Abstract
MicroRNAs (miRs) are small non-coding RNAs implicated mainly in post-transcriptional gene silencing by interacting with the unstranslated region of the transcript. miR-210 represents a major hypoxia-inducible miRs, also known as hypoxamirs, which is ubiquitously expressed in a wide range of cells, serving versatile functions. This review article summarizes the current progress on biogenesis of miR-210 and its physiological roles including arrest of cell proliferation, repression of mitochondrial respiration, arrest of DNA repair, vascular biology, and angiogenesis. Given the fact that miR-210 is aberrantly expressed in a number of diseases such as tumor progression, myocardial infarction and cutaneous ischemic wounds, miR-210 could serve as an excellent candidate for prognostic purposes and therapeutic intervention. With the advancement of computational prediction, high-throughput target validation methodology, sequencing, proteomic analysis and microarray, it is anticipated that more down-stream targets of miR-210 and its-associated biological consequences under hypoxia will be unveiled establishing miR-210 as a major hub in the biology of hypoxia-response.
Keywords: miR-210, hypoxamiRs, microRNAs, tissue repair
INTRODUCTION
MicroRNAs (miRs) are small RNAs consisting of around 22 nucleotides. Unlike protein-coding genes, miRs exhibit extraordinary gene regulatory functions, silencing gene expression via interaction with the 3′ unstranslated region (3′ UTR) of the transcript [3,8,12,44,74,81,82,85,87,88,91,90]. The biogenesis of miRs is a highly-orchestrated process which essentially requires the co-ordination of ribonucleases, RNA-binding proteins and the miR gene itself [93]. Transcription and appropriate truncation of the nucleic acid, which is known as miR maturation, are vital for the synthesis of correct miR strand. Certain key proteins such as Drosha, DiGeorge critical region 8 (DGCR8), exportin-5, Dicer and Ago2 are critical for this process. Any disruption or inactivation of these molecules results in pathological outcomes or developmental defects [56,101]. The details of miR maturation have been reviewed elsewhere and will not be discussed in this article [91,93].
Over decades, induction of protein coding genes by low oxygen has dominated the focal point of hypoxia research. One of the most sensitive physiological sensors of hypoxia is hypoxia inducible factor (HIF). HIFs control the cellular response to hypoxia by regulating genes that are involved in metabolism, angiogenesis, erythropoiesis, cell proliferation, differentiation and apoptosis. Although three isoforms of HIF have been identified, HIF1α and HIF2α are the most dominant sensors of hypoxia [48]. When oxygen tension falls below the normoxic setpoint for any given tissue [50], HIF1α is stabilized and binds to its more constitutive partner HIF1β, and this complex regulates the expression of downstream genes [48]. HIF transactivates a wide variety of genes involved in the hypoxia response, some of the most noted ones being erythropoietin (EPO), vascular endothelial growth factor (VEGF) and glucose transporter 1 (GLUT1) [72]. More recently, however, the study of gene regulation promoted by a low oxygen microenvironment has received increased attention. The regulation is under the control of specific hypoxia-inducible miRs also termed as “hypoxamiRs”. Some of the reported hypoxamiRs are summarized in Table 1.
Table 1.
Summary of hypoxamiRs
| Name of microRNAs | Reference | |
|---|---|---|
| Hypoxia up-regulated miRs | miR-21, -23a, -23b, -24, -26a, -26b, -27a, -30b, -93, -103, -103, -106a, -107, -125b, -181a, -181b, -181c, -192, -195, -210, -213, -429, -498, -572, -563, -637 and -628 | [25, 41, 45, 83] |
| Hypoxia down-regulated miRs | miR- 15b, -16, -19a, -20a, -20b, -29b, -30b, -30e-5p, -101, -122a, -141, -186, -195, -197, -200b, -224, -320, -374, -422b, -424, and -565 | [12, 41, 78] |
mIR-210: GENETIC LOCUS AND PROMOTER
MiR-210 is a master hypoxamir which is induced under hypoxia in wide range of primary and transformed cells [20]. The stem–loop of miR-210 is located in an intron of a noncoding RNA, which is transcribed from AK123483 on chromosome 11p15.5 [41]. MiR-210 is regulated by both HIF1α [9,40,41] and HIF2α [104]. HIF1α directly binds to a hypoxia responsive element (HRE) on the proximal miR-210 promoter, located 400 bp upstream of the structure [41]. The HRE of miR-210 promoter is highly conserved across species, suggesting that HIF is phylogenetically conserved in regulation of miR-210 de novo synthesis under hypoxia. Apart from HIF, nuclear factor κB (NFκB), a hypoxia-sensitive transcription factor [18], is also responsible for miR-210 induction in responsive to hypoxia. Mapping of 200-bp core promoter region immediately upstream of miR-210 stem-loop structure indicated a conserved κB binding site [103]. Chromatin immunoprecipitation, promoter luciferase assay, gene knockdown studies revealed that NFκB p50 can physically interact with and transactivate miR-210 promoter under hypoxia [103]. Recent study also reported that Akt activation facilitates the hypoxia-associated accumulation of miR-210 in a HIF-independent manner [69], indicating that multiple circuits of signaling can switch on miR-210 in responsive to low oxygen condition.
MIR-210 AND CELL GROWTH ARREST
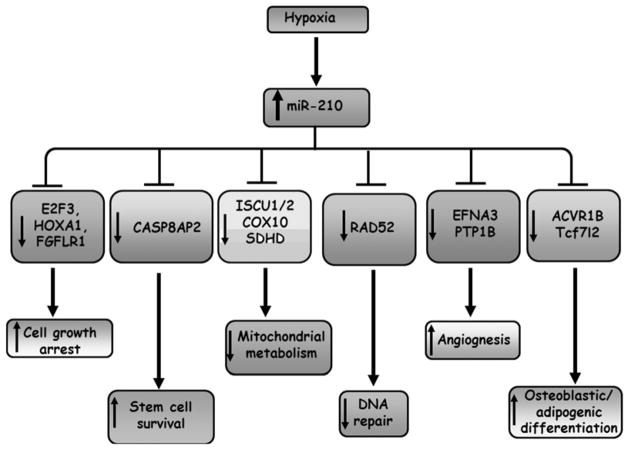
MiR-210 inhibits cell proliferation by targeting proteins that are crucial for cell cycle progression. A number of studies reported that hypoxia-driven miR-210 directly targets E2F3 in a wide variety of cells such as keratinocytes [8], ovarian cancer cells [31], and human embryonic kidney (HEK) cells [70]. E2F3 belongs to the E2F family transcription factor involved in regulation of cell proliferation, differentiation and apoptotic response [35,58]. It is well-documented that E2F3 promotes cell proliferation by allowing the cell cycle progression from G1 to S phase and the initiation of DNA replication [22,57,96]. Recently, miR-210 has been shown to target two other proteins, namely fibroblast growth factor receptor like 1 (FGFRL1) [40,95] and homeobox A1 (HOXA1) [40] to modulate cell proliferation. FGFRL1 is the fifth FGFR family [100] which contains similar structure of extracellular-transmembrane domain to other FGFR family members but lacks the intracellular protein tyrosine domain [92]. FGFRL1 promotes proliferation in esophageal squamous cell carcinoma cells by facilitating cell cycle progression [95]. More importantly, over-expression of FGFRL1 significant rescued the growth inhibitory effect of miR-210 in vitro [95] and in vivo tumor xenograft [40], suggesting that miR-210 inhibits cell proliferation via a FGFRL1-dependent mechanism. HOXA1 is one of the members of the homeobox protein cluster A which is essential in patterning the early hindbrain along the anterior-posterior axis during development [29]. It is highly expressed in a wide variety of cells including mammary epithelial cells, esophageal squamous cells and cervical cancer cells. E-cadherin signaling induces HOXA1 expression, which subsequently promotes anchorage-dependent growth [102]. Forced over-expression of HOXA1 induced activation of p44/42 MAP kinase, supporting cell proliferation [66]. Over-expression of HOXA1 reversed growth inhibitory effect of miR-210 [40], indicating that miR-210-dependent growth inhibitory effect is, at least partially, due to direct silencing of HOXA1.
Expression profile of miR-210 targets are different in healthy versus cancer cells. Expression of some targets is restricted to transformed cells. Besides, some of the miR-210 target proteins serve different functions in different cell type. In cancer cells, miR-210 may support cell proliferation. miR-210 targets MNT, a Myc-antagonist, promoting cell cycle progression in transformed cells such as colon cancer cells and cervical cancer cells [104]. MNT competes with Myc for its binding partner myc-associated factor X and the Enhancer Box sequences to inhibit transactivation of genes that control cell cycle progression [42].
MIR-210 SUPPORTS STEM CELL SURVIVAL
miR-210 supports stem cell survival under hypoxic condition [51]. Episodes of ischemic preconditioning (IP) enhanced mesenchymal stem cells (MSCs) survival under anoxic condition, with the concomitant elevation of miR-210. Interestingly, the cytoprotective effect of IP could be reversed by anti-miR-210. Studies dissecting mechanistic insight revealed that miR-210 promotes stem cell survival via targeting caspase-8-associated protein 2 (CASP8AP2) [51], or its human homologue FLICE-associated protein homolog (FLASH), a protein that facilitates Fas-induced apoptosis [43].
MIR-210 REPRESSES MITOCHONDRIAL METABOLISM
When oxygen is available on a limited basis, metabolic shift from mitochondria respiration to glycolysis takes place, generating 2 mole of ATP (instead of 38 moles in normoxia) per one mole of glucose [41]. Accumulating evidence reveal that miR-210 inhibits mitochondrial metabolism by targeting a number of proteins that are crucial for normal TCA cycle. MiR-210 delivery alone under normal oxygen condition was potent enough to inhibit mitochondrial energy production [11], impair the oxygen consumption [11], induce lactate accumulation [15,27], alter mitochondrial membrane potential [77] and disrupt mitochondrial structure [77]. The iron-sulfur cluster assembly homologue (ISCU) 1/2 is one of the direct targets of miR-210. ISCU1/2 expression is negatively correlated with miR-210 level in wide variety of cells such as human pulmonary endothelial cells [11], breast cancer cells [27], colon cancer cells [27], and trophoblasts [55]. ISCU1/2 catalyzes the assembly of [4Fe-4S] and [2Fe-2S] iron-sulfur clusters. FeS clusters serve as prosthetic groups of the flavoproteins in electron transport chain, enabling the oxidation-reduction reactions in mitochondrial respiration and energy production [80]. Fe-S cluster is critical for the enzymatic activity of aconitase, a stereo-specific isomerization of citrate to isocitrate which fuels the TCA-cycle [4,80]. Constitutive expression of miR-210-resistant form of ISCU1/2 (devoid of 3′UTR region) partially reversed miR-210-dependent inhibition of mitochondrial respiration activity [15], indicating that miR-210 targets ISCU1/2 to suppress mitochondrial functions during hypoxia. MiR-210 not only targets ISCU1/2 but also regulates cytochrome c oxidase assembly protein (COX10) [15] and succinate dehydrogenase subunit D (SDHD) [77], repressing mitochondrial respiration. COX10 encodes the enzyme protoheme: heme O farnesyl transferase that facilitates the biosynthesis of heme-α, a vital component for the terminal enzyme of the respiratory chain cytochrome c oxidase [75]. Loss of COX10 inhibits the activity of mitochondrial complex I and complex IV [21]. SDHD is one of the subunits of the inner mitochondrial enzyme succinate dehydrogenase or succinate-coenzyme Q reductase (SQR) which catalyzes the oxidation of succinate (coupled to reduction of ubiquinone) during mitochondrial respiration [76]. In this regard, hypoxia-dependent elevation of miR-210 serves as a potent inhibitor of mitochondrial metabolism by targeting TCA cycle and electron transport chain activity. MiR-210-dependent acute transient down-regulation of mitochondrial respiration is on one hand important to enable the cells to ‘hang in there’ as the cells are less sensitive to oxygen for energy production under hypoxic environment. On the other hand it is in conflict with energy demanding processes such as tissue repair. If the inhibition of mitochondrial respiration is prolonged, cell death may ensue because of energy starvation. Interestingly, hypoxia-inducible miR-210 remains elevated even after return to normoxic environment for a day [25], suggesting that miR-210 induces a long-lasting inhibitory effect on mitochondrial metabolism even in the presence of sufficient oxygen. Strategies to antagonize the persistent miR-210 up-regulation during re-oxygenation phase would help to re-establish normal mitochondrial respiration and direct the cells toward an effective energy metabolism status.
MIR-210 STALLS DNA REPAIR
DNA damage is induced under normal metabolic conditions and some environmental factors including UV and radiation. It is expected that over a million DNA lesions per cell take place in a day [68], leading to severe detrimental consequences including cell senescence and tumor transformation. DNA repair is particularly important to ensure the genetic material remains intact throughout the life. Recently, it was reported that miR-210 can silence the DNA repair system via targeting the enzyme RAD52 [17]. RAD52 is a protein that fixes DNA double-strand break repair, repairs single-stranded DNA gaps and facilitates RAD51-mediated strand invasion during homologous recombination [67,99]. miR-210 directly binds to the 3′UTR of RAD52 to induce translational repression [17]. Hypoxia-dependent down-regulation of RAD52 can be rescued by treatment with anti-miR-210 [17], suggesting that RAD52 is down-regulated via a miR-210 dependent mechanism under low oxygen environment. Shutdown of the DNA repair under low oxygen condition might be critical for ATP conservation for cell preservation (‘hang in there’ response) during acute hypoxic condition [17]. However, such shutdown is in direct conflict with tissue repair. Chronic hypoxia (thus subsequently leading to sustained miR-210 level) substantially arrests DNA repair mechanism and induce genetic instability, resulting in either cell senescence or conferring a mutation phenotype during tumor transformation.
MIR-210 INDUCES ANGIOGENESIS
Cells respond to hypoxic challenge by up-regulation of genes that are essential for endothelial cells laying new blood vessel which would help to correct hypoxia and ensure survival. While the majority of studies focus on the regulation of VEGF, which is a potent pro-angiogenic factor to support sprouting of endothelial cells, current progress in miR biology has shed some light on the involvement of miR-210 in regulating angiogenesis under low oxygen environment. Fasanaro et al., first reported that hypoxia-driven miR-210 supports angiogenic response in endothelial cells [25]. These effects were, at least partially due to the down-regulation of ephrin-A3 (EFNA3), an ephrin family member involving vascular development [25,39]. Over-expression of EFNA3 significantly blocked the pro-angiogenic effect of miR-210 or hypoxia preconditioning [25]. Apart from EFNA3, miR-210 directly targets protein-tyrosine phosphatase 1B (PTP1B) [26,39], which negatively regulates VEGF signaling by de-phosphorylation of VEGFR2 in endothelial cells [71]. Elevation of miR-210 leads to repression of PTP1B, allowing successful VEGF signaling to proceed under hypoxia. The pro-angiogenic effect of miR-210 was evaluated in myocardial infarction, as evidenced by improved endothelial cell survival after delivery of miR-210 in the heart [39]. The involvement of miR-210 in regulation of pathophysiological angiogenesis has also been demonstrated in ischemic renal ischemia/reperfusion (I/R) injury, indicating that miR-210 induction is necessary to drive the expression of VEGF and VEGFR2 in endothelial cells [61]. In this regard, elevation of miR-210 supports angiogenic response and facilitates microcirculation under both physiological as well as pathophysiological conditions.
MIR-210 SUPPORTS CELL DIFFERENTIATION
Oxygen tension represents an important microenvironmental cue that directs the cell differentiation program towards lineage commitment [36,60,79]. In general, stem cells tend to retain their pluripotency and undifferentiated state under hypoxia while certain progenitor cells exhibit either accelerated or arrested differentiation program depending on the cell type. A number of investigations revealed that miR-210 supports cell differentiation. Bianchi et al., reported that mithramycin, a DNA-binding drugs which promote erythroid differentiation, induced the expression of miR-210 in erythroid progenitor cells [7], with the concomitant expression of erythroid marker γ-globin. On the other hand, miR-210 promotes bone morphogenic protein (BMP)-induced osteoblastic differentiation via targeting activin receptor type 1 B (ACVR1B) [65]. ACVR1B transmits signal from activin via Smad 2/3. Inhibition of Smad 2/3 leads to activation of Smad 1/5/8, resulting in promotion of differentiation of osteoblast to osteoclast [65]. In this regard, hypoxia-inducible miR-210 down-regulates ACVR1B, shutting down Smad 2/3 signaling, and promoting Smad 1/5/8-dependent osteoblastic differentiation [65]. In addition, miR-210 is strongly induced during adipogenesis [78]. Delivery of miR-210 markedly promoted lipogenesis while anti-miR-210 treatment significantly impaired the expression of lithium-induced adipogenic markers [78]. The pro-adipogenic response may be attributed by direct targeting of Wnt signaling mediator Transcription factor 7-like 2 (Tcf7l2). Given the fact that hypoxia promotes adipogenic differentiation [45], it is anticipated that a low oxygen environment fosters the accumulation of miR-210, which in turn down-regulates Tcf7l2 and subsequently induces adipogenesis. Figure 1 summarizes the major biological significance of miR-210 elevation and its corresponding targets under hypoxic condition.
Figure 1.
Summary of miR-210 targets and their biological consequences under hypoxia
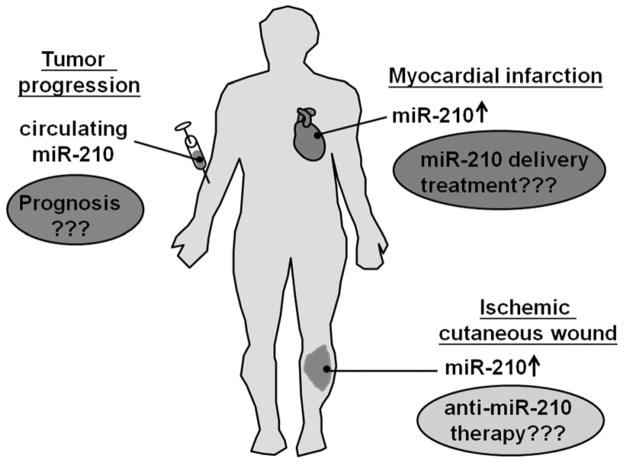
MIR-210 AS A PROGNOSTIC BIOMARKER
The average half-life of miRs is around 5 days, 10 times more than that of regular mRNA [28]. These small RNAs are extremely stable and are resistant to degradation by RNase A [14], high temperature, extreme pH and freeze-and-thaw cycle [59]. The extraordinary stability of miR makes it suitable to serve as a biomarker of certain diseases for prognostic purpose. The expression of miR-210 is elevated in human solid tumors, including glioma [64], head and neck carcinoma [40], lung adenocarcinoma [16], late stage small cell lung cancer [77], malignant melanoma [83], and pancreatic ductal adenocarcinomas [32]. Aberrant miR-210 expression is not only present in the solid tumor or injured organs but also being secreted into circulation which can be detected in the plasma of patients. Elevated circulating level of miR-210 can serve as a marker of diffuse large B-cell lymphoma [54], pancreatic ductal carcinoma [37,97], malignant solitary pulmonary nodules [89] and acute renal injury [62]. Recently, Lorenzen et al., reported that miR-210 can also be detected in the urine from normal individual and renal allograft recipients [63], suggesting a wide variety of miR-210 source for prognosis. To date, a number of studies have worked on the prognostic power of miR-210 expression in different diseases. High levels of miR-210 were associated with disease recurrence and short overall survival in in head and neck squamous cell carcinoma [30]. High miR-210 expression was also associated with a lower relative risk (RR) of tumor-related death compared with the intermediate expression of miR-210 expression in soft-tissue sarcoma patients [33].
MIR-210 DELIVERY OR ANTI-MIR-210: POTENTIAL THERAPY OF ISCHEMIC DISORDERS?
Given the fact that miR-210 exerts versatile effects on cellular functions and its deregulation under pathological conditions, strategies targeting correction of aberrantly expressed miR-210 might open up a new therapeutic avenue to a wide range of diseases such as ischemic disorders and tumor progression. The employment of miR-210 mimic delivery or anti-miR-210 therapy depends on whether miR-210 is insufficient or over-produced, respectively, under the corresponding disease state. Recently, it has been reported that intramyocardial injection of non-viral vector minicircle DNA carrying miR-210 precursor can stably transduce miR-210 expression for at least 8 weeks in the heart [39]. More importantly, this strategy improved cardiac function, reduced the infarct size and rectified angiogenesis after myocardial infarction [39], indicating miR-210 delivery might serve as a therapeutic approach in ischemic heart disease. On the other hand, aberrantly up-regulated miR-210 level can be suppressed by delivery of anti-miR-210 strategy using antagomir (with 2′O-methylation and phosphothioates) or lock-nucleic acid (LNA) (with extra bridge connecting 2′ oxygen and 4′ carbon on ribose moiety). Stoffel and colleagues first reported that murine endogenous miRs could be silenced by bolus intravenous injection of antagomir in wide range of tissues [53]. The anti-miR in vivo study was further extended to non-human primates. Acute administration of unconjugated LNA-modified oligonucleotide against miR-122, a miR that regulates cholesterol biosynthesis, effectively down-regulated hepatic miR-122 in African green monkeys, which was accompanied by a decrease in plasma cholesterol in a dose-dependent manner [24].
Our group reported that ischemic cutaneous wounds exhibit elevated miR-210 expression, which was associated with the down-regulation of E2F3 and impairment in keratinocyte proliferation and wound re-epithelialization [8]. Besides arrest of keratinocyte proliferation, elevated miR-210 may complicate wound closure by repressing mitochondrial respiration and silencing DNA repair. Wound healing is a energy-demanding process [86]. Energy supply, as ATP, is required to fuel the growth of new tissue. Indeed, extracellular ATP supports wound healing response by a number of mechanisms including epidermal growth factor (EGF) receptor transactivation and NADPH oxidase activation [86]. Limited ATP generation is therefore in direct conflict with wound healing. Disruption of DNA repair system by elevated miR-210 in ischemic wounds is yet another roadblock as excessive DNA damage leads to cell senescence, which blunts the healing response. Angiogenesis is important but not in most cases not singularly sufficient to drive wound healing [46]. Clinical experience show that successful re-vascularization failed to heal ischemic lower extremity wounds [1,2,5,6,10,13,19,23,47,49,73,84,94]. Thus, elevation of miR-210 in ischemic wounds hurts wound closure by inhibiting keratinocyte proliferation, disrupting mitochondrial metabolism and compromising DNA repair despite pro-angiogenic effects. Delivery of anti-miR-210 should be tested for its efficacy to improve cutaneous wound outcomes.
PERSPECTIVE AND CONCLUDING REMARKS
miR-210 is steadily establishing itself as a major hypoxia-response factor that regulates several key aspects of health and disease. In silico prediction algorithms including Targetscan [38], MiRanda [38], Pictar [52], miRBase Target Database [34] and miRDB [98] represent powerful tools in the search for novel direct targets of miR-210. One of the limitations of these approaches is the possibility of raising false positive prediction because of the short seed sequence (7–8 nt). Experimental validation of specific miR-binding to 3′UTR is necessary to confirm the induction of translational repression. Recently, the employment of RNA induced silencing complex (RISC) immunoprecipitation, a robust high-throughput approach to biologically validate the enrichment of transcript in RISC complex in responsive to miR-210 over-expression, has unveiled a number of novel miR-210 targets that are of significance in ischemic diseases [26,39,40]. Strategies adopting combined approaches including in silico prediction, RISC immunoprecipitation, proteomic analysis, microarray analysis, would help to dissect the biological consequences of miR-210 and its associated target under diseases state. As it relates to regulating biological functions, miR-210 serves as a potent maestro in fine-tuning hypoxia response. Apart from prognostic value, miR-210 may serve as a target for therapeutic purpose in treating ischemic disorders such as myocardial infarction and cutaneous ischemic wounds (Figure 2).
Figure 2.
miR-210 serves as prognostic and therapeutic targets
Acknowledgments
This work was supported by NIH awards GM 077185, GM069589, and HL073087 to C.K.S.
Abbreviation used
- ACVR1B
activin receptor type 1 B
- CASP8AP2
caspase-8-associated protein 2
- COX10
cytochrome c oxidase assembly protein
- DGCR8
DiGeorge critical region 8
- EFNA3
ephrin-A3
- FGFRL1
fibroblast growth factor receptor like 1
- FLASH
FLICE-associated protein homolog
- HIF
hypoxia inducible factor
- HOXA1
homeobox A1
- HRE
hypoxia responsive element
- IP
ischemia preconditioning
- I/R
ischemia/reperfusion
- ISCU
Iron-sulfur cluster assembly homologue
- LNA
lock-nucleic acid
- MSCs
mesenchymal stem cells
- miR
MicroRNAs
- 3′UTR
3′ unstranslated region
- NFκB
nuclear factor κB
- PTP1B
phosphatase 1B
- RISC
RNA induced silencing complex
- SDHD
succinate dehydrogenase subunit D
- Tcf7l2
Transcription factor 7-like 2
- VEGF
vascular endothelial growth factor
Biographies

Professor Chandan Sen is the Executive Director of the Ohio State University (OSU) Comprehensive Wound Center and Associste Dean (Research) at OSU Medical Center. Professor Sen obtained his PhD in Physiology from University of University of Kuopio, Finland. He was appointed to his present position at the OSU in 2004. His research interests focuses on tissue injury and repair. Professor Sen is the Editor-in-Chief of the journal, Antioxidants & Redox Signaling (IF: 8.209) and the Wound Healing Society Yearbook Advances in Wound Care. He has published over 250 papers and is cited more than 900 times annually.

Dr Yuk Cheung Chan obtained his BSc in Biochemistry (2004) and PhD in Physiology (2008) from the Chinese University of Hong Kong, China. He is currently a postdoctoral researcher in the department of surgery, OSU. His research focuses on studying the novel role of microRNAs in regulation of endothelial cell biology, and its association with cutaneous wound angiogenesis.

Jaideep Banerjee completed his Bachelor of Science in Physiology and Master of Science in Molecular Biology from the University of Calcutta, India. He is currently in the Molecular Cell and Developmental Biology PhD program at OSU. His research interests include the role of microRNAs in tissue injury and repair, primarily in wound healing and heart failure models.

Sang-Yong Choi, M.S. attended Yonsei University in South Korea where he double-majored in Biology and Food & Nutrition (2006) and obtained M.S. in Food & Nutrition (2008). Mr Choi is a graduate student in an Interdisciplinary Ph.D. program in Nutrition at OSU.
REFERECES
- 1.Abou-Zamzam AM, Jr, Moneta GL, Lee RW, Nehler MR, Taylor LM, Jr, Porter JM. Peroneal bypass is equivalent to inframalleolar bypass for ischemic pedal gangrene. Arch Surg. 1996;131:894–898. doi: 10.1001/archsurg.1996.01430200104018. discussion 898–899. [DOI] [PubMed] [Google Scholar]
- 2.Andros G, Harris RW, Dulawa LB, Oblath RW, Salles-Cunha SX. The need for arteriography in diabetic patients with gangrene and palpable foot pulses. Arch Surg. 1984;119:1260–1263. doi: 10.1001/archsurg.1984.01390230032007. [DOI] [PubMed] [Google Scholar]
- 3.Banerjee J, Chan YC, Sen CK. MicroRNAs in skin and wound healing. Physiol Genomics. 2011;43:543–556. doi: 10.1152/physiolgenomics.00157.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beinert H, Kennedy MC. Aconitase, a two-faced protein: enzyme and iron regulatory factor. FASEB J. 1993;7:1442–1449. doi: 10.1096/fasebj.7.15.8262329. [DOI] [PubMed] [Google Scholar]
- 5.Berceli SA, Chan AK, Pomposelli FB, Jr, Gibbons GW, Campbell DR, Akbari CM, Brophy DT, LoGerfo FW. Efficacy of dorsal pedal artery bypass in limb salvage for ischemic heel ulcers. J Vasc Surg. 1999;30:499–508. doi: 10.1016/s0741-5214(99)70077-7. [DOI] [PubMed] [Google Scholar]
- 6.Bergamini TM, George SM, Jr, Massey HT, Henke PK, Klamer TW, Lambert GE, Jr, Banis JC, Jr, Miller FB, Garrison RN, Richardson JD. Pedal or peroneal bypass: which is better when both are patent? J Vasc Surg. 1994;20:347–355. doi: 10.1016/0741-5214(94)90132-5. discussion 355–346. [DOI] [PubMed] [Google Scholar]
- 7.Bianchi N, Zuccato C, Lampronti I, Borgatti M, Gambari R. Expression of miR-210 during erythroid differentiation and induction of gamma-globin gene expression. BMB Rep. 2009;42:493–499. doi: 10.5483/bmbrep.2009.42.8.493. [DOI] [PubMed] [Google Scholar]
- 8.Biswas S, Roy S, Banerjee J, Hussain SR, Khanna S, Meenakshisundaram G, Kuppusamy P, Friedman A, Sen CK. Hypoxia inducible microRNA 210 attenuates keratinocyte proliferation and impairs closure in a murine model of ischemic wounds. Proc Natl Acad Sci U S A. 2010;107:6976–6981. doi: 10.1073/pnas.1001653107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Camps C, Buffa FM, Colella S, Moore J, Sotiriou C, Sheldon H, Harris AL, Gleadle JM, Ragoussis J. hsa-miR-210 Is induced by hypoxia and is an independent prognostic factor in breast cancer. Clin Cancer Res. 2008;14:1340–1348. doi: 10.1158/1078-0432.CCR-07-1755. [DOI] [PubMed] [Google Scholar]
- 10.Carsten CG, 3rd, Taylor SM, Langan EM, 3rd, Crane MM. Factors associated with limb loss despite a patent infrainguinal bypass graft. Am Surg. 1998;64:33–37. discussion 37–38. [PubMed] [Google Scholar]
- 11.Chan SY, Zhang YY, Hemann C, Mahoney CE, Zweier JL, Loscalzo J. MicroRNA-210 controls mitochondrial metabolism during hypoxia by repressing the iron-sulfur cluster assembly proteins ISCU1/2. Cell Metab. 2009;10:273–284. doi: 10.1016/j.cmet.2009.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan YC, Khanna S, Roy S, Sen CK. miR-200b targets Ets-1 and is down-regulated by hypoxia to induce angiogenic response of endothelial cells. J Biol Chem. 2011;286:2047–2056. doi: 10.1074/jbc.M110.158790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang BB, Paty PS, Shah DM, Kaufman JL, Leather RP. Results of infrainguinal bypass for limb salvage in patients with end-stage renal disease. Surgery. 1990;108:742–746. discussion 746–747. [PubMed] [Google Scholar]
- 14.Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, Guo J, Zhang Y, Chen J, Guo X, Li Q, Li X, Wang W, Wang J, Jiang X, Xiang Y, Xu C, Zheng P, Zhang J, Li R, Zhang H, Shang X, Gong T, Ning G, Zen K, Zhang CY. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 15.Chen Z, Li Y, Zhang H, Huang P, Luthra R. Hypoxia-regulated microRNA-210 modulates mitochondrial function and decreases ISCU and COX10 expression. Oncogene. 2010;29:4362–4368. doi: 10.1038/onc.2010.193. [DOI] [PubMed] [Google Scholar]
- 16.Cho WC, Chow AS, Au JS. Restoration of tumour suppressor hsa-miR-145 inhibits cancer cell growth in lung adenocarcinoma patients with epidermal growth factor receptor mutation. Eur J Cancer. 2009;45:2197–2206. doi: 10.1016/j.ejca.2009.04.039. [DOI] [PubMed] [Google Scholar]
- 17.Crosby ME, Kulshreshtha R, Ivan M, Glazer PM. MicroRNA regulation of DNA repair gene expression in hypoxic stress. Cancer Res. 2009;69:1221–1229. doi: 10.1158/0008-5472.CAN-08-2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cummins EP, Taylor CT. Hypoxia-responsive transcription factors. Pflugers Arch. 2005;450:363–371. doi: 10.1007/s00424-005-1413-7. [DOI] [PubMed] [Google Scholar]
- 19.Darling RC, 3rd, Chang BB, Paty PS, Lloyd WE, Leather RP, Shah DM. Choice of peroneal or dorsalis pedis artery bypass for limb salvage. Am J Surg. 1995;170:109–112. doi: 10.1016/s0002-9610(99)80266-9. [DOI] [PubMed] [Google Scholar]
- 20.Devlin C, Greco S, Martelli F, Ivan M. miR-210: More than a silent player in hypoxia. IUBMB Life. 2011;63:94–100. doi: 10.1002/iub.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Diaz F, Fukui H, Garcia S, Moraes CT. Cytochrome c oxidase is required for the assembly/stability of respiratory complex I in mouse fibroblasts. Mol Cell Biol. 2006;26:4872–4881. doi: 10.1128/MCB.01767-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dirks PB, Rutka JT, Hubbard SL, Mondal S, Hamel PA. The E2F-family proteins induce distinct cell cycle regulatory factors in p16-arrested, U343 astrocytoma cells. Oncogene. 1998;17:867–876. doi: 10.1038/sj.onc.1202008. [DOI] [PubMed] [Google Scholar]
- 23.Elliott BM, Robison JG, Brothers TE, Cross MA. Limitations of peroneal artery bypass grafting for limb salvage. J Vasc Surg. 1993;18:881–888. doi: 10.1067/mva.1993.49636. [DOI] [PubMed] [Google Scholar]
- 24.Elmen J, Lindow M, Schutz S, Lawrence M, Petri A, Obad S, Lindholm M, Hedtjarn M, Hansen HF, Berger U, Gullans S, Kearney P, Sarnow P, Straarup EM, Kauppinen S. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 25.Fasanaro P, D’Alessandra Y, Di Stefano V, Melchionna R, Romani S, Pompilio G, Capogrossi MC, Martelli F. MicroRNA-210 modulates endothelial cell response to hypoxia and inhibits the receptor tyrosine kinase ligand Ephrin-A3. J Biol Chem. 2008;283:15878–15883. doi: 10.1074/jbc.M800731200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fasanaro P, Greco S, Lorenzi M, Pescatori M, Brioschi M, Kulshreshtha R, Banfi C, Stubbs A, Calin GA, Ivan M, Capogrossi MC, Martelli F. An integrated approach for experimental target identification of hypoxia-induced miR-210. J Biol Chem. 2009;284:35134–35143. doi: 10.1074/jbc.M109.052779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Favaro E, Ramachandran A, McCormick R, Gee H, Blancher C, Crosby M, Devlin C, Blick C, Buffa F, Li JL, Vojnovic B, Pires das Neves R, Glazer P, Iborra F, Ivan M, Ragoussis J, Harris AL. MicroRNA-210 regulates mitochondrial free radical response to hypoxia and krebs cycle in cancer cells by targeting iron sulfur cluster protein ISCU. PLoS One. 2010;5:e10345. doi: 10.1371/journal.pone.0010345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gantier MP, McCoy CE, Rusinova I, Saulep D, Wang D, Xu D, Irving AT, Behlke MA, Hertzog PJ, Mackay F, Williams BR. Analysis of microRNA turnover in mammalian cells following Dicer1 ablation. Nucleic Acids Res. 2011;39:5692–5703. doi: 10.1093/nar/gkr148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gavalas A, Studer M, Lumsden A, Rijli FM, Krumlauf R, Chambon P. Hoxa1 and Hoxb1 synergize in patterning the hindbrain, cranial nerves and second pharyngeal arch. Development. 1998;125:1123–1136. doi: 10.1242/dev.125.6.1123. [DOI] [PubMed] [Google Scholar]
- 30.Gee HE, Camps C, Buffa FM, Patiar S, Winter SC, Betts G, Homer J, Corbridge R, Cox G, West CM, Ragoussis J, Harris AL. hsa-mir-210 is a marker of tumor hypoxia and a prognostic factor in head and neck cancer. Cancer. 2010;116:2148–2158. doi: 10.1002/cncr.25009. [DOI] [PubMed] [Google Scholar]
- 31.Giannakakis A, Sandaltzopoulos R, Greshock J, Liang S, Huang J, Hasegawa K, Li C, O’Brien-Jenkins A, Katsaros D, Weber BL, Simon C, Coukos G, Zhang L. miR-210 links hypoxia with cell cycle regulation and is deleted in human epithelial ovarian cancer. Cancer Biol Ther. 2008;7:255–264. doi: 10.4161/cbt.7.2.5297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Greither T, Grochola LF, Udelnow A, Lautenschlager C, Wurl P, Taubert H. Elevated expression of microRNAs 155, 203, 210 and 222 in pancreatic tumors is associated with poorer survival. Int J Cancer. 2010;126:73–80. doi: 10.1002/ijc.24687. [DOI] [PubMed] [Google Scholar]
- 33.Greither T, Wurl P, Grochola L, Bond G, Bache M, Kappler M, Lautenschlager C, Holzhausen HJ, Wach S, Eckert AW, Taubert H. Expression of microRNA 210 associates with poor survival and age of tumor onset of soft-tissue sarcoma patients. Int J Cancer. 2011 doi: 10.1002/ijc.26109. [DOI] [PubMed] [Google Scholar]
- 34.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.He Y, Armanious MK, Thomas MJ, Cress WD. Identification of E2F-3B, an alternative form of E2F-3 lacking a conserved N-terminal region. Oncogene. 2000;19:3422–3433. doi: 10.1038/sj.onc.1203682. [DOI] [PubMed] [Google Scholar]
- 36.Heinis M, Simon MT, Ilc K, Mazure NM, Pouyssegur J, Scharfmann R, Duvillie B. Oxygen tension regulates pancreatic beta-cell differentiation through hypoxia-inducible factor 1alpha. Diabetes. 2010;59:662–669. doi: 10.2337/db09-0891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ho AS, Huang X, Cao H, Christman-Skieller C, Bennewith K, Le QT, Koong AC. Circulating miR-210 as a Novel Hypoxia Marker in Pancreatic Cancer. Transl Oncol. 2010;3:109–113. doi: 10.1593/tlo.09256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hsu PW, Lin LZ, Hsu SD, Hsu JB, Huang HD. ViTa: prediction of host microRNAs targets on viruses. Nucleic Acids Res. 2007;35:D381–385. doi: 10.1093/nar/gkl1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hu S, Huang M, Li Z, Jia F, Ghosh Z, Lijkwan MA, Fasanaro P, Sun N, Wang X, Martelli F, Robbins RC, Wu JC. MicroRNA-210 as a novel therapy for treatment of ischemic heart disease. Circulation. 2010;122:S124–131. doi: 10.1161/CIRCULATIONAHA.109.928424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang X, Ding L, Bennewith KL, Tong RT, Welford SM, Ang KK, Story M, Le QT, Giaccia AJ. Hypoxia-inducible mir-210 regulates normoxic gene expression involved in tumor initiation. Mol Cell. 2009;35:856–867. doi: 10.1016/j.molcel.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang X, Le QT, Giaccia AJ. MiR-210--micromanager of the hypoxia pathway. Trends Mol Med. 2010;16:230–237. doi: 10.1016/j.molmed.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hurlin PJ, Huang J. The MAX-interacting transcription factor network. Semin Cancer Biol. 2006;16:265–274. doi: 10.1016/j.semcancer.2006.07.009. [DOI] [PubMed] [Google Scholar]
- 43.Imai Y, Kimura T, Murakami A, Yajima N, Sakamaki K, Yonehara S. The CED-4-homologous protein FLASH is involved in Fas-mediated activation of caspase-8 during apoptosis. Nature. 1999;398:777–785. doi: 10.1038/19709. [DOI] [PubMed] [Google Scholar]
- 44.Ioshikhes I, Roy S, Sen CK. Algorithms for mapping of mRNA targets for microRNA. DNA Cell Biol. 2007;26:265–272. doi: 10.1089/dna.2006.0566. [DOI] [PubMed] [Google Scholar]
- 45.Itoigawa Y, Kishimoto KN, Okuno H, Sano H, Kaneko K, Itoi E. Hypoxia induces adipogenic differentitation of myoblastic cell lines. Biochem Biophys Res Commun. 2010;399:721–726. doi: 10.1016/j.bbrc.2010.08.007. [DOI] [PubMed] [Google Scholar]
- 46.Jang YC, Arumugam S, Gibran NS, Isik FF. Role of alpha(v) integrins and angiogenesis during wound repair. Wound Repair Regen. 1999;7:375–380. doi: 10.1046/j.1524-475x.1999.00375.x. [DOI] [PubMed] [Google Scholar]
- 47.Johnson BL, Glickman MH, Bandyk DF, Esses GE. Failure of foot salvage in patients with end-stage renal disease after surgical revascularization. J Vasc Surg. 1995;22:280–285. doi: 10.1016/s0741-5214(95)70142-7. discussion 285–286. [DOI] [PubMed] [Google Scholar]
- 48.Kaelin WG, Jr, Ratcliffe PJ. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol Cell. 2008;30:393–402. doi: 10.1016/j.molcel.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 49.Karp NS, Kasabian AK, Siebert JW, Eidelman Y, Colen S. Microvascular free-flap salvage of the diabetic foot: a 5-year experience. Plast Reconstr Surg. 1994;94:834–840. doi: 10.1097/00006534-199411000-00013. [DOI] [PubMed] [Google Scholar]
- 50.Khanna S, Roy S, Maurer M, Ratan RR, Sen CK. Oxygen-sensitive reset of hypoxia-inducible factor transactivation response: prolyl hydroxylases tune the biological normoxic set point. Free Radic Biol Med. 2006;40:2147–2154. doi: 10.1016/j.freeradbiomed.2006.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim HW, Haider HK, Jiang S, Ashraf M. Ischemic preconditioning augments survival of stem cells via miR-210 expression by targeting caspase-8-associated protein 2. J Biol Chem. 2009;284:33161–33168. doi: 10.1074/jbc.M109.020925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, Rajewsky N. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 53.Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 54.Lawrie CH, Gal S, Dunlop HM, Pushkaran B, Liggins AP, Pulford K, Banham AH, Pezzella F, Boultwood J, Wainscoat JS, Hatton CS, Harris AL. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol. 2008;141:672–675. doi: 10.1111/j.1365-2141.2008.07077.x. [DOI] [PubMed] [Google Scholar]
- 55.Lee DC, Romero R, Kim JS, Tarca AL, Montenegro D, Pineles BL, Kim E, Lee J, Kim SY, Draghici S, Mittal P, Kusanovic JP, Chaiworapongsa T, Hassan SS, Kim CJ. miR-210 targets iron-sulfur cluster scaffold homologue in human trophoblast cell lines: siderosis of interstitial trophoblasts as a novel pathology of preterm preeclampsia and small-for-gestational-age pregnancies. Am J Pathol. 2011;179:590–602. doi: 10.1016/j.ajpath.2011.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee EJ, Baek M, Gusev Y, Brackett DJ, Nuovo GJ, Schmittgen TD. Systematic evaluation of microRNA processing patterns in tissues, cell lines, and tumors. RNA. 2008;14:35–42. doi: 10.1261/rna.804508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Leone G, DeGregori J, Yan Z, Jakoi L, Ishida S, Williams RS, Nevins JR. E2F3 activity is regulated during the cell cycle and is required for the induction of S phase. Genes Dev. 1998;12:2120–2130. doi: 10.1101/gad.12.14.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Leone G, Nuckolls F, Ishida S, Adams M, Sears R, Jakoi L, Miron A, Nevins JR. Identification of a novel E2F3 product suggests a mechanism for determining specificity of repression by Rb proteins. Mol Cell Biol. 2000;20:3626–3632. doi: 10.1128/mcb.20.10.3626-3632.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li J, Smyth P, Flavin R, Cahill S, Denning K, Aherne S, Guenther SM, O’Leary JJ, Sheils O. Comparison of miRNA expression patterns using total RNA extracted from matched samples of formalin-fixed paraffin-embedded (FFPE) cells and snap frozen cells. BMC Biotechnol. 2007;7:36. doi: 10.1186/1472-6750-7-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lin Q, Lee YJ, Yun Z. Differentiation arrest by hypoxia. J Biol Chem. 2006;281:30678–30683. doi: 10.1074/jbc.C600120200. [DOI] [PubMed] [Google Scholar]
- 61.Liu F, Lou YL, Wu J, Ruan QF, Xie A, Guo F, Cui SP, Deng ZF, Wang Y. Upregulation of MicroRNA-210 Regulates Renal Angiogenesis Mediated by Activation of VEGF Signaling Pathway under Ischemia/Perfusion Injury in vivo and in vitro. Kidney Blood Press Res. 2011;35:182–191. doi: 10.1159/000331054. [DOI] [PubMed] [Google Scholar]
- 62.Lorenzen JM, Kielstein JT, Hafer C, Gupta SK, Kumpers P, Faulhaber-Walter R, Haller H, Fliser D, Thum T. Circulating miR-210 predicts survival in critically ill patients with acute kidney injury. Clin J Am Soc Nephrol. 2011;6:1540–1546. doi: 10.2215/CJN.00430111. [DOI] [PubMed] [Google Scholar]
- 63.Lorenzen JM, Volkmann I, Fiedler J, Schmidt M, Scheffner I, Haller H, Gwinner W, Thum T. Urinary miR-210 as a Mediator of Acute T-Cell Mediated Rejection in Renal Allograft Recipients. Am J Transplant. 2011;11:2221–2227. doi: 10.1111/j.1600-6143.2011.03679.x. [DOI] [PubMed] [Google Scholar]
- 64.Malzkorn B, Wolter M, Liesenberg F, Grzendowski M, Stuhler K, Meyer HE, Reifenberger G. Identification and functional characterization of microRNAs involved in the malignant progression of gliomas. Brain Pathol. 2010;20:539–550. doi: 10.1111/j.1750-3639.2009.00328.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mizuno Y, Tokuzawa Y, Ninomiya Y, Yagi K, Yatsuka-Kanesaki Y, Suda T, Fukuda T, Katagiri T, Kondoh Y, Amemiya T, Tashiro H, Okazaki Y. miR-210 promotes osteoblastic differentiation through inhibition of AcvR1b. FEBS Lett. 2009;583:2263–2268. doi: 10.1016/j.febslet.2009.06.006. [DOI] [PubMed] [Google Scholar]
- 66.Mohankumar KM, Xu XQ, Zhu T, Kannan N, Miller LD, Liu ET, Gluckman PD, Sukumar S, Emerald BS, Lobie PE. HOXA1-stimulated oncogenicity is mediated by selective upregulation of components of the p44/42 MAP kinase pathway in human mammary carcinoma cells. Oncogene. 2007;26:3998–4008. doi: 10.1038/sj.onc.1210180. [DOI] [PubMed] [Google Scholar]
- 67.Mortensen UH, Lisby M, Rothstein R. Rad52. Curr Biol. 2009;19:R676–677. doi: 10.1016/j.cub.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 68.Muniappan BP, Thilly WG. The DNA polymerase beta replication error spectrum in the adenomatous polyposis coli gene contains human colon tumor mutational hotspots. Cancer Res. 2002;62:3271–3275. [PubMed] [Google Scholar]
- 69.Mutharasan RK, Nagpal V, Ichikawa Y, Ardehali H. microRNA-210 is upregulated in hypoxic cardiomyocytes through Akt- and p53-dependent pathways and exerts cytoprotective effects. Am J Physiol Heart Circ Physiol. 2011;301:H1519–1530. doi: 10.1152/ajpheart.01080.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nakada C, Tsukamoto Y, Matsuura K, Nguyen TL, Hijiya N, Uchida T, Sato F, Mimata H, Seto M, Moriyama M. Overexpression of miR-210, a downstream target of HIF1alpha, causes centrosome amplification in renal carcinoma cells. J Pathol. 2011;224:280–288. doi: 10.1002/path.2860. [DOI] [PubMed] [Google Scholar]
- 71.Nakamura Y, Patrushev N, Inomata H, Mehta D, Urao N, Kim HW, Razvi M, Kini V, Mahadev K, Goldstein BJ, McKinney R, Fukai T, Ushio-Fukai M. Role of protein tyrosine phosphatase 1B in vascular endothelial growth factor signaling and cell-cell adhesions in endothelial cells. Circ Res. 2008;102:1182–1191. doi: 10.1161/CIRCRESAHA.107.167080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nakayama K. Cellular signal transduction of the hypoxia response. J Biochem. 2009;146:757–765. doi: 10.1093/jb/mvp167. [DOI] [PubMed] [Google Scholar]
- 73.Neville RF, Attinger CE, Bulan EJ, Ducic I, Thomassen M, Sidawy AN. Revascularization of a specific angiosome for limb salvage: does the target artery matter? Ann Vasc Surg. 2009;23:367–373. doi: 10.1016/j.avsg.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 74.Park HA, Kubicki N, Gnyawali S, Chan YC, Roy S, Khanna S, Sen CK. Natural vitamin E alpha-tocotrienol protects against ischemic stroke by induction of multidrug resistance-associated protein 1. Stroke. 2011;42:2308–2314. doi: 10.1161/STROKEAHA.110.608547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pecina P, Houstkova H, Hansikova H, Zeman J, Houstek J. Genetic defects of cytochrome c oxidase assembly. Physiol Res. 2004;53 (Suppl 1):S213–223. [PubMed] [Google Scholar]
- 76.Piruat JI, Pintado CO, Ortega-Saenz P, Roche M, Lopez-Barneo J. The mitochondrial SDHD gene is required for early embryogenesis, and its partial deficiency results in persistent carotid body glomus cell activation with full responsiveness to hypoxia. Mol Cell Biol. 2004;24:10933–10940. doi: 10.1128/MCB.24.24.10933-10940.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Puissegur MP, Mazure NM, Bertero T, Pradelli L, Grosso S, Robbe-Sermesant K, Maurin T, Lebrigand K, Cardinaud B, Hofman V, Fourre S, Magnone V, Ricci JE, Pouyssegur J, Gounon P, Hofman P, Barbry P, Mari B. miR-210 is overexpressed in late stages of lung cancer and mediates mitochondrial alterations associated with modulation of HIF-1 activity. Cell Death Differ. 2011;18:465–478. doi: 10.1038/cdd.2010.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Qin L, Chen Y, Niu Y, Chen W, Wang Q, Xiao S, Li A, Xie Y, Li J, Zhao X, He Z, Mo D. A deep investigation into the adipogenesis mechanism: profile of microRNAs regulating adipogenesis by modulating the canonical Wnt/beta-catenin signaling pathway. BMC Genomics. 2010;11:320. doi: 10.1186/1471-2164-11-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rogers HM, Yu X, Wen J, Smith R, Fibach E, Noguchi CT. Hypoxia alters progression of the erythroid program. Exp Hematol. 2008;36:17–27. doi: 10.1016/j.exphem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rouault TA, Tong WH. Iron-sulfur cluster biogenesis and human disease. Trends Genet. 2008;24:398–407. doi: 10.1016/j.tig.2008.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Roy S, Khanna S, Hussain SR, Biswas S, Azad A, Rink C, Gnyawali S, Shilo S, Nuovo GJ, Sen CK. MicroRNA expression in response to murine myocardial infarction: miR-21 regulates fibroblast metalloprotease-2 via phosphatase and tensin homologue. Cardiovasc Res. 2009;82:21–29. doi: 10.1093/cvr/cvp015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Roy S, Sen CK. MiRNA in innate immune responses: novel players in wound inflammation. Physiol Genomics. 2011;43:557–565. doi: 10.1152/physiolgenomics.00160.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Satzger I, Mattern A, Kuettler U, Weinspach D, Voelker B, Kapp A, Gutzmer R. MicroRNA-15b represents an independent prognostic parameter and is correlated with tumor cell proliferation and apoptosis in malignant melanoma. Int J Cancer. 2010;126:2553–2562. doi: 10.1002/ijc.24960. [DOI] [PubMed] [Google Scholar]
- 84.Seeger JM, Pretus HA, Carlton LC, Flynn TC, Ozaki CK, Huber TS. Potential predictors of outcome in patients with tissue loss who undergo infrainguinal vein bypass grafting. J Vasc Surg. 1999;30:427–435. doi: 10.1016/s0741-5214(99)70069-8. [DOI] [PubMed] [Google Scholar]
- 85.Sen CK. MicroRNAs as new maestro conducting the expanding symphony orchestra of regenerative and reparative medicine. Physiol Genomics. 2011;43:517–520. doi: 10.1152/physiolgenomics.00037.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sen CK. Wound healing essentials: let there be oxygen. Wound Repair Regen. 2009;17:1–18. doi: 10.1111/j.1524-475X.2008.00436.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sen CK, Gordillo GM, Khanna S, Roy S. Micromanaging vascular biology: tiny microRNAs play big band. J Vasc Res. 2009;46:527–540. doi: 10.1159/000226221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sen CK, Roy S. miRNA: licensed to kill the messenger. DNA Cell Biol. 2007;26:193–194. doi: 10.1089/dna.2006.0567. [DOI] [PubMed] [Google Scholar]
- 89.Shen J, Liu Z, Todd NW, Zhang H, Liao J, Yu L, Guarnera MA, Li R, Cai L, Zhan M, Jiang F. Diagnosis of lung cancer in individuals with solitary pulmonary nodules by plasma microRNA biomarkers. BMC Cancer. 2011;11:374. doi: 10.1186/1471-2407-11-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Shilo S, Roy S, Khanna S, Sen CK. Evidence for the involvement of miRNA in redox regulated angiogenic response of human microvascular endothelial cells. Arterioscler Thromb Vasc Biol. 2008;28:471–477. doi: 10.1161/ATVBAHA.107.160655. [DOI] [PubMed] [Google Scholar]
- 91.Shilo S, Roy S, Khanna S, Sen CK. MicroRNA in cutaneous wound healing: a new paradigm. DNA Cell Biol. 2007;26:227–237. doi: 10.1089/dna.2006.0568. [DOI] [PubMed] [Google Scholar]
- 92.Sleeman M, Fraser J, McDonald M, Yuan S, White D, Grandison P, Kumble K, Watson JD, Murison JG. Identification of a new fibroblast growth factor receptor, FGFR5. Gene. 2001;271:171–182. doi: 10.1016/s0378-1119(01)00518-2. [DOI] [PubMed] [Google Scholar]
- 93.Slezak-Prochazka I, Durmus S, Kroesen BJ, van den Berg A. MicroRNAs, macrocontrol: regulation of miRNA processing. RNA. 2010;16:1087–1095. doi: 10.1261/rna.1804410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Treiman GS, Oderich GS, Ashrafi A, Schneider PA. Management of ischemic heel ulceration and gangrene: An evaluation of factors associated with successful healing. J Vasc Surg. 2000;31:1110–1118. doi: 10.1067/mva.2000.106493. [DOI] [PubMed] [Google Scholar]
- 95.Tsuchiya S, Fujiwara T, Sato F, Shimada Y, Tanaka E, Sakai Y, Shimizu K, Tsujimoto G. MicroRNA-210 regulates cancer cell proliferation through targeting fibroblast growth factor receptor-like 1 (FGFRL1) J Biol Chem. 2011;286:420–428. doi: 10.1074/jbc.M110.170852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Vigo E, Muller H, Prosperini E, Hateboer G, Cartwright P, Moroni MC, Helin K. CDC25A phosphatase is a target of E2F and is required for efficient E2F-induced S phase. Mol Cell Biol. 1999;19:6379–6395. doi: 10.1128/mcb.19.9.6379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang J, Chen J, Chang P, LeBlanc A, Li D, Abbruzzesse JL, Frazier ML, Killary AM, Sen S. MicroRNAs in plasma of pancreatic ductal adenocarcinoma patients as novel blood-based biomarkers of disease. Cancer Prev Res (Phila) 2009;2:807–813. doi: 10.1158/1940-6207.CAPR-09-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wang X. miRDB: a microRNA target prediction and functional annotation database with a wiki interface. RNA. 2008;14:1012–1017. doi: 10.1261/rna.965408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.West SC. Molecular views of recombination proteins and their control. Nat Rev Mol Cell Biol. 2003;4:435–445. doi: 10.1038/nrm1127. [DOI] [PubMed] [Google Scholar]
- 100.Wiedemann M, Trueb B. Characterization of a novel protein (FGFRL1) from human cartilage related to FGF receptors. Genomics. 2000;69:275–279. doi: 10.1006/geno.2000.6332. [DOI] [PubMed] [Google Scholar]
- 101.Yi R, Pasolli HA, Landthaler M, Hafner M, Ojo T, Sheridan R, Sander C, O’Carroll D, Stoffel M, Tuschl T, Fuchs E. DGCR8-dependent microRNA biogenesis is essential for skin development. Proc Natl Acad Sci U S A. 2009;106:498–502. doi: 10.1073/pnas.0810766105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhang X, Emerald BS, Mukhina S, Mohankumar KM, Kraemer A, Yap AS, Gluckman PD, Lee KO, Lobie PE. HOXA1 is required for E-cadherin-dependent anchorage-independent survival of human mammary carcinoma cells. J Biol Chem. 2006;281:6471–6481. doi: 10.1074/jbc.M512666200. [DOI] [PubMed] [Google Scholar]
- 103.Zhang Y, Fei M, Xue G, Zhou Q, Jia Y, Li L, Xin H, Sun S. Elevated levels of hypoxia-inducible microRNA-210 in preeclampsia: new insights into molecular mechanisms for the disease. J Cell Mol Med. 2011 doi: 10.1111/j.1582-4934.2011.01291.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhang Z, Sun H, Dai H, Walsh RM, Imakura M, Schelter J, Burchard J, Dai X, Chang AN, Diaz RL, Marszalek JR, Bartz SR, Carleton M, Cleary MA, Linsley PS, Grandori C. MicroRNA miR-210 modulates cellular response to hypoxia through the MYC antagonist MNT. Cell Cycle. 2009;8:2756–2768. doi: 10.4161/cc.8.17.9387. [DOI] [PubMed] [Google Scholar]