fumigatus A1163 | 11 | 2 | 13 | 13 | 1 | 14F | 1 | 5 | 1 | 5 | 12 | 2 | 14 |
| fumigatus Af293 | 13 | 0 | 13 | 13 | 2 | 15F | 0 | 6 | 3 | 4 | 13 | 2 | 15 |
| fumigatus | – | – | – | – | – | – | 0 | 8 | 0 | 0 | 8 | 0 | 8 |
| nidulans | 26 | 1 | 27 | 27 | – | 27N | 0 | 14 | 3 | 13 | 30 | 1 | 31 |
| niger CBS 513.88 | 13 | 0 | 13 | 34 | 7 | 41P | 1 | 5 | 7 | 25 | 38 | 6 | 44 |
| oryzae | 29 | 0 | 29 | 32 | – | 32R | 1 | 11 | 7 | 10 | 29 | 2 | 31 |
| terreus NIH2624 | – | – | – | – | – | – | 2 | 10 | 4 | 13 | 29 | 1 | 30 |
| terreus | – | – | – | – | – | – | 1 | 3 | 1 | 1 | 6 | 0 | 6 |
| N. fischeri | 15 | 2 | 17 | 17 | 1 | 18F | 2 | 6 | 0 | 7 | 15 | 2 | 17 |
Notes: Numbers include putative genes and are intended as upper bounds. The numbers for the CADRE database were obtained from release 3. For terreus, the database (as of April 2012) did not provide accurate information on PKS genes. Superscripts indicate reference sources: F, Ref. 34; N, Ref. 30; P, Ref. 32; R, Ref. 33; Y, Ref. 31.
Obligate plant pathogens such as Botryotinia, Cochliobolus, and Fusarium (Gibberella) species, all of which, like Aspergillus, are in Pezizomycotina, also harbor many PKS genes. Although few species from these genera have been fully sequenced,36 a comparative study of Fusarium species in Sordariomycetes identified around 15 PKS genes in each species.37 Similarly, Botryotinia fuckeliana in Leotiomycetes had 20-, and Cochliobolus heterostrophus in Dothideomycetes had 25 PKS genes.38 These numbers indicate that saprotrophic Aspergilli and obligate plant pathogens contain a comparable number of iPKS genes and that their number is not directly associated with the fungal lifestyle. This observation is supported by the earlier finding that Actinomycetes bacteria harbor similar numbers of PKS genes, ie, around 20 in Streptomyces coelicolor and 30 in S. avermitilis.39,40 The distribution of PKS types varies extensively inside the Aspergillus genus; saprotrophic species such as clavatus, fumigatus, and N. fischeri do not share many orthologs and even in flavus and oryzae more than 10% of the PKS genes are different (see below). The variation in PKS genes is attributable to subtelomeric rearrangements and their evolutionary history requires a detailed analysis of synteny blocks.
The numbers in Table 2 are not definitive. The statistics are intended as upper bounds and may change with the counting method used. For example, for fumigatus and oryzae, we obtained some information for unknown strains. A small number of orphan data in oryzae (2 genes) were integrated into the data of the sequenced RIB40 strain. On the other hand, we treated the orphan data in fumigatus independently because the remaining data were separated into two sequenced strains (A1163 and Af293) with different PKS composition.
Domain compositions
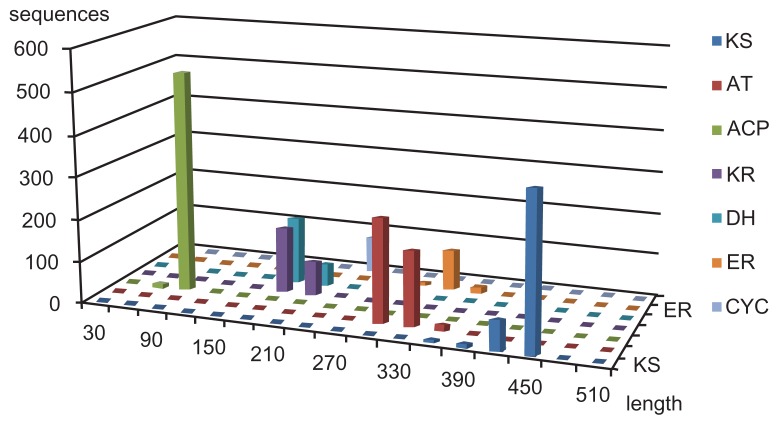
Domains were manually assigned by referencing computational predictions such as MAPSI, ITERDB, and CDD as described in the Methods section. The MAPSI tool can assign 7 domains (KS, AT, ACP, KR, DH, ER, and CYC) and CDD can assign more than 20 different domains. Information on predicted domains for all sequences is freely accessible through our wiki-based database at http://metabolomics.jp/wiki/Category:PK by following links at the top of the page. The classification of PKS types was based on the literature and on the database annotation as described (Table 1). The 6-MSAS type was assigned for small genes with the domain structure KS-AT-DH- KR-ACP.15,16 Although the assignment may look simplistic, our phylogenetic analysis supports this definition (see the concise phylogenetic trees shown in Figs. 1 and 2). The remaining iPKS genes were assigned to the nonreducing (NR)-, partially reducing (PR)-, or reducing (R) type depending on the existence of reducing domains and literature information, if any (see Methods). Since the number of functionally identified genes is small, their categorization largely depends on the predictions by the MAPSI tool which is specialized for type I PKS. We listed results of both MAPSI and CDD in our website entries and added more entries if literature information was available. The statistics of the MAPSI output are shown in Fig. 3. Among the 7 assigned domains, KS was the longest (422 aa ± 21 SD), followed by ER (320 aa ± 9 SD), and AT (300 aa ± 13 SD). The number of the inessential ER domain is only one quarter of KS. Some reducing PKS genes in bacteria did not have an ER domain; such exceptions were rare in fungi.
Figure 1.
The phylogenetic tree of the KS domain for representative sequences (ie, the concise tree) computed with the maximum parsimony method.
Notes: There are three large categories: R-PKS, bacterial PKS, and NR-PKS. R-PKS is separated into many small groups. Bacterial PKS contains the Nested Fungal Clade (NFC; see text). NR-PKS contains three large groups. The tree topology is slightly different from the full tree containing 471 sequences due to the selection of sequences. Also see the Methods section for the selection method of sequences.
Figure 2.
Overview of the KS domain phylogeny.
Figure 3.
Statistics of the number of domains and their length predicted by the MAPSI tool.
Notes: The horizontal axis is the amino acid length and the vertical axis is the number of sequences. The KS domain is the longest and also the best conserved.
The statistics of the domain length justify our decision to consider only sequences of more than 800 aa as residues for PKS. In fact, the CADRE database contains shorter PKS genes, but their domains are partial and probably not functional. Such genes were excluded from our analysis and from Table 2.
Since some PKS genes exploit external genes, the assigned types may not always correspond to the chemical type of their products. For example, the lovastatin synthase in terreus requires an external lovC gene with an ER domain. Such limitations of the computational tools can be assessed by cross comparisons. We found that MAPSI cannot assign ME or TE domains and tends to ignore domains after the first appearance of ACP. Especially the Rossmannfold NAD(P)(+)-binding domain in the C-terminal of PKS tends to be misidentified as a KR- rather than a TE domain. Also, many ME domains are not functional and their verification is difficult. For these reasons we used the CDD output only as a reference to check for the possible existence of ME-, TE-, and other domains. Nevertheless, we identified several global characteristics. First, close to 30% of all PKS genes show the simple KS-AT-ACP-(ACP)-(CYC) composition for NR-PKS. A. flavus and nidulans contain more NR-PKS genes than do others, while niger contains the largest number of R-PKS genes. Second, the number of NRPS-PKS differs greatly among species. While niger contains as many as 7, terreus and nidulans has only one. Since the annotation for NRPS-PKS is based on literature sources and not on predictions, the difference may reflect the extent of research in each species. Still, the large difference suggests species-specific biases, a hypothesis that is supported by the observation that extremely close relatives such as oryzae and flavus yielded different statistics with respect to the category of PKS genes.
Phylogenomic analysis of KS domains
The KS region is the best conserved domain. Phylogenetic estimates on KS only coincide with results that are based on combined KS and AT domains. The only exceptions are PKS genes from the protozoa Cryptosporidium parvum; they manifest a non-standard evolutionary trace for the AT domain.18 Figs. 1 and 2 provide a concise view of our phylogenetic analysis on KS domains; Supplementary Fig. S1 shows details.
Phylogenetically, NR-PKS, (P)R-PKS, and 6-MSAS types clustered well.38 The NR-PKS type is adjacent to bacterial PKS genes, many of which are involved in phenolpthiocerol synthesis. There are two fungal clades deep inside bacterial PKS (called the nested fungal clade or NFC); one is for NRPS-PKS and the other, larger one, for 6-MSAS. The two NFCs were statistically supported to have originated in actinobacteria and to have been horizontally transferred later into ascomycete fungi.16,38,41
The NR-PKS type is separated into three classes (I–III in Fig. 1). The NR-PKS I type contains genes for aflatoxins, melanins, pigments, and naphthopyrones. NR-PKS III type contains one gene for citrinin (pksCT in Monascus purpureus), but the function of most other genes remains unknown. NR-PKS II type is a small class of genes of unknown function. The higher relationship among NR-PKS, R-PKS, 6-MSAS, and bacterial PKS types was different from that reported in earlier studies,17,38 as was the topological position of FAS (Figs. 1 and 2). The higher part was susceptible to the number of genes and also the clustering method. In our analysis, the NJ method tended to place the outgroup FAS between R-PKS and NR-PKS, whereas the MP method did not. The relationship within each gene-type was much better conserved in all phylogenomic studies.17–19,38,41
In contrast to the well-clustered NR-PKS genes, R-PKS and 6-MSAS types were separated into many small groups. The R-PKS type contains the genes for fumonisin, lovastatin, and t-toxin but few genes have been experimentally verified. A recent excellent work identified a number of new products for NR-PKS genes in nidulans;10 such identification would be more difficult for reducing PKS genes. An interesting finding was polyphyly of the NRPS-PKS type; this is in contrast with observations derived from much wider studies focusing on NRPS.41,42 In our study, the KS sequences for NRPS-PKS co-occurred with those for 6-MSAS type in clades for equisetin, pseurotin A, and cyclopiazonic acid.43 While this observation may be attributable to incomplete experimental evidence or to incomplete resolution of the KS phylogeny, it points to an evolutionary and mechanistic relationship of the hybrid genes with the 6-MSAS type. This also justifies our curation process in which we did not annotate hybrid PKS only from sequence similarity. Among such hybrid genes, NRPS7/PKS24 in Cochliobolus heterostrophus (AAR90278) drew significant attention as the model HGT target.41,44 This hybrid gene is similar to a putative PR-PKS gene in Chaetomium globosum (XP_001224327) and a NRPS-PKS gene in niger (AM270324). All three sequences are grouped inside the NFC. Lawrence et al44 reconfirmed the simultaneous transfer of the NRPS and PKS domains for the smaller NFC in the early evolutionary phase of Pezizomycotina (euascomycota), ie, after divergence of the saccharomyces group. The other, larger NFC is expected to be equally ancient; it includes pksPN (AAP33839) for ochratoxin A (OTA) biosynthesis in Penicillium nordicum and a putative aflatoxin gene pksL2 (AAC23536) in parasiticus. The involvement of pksL2 in aflatoxin biosynthesis is esoteric because all aflatoxin-related genes belong to the NR-PKS group. Additional evidence is required to detect its true function. On the other hand, investigations into pksPN are a worthwhile challenge. The OTA-PKS of P. nordicum is most similar to the 6-MSAS type and requires an NRPS to catalyze the ligation of phenylalanine. However, a different biosynthetic pathway may exist in ochratoxigenic Aspergillus species.45 Our phylogenetic analysis based on the KS domain indicates such an alternative: the OTA-PKS of A. ochraceus is similar to R-PKS, not 6-MSAS. Earlier studies of ochraceus also support such alternative routes using different domains.46,47 The simultaneous transfer of NRPS and PKS, and the subsequent neofunctionalization as OTA synthase is an intriguing hypothesis that we intend to test in the future. As more sequences are obtained, similar analyses can be performed for equisetin-, pseurotin A-, and cyclopiazonic acid-related genes in Aspergilli to clarify the origin of NRPS-PKS hybrids in relation to 6-MSAS types.
Orthologous genes
Phylogenetic analysis yields information on orthologous genes. We used ‘orthology’ loosely to describe genes with identical domain structures with highly similar KS sequences. Such orthologs are expected to synthesize polyketides with identical backbones, although their structural modifications may vary depending on the external modifier genes or ecological usage. Surprisingly, only two orthologous genes were conserved by all sequenced Aspergilli (Table 3), ie, the R-PKS gene represented by the UniRef identifiers Q8TGD6 and A1CVN0, and the 6-MSAS type gene, including the lovB-like gene in fumigatus (XP_751268; UniRef50_Q4WLD4). The former orthologous class encodes the conidial pigment emodin. The latter is a less conserved 6-MSAS type and its final product is unknown. Several other orthologs were shared among multiple but not all species (Table 3). For example, the R-PKS V group for lovastatin and t-toxin biosynthesis does not include fumigatus and N. fischeri but includes clavatus (Table 3). The NR-PKS I(7) group for the biosynthesis of naphthopyrone pigment was highly conserved (more than 50% of amino acids) but terreus lacks this gene. Such sporadic distribution suggests a significant shuffling of PKS genes without severe environmental selection pressure. This hypothesis is supported by the observation that in certain species such conserved groups always contain duplicate genes (paralogs). One evolutionarily recent example is the pseurotin A gene in fumigatus (UniRef50_Q4WAZ9) in the NRPS-PKS type. Another recent event is the presence of lovB genes in terreus (UniRef50_Q0C8M3). Although not highly similar, nidulans has three copies of 6-MSAS type genes of unknown function (Uni-Ref50_C5FNM3). Such distant duplications are most evident in niger, in which at least four duplicates are present (UniRef50_A2QDU0, G7XYT5, A2R4Z4, A2QW54) in 8 chromosomes (nidulans, fumigatus, flavus, and oryzae have 8 chromosomes in haploid cells). Frequent duplication and the absence of conserved orthologs both support active genome rearrangement in Aspergilli. This observation is in good contrast with findings in Fusarium species, which lack apparent PKS duplications, presumably due to repeat-induced point mutations (RIP).37
Table 3.
Well conserved orthologous genes in Aspergillus species.
| Type | clavatus | flavus | fumigatus A1163 | fumigatus Af293 | nidulans | niger | oryzae | terreus | fischeri |
|---|---|---|---|---|---|---|---|---|---|
| 6-MSAS I | A1CLJ5 | B8NPY7 | Q4WLD4 | Q4WLD4 | Q5BCE6 | An11g09720 An04g04340 |
AO090102000166 | Q0C9L7 | Q4WLD4 |
| R-PKS I(1) | Q4WAY3 | Q4WAY3 | Q5AX96 | AO090011000015 | Q0CNL6 |
Q0CNL6 A1DA81 |
|||
| R-PKS V (lovastatin and t-toxin) | C5FYS0 A1CD09 |
B8NFE8 |
Q5BEJ4 B0XZN7 Q5AY39 |
An09g01930 | AO090701000826 |
Q8J0F5 Q0C8L6 Q0CF75 |
|||
| R-PKS IV | P0CJ32 | P0CJ32 | P85915 | P85915 | AO090009000052 | Q0CB46 | P85915 | ||
| bacMSAS 6-MSAS (NFC) | P22367 | B8NYX0 | An10g00140 | AO090206000074 |
P22367 P22367 |
||||
| NR-PKS I(2) | Q8TGD6 | P0C8J3 |
Q8TGD6 B0Y591 |
Q8TGD6 A1CVN0 |
A1CVN0 | An04g09530 | AO090023000444 |
Q8TGD6 Q8TGD6 A1CVN0 |
Q8TGD6 A1CVN0 A1CVN0 |
| NR-PKS I(7) Pigment-I |
Q03149 | Q03149 | Q03149 | Q03149 |
Q03149 Q03149 |
An09g05730 | Q03149 | Q03149 | |
| NR-PKS I(3) | Q2U886 | Q4WA61 | Q4WA61 |
Q2U886 D7PHZ2 |
D7PHZ2 | Q2U886 | Q4WA61 |
Notes: This putative orthology was determined by the KS sequences and not by their gene structures. Large groups covering more than 5 species with more than 75 bootstrapping values are listed with duplicate genes. The 6-digit alphanumeric codes are UniRef50 identifiers (as of April 2012). The longer codes for niger and oryzae are standard genomic IDs provided by their sequencing communities, which are also accessible at our website. Bold identifiers are referred to in the main text.
Flexible rearrangements are also evidenced upon comparison of close pairs, ie, flavus and oryzae, or the two strains of fumigatus (Af293 and A1163). The industrially important oryzae and the pathogenic and toxic flavus both belong to section Flavi and are indistinguishable by DNA reassociation kinetics (Cot analysis) or from ribosomal DNA fragments.48 In fact, oryzae is considered a domesticated ecotype of an aflatoxin-producing, ancestral flavus strain.33 In the course of domestication the genome size of oryzae increased, the organism lost the ability to produce secondary metabolites including aflatoxin, and it acquired the ability to use carbohydrate(s). The number of PKS genes changed accordingly. Of the 31 (32) PKS genes in oryzae (flavus), 27 showed clear orthology with short evolutionary distances. In other words, more than 10% are functionally different. Since the total number of PKS genes is not different in oryzae and flavus, the seemingly unrelated genes may share common origins. However, the elucidation of this issue requires detailed information at a chromosomal level (gene clusters) and is beyond the scope of this work. Likewise, the two fumigatus strains show some differences. Two PKS genes were not orthologous (XP 749851 and XP 748578 in Af293; EDP53389 and EDP49937 in A1163) and the degree of orthology was also often not close enough for the remaining genes (Supplementary Fig. S1). A similar observation with respect to differing numbers of PKS genes was recently reported within the niger strains.49
Web-based database system
When sequences of more than 400 genes are involved, it is difficult to obtain detailed information for each sequence from published research only. Most genomic information is available on the Internet, eg, from NCBI and EBI repositories. Thus, a web-based database is the best interface to fully utilize the results of phylogenomic analyses. We created a website for PKS information in Aspergilli; users have full access to sequence data and experimental, referenced annotations and classifications. For the best results, the website is maintained by a wiki-based system and each entry can be asynchronously updated through a web browser by registered users. As more systematic studies on iPKS genes are presented,10,35 we expect to elaborate our analysis through such rapidly accumulating information. To maintain the quality of academic information we do not allow publicly free edits. Collaborating users are requested to contact the author (M.A.) to obtain a login ID.
Supplementary Data
Supplementary Sequence Data for 471 genes: Information on predicted domains and species.
Supplementary FASTA format of the 43 KS clusters: Information on aligned groups generated by the Geneious software program (basic version v5.63).
Acknowledgements
We thank Prof. Isao Fujii (Iwate Medical University, Japan) for comments and kind suggestions on our manuscript. We also thank Ms. Ursula Petralia for editing our manuscript.
Footnotes
Funding Sources
This research was supported by a MEXT Grant-in-Aid for Scientific Research on Innovative Areas “Biosynthetic Machinery” 11001359. The early part of the research was carried out at The University of Tokyo when SHL was a visiting student in Japan. His sojourn was financially supported by the National Security Council of Taiwan under Grant No. NSC096- 2917-I-007-005 and by the Elite Scholarship Program, Ministry of Education, Taiwan.
Competing Interests
MA’s institution received a grant for JST-NSF joint research on metabolomics. Other authors disclose no potential conflicts of interest.
Author Contributions
Conceived and designed the experiments: SHL, MA. Analyzed the data: SHL, MY, MA. Supervised the data analysis of SHL: PCL, CYT, MA. Wrote the first draft of the manuscript: SHL, MA. Agreed with manuscript results and conclusions: SHL, PCL, CYT, MA. Made critical revisions and approved final version: SHL, MA. All authors reviewed and approved the final manuscript.
Disclosures and Ethics
As a requirement of publication, the author(s) have provided to the publisher signed confirmation of compliance with legal and ethical obligations including but not limited to the following: authorship and contributorship, conflicts of interest, privacy and confidentiality, and (where applicable) protection of human and animal research subjects. The authors have read and confirmed their agreement with the ICMJE authorship and conflict of interest criteria. The authors have also confirmed that this article is unique and not under consideration or published in any other publication, and that they have permission from rights holders to reproduce any copyrighted material. Any disclosures are made in this section. The external blind peer reviewers report no conflicts of interest.
References
- 1.Cavicchioli R, editor. Archaea: Molecular and cellular biology. Washington DC: American Society for Microbiology Press; 2007. [Google Scholar]
- 2.Staunton J, Weissman KJ. Polyketide biosynthesis: A millennium review. Nat Prod Rep. 2001;18:380–416. doi: 10.1039/a909079g. [DOI] [PubMed] [Google Scholar]
- 3.Shen B. Polyketide biosynthesis beyond the type I, II and III polyketide synthase paradigms. Curr Opin Chem Biol. 2003;7:285–95. doi: 10.1016/s1367-5931(03)00020-6. [DOI] [PubMed] [Google Scholar]
- 4.Li Y, Xu W, Tang Y. Classification, prediction, and verification of the regioselectivity of fungal polyketide synthase product template domains. J Biol Chem. 2010;285:22764–73. doi: 10.1074/jbc.M110.128504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gaisser S, Trefzer A, Stockert S, Kirschning A, Bechthold A. Cloning of an avilamycin biosynthetic gene cluster from Streptomyces viridochromogenes Tu57. J Bacteriol. 1997;179:6271–8. doi: 10.1128/jb.179.20.6271-6278.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Whitwam RE, Ahlert J, Holman TR, Ruppen M, Thorson JS. The gene calC encodes for a non-heme iron metalloprotein responsible for calicheamicin self-resistance in Micromonospora. J Am Chem Soc. 2000;122:1556–7. [Google Scholar]
- 7.Tokuoka M, Seshime Y, Fujii I, Kitamoto K, Takahashi T, Koyama Y. Identification of a novel polyketide synthase-nonribosomal peptide synthetase (PKS-NRPS) gene required for the biosynthesis of cyclopiazonic acid in Aspergillus oryzae. Fungal Genet Biol. 2008;45:1608–15. doi: 10.1016/j.fgb.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 8.Varga J, Samson RA, editors. Aspergillus in the genomic era. The Netherlands: Wageningen Academic Publishers; 2008. [Google Scholar]
- 9.Fujii I. Functional analysis of fungal polyketide biosynthesis genes. J Antibiotics. 2010;63:207–18. doi: 10.1038/ja.2010.17. [DOI] [PubMed] [Google Scholar]
- 10.Ahuja M, Chiang YM, Chang SL, et al. Illuminating the diversity of aromatic polyketide synthases in Aspergillus nidulans. J Am Chem Soc. 2012;134:8212–21. doi: 10.1021/ja3016395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Machida M, Yamada O, Gomi K. Genomics of Aspergillus oryzae: Learning from the history of koji mold and exploration of its future. DNA Res. 2008;15:173–83. doi: 10.1093/dnares/dsn020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mabey GJ, Cooley J, Bowyer P. CADRE: The Central Aspergillus Data REpository 2012. Nucleic Acids Res. 2012;40:D660–6. doi: 10.1093/nar/gkr971. The database is available at: http://www.cadre-genomes.org.uk. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arnaud MB, Chibucos MC, Costanzo MC, et al. The Aspergillus Genome Database, a curated comparative genomics resource for gene, protein and sequence information for the Aspergillus research community. Nucleic Acids Res. 2010;38:D420–7. doi: 10.1093/nar/gkp751. The database is available at: http://www.aspgd.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Machida M, Gomi K, editors. Aspergillus: Molecular Biology and Genomics. Norfolk UK: Caister Academic Press; 2010. [Google Scholar]
- 15.Schmitt I, Kautz S, Lumbsch HT. 6-MSAS-like polyketide synthase genes occur in lichenized ascomycetes. Mycol Res. 2008;112:289–96. doi: 10.1016/j.mycres.2007.08.023. [DOI] [PubMed] [Google Scholar]
- 16.Schmitt I, Lumbsch HT. Ancient horizontal gene transfer from bacteria enhances biosynthetic capabilities of fungi. PLoS One. 2009;4:e4437. doi: 10.1371/journal.pone.0004437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Varga J, Rigó K, Kocsubé S, Farkas B, Pál K. Diversity of polyketide synthase gene sequences in Aspergillus species. Res Microbiol. 2003;154:593–600. doi: 10.1016/S0923-2508(03)00169-4. [DOI] [PubMed] [Google Scholar]
- 18.Castoe TA, Stephens T, Noonan BP, Calestani C. A novel group of type I polyketide synthases (PKS) in animals and the complex phylogenomics of PKSs. Gene. 2007;392:47–58. doi: 10.1016/j.gene.2006.11.005. [DOI] [PubMed] [Google Scholar]
- 19.John U, Beszteri B, Derelle E, et al. Novel insights into evolution of protistan polyketide synthases through phylogenomic analysis. Protist. 2008;159:21–30. doi: 10.1016/j.protis.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 20.Valarmathi R, Hariharan GN, Venkataraman G, Parida A. Characterization of a non-reducing polyketide synthase gene from lichen Dirinaria applanata. Phytochem. 2009;70:721–9. doi: 10.1016/j.phytochem.2009.04.007. [DOI] [PubMed] [Google Scholar]
- 21.Fisch KM, Gillaspy AF, Gipson M, et al. Chemical induction of silent biosynthetic pathway transcription in Aspergillus niger. J Ind Microbiol Biotechnol. 2009;36(9):1199–213. doi: 10.1007/s10295-009-0601-4. [DOI] [PubMed] [Google Scholar]
- 22.Yadav G, Gokhale RS, Mohanty D. Computational approach for prediction of domain organization and substrate specificity of modular polyketide synthases. J Mol Biol. 2003;328:335–63. doi: 10.1016/s0022-2836(03)00232-8. [DOI] [PubMed] [Google Scholar]
- 23.Ansari MZ, Yadav G, Gokhale RS, Mohanty D. NRPS-PKS: A knowledge-based resource for analysis of NRPS/PKS megasynthases. Nucleic Acids Res. 2004;32:W405–13. doi: 10.1093/nar/gkh359. Tools are available at: http://www.nii.ac.in/~pksdb/sbspks/master.html. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tae H, Sohng JK, Park K. MapsiDB: An integrated web database for type I polyketide synthases. Bioprocess Biosyst Eng. 2009;32:723–7. doi: 10.1007/s00449-008-0296-3. The database is available at: http://gate.smallsoft.co.kr:8008/~hstae/pks/mapsidb/ [DOI] [PubMed] [Google Scholar]
- 25.Marchler-Bauer A, Panchenko AR, Shoemaker BA, Thiessen PA, Geer LY, Bryant SH. CDD: A database of conserved domain alignments with links to domain three-dimensional structure. Nucleic Acids Res. 2002;30(1):281–3. doi: 10.1093/nar/30.1.281. The database is available at: http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. Mega5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–9. doi: 10.1093/molbev/msr121. The software is available at: http://www.megasoftware.net/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chevenet F, Brun C, Banuls AL, Jacq B, Christen R. TreeDyn: Towards dynamic graphics and annotations for analyses of trees. BMC Bioinformatics. 2006;7:439. doi: 10.1186/1471-2105-7-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang Y, Niu B, Gao Y, Fu L, Li W. CD-HIT Suite: A web server for clustering and comparing biological sequences. Bioinformatics. 2010;26:680–2. doi: 10.1093/bioinformatics/btq003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suzek BE, Huang H, McGarvey P, Mazumder R, Wu CH. UniRef: Comprehensive and non-redundant UniProt reference clusters. Bioinformatics. 2007;23:1282–8. doi: 10.1093/bioinformatics/btm098. [DOI] [PubMed] [Google Scholar]
- 30.Nierman WC, Pain A, Anderson MJ, et al. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature. 2005;438:1151–6. doi: 10.1038/nature04332. [DOI] [PubMed] [Google Scholar]
- 31.Payne GA, Nierman WC, Wortman JR, et al. Whole genome comparison of Aspergillus flavus and A. oryzae. Med Mycol. 2006;44(s1):9–11. doi: 10.1080/13693780600835716. [DOI] [PubMed] [Google Scholar]
- 32.Pel HJ, de Winde JH, Archer DB, et al. Genome sequencing and analysis of the versatile cell factory Aspergillus niger cbs 513.88. Nat Biotechnol. 2007;25(2):221–31. doi: 10.1038/nbt1282. [DOI] [PubMed] [Google Scholar]
- 33.Rokas A, Payne G, Fedorova ND, et al. What can comparative genomics tell us about species concepts in the genus Aspergillus? Stud Mycol. 2007;59:11–7. doi: 10.3114/sim.2007.59.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fedorova ND, Khaldi N, Joardar VS, et al. Genomic islands in the pathogenic filamentous fungus Aspergillus fumigatus. PLoS Genet. 2008;4(4):e1000046. doi: 10.1371/journal.pgen.1000046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nielsen ML, Nielsen JB, Rank C, et al. A genome-wide polyketide synthase deletion library uncovers novel genetic links to polyketides and meroterpenoids in Aspergillus nidulans. FEMS Microbiol Lett. 2011;321(2):157–66. doi: 10.1111/j.1574-6968.2011.02327.x. [DOI] [PubMed] [Google Scholar]
- 36.Cuomo CA, Güldener U, Xu JR, et al. The Fusarium graminearum genome reveals a link between localized polymorphism and pathogen specialization. Science. 2007;317:1400–2. doi: 10.1126/science.1143708. [DOI] [PubMed] [Google Scholar]
- 37.Brown DW, Butchko RA, Baker SE, Proctor RH. Phylogenomic and functional domain analysis of polyketide synthases in Fusarium. Fungal Biol. 2012;116(2):318–31. doi: 10.1016/j.funbio.2011.12.005. [DOI] [PubMed] [Google Scholar]
- 38.Kroken S, Glass NL, Taylor JW, Yoder OC, Turgeon BG. Phylogenomic analysis of type I polyketide synthase genes in pathogenic and saprobic ascomycetes. Proc Natl Acad Sci U S A. 2003;100:15670–5. doi: 10.1073/pnas.2532165100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bentley SD, Chater KF, Cerdeno-Tárraga AM, et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2) Nature. 2002;417:141–7. doi: 10.1038/417141a. [DOI] [PubMed] [Google Scholar]
- 40.Ikeda H, Ishikawa J, Hanamoto A, et al. Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat Biotechnol. 2003;21:526–31. doi: 10.1038/nbt820. [DOI] [PubMed] [Google Scholar]
- 41.Bushley KE, Turgeon BG. Phylogenomics reveals subfamilies of fungal nonribosomal peptide synthases and their evolutionary relationships. BMC Evol Biol. 2010;10:26. doi: 10.1186/1471-2148-10-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cramer RA, Jr, Stajich JE, Yamanaka Y, Dietrich FS, Steinbach WJ, Perfect JR. Phylogenomic analysis of non-ribosomal peptide synthetases in the genus Aspergillus. Gene. 2006;383:24–32. doi: 10.1016/j.gene.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 43.Maiya S, Grundmann A, Li X, Li SM, Turner G. Identification of a hybrid PKS/NRPS required for pseurotin A biosynthesis in the human pathogen Aspergillus fumigatus. Chembiochem. 2007;8:1736–43. doi: 10.1002/cbic.200700202. [DOI] [PubMed] [Google Scholar]
- 44.Lawrence DP, Kroken S, Pryor BM, Arnold AE. Interkingdom gene transfer of a hybrid NRPS/PKS from bacteria to filamentous ascomycota. PLoS One. 2011;6:e28231. doi: 10.1371/journal.pone.0028231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huffman J, Gerber R, Du L. Recent advancements in the biosynthetic mechanisms for polyketide-derived mycotoxins. Biopolymers. 2010;93:764–76. doi: 10.1002/bip.21483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.O’Callaghan J, Dobson ADW. Phylogenetic analysis of polyketide synthase genes from Aspergillus ochraceus. Mycotoxin Res. 2006;22:125–33. doi: 10.1007/BF02956776. [DOI] [PubMed] [Google Scholar]
- 47.Atoui A, Dao HP, Mathieu F, Lebrihi A. Amplification and diversity analysis of ketosynthase domains of putative polyketide synthase genes in Aspergillus ochraceus and Aspergillus carbonarius producers of ochratoxin A. Mol Nutr Food Res. 2006;50(6):488–93. doi: 10.1002/mnfr.200500165. [DOI] [PubMed] [Google Scholar]
- 48.Lee CZ, Liou GY, Yuan GF. Comparison of Aspergillus flavus and Aspergillus oryzae by amplified fragment length polymorphism. Bot Bull Acad Sin. 2004;45:61–8. [Google Scholar]
- 49.Ferracin LM, Fier CB, Vieira ML, et al. Strain-specific polyketide synthase genes of Aspergillus niger. Intern J Food Microbiol. 2012;155:137–45. doi: 10.1016/j.ijfoodmicro.2012.01.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.