Abstract
Dendritic cells (DCs) play a critical role in orchestrating the host responses to a wide variety of foreign antigens and are essential in maintaining immune tolerance. Distinct biomaterials have been shown to differentially affect the phenotype of DCs, which suggested that biomaterials may be used to modulate immune response towards the biologic component in combination products. The elucidation of biomaterial property-DC phenotype relationships is expected to inform rational design of immuno-modulatory biomaterials. In this study, DC response to a set of 12 polymethacrylates (pMAs) was assessed in terms of surface marker expression and cytokine profile. Principal component analysis (PCA) determined that surface carbon correlated with enhanced DC maturation, while surface oxygen was associated with an immature DC phenotype. Partial square linear regression, a multivariate modeling approach, was implemented and successfully predicted biomaterial-induced DC phenotype in terms of surface marker expression from biomaterial properties with R2prediction = 0.76. Furthermore, prediction of DC phenotype was effective based on only theoretical chemical composition of the bulk polymers with R2prediction = 0.80. These results demonstrated that immune cell response can be predicted from biomaterial properties, and computational models will expedite future biomaterial design and selection.
Keywords: dendritic cells, polymethacrylate, combinatorial library, material properties, principal component analysis, partial linear squares regression
Introduction
DCs are the most potent antigen presenting cells (APCs) that bridge innate and adaptive immunity. They are sparsely but widely distributed APCs of the hematopoietic origin and differentiate from CD34+ hematopoietic stem cells or CD14+ monocytes [1]. In their immature state, DCs act as sentinels that are specialized in the uptake, transport, processing and presentation of antigens to T cells. Using pattern recognition receptors (PRRs), these immature DCs (iDCs) constantly sample the environment for potentially dangerous pathogens and antigens by recognizing pathogen-associated molecular patterns (PAMPs), conserved motifs derived from pathogens [2,3]. DCs can also recognize and respond to “danger signals”, tissue fragments and intracellular molecules resulting from necrotic tissues [4], Upon stimulation with PAMPs or inflammatory cytokines, DCs internalize, process, and present the antigens to T-cells by via major histocompatibility complex (MHC) molecules, while DCs migrate to the lymph nodes and become phenotypically mature [5]. The maturation of DCs is a continuous process initiated in the periphery upon the encounter of antigens and the balance of pro- and anti-inflammatory cytokines and completed during the DC-T cell interaction in the secondary lymphoid organs [5,6]. DCs are not only critical in mediating T cell polarization for optimal immune response [7–9], but they also actively maintain immunological tolerance towards self or non-harmful antigens by actively inducing the formation of regulatory T (Treg)-cells [10].
Biomaterials commonly used in combination products were previously shown to have differential effects on DC phenotype in vitro. Specifically, poly(lactic-co-glycolic acid) (PLGA) and chitosan films promote DC maturation, agarose films maintain an iDC phenotype, and hyaluronic acid films promoted an even more immature phenotype [11,12]. In addition, PLGA, but not agarose, enhanced the humoral immune response towards a co-delivered antigen in vivo in a murine model through a biomaterial adjuvant effect [13,14]. These studies suggested that biomaterials can be used to modulate DC phenotype, thereby controlling associated in vivo host immune response to the biologic components in combination products such as tissue-engineered constructs and vaccines. However, the biomaterial systems used previously were not sufficient in deducing which material properties determined the distinct DC response. Therefore, deriving the correlations between DC phenotype and material properties using well-characterized material systems is expected to provide guidelines for immuno-modulatory biomaterial design for both tissue engineering and vaccine delivery applications.
Combinatorial and computational approaches in biomaterial design can potentially accelerate the discovery of new biomaterials and increase the diversity of promising polymeric structures for biomedical uses [15,16]. Quantitative structure-property relationship (QSPR) modeling has long been used in drug discovery [17]. This approach was translated into predicting biological response to polymeric biomaterials. For instance, computation models, including artificial neural network (ANN), surrogate modeling, and partial linear squares regression (PLSR), were developed to successfully predict fibrinogen adsorption, rat lung fibroblast growth, and/or metabolic activity after culture on a combinatorial library of 112 polyarylates from a number of material descriptor, including hydrophobicity, glass transition temperature (Tg), and chemical structure [18–21]. Furthermore, computational modeling has been used successfully to create a large virtual library of 40,000 pMAs by the Kohn laboratory with predictable fibrinogen adsorption, fibroblast attachment, and fibroblast growth based on a selection of material descriptors [22]. These models allow for rational design of biomaterials before any polymers are synthesized or any biological experiments are performed and are expected to unprecedentedly expedite the discovery of biomaterials and advance the field of biomaterials.
Previously, a differential equation-based model has been developed to describe primary macrophage fusion in response to different common biomaterials [23]. However, this model did not directly predict host response based on material properties as the predictors. Thus far, no computational models have been developed to describe or predict the response of human primary immune cells to biomaterial properties, which is of outstanding clinical relevance. In this report, the biomaterial-mediated DC phenotype was assessed for a selection of the pMAs developed in Kohn laboratory. This selected set of pMAs was previously shown to induce a wide range of biological response in terms of fibrinogen adsorption, fibroblast adhesion and growth [22]. In this report, principal component analysis (PCA) was used to identify correlations of DC phenotypic variables with pMA material properties. Furthermore, PLSR models were used to predict DC response from different sets of material property predictors.
Materials and Methods
Synthesis pMA combinatorial library
pMAs (Table 1) were prepared using chain-growth polymerization via free radical solution (FRS) and reversible addition-fragmentation transfer (RAFT) [24]. All chemicals were high purity, reagent-grade, or HPLC-grade and used as received except as noted: (i) AIBN was recrystallized from MeOH, (ii) Monomers (Fig. 1) were purified through a column of alumina to remove inhibitors, and (iii) Solvents, solutions, and monomers were degassed with nitrogen or argon before use in free radical or RAFT polymerizations.
Table 1.
List of pMAs that were used for the training set
| pMA# | Name | Abbreviation |
|---|---|---|
| 1 | Ethylene Glycol Methyl Ether MA | EGMA |
| 2 | Hydroxypropyl MA | HP |
| 3 | Isobornyl MA | Isobornyl |
| 4 | Methyl-EGMA | Me-EGMA |
| 5 | Methyl-hydroxypropyl MA | Me-HP |
| 6 | Methyl-tetrahydrofurfuryl MA | Me-THFF |
| 7 | Ethyl-Benzyl-TEGMA | E-B-TEGMA |
| 8 | Isobutyl-Benzyl-THFF MA | I-B-THFF |
| 9 | Lauryl-Isobornyl-Hydroxyethyl MA | L-I-HE |
| 10 | nButyl-Cyclohexyl-Undecyl MA | nB-C-Undecyl |
| 11 | Octyl-Isobornyl-Hydroxypropyl MA | O-I-HP |
| 12 | HEMA | HEMA |
Fig. 1.
Chemical structure of the pMA monomers
The polymerization was carried out using an automated parallel synthesizer. Briefly the synthesizer was inertized by five cycles of evacuation under vacuum at 120°C and degassed monomers (single or multiple monomers in desired mole ratios for desired target ratios), stock solutions of either AIBN (for FRS) or a cosolution of AIBN and 2-cyanoprop-2-yl dithiobezoate (for RAFT) and solvents were charged to the reactors that were vortexed at 600 rpm at 70°C for 6 hours (FRS) or 20 hrs (RAFT) under Argon. More than 90 reactions were carried out in a single run. The reactors were cooled to room temperature and the polymers were precipitated manually and dried under vacuum for more than 24 hours at 60°C. The pMAs used in this study were selected from more than 150 members of the pMA library that were synthesized using this method and consisted of homo-, co- and ter-polymers. Proton NMR was used to determine the composition and gel permeation chromatography for molecular weight determination [24]. Table 1 lists the pMAs and the corresponding abbreviations used in PCA and PLSR plots.
Coating of pMAs in 96-well plate
Each of the pMAs were dissolved 0.5% (w/v) in tetrahydrafuran (THF; Sigma, St. Louis, MO), and the solutions were used to fill the wells of a 96-well polypropylene (PP) plate (Corning, Corning, NY) to ensure that the walls of the wells were coated. The filled plate was then transferred into an Isotemp vacuum oven (Fisher Scientific). The temperature of the oven was increased 10°C/h from 40°C to 80°C. Vacuum was generated at 80°C, and the solutions were dried under vacuum for 5 days until uniform coatings were formed. Uniform coating of the wells, including the walls, was inspected by mixing the pMA solution with fluorescein to check for homogeneous fluorescence.
Synthesis and coating of terpolymer combinatorial library
The synthesis of terpolymers (Table 2) has been described previously [25]. The monomers (Fig. 2) hydroxyethyl methacrylate (ophthalmic grade) (HEMA or H), 2-ethylhexyl acrylate (EHA or A), triethyleneglycol monomethylether monomethacrylate (TEGMA or T), N-ispropylacryamide (NIPAAM or N), glycidylmethacrylate (GMA or G), and azobisisobutyronitrile (AIBN) were purchased from either Sigma-Aldrich or Polysciences and used as received. The polymers were synthesized by AlBN-initiated radical polymerization of the three monomers in DMF at 70°C. A typical reaction such as the synthesis of 40%HEMA-co-35%TEGMA-co-25%GMA was carried out as follows: HEMA (0.78 mL, 0.006 mols), TEGMA (1.29 mL, 0.005 mols), GMA (0.53 mL, 0.004 mols), and AIBN (2.5mg, 0.015 mmol) were taken place in a round bottomed flask. Dimethylformamide (DMF) (12 mL) was added and the reaction was purged by a stream of nitrogen. The reaction was heated at 70°C for 6 h with rapid stirring. The polymer was precipitated in diethyl ether. The obtained polymer was re-dissolved and precipitated 2×, after which the polymer was dried for 2 days under vacuum. Terpolymer coating procedure was carried out the same way as pMAs. The terpolymers were used as the prediction set.
Table 2.
List of terpolymers that were used for the prediction set
| terpolymer # | Abbreviation | Composition |
|---|---|---|
| 1 | 2A | A55T20G25 — 55%A-co-20%T-co-25%GMA |
| 2 | 2B | A40T35G25 — 40%A-co-35%T-co-25%GMA |
| 3 | 2D | A10T65G25 — 10%A-co-65%T-co-25%GMA |
| 4 | 5B | H40T35G25 — 40%H-co-35%T-co-25%GMA |
| 5 | 5C | H25T50G25 — 25%H-co-50%T-co-25%GMA |
| 6 | 5D | H10T65G25 — 10%H-co-65%T-co-25%GMA |
| 7 | 6A | A55H20G25 — 55%A-co-20%H-co-25%GMA |
| 8 | 6B | A40H35G25 — 40%A-co-35%H-co-25%GMA |
| 9 | 6C | A25H50G25 — 25%A-co-50%H-co-25%GMA |
| 10 | HEMA | 100% H |
Fig. 2.
Chemical structure of the monomers that made up the terpolymer prediction set.
Surface roughness and surface area measurements
All surface roughness was measured in pMA-coated 96-well wells after cutting off the walls of the wells. Line roughness (Ra) of the pMA coatings were measured by Wyko optical profilometer (Veeco, Plainview, NY) with a 5× objective. Surface area, surface roughness (Sa), and other surface roughness variables (Table S2) were measured by LEXT OLS4000 3D material confocal microscope (Olympus, Center Valley, PA) using a 20× objective in an organic cleanroom in the Marcus Nanotechnology Building at Georgia Tech. Two measurements were performed on each of three coatings for each pMA. The value for each pMA was the average of the six measurements.
Water-air contact angle and glass transition temperature (Tg) measurements
Polymers were coated on coverslips by solvent casting for contact angle measurements at Rutgers University. Contact angles were determined on a Ramé-Hart goniometer equipped with a camera and Droplmage software. A drop of deionized water was placed on the polymer film and the contact angle measured within 1 – 3 s. Tg was measured by differential scanning calorimetry (2910 Modulated DSC, TA Instruments).
Surface chemical composition measurements by X-ray photoelectron spectroscopy (XPS)
XPS was performed on pMA-coated 96-well wells after cutting off the walls of the wells at the University of Toronto. The samples were analyzed on the Thermo Scientific K-Alpha XPS spectrometer (ThermoFisher, E. Grinstead, UK). Measurements were obtained at a take-off angle (relative to the surface) of 90°. A monochromatic Al Kα X-ray source was used with a spot area of 400 μm. Charge compensation was necessary and was provided using the flood gun supplied with the instrument. Survey spectra were obtained at low energy resolution (pass energy = 150 eV) in a scanned mode. Quantification was obtained from low resolution spectra acquired in a snapshot mode. The C 1s spectrum was also recorded at high resolution (PE = 25 eV) in a scanned mode. All data processing was performed using the software package supplied with the instrument (Avantage). The high resolution peak fitting was independently performed in Georgia Tech, which included the basic C-C, O-C=O, and C-0 bonds (denoted as GT), and in University of Toronto, which included beta carbons and slight C=O contamination in addition to the three basic bonds (denoted as UT). Theoretical values were calculated based on polymer structures and molar ratios determined by NMR.
Human dendritic cell culture
Human blood was collected from healthy donors with informed consent and heparinized (333 U/ml blood) (Abraxis Pharmaceutical Products, Schaumburg, IL) at the Student Health Center Phlebotomy Laboratory, in accordance with protocol H10011 of the Institutional Review Board of Georgia Institute of Technology. Dendritic cells were derived from human peripheral blood mononuclear cells (PBMCs) using a previously described method [26] with some modifications [27]. Briefly, the collected blood was diluted 1:1 in Mg2+- and Ca2+-free phosphate buffer saline (D-PBS; Invitrogen, Carlsbad, CA), and the PBMCs were isolated by differential centrifugation using lymphocyte separation medium (Cellgro MediaTech, Herndon, VA). After the lysis of residual erythrocytes with RBC lysis buffer (155 mM NH4CI, 10 mM KHCO3, 0.1 mM EDTA), the PBMCs were washed with D-PBS. Ten milliliters of PBMCs were plated in a Primaria 100 × 20 mm2 tissue-culture dish (Becton Dickinson, Franklin Lakes, NJ) at a concentration of 5 × 106 cells/ml in DC media [RPMI-1640 (Invitrogen), 10% heat inactivated FBS (Cellgro MediaTech) and 100 U/ml of penicillin/streptomycin (Cellgro MediaTech)]. After 2 h of incubation at 95% relative humidity and 5% CO2 at 37°C for the selection of adherent monocytes, the dishes were washed three times with warm DC media to remove non-adherent cells. The remaining adherent monocytes were incubated with 10 ml/plate fresh warm DC media, supplemented with 1000 U/ml GM-CSF and 800 U/ml IL-4 (PeproTech, Rocky Hill, NJ), for 5 days to induce the differentiation of monocytes into iDCs.
Exposure of DCs to coated pMAs or terpolymers in 96-well plate
On day 5 of culture, loosely adherent and non-adherent cells containing iDCs were harvested and resuspended in DC media with 1000 U/ml GM-CSF and 800 U/ml IL-4 at 5 × 105 DCs/ml. 150 μl of cell suspension (7.5 × 104 DCs) was plated on pMA coatings in quadruplicate in the wells of a 96-well PP plate. The extent of DC maturation was compared to the reference controls cultured on TCPS 96-well plate in parallel: untreated iDCs for the negative reference control and lipopolysaccharide (LPS) (1 μg/ml; E. coli 055:B5; Sigma)-treated mDCs for the positive reference control. Four of the donors used in the pMA library experiment were used again in the terpolymer library experiment on different days.
Maturation analysis with 96-well filter plate-based high throughput (HTP) method
Differentially-treated and reference control DCs were harvested after 24 h for analysis using a HTP method previously described [27]. Briefly, all treated DCs and controls were transferred to a black 96-well filter plate, and the supernatants were immediately collected into a 96-well plate through the filters by stacking the filter plate on top of the collection plate and centrifuging at 250 × g for 2 min. A portion of the supernatants were used immediately for cytotoxicity assessment by measuring glucose-6-phosphate dehydrogenase (G6PD) release from damaged cells (described in the following section), while the remaining portion was stored at −80°C for multiplex cytokine profiling. The cells retained in the wells were assessed for maturation phenotype by immunostaining using antibodies anti-CD86-PE and anti-DC-SIGN-FITC. CD86 is a costimulatory molecule that is upregulated upon DC maturation, and DC-SIGN an endocytic receptor that is downregulated upon LPS-stimulated maturation. The fluorescent intensities were measured with a Tecan Infinite F500 microplate reader, and the ratio of CD86/DC-SIGN, a cell number independent metric named “maturation factor (MF)”, was used to represent DC maturation.
Cytotoxicity and Endotoxin Assessment
Biomaterial-induced cytotoxicity was assessed by the release of glucose-6-phosphate dehydrogenase (G6PD) into the media from cells cultured with or without biomaterials according to the manufacturer's protocol. G6PD is a cytosolic enzyme that is released from damaged or dead cells, and its relative quantity is directly correlated to cytotoxicity and was measured using the Vybrant Cytotoxicity Assay (Molecular Probes, Eugene, OR) as described previously [27]. Briefly, 50 μl of the supernatants were assayed immediately, and the fluorescent intensity was compared to that of the medium from lysed cells. The fluorescence signals were measured after 30 min incubation at 37°C with excitation and emission filters 535/25 and 590/20, respectively. The endotoxin contents of the pMA coatings were measured using a chromogenic substrate (QCL-1000 LAL assay, Lonza) and determined to be less than 0.1 EU/mL, which is well below the FDA limit of 0.5 EU/mL.
Multiplex cytokine profiling
The supernatants collected from the cell culture media in the presence of pMAs or controls were stored at −80°C and were thawed only once for multiplex cytokine analysis as described previously [28]. The concentrations of cytokines and chemokines in the cell culture supernatants were quantified using Bio-Plex suspension array systems (Bio-Rad, Hercules, CA) following the manufacturer's instructions. Briefly, beads conjugated with capture antibodies for the target proteins were thoroughly mixed and incubated with standards or supernatants for 30 min in the wells of a 96-well MultiScreenHTS-BV filter plate (Millipore, Billerica, MA). After three washes using a vacuum manifold (Pall Life Science, Ann Arbor, MI), the beads were incubated with biotinylated detection antibodies for 30 min, washed, incubated with streptavidin-PE for 10 min, and then washed. Finally, the beads were analyzed using a Bio-Plex 200 instrument with Bio-Plex Manager 4.0 software. For the pMA training set, through six independent experiments each with a different donor, cytokine analysis was performed for pro-inflammatory cytokines (IL-1β, IL-12p70, IL-15, IL-18, and TNF-α), anti-inflammatory cytokines (IL-1ra and IL-10), a pleiotropic cytokine (IL-16), and chemokines (IL-8, MCP-1, and MIP-1α). For the terpolymer prediction set, cytokine analysis was performed for IL-1β, IL-ra, IL-8, IL-16, MCP-1, and TNF-α.
Statistical analysis
To observe any significant differences between all sample groups in pairs, a pair-wise two-way ANOVA followed by Tukey post test was performed using the GraphPad Prism 5 software (La Jolla, CA), and the p-value equal to or less than 0.05 was considered significant.
Principal component analysis (PCA)
PCA was performed on the DC phenotype and pMA material property dataset to draw correlations between DC response and material properties. PCA is a dimension reduction technique, which finds a few principal components (PCs) (new axes in the PC space) that represent dimensions with maximal variability and highlight the global covariance patterns of the variables [29]. The data matrix X ε R72×29 consisted of both phenotypic and material variables (29 variables). Phenotypic variables included MF (CD86/DC-SIGN) obtained in the HTP assay and production levels of seven cytokines and chemokines (IL-1β, IL-1ra, IL-8, IL16, MCP-1, MIP-1α, and TNF-α). Values of IL-10, IL-12p70, IL15, and IL-18 measurements were very close to the detection limit or could not be detected and therefore were not included in the analysis. Material property variables included air-water contact angle (θ or Theta), glass transition temperature (Tg), line roughness, surface roughness, and surface chemical composition by XPS (Table S1). The 72 observations included six independent experiments with 12 different pMAs. PCA algorithm was applied to the data matrix to extract the latent correlations among the variables. All variables were pre-processed by mean centering and unit-variance scaling [30,31]. Based on the distribution of the variables, some were log-transformed to ensure Gaussian distributions of data. PCA was performed using the software SIMCA P+ (Umetrics, Malmö, Sweden). The performance of a PCA model can be summarized by two primary quantitative measures: goodness of fit of the model to the current dataset as given by R2 and goodness of prediction of the model for predicting outcomes of future experiments as given by Q2, a measure of the cumulative fraction of the total variation of the X block that can be predicted by all PCs. A high R2 is required for a high Q2, and a Q2 of > 0.5 is considered good [32].
Where SSXtot.corr. is the total sum of squares of X matrix (i.e. total variation in the X block), RSS is the fitted residual sum of squares, and PRESS is the predictive residual sum of squares calculated from cross-validation and is defined as
where xik is the experimental values of the variables, is the predicted values of the variables from the reduced models during cross validation, l is the row position of data matrix, and k is the column position of the data matrix. The suitable number of PCs is determined by the optimal balance between fit and predictive ability (see reference [32–34] for details).
Partial Least Square Regression (PLSR) Modeling
PLSR is a powerful computational method that expresses a set of dependent variables (outcomes) in terms of linear combinations (principal components) of the independent variables (predictors). PLSR is very similar to PCA with an added algorithm for maximizing the correlations between the X (independent or predictor variable) and Y (dependent or outcome variable) blocks in the PC space. The dataset used for PLSR included all the variables in PCA as well as additional roughness variables and was divided into two matrices: X ε R72×119, consisting of the material property measurements as the predictor variables, and Y ε R72×1, containing only one DC response variable, MF. The material variables were copied six times for the six donor trials as they were the same for each donor. Table S2 summarizes the variables included for each observation. The performance of a PLSR model can be determined by R2Y and Q2Y. A Q2Y of > 0.5 is considered good [32]. Analogous to PCA,
where SSYtot.corr is the total sum of squares of Y matrix (i.e. total variation in the Y block), RSS is the fitted residual sum of squares, and PRESS is defined as:
where Yim is the experimental values of the Y variables, is the predicted values of the variables from the reduced models during cross validation, i is the row position of data matrix, and m is the column position of the data matrix. The PCs derived from the PLSR model result in linear combinations of the predictor variables optimized for the maximum covariance with the dependent DC phenotypic variable. From this initial model, a pruning step was performed to remove variables with low variance influence on projection (VIP < 0.7) and low reliability as determined by jack-knifing [34]. PLSR modeling was performed using SIMCA P+.
The model performance was determined by how well the predicted values matched the observed values by evaluating the regression coefficient, R2prediction. Because with more explanatory terms in a model, the model fit generally tends to improve with an inflated R2prediciton, the adjusted R2prediction () is also presented here to penalize the use of more PCs in the models. is defined as:
where n is the sample size, which is 72 (6 donors × 12 treatments) in all the models, and p is the number of explanatory terms, which is equivalent to the number of principal components necessary for the model. In addition, CV-ANOVA (cross validation-ANOVA) was performed for significance testing of the models. This technique results in F-statistcs and p-values, which are useful for evaluating the significance of the model, i.e. whether the model is developed merely by chance [35].
Results
pMAs-induced DC responses
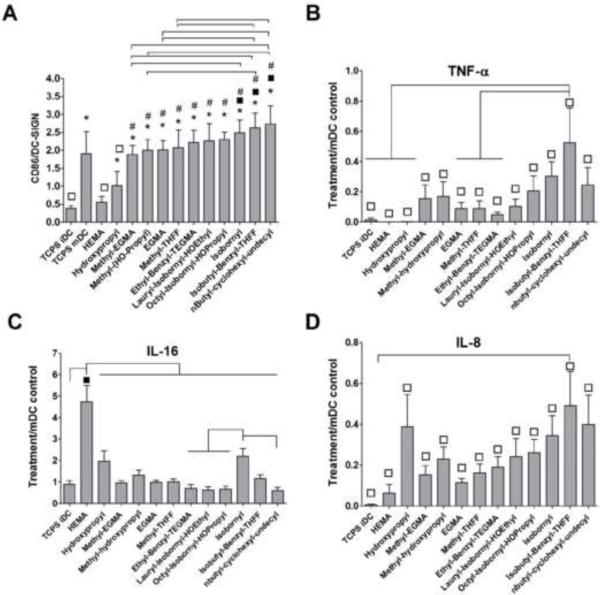
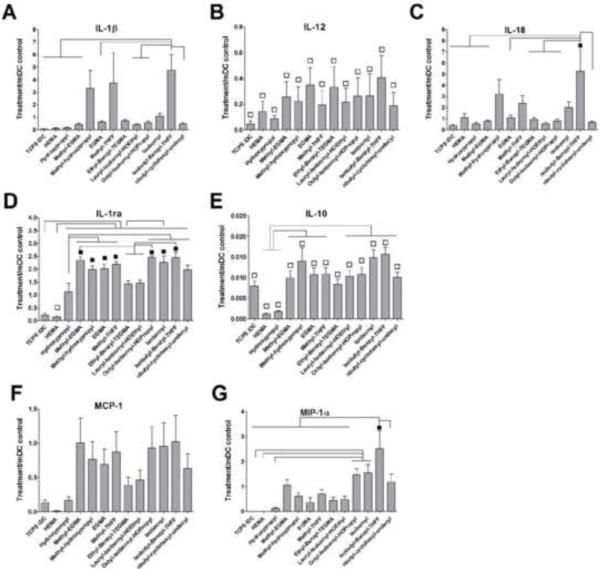
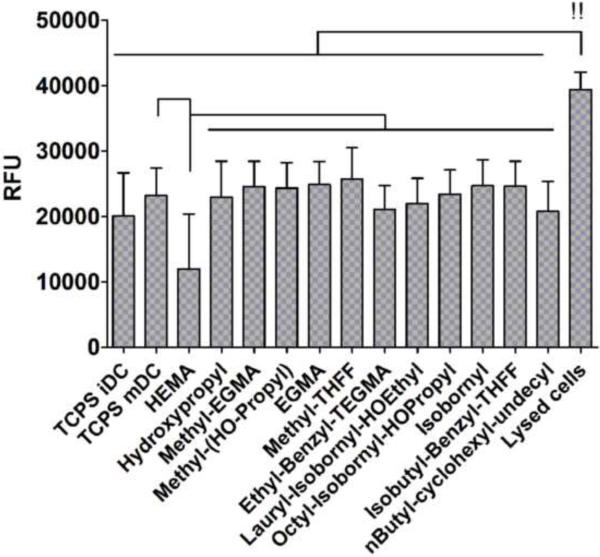
The results of material characterization of the twelve pMAs are summarized in Table 3. DCs responded differentially to this set of polymers. There was a trend of increasing DC maturation as shown by the metric CD86/DC-SIGN for the ordering of polymers used to treat DCs, as represented on the x-axis (Fig. 3A). When keeping the ordering of polymers the same, the release of pro-inflammatory cytokine, TNF-α (Fig. 3B), and chemokine, IL-8 (Fig. 3D) by treated DCs appeared to follow the same trend with pHEMA inducing the lowest amount of TNF-α and IL-8 production in DCs. In contrast, pHEMA induced the highest amount of IL-16 secretion from DCs, which was statistically different from iDC and mDC reference controls as well as all the other pMA treatments examined (Fig. 3C). Furthermore, the pMAs induced differential levels of cytokine secretion. In particular, poly(isobutyl-benzyl-THFF)MA [p(l-B-THFF)MA] induced the highest levels of TNF-α (Fig. 3B) and IL-8 (Fig. 3D) relative to all the other pMAs, while poly(isobornyl)MA induced the most IL-16 after pHEMA (Fig. 3C). Other cytokines and chemokines assayed did not appear to follow as a clear trend (Fig. 4), but they induced differential levels of IL-1β (Fig. 4A), IL-1ra (Fig. 4D), IL-10 (Fig. 4E), IL-18 (Fig. 4C), and MIP-1α (Fig. 4G) production. Generally, the pMAs on the left of the plots induced low amounts of the cytokines or chemokines from treated DCs, while the pMAs on the right induced relatively higher amounts. Again, p(l-B-THFF)MA induced the highest amounts of pro-inflammatory cytokines and chemokine such as IL-1β (Fig. 4A), IL-18 (Fig. 4C), and MIP-1α (Fig. 4G). The pMAs did not induce significant cytotoxicity in treated DCs. Interestingly, pHEMA induced lower cell death as compared to all the other pMAs (Fig. 5).
Table 3.
Summary of material property measurements for the 12 pMAs (the training set). The pMAs are ordered the same way as in Figure 6–3.
| HEMA | HP | Me-EGMA | Me-HP | EGMA | Me-THFF | E-B-TEGMA | L-I-HE | O-I-HP | Isobornyl | I-B-THFF | nB-C-Undecyl | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Theta | 69.5 | 71.2 | 17.2 | 73.8 | 21.5 | 70.5 | 20.8 | 29.0 | 62.8 | 29.2 | 28.7 | 39.0 |
| Tg | 87.6 | 83.8 | 55.8 | 90.7 | 15.2 | 91.3 | 1.6 | −7.7 | 27.3 | 130.2 | 30.9 | 148.9 |
| Si2p (E) | 0.1 | 0.3 | 2.0 | 0.1 | 0.5 | 0.2 | 2.2 | 1.0 | 0.5 | 0.4 | 0.3 | 3.0 |
| Cls (E) | 84.6 | 78.7 | 75.3 | 91.5 | 91.7 | 95.7 | 78.4 | 87.0 | 86.3 | 90.4 | 91.5 | 81.2 |
| Ols (E) | 15.3 | 21.0 | 22.7 | 8.4 | 7.8 | 4.1 | 19.4 | 12.1 | 13.2 | 9.2 | 8.2 | 15.8 |
| Cls (T) | 66.7 | 70.0 | 75.4 | 72.5 | 70.0 | 72.5 | 78.3 | 87.0 | 85.1 | 87.5 | 78.5 | 87.8 |
| Ols (T) | 33.3 | 30.0 | 24.6 | 27.5 | 30.0 | 27.5 | 21.7 | 13.0 | 14.9 | 12.5 | 21.5 | 12.2 |
| C-C (UT) | 63.9 | 62.3 | 54.9 | 71.7 | 82.2 | 68.5 | 64.7 | 76.8 | 64.5 | 74.5 | 84.0 | 75.8 |
| CO (UT) | 20.8 | 14.7 | 22.8 | 10.7 | 7.7 | 11.8 | 19.3 | 10.0 | 17.3 | 5.9 | 6.6 | 11.4 |
| O-C=O (UT) | 5.6 | 8.1 | 11.8 | 4.8 | 2.6 | 5.6 | 7.0 | 4.3 | 9.1 | 2.6 | 3.3 | 3.9 |
| Beta C | 9.7 | 11.5 | 10.5 | 12.8 | 7.5 | 13.1 | 9.0 | 9.0 | 7.6 | 14.9 | 6.1 | 4.9 |
| C=O | 0.0 | 3.4 | 0.0 | 0.0 | 0.0 | 1.1 | 0.0 | 0.0 | 1.6 | 2.0 | 0.0 | 4.0 |
| C-C (GT) | 73.6 | 76.4 | 64.9 | 84.9 | 88.9 | 75.5 | 73.8 | 89.2 | 70.8 | 87.9 | 91.3 | 90.3 |
| CO (GT) | 19.8 | 13.8 | 18.5 | 8.0 | 7.3 | 18.9 | 19.0 | 5.0 | 19.5 | 7.0 | 5.9 | 3.6 |
| O-C=O (GT) | 6.6 | 9.8 | 16.6 | 7.1 | 3.8 | 5.8 | 7.2 | 5.8 | 9.7 | 5.2 | 2.8 | 6.1 |
| C-C(T) | 0.50 | 0.57 | 0.51 | 0.58 | 0.43 | 0.62 | 0.58 | 0.85 | 0.83 | 0.86 | 0.73 | 0.86 |
| CO (T) | 0.33 | 0.29 | 0.33 | 0.27 | 0.43 | 0.14 | 0.31 | 0.09 | 0.10 | 0.07 | 0.12 | 0.07 |
| O-C=O (T) | 0.17 | 0.14 | 0.17 | 0.16 | 0.14 | 0.24 | 0.11 | 0.06 | 0.08 | 0.07 | 0.16 | 0.07 |
| Ra | 0.23 | 0.84 | 0.44 | 0.25 | 0.59 | 1.05 | 0.13 | 0.45 | 0.24 | 0.13 | 0.81 | 0.44 |
| Sq | 0.85 | 0.70 | 0.94 | 0.37 | 0.55 | 0.51 | 0.31 | 0.33 | 0.23 | 0.72 | 0.30 | 0.30 |
| Ssk | 1.49 | 0.07 | 1.37 | −0.13 | 0.51 | 5.45 | −0.53 | 3.68 | 1.84 | −1.02 | 2.29 | 4.89 |
| Sku | 22.17 | 10.96 | 29.49 | 16.1 | 47.70 | 134.5 | 30.64 | 129.5 | 48.1 | 7.91 | 66.83 | 111.96 |
| Sp | 10.89 | 9.06 | 12.35 | 7.26 | 12.67 | 13.46 | 6.16 | 9.10 | 5.71 | 6.46 | 6.98 | 8.98 |
| Sv | 4.13 | 4.92 | 8.39 | 3.13 | 8.96 | 5.47 | 4.31 | 3.94 | 1.74 | 6.87 | 4.06 | 3.56 |
| Sz | 15.02 | 13.98 | 20.74 | 10.4 | 21.63 | 18.93 | 10.46 | 13.04 | 7.45 | 13.33 | 11.04 | 12.54 |
| Sa | 0.66 | 0.53 | 0.63 | 0.27 | 0.34 | 0.31 | 0.20 | 0.20 | 0.17 | 0.54 | 0.21 | 0.18 |
| Sk | 2.05 | 1.45 | 1.52 | 0.73 | 0.82 | 0.87 | 0.51 | 0.54 | 0.53 | 1.40 | 0.64 | 0.54 |
| Spk | 0.90 | 0.57 | 1.51 | 0.35 | 0.52 | 0.68 | 0.22 | 0.35 | 0.25 | 0.39 | 0.32 | 0.39 |
| Svk | 0.67 | 0.97 | 1.19 | 0.60 | 1.13 | 0.55 | 0.64 | 0.51 | 0.22 | 1.28 | 0.32 | 0.31 |
| SMrl | 11.17 | 9.62 | 13.62 | 10.1 | 9.92 | 10.44 | 9.47 | 10.21 | 10.1 | 7.85 | 10.16 | 10.62 |
| SMr2 | 89.95 | 85.03 | 87.48 | 86.5 | 86.15 | 87.60 | 87.68 | 88.30 | 88.7 | 82.45 | 89.23 | 89.89 |
| Sxp | 1.73 | 1.30 | 1.52 | 0.63 | 0.70 | 0.75 | 0.43 | 0.45 | 0.45 | 1.24 | 0.55 | 0.45 |
| Vvv | 0.09 | 0.10 | 0.12 | 0.06 | 0.10 | 0.06 | 0.06 | 0.04 | 0.03 | 0.13 | 0.03 | 0.03 |
| Vvc | 1.01 | 0.71 | 0.90 | 0.35 | 0.40 | 0.43 | 0.24 | 0.26 | 0.25 | 0.65 | 0.31 | 0.26 |
| Vmp | 0.05 | 0.03 | 0.07 | 0.02 | 0.03 | 0.04 | 0.01 | 0.02 | 0.01 | 0.02 | 0.02 | 0.02 |
| Vmc | 0.72 | 0.59 | 0.62 | 0.28 | 0.31 | 0.32 | 0.19 | 0.19 | 0.19 | 0.59 | 0.23 | 0.19 |
| Sal | 146.1 | 5.61 | 82.17 | 70.5 | 8.29 | 59.30 | 4.14 | 25.59 | 70.9 | 3.10 | 51.71 | 72.18 |
| Str | 0.45 | 0.59 | 0.38 | 0.64 | 0.54 | 0.56 | 0.85 | 0.61 | 0.59 | 0.82 | 0.65 | 0.53 |
Fig. 3.
DC responded differential to the pMAs. A) Maturation factor (CD86/DC-SIGN). Release of B) pro-inflammatory cytokine TNF-α, C) anti-inflammatory cytokine IL-16, and D) chemokine IL-8 was represented by the fold change against iDC. Mean ± SEM; n = 6 donors. *: p<0.05 different from iDC; #: p<0.05 different from HEMA and hydroxypropyl; ■: p<0.05 higher than mDC; □: p<0.05 lower than mDC; brackets: p<0.05 different between treatments
Fig. 4.
Cytokine and chemokine profiles induced by pMA treatments of DCs. This set of cytokines and chemokines analyzed did not follow as clear trend of increasing DC maturation along the same ordering of polymers listed in the x-axis. A) IL-1β, B) IL-12p70, and C) IL-18 are pro-inflammatory cytokines; D) IL-1ra and E) IL-10 are anti-inflammatory cytokines; F) and G) are chemokines. Mean ± SEM; n = 6 donors. ■: p<0.05 higher than mDC; □: p<0.05 lower than mDC; brackets: p<0.05 different between treatments.
Fig. 5.
pMAs did not induce significant cytotoxicity of DCs. Mean ± SEM; n = 5 donors. Brackets: p < 0.05 between treatments. !! indicates the signal was beyond upper limit for this assay.
Correlations between DC phenotype and material properties
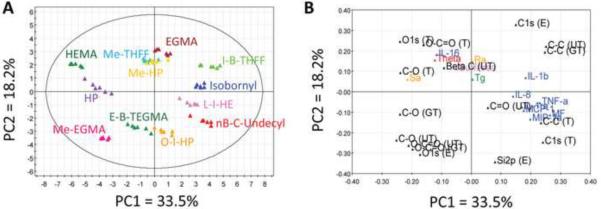
A five-component PCA model was determined by cross-validation to be the most optimal for representing this dataset with R2 = 0.78 and Q2 = 0.61, meaning that this model can capture 78% of the information in the original data space with good predictability. The five components could individually capture 33.5%, 18.2%, 10.9%, 9.6%, and 6.1% of data information, respectively. No major outliers were identified by the Hotelling's T2 statistic, which is a multivariate generalization of the Student's T distribution. The score plots showed a wide spread nature of the projection of the observations, indicating that the pMAs induced a wide range of DC responses (Fig. 6A). Furthermore, pMAs that induced low DC maturation in experiments, such as pHEMA and pHPMA, segregated to the left of the PC1, while pMAs that induced high DC maturation, such as p(l-B-THFF)MA and p(nB-C-Undecyl)MA, located to the right of PC1, suggesting that PC1 can be roughly defined as the “maturation” axis.
Fig. 6.
Score and loading plots showing the projection of the treatments and variables on the PC space. PC1 captures 33.5% and PC2 captures 18.2% of the data, which together represent >50% of the original data information. A) Score plot shows the projection of the pMA treatments, each with six data points obtained from six independent experiments with different donors. B) Loading plots shows the projection of the variables on the PC space. See text for detailed interpretation of the plots. The following color code is used for the loading plot: Blue: phenotypic variables; black: chemical composition; red: contact angle; orange: roughness; green: Tg; pink: surface area. The interpretation of the combination of PC1 with other PCs resulted in similar conclusion; therefore these plots are omitted for simplicity.
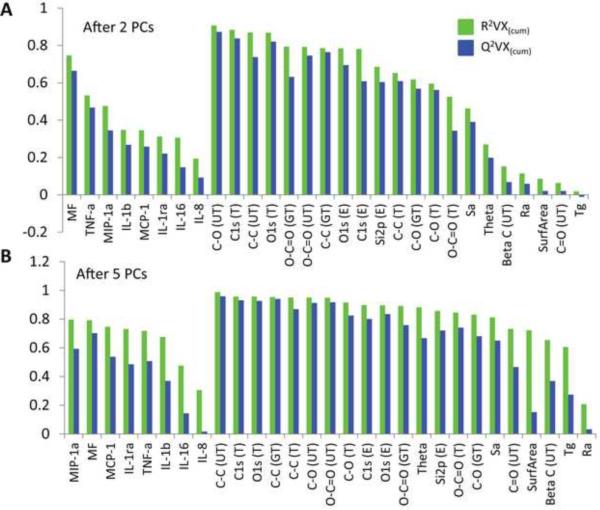
The loading plot represents how the material properties and DC phenotypes correlate to each other (Fig. 6B). First, all the phenotypic variables measured strongly clustered and mostly located in a quadrant diagonally opposite from IL-16. Overall, the theoretical values of C-C, C-O, and O-C=O bond composition mostly clustered with experimental values (both fitted in U of Toronto and Georgia Tech), and they were situated away from DC maturation variables and were associated with an immature DC phenotype. Although there was difference in the projection of theoretical and experimental values of C1s onto the other PCs, these variables were always associated with the maturation phenotypic variations along PC1 (data not shown). Most importantly, the projection of O-containing bonds in the loading plots was always diagonally opposite to that of C only bonds (Fig. 6B). In addition, contact angle and surface roughness (Sa) were located away from the maturation variables. Interestingly, line roughness (Ra), Tg, and surface area were consistently situated close to the origin of the plots except that Tg had influence on PC3, and surface area had influence on PC3 and PC5, neither of which captured more than 10% of data information (data not shown). The overall results were very similar regardless of the combinations of PCs. Although the model was best fitted with five PCs, MF and most of the surface chemical compositions could be well modeled by only two PCs (Fig. 7A). However, after five PCs were applied (Fig. 7B), many of the cytokines and chemokines, along with Tg, Ra, and surface area, were still poorly modeled, with low R2 or large difference between R2 and Q2 (an indicator of poor model performance when R2 − Q2> 0.3 [32]).
Fig. 7.
Cumulative R2 and Q2 after (A) 2 PCs or (B) 5 PCs for the PCA model.
Prediction of DC maturation levels from material properties
After pruning the initial PLSR model to remove variables of low importance and reliability (e.g. C=O contamination, Tg, Ra, surface area, etc.), the resulting X ∈ R72×91 data matrix was fitted with a two-component model with R2Y = 0.63 and Q2 = 0.58. CV-ANOVA determined that the model is significant with p-value = 4.41 ×10−12 (Table 4). Consistent with PCA, the score plot segregated pMAs that induced less DC maturation to the left of PC1, but segregated pMAs that induced high DC maturation to the right of the PC1; therefore, PC1 could also be defined as the “maturation axis” for this PLSR model (Fig. 8A). PC1 could correlate material properties to MF with R2Y = 0.52 and Q2 = 0.50, while PC2 could draw additional correlation with R2Y = 0.12 and Q2 = 0.17.
Table 4.
Summary of model performance (all models have undergone pruning steps except for the model based on only theoretical or experimental XPS)
| Predictors Used in Model | R2Y | Q2 | R2prediction | F | p-value | |
|---|---|---|---|---|---|---|
| Full set of material properties | 0.63 | 0.58 | 0.76 | 0.75 | 23.3 | 4.4×10−12 |
| Theoretical XPS only | 0.61 | 0.58 | 0.80 | 0.79 | 14.4 | 2.3×10−10 |
| Experimental XPS only (both GT and UT) | 0.21 | 0.19 | 0.26 | 0.25 | 8.3 | 5.8×10−4 |
| Experimental XPS without beta carbon fitting only (GT only) | 0.19 | 0.17 | 0.24 | 0.23 | 7.2 | 1.5×10−3 |
| Experimental XPS with beta carbon fitting only (UT only) | 0.20 | 0.19 | 0.32 | 0.31 | 7.9 | 8.5×10−4 |
Fig. 8.
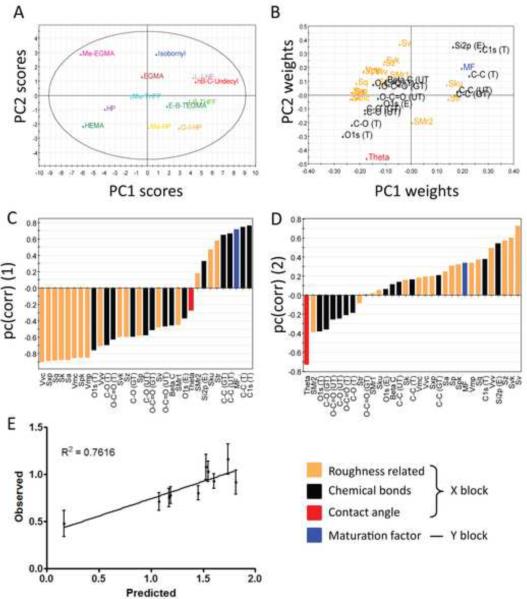
Maturation factor prediction from surface material properties using a PLSR model. A) Score plot showing the projection of the pMAs onto the PC space. B) Loading plot showing the projection of the material properties (predictor variables) onto the PC space. C–D) loading weight of the variables in PC1 (C) and PC2 (D). E) Prediction of MF induced by a different set of acrylate- and methacrylate-based polymers in six independent experiments with different donors. The error bars indicate the standard errors of the experimental values.
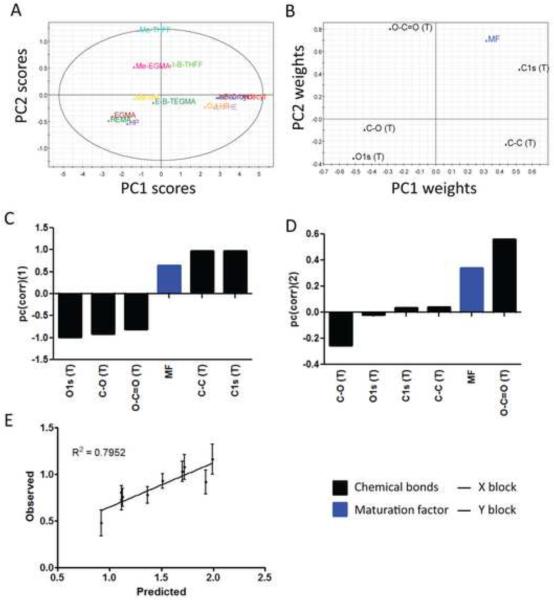
Predictor variables that were highly correlated or anti-correlated with the outcome variable, MF, could be visualized by their relative placement in the loading plot (Fig. 8B). Again, surface carbon, along with Si contamination, was strongly correlated to DC maturation, MF, whereas surface oxygen was strongly anti-correlated to DC maturation. The analysis of the loading weight of the variables (w*c) and the VIP underscored the importance of surface chemical composition, particularly the theoretical values, on predicting DC responses (Table S3–4). Specifically, among chemical composition variables, C-C (T), C1s (T), and MF had the strongest weight along positive PC1, while O1s (T), C-O (T), and O-C=O (T) had the strongest weight along negative PC1 (Fig. 8C), where “T” indicates theoretical values. As expected, the C bonds were positively weighted on PC1, while O-containing bonds were negatively weighted on PC1. Interestingly, most of the roughness structures were negatively weighted on PC1 (Fig. 8C). In contrast, contact angle (Theta) was weighted negatively and most of the roughness variables positively on PC2 (Fig. 8D).
The robustness of this model was tested by applying it to predict the MF levels in DCs induced by the prediction set, which was derived from the independent experiments using a different set of acrylate-and methacrylate-based terpolymers averaged from six donors. Linear regression on the prediction resulted in R2prediction = 0.76 (Fig. 8E) and (Table 4), indicating that the model was able to predict future experiments well, at least with methacrylate and acrylate-based polymers. The detailed phenotypic analysis and material characterization are described in Appendix 6.
Interestingly, by removing the experimental XPS values either obtained with (in UT) or without beta carbon fitting (in GT) still resulted in strong predictive models with R2prediction = 0.71 and if beta carbon fitting is included (i.e. remove XPS values at GT) (Fig. S1A) or with R2prediction = 0.77 and if beta carbon is not included (i.e. remove XPS values at UT) (Fig. S1B, Table).
Analogous models were constructed to predict the cytokine levels and were found to have R2Y < 0.55 and Q2 < 0.4. These models were also unable to predict future experiments with low R2prediction < 0.3. Due to the high correlation among the maturation variables as shown by the PCA model, a full Y block containing all these variables were expected to be reduced to a few dimensions. Therefore, a PLSR model with a full Y-block: Y ∈ R72×8 was also constructed with R2Y = 0.37 and Q2 = 0.33 (3 PCs), which was still effective in predicting MF (R2prediction = 0.71) but ineffective in predicting cytokine profile outcomes in future experiments with R2prediction < 0.15.
Prediction of DC maturation levels from theoretical chemical composition
A PLSR model was built using only the theoretical values of chemical compositions of the pMAs. A three-component model was fitted for this X ∈ R72×5 data matrix with R2Y = 0.61 and Q2 = 0.58 during cross-validation (Fig. 9) and a p-value = 2.3×10−10 based on CV-ANOVA (Table 4). PC1 can correlate material properties to MF with R2Y = 0.43 and Q2 = 0.41, PC2 can draw additional correlation with R2Y = 0.12 and Q2 = 0.18, and PC3 can capture the remaining covariance with R2Y = 0.07 and Q2 = 0.13. Similar to the previous PLSR model, PC1 could be used to roughly segregate the effects of the materials that induced differential levels of DC maturation (Fig. 9). Consistent with both PCA and previous PLSR models, surface carbon (C1s) was strongly associated with DC maturation by locating the same quadrant as MF, while surface oxygen was found to associate with an immature DC phenotype by situating oppositely to MF. In addition, surface carbon was strongly positively weighted in PC1, but surface oxygen was strongly negatively weighted in PC1 (Fig. 9C). In contrast, C1s and O-C=O along with MF had positive influence on PC2, C-C and O1s had negative influence on PC2, and C-O had little influence on PC2 (Fig. 9D). All five variables had strong influence on the model (Table S5–6). This simple model could effectively predict the MF levels in DCs induced by the prediction set (Table 2) with R2prediction = 0.80 and , indicating strong predictive robustness (Fig. 9E, Table 4).
Fig. 9.
Maturation factor prediction from theoretical surface chemical composition alone using a PLSR model. A) Score plot showing the projection of the pMAs onto the PC space. B) Loading plot showing the projection of the material properties (predictor variables) onto the PC space. C–D) loading weight of the variables in PC1 (C) and PC2 (D). E) Prediction of MF induced by a different set of acrylate- and methacrylate-based polymers in six independent experiments with different donors. The error bars indicate the standard errors of the experimental values (n=6).
Similar to the previous PLSR model, analogous models aimed at predicting the cytokine levels had low model performance with R2Y < 0.45 and Q2 < 0.4 and were unable to predict future experiments with low R2Prediction < 0.15. In addition, a PLSR model with a full Y-block; Y ∈ R72×8 also resulted in a poorly performing model R2Y = 0.37 and Q2 < 0.33 (3 PCs), which, consistent with previous PLSR model, was still effective in predicting MF with R2prediction = 0.79 but ineffective in predicting cytokine profile outcomes in future experiments with R2prediction < 0.1.
In contrast, when experimental XPS values were used alone, the predictability of the models was much poorer. PLSR models derived from a) both experimental XPS values analyzed with (in UT) or without (in GT) beta carbon fitting, b) only experimental XPS values with carbon fitting, and c) only experimental XPS values without carbon fitting all performed poorly with R2Y ≅ 0.2 and Q2 < 0.2. These models could predict MF with R2preciiction ≤ 0.32 and with p-values at least five magnitudes lower than the model developed using only theoretical values (Table 4). These models were developed with the removal of Si contamination and C=O contamination on the surfaces because the effects of these minute contamination were amplified in PLSR modeling, where each variable is given equal weight to the problem at hand. When Si and C=O were maintained in the models, although the model fit would be improved due to the addition of two more PCs, the resulting models made even poorer prediction for MF. Alternatively, a multiplier of smaller than 1 could be manually set for variables that are not expected to contribute significantly to the modeling due to their small quantities. This approach did not result in improved models.
Discussion
The phenotype of DCs was differentially modulated by the 12 pMAs. The pMAs induced a wide range of DC response as represented by the MF metric (Fig. 3). Interestingly, three of the pMAs induced higher level of DC maturation upon treatment represented by this metric as compared to LPS stimulation (TCPS mDC) (Fig. 3A). When the ordering of pMAs was kept consistent throughout the other plots, the production of TNF-α and IL-8 by DCs induced by the pMAs appeared to follow roughly the same trend with pHEMA inducing the least amount of TNF-α and IL-8 release by DCs (Fig. 3B,D). In contrast, pHEMA induced the highest amount of IL-16 production that is significantly different from iDCs, mDCs and all other pMA treatments (Fig. 3C). TNF-α is a pro-inflammatory cytokine, and IL-8 is a chemokine, both of which are released upon DC maturation. IL-16 has been shown to be a pleiotropic cytokine that can have both pro- and anti-inflammatory properties. The functions of IL-16 are likely dependent on the presence of surrounding cell types and cytokines in the microenvironment [28]. Because pHEMA induced very low maturation marker expression and low secretion of cytokines and chemokines that are typically elevated in inflammation, the pHEMA-treated DCs likely produced IL-16 as a part of anti-inflammatory response.
The production of the other seven cytokines by DCs did not follow a clear trend based on the same ordering of pMAs in the x-axis (Fig. 4), but the low amounts of cytokine release are clustered to the left while the higher amounts clustered to the right, which is consistent with the DC maturation represented by MF. Particularly, this set of pMAs induced differential levels of IL-1β (Fig. 4A), IL-1ra (Fig, 4D), IL-10 (Fig. 4E), IL-18 (Fig. 4C), and MIP-1α (Fig. 4G) secretion. Most notably, besides TNF-α (Fig. 3B) and IL-8 (Fig. 3D), p(I-B-THFF)MA induced the highest levels of IL-1β, IL-18, and MIP-1α production by treated DCs relative to all other pMA treatments. Since p(I-B-THFF)MA-treated DCs also expressed high level of MF, it can be concluded that p(I-B-THFF)MA potentially induced the highest level of DC maturation among these pMAs examined. Anti-inflammatory cytokines IL-1ra and IL-10 increased with enhanced DC maturation as well. Most likely, these anti-inflammatory cytokines have been naturally up-regulated as a negative feedback to modulate the inflammatory response [36], Interestingly, pHEMA induced very low levels of all these cytokines and chemokines; in some cases, the levels were even lower than those induced by iDCs. IL-15 was only detectable in one of the six donors and the data were not shown.
pHEMA, while least activating on DCs, supported the least Fgn adsorption (Fig. S2A). In contrast, DC-stimulating pMAs such as p(isobornyl)MA or p(I-B-THFF)MA supported the most Fgn adsorption. However, no clear correlations exist between DC response and amount of Fgn adsorption (Fig. S2A). Previously, Fgn-coated beads were shown to induce secretion of inflammatory cytokines/chemokines such as IL-6, IL-8, MIP-1β, and MCP-1 [37], which is consistent with the results herein that some of the most activating pMAs also adsorbed the most Fgn. The lack of matching trends between DC response and Fgn adsorption could be due to the change of protein conformation or orientation on some of these pMA surfaces. Many studies have suggested that adsorbed proteins might acquire enhanced or impaired functions, most likely due to conformational changes of the adsorbed proteins [38].
PCA was applied to the dataset that contains both the DC phenotype and material property information to draw correlations among the variables. PCA reduced the dimensions of the dataset into lower dimension space to facilitate the analysis of latent relationships. Most of the cytokine variables were log-transformed due to the multi-magnitude differences induced by the pMA treatments. A five-component PCA model was the most optimal to represent the original dataset in the new principal component space with R2 = 0.78 and Q2 = 0.61. These values are excellent for this dataset, which contains many phenotypic and material property variables (29 variables) and large donor-to-donor variations, particularly in the cytokine profiles. pMAs were spread along PC1 in the score plot roughly based on their effects on DC maturation, so PC1 was coarsely defined as the “maturation axis” (Fig. 6A). PCA allows the multi-dimensional data to be projected onto this reduced PC space for easy visualization of the correlations among the variables.
The loading plots represent the correlations among the variables (Fig. 6B). Consistent with the experimental results, all the phenotypic variables associated with DC maturation formed a cluster and located diagonally opposite from IL-16 (Fig. 6B). The strong association of the phenotypic variables suggests that high redundancy exists, and potentially only a few of these variables need to be assayed to obtain general phenotypic information about DCs. Overall, all the carbon bonds located at positive PC1 but the oxygen bonds at negative PC1 (Fig. 6B). As expected, theoretical XPS values mostly clustered with experimental values regardless of whether beta carbons were taken into account during peak fitting. Material properties such as Ra, Tg and surface area located primarily close to the origin of the loading plots, indicating that these variables are not important in predicting DC response. In contrast, high Sa is associated with lower DC maturation in this pMA library (Fig. 6B). Sa is computed across the entire area in the field of view, while Ra only measures the roughness along some arbitrary line across a surface. For a uniform surface, Ra and Sa are expected to be very similar, but this may not be the case for a heterogeneous surface. Since the pMAs were solvent cast into the PP plates, the formation of the films was not homogeneous, and therefore Sa should be a better roughness variable for such surfaces. Generally speaking, the carbon bonds located in a quadrant was diagonally opposite from the oxygen bonds in all the loading plots (Fig. 6B), indicating that surface carbon and oxygen anti-correlated each other in terms of inducing DC maturation. Interestingly, increase in contact angle was associated with low DC maturation for this set of pMAs. It is important to note that the range of contact angle for this set of polymers was 17.2 – 71.2°. Therefore, the conclusion drawn in the PCA model is only valid for this range of contact angles. The higher contact angles (close to 71.2°) are similar to that of TCPS [39], which is known to be suitable for most cell cultures and may be approximately the optimal contact angles for maintaining DC immature state. However, close inspection of Table 3 indicated that no strong correlation exists between DC maturation and contact angle, but pMAs with higher contact angles do group towards the left of Table 3 with pMAs such as pHEMA and pHPMA that caused low DC maturation.
Fig. 7 indicated that although the model was best fitted with five PCs, the variations in MF and most of the surface chemical composition variables were well-captured by the first two PCs (Fig. 7A). In contrast, most of the cytokines and chemokines were still poorly modeled after five PCs (Fig. 7B), presumably due to the large high donor-to-donor variations that introduced significant noise to model.
Subsequently, predictive models were constructed in an attempt to predict DC phenotype using PLSR. PLSR is very similar to PCA; the major difference is that in finding the optimal PCs to describe the variations in the X-block, the algorithm also maximizes the correlations between the X- and Y-blocks, and the resulting model can be used to predict outcomes of future experiments. Because MF was determined from PCA as the best DC phenotypic variable that could be modeled (Fig. 7), a PLSR was first developed to correlate material properties and MF variations. To begin the modeling, all material variables were included into the X-block. From this initial model, pruning steps were implemented to exclude variables that have little importance or reliability to reduce the X-block, including C=O contamination, Tg, Ra, surface area, and a few others. Such procedures were successful in creating a model with R2Y = 0.63 and Q2 = 0.58 with model significance of p-value = 4.4×10−12, which was able to predict future experiment with a high R2prediction = 0.76 and (Fig. 8E and Table 4). As expected, the score (Fig. 8A) and loading (Fig. 8B) plots resulted in similar conclusions as in the PCA model, with MF being the most strongly associated with surface carbon due to its proximity with carbon bonds in the loading plot (Fig. 8B). In addition, MF was anti-correlated with surface oxygen by locating at opposite quadrants (Fig. 8B).
Interestingly, theoretical values for XPS were weighted more heavily to the PC space (Fig. 8C–D) with higher VIP (Table S3 – 4) as compared to the experimental values. A possible explanation is that XPS has large inherent errors (30 – 50%), which affects the projection of these variables onto the PC space. Since this set of pMA films was prepared from bulk materials without further modification, the surface chemical composition should be similar to the bulk composition unless serious contamination is present. The theoretical values might actually reflect a more accurate estimation of the surface composition, while the experimental values contained extra instrumental errors that affect their projection onto the model. However, XPS was still valuable to ensure that the material surfaces were not contaminated during regular storage. If surface modification was implemented, XPS must be performed to obtain a better estimation of surface chemical composition for model development.
After removing either set of experimental XPS values, the model was improved to have prediction regression coefficient of R2prediction = 0.71 and (if the set of experiment XPS values without beta carbon fitting [GT] was removed) and R2prediction = 0.77 and (if the set of experimental XPS values with beta carbon fitting [UT] was removed), respectively. This further supports the hypothesis that experimental XPS values could deteriorate the predictive power of this PLSR model, and that the theoretical values of surface composition may be the most informative. However, experimental XPS is expected to be necessary to determine the actual surface composition if surface modification is performed on the materials.
Analogous models for cytokine and chemokine level prediction were found to be poor in performance with R2Y < 0.55 and Q2<0.4 and were not able to predict outcomes in future experiments, likely due to the high donor-to-donor variations in cytokine profiles.
PCA indicated that a strong covariance exists among the phenotypic variables that are associated with DC maturation, suggesting their strong redundancy and that a Y-block that contains all the DC response variables would be suitable (because it can be reduced to lower dimensions). However, a PLSR model built with a full Y-block resulted in a poor model (R2Y = 0.37, Q2 = 0.33). Although this model has poor predictive power for the cytokines and chemokines, it was quite effective in predicting MF (R2prediction = 0.71). Again, this confirms that MF was the only predictable phenotypic variable among those assayed.
Since theoretical XPS values appeared to be the most important in predicting DC response for the pMA library, a simple model with only these variables as the X-block was developed (Fig. 9). The resulting three-component model had fair performance with R2Y = 0.61 and Q2 = 0.58 with model significance of p-value = 2.3×10−10. This model could predict MF levels in future experiments with R2prediction = 0.80 and (Fig. 9E and Table 4). Similar to the previous PLSR model, cytokine and chemokine levels were not predictable in analogous models. In addition, a model with a full Y-block was able to predict MF (R2prediction = 0.79) but not cytokines and chemokines.
When experimental XPS values were used alone to construct the model, the resulting models performed prediction poorly (Table 4). If both sets of experimental XPS values (i.e. experimental XPS values with beta fitting [UT] and without beta fitting [GT]) were used, the model could only predict future outcomes at R2prediction = 0.26 and . If the set of XPS values without beta carbon (fitted in GT) was used only, the predictability was merely R2prediction = 0.24 and . If the set of XPS values with beta carbon (fitted in UT) was used only, the predictability was improved to R2prediction = 0.32 and . Furthermore, the p-values for these models were at least five magnitudes lower than the previous models. Therefore, at least in this study, theoretical XPS values were the most informative for the prediction of DC response based on material characteristics.
We hypothesized that the importance of material chemistry on dendritic cell response is because chemistry contributes to the fundamental differences among the pMAs. The change of chemistry affects many other bulk properties of polymers such as glass transition temperature and wettability. Previous research has demonstrated that ECM proteins adsorbed on tissue-culture polystyrene affected DC morphology, cytokine production, and allstimulatory capacity. Furthermore, self-assembled monolayers presenting -CH3, -OH, -COOH, or -NH2 presented different profiles of carbohydrate ligands of C-type lectin receptors [40]. Therefore, the chemical differences resulted in differences in surface properties, which in turn directed the presentation, orientation, or conformation of adsorbed proteins. These “biomaterial-associated molecular patterns” [41] modulated DC response based on the pMAs used to treat the cells.
Although biomaterial chemistry was shown in this study to be the most critical material property for predicting DC response in vitro, many previous studies have also shown that despite the differential levels of protein adsorption and short-term leukocyte infiltration induced by varied surface chemistries, similar long term inflammatory outcome of fibrous capsule formation was achieved in vivo. For example, polyethylene terephthalate (PET) surfaces functionalized with -OH, -NH2, or -CF3 induced differential adsorption and denaturation of fibrinogen. Although these materials also induced different numbers of total adherent phagocytes depending on the functional group (-NH2> -CF3> -OH) after short term implantation, the chronic fibrotic outcome was similar despite the different chemistries [42]. In another study, tetrafluoroethylene-hexafluoropropylene-copolymer (FEP) films were plasma polymerized with tetraglyme to minimize fibrinogen adsorption on the surfaces. However, in vivo studies showed that FEP films with or without polymerized tetraglyme resulted in similar levels of fibrous encapsulation [43]. However, Thevenot et al. hypothesized that the ineffectiveness of surface chemistry on long-term in vivo fibrotic reactions might be due to inefficient interactions between the cells and surface functional groups [44]. By enhancing the cell-surface interaction using microspheres with different functionalities, Kamath et al. showed surfaces with -OH and -NH2 groups induced stronger fibrotic reaction in comparison to -CFx and -COOH surfaces [45]. Therefore, in order to translate the in silico predictions into in vivo host response, biomaterials that cause extremes of DC phenotype should be selected based on the predictions generated by the computational model. Then, these biomaterials would first be used to validate the induced DC phenotype upon treatment in vitro. Subsequently, scaffolds or microspheres of these biomaterials would be created to maximize the interactions between the host and the material for optimal biomaterial effects in vivo. These biomaterial scaffolds or microspheres with optimal architecture for host interaction are expected to differentially modulate host response against the implanted devices according the biomaterial chemistry, which exerts its effects on host response through the adsorbed layer of proteins.
The 12 pMAs used in this study were selected from a larger library of pMAs that were used for computational modeling of Fgn adsorption and fibroblast attachment and proliferation [46]. Different sets of material descriptors were necessary for the prediction of Fgn adsorption or fibroblast response on different sets of pMAs. Therefore, a different set of material descriptors may be required to optimize the prediction accuracy of a certain biological response for a particular group of polymers. In this study, simple chemistry information of the pMAs was sufficient for DC response prediction. However, it would be of interest to test the model with a larger set of materials to determine its reproducibility. Due to the large variations in the biological responses of human primary DCs, it is important to consistently produce polymer coatings for biological experiments for any modeling efforts.
Altogether, the results reported herein demonstrated co-variations exist between DC response and material properties as demonstrated by PCA with surface chemistry being the most influential in resultant DC response. More importantly, DC response can be predicted by basic material properties. However, not all phenotypic variables are predictable. In this study, MF was a predictable variable, while none of the cytokines and chemokines was predictive. A potential explanation for the lack of predictability of cytokines and chemokines could be that these molecules are the most downstream in the inflammatory response and are subjected to very complex post-transcriptional control [47] as well as degradation upon release into the culture medium. Large donor-to-donor variations might be caused by the individuals' unique network of mechanisms that regulate cytokine release. These non-specific variations cannot be projected onto the PLSR models well and distort the model; therefore, they cannot be used for future predictions.
This study also demonstrated that an overall DC response represented by MF can be predicted using only theoretical chemical composition values. This has very significant implications for future biomaterial design. Researcher may potentially design a polymer based on the optimal ratios of chemical bonds to achieve a certain target DC phenotype and save many research dollars that would otherwise be spent on the testing of unnecessary polymer formulations. It is important to note that these models did not differentiate aromatic rings from single C-C bonds (as in XPS). A model with more complex chemical structure information is expected to have more predictive power.
Several limitations of the models exist. First, as seen by the slope ≠ 1 in the plots in Fig. 8E, 9E, and S1, the models were able to differentiate the relative DC response that can be induced by a set of polymers, but they do not suggest a particular level of DC response. Therefore, these models are the most useful for theoretically predicting the effect of a set of polymers on DCs, from which the researcher can choose materials that induce low, medium, or high DC maturation and perform further experimentation to verify the prediction. Second, the models were tested on acrylate- and methacrylate-based materials that contain only carbon and oxygen with strong predictive robustness. The models were not as effectively in predicting DC response to PP surface (data not shown) or materials from a different category. A model can only predict what it is trained on. Therefore, predictive PLSR models are expected to be required for different classes of polymers, although a model developed with very large dataset composed of different classes of materials may be explored for its predictive power. Third, this model did not take into account other important material properties such as polymer swelling or polymer mechanical properties, which could be an important material property that directs DC response. It is noteworthy that pHEMA did noticeably absorb cell culture medium to a greater extent than other pMAs, which could have contributed to its non-inflammatory properties. However, polymer swelling is often measured using bulk materials and the results might not be applicable to coated thin films. More reliable evaluation methods for the swelling of polymer thin film should be employed. Finally, immune response is very complex with different polarization, including Th1, Th2, Th17, and Treg [8,48–50]; therefore, DC response based on surface marker expression can only capture a part of the response.
Despite the limitations, these predictive models can assist future biomaterial design for the applications in tissue engineering, vaccine delivery, or cancer therapy, where effective and appropriate immune response is required. Potential immune response approximated by DC maturation factor can be predicted based on polymer formulation before any experimentation is performed, which is expected to expedite the advances in immuno-modulatory biomaterial design.
Conclusion
Different pMAs were shown to induce differential DC phenotype upon treatment. PCA was used to determine the co-variations between material properties and DC response. Furthermore, the phenotypic response based on maturation factor (based on the ratio of CD86/DC-SIGN surface expression) could be predicted from the properties of the materials used to treat the cells. Interestingly, theoretical values of chemical composition of the bulk materials were sufficient in the prediction, suggesting that DC response can be predicted from polymers prior to lengthy polymer synthesis and material characterization procedures. However, cytokine and chemokine levels were not successfully predicted by the model, presumably due to the large donor-to-donor variation in their production by DCs.
Supplementary Material
Prediction results using PLSR models similar to first PLSR model built with surface material properties but with high resolution experimental surface chemical composition fitted (A) at GT (no beta carbons and C=O) removed from the X-block, and (B) at UT (with beta carbons and C=O) removed from the X-block. After pruning steps, prediction of MF induced by the prediction set terpolymers in six independent experiments with different donors was determined. The error bars indicate the standard errors of the experimental values (n = 6).
Fgn adsorption (A) and fibroblast proliferation (B) on the pMAs. The Fgn adsorption on each pMA was normalized by that on the control polypropylene surface (Mean ± SD; n = 14 wells). Fibroblast proliferation on each pMA was normalized by that on the control TCPS surface and is represented in terms of relative proliferation index. Detailed methods are published in [46]. The corresponding CD86/DC-SIGN ratios (solid squares, Mean ± SEM) are also shown for comparison. Brackets: p < 0.05 different between treatments for fibrinogen adsorption (A) or relative proliferation index (B).
Acknowledgement
The authors thank Dr. Melissa L. Kemp of Georgia Institute of Technology for helpful discussion on multivariate analysis and critical review of the manuscript, Dr. Manu O. Platt of Georgia Institute of Technology for access to the Simca P+ software, and Dr. Rana Sodhi for performing the XPS measurements and analysis. This research was supported by the National Institutes of Health grants EB004633 (JEB), and the 96-well pMA testbed was developed under NIH grant EB001046 (RESBIO) at NJ Center for Biomaterials, Rutgers University.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Shortman K, Naik SH. Steady-state and inflammatory dendritic-cell development. Nat Rev Immunol. 2007;7:19–30. doi: 10.1038/nri1996. [DOI] [PubMed] [Google Scholar]
- 2.Trinchieri G, Sher A. Cooperation of toll-like receptor signals in innate immune defence. Nat Rev Immunol. 2007;7:179–190. doi: 10.1038/nri2038. [DOI] [PubMed] [Google Scholar]
- 3.Janeway CA, Jr, Medzhitov R. Innate immunity: lipoproteins take their toll on the host. Curr Biol. 1999;9:R879–R882. doi: 10.1016/s0960-9822(00)80073-1. [DOI] [PubMed] [Google Scholar]
- 4.Matzinger P. The danger model: a renewed sense of self. Science. 2002;296:301–305. doi: 10.1126/science.1071059. [DOI] [PubMed] [Google Scholar]
- 5.Banchereau J, Briere F, Caux C, Davoust J, Lebecque S, Liu Y-J, et al. Immunobiology of dendritic cells. Annu Rev Immunol. 2000;18:767–811. doi: 10.1146/annurev.immunol.18.1.767. [DOI] [PubMed] [Google Scholar]
- 6.Jarrossay D, Napolitani G, Colonna M, Sallusto F, Lanzavecchia A. Specialization and complementarity in microbial molecule recognition by human myeloid and plasmacytoid dendritic cells. EurJ Immunol. 2001;31:3388–3393. doi: 10.1002/1521-4141(200111)31:11<3388::aid-immu3388>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 7.Kapsenberg ML. Dendritic-cell control of pathogen-driven T-cell polarization. Nat Rev Immunol. 2003;3:984–993. doi: 10.1038/nri1246. [DOI] [PubMed] [Google Scholar]
- 8.Kalinski P, Hilkens CMU, Wierenga EA, Kapsenberg ML. T-cell priming by type-1 and type-2 polarized dendritic cells: the concept of a third signal. Immunol Today. 1999;20:561–567. doi: 10.1016/s0167-5699(99)01547-9. [DOI] [PubMed] [Google Scholar]
- 9.Langenkamp A, Messi M, Lanzavecchia A, Sallusto F. Kinetics of dendritic cell activation: impact on priming of TH1, TH2 and nonpolarized T cells. Nat Immunol. 2000;1:311–316. doi: 10.1038/79758. [DOI] [PubMed] [Google Scholar]
- 10.Steinman RM, Hawiger D, Nussenzweig MC. Tolerogenic dendritic cells. Annu Rev Immunol. 2003;21:685–711. doi: 10.1146/annurev.immunol.21.120601.141040. [DOI] [PubMed] [Google Scholar]
- 11.Babensee JE, Paranjpe A. Differential levels of dendritic cell maturation on different biomaterials used in combination products. J Biomed Mater Res A. 2005;74A:503–510. doi: 10.1002/jbm.a.30429. [DOI] [PubMed] [Google Scholar]
- 12.Yoshida M, Babensee JE. Differential effects of agarose and poly(lactic-co-glycolic acid) on dendritic cell maturation. J Biomed Mater Res A. 2006;79A:393–408. doi: 10.1002/jbm.a.30798. [DOI] [PubMed] [Google Scholar]
- 13.Bennewitz NL, Babensee JE. The effect of the physical form of poly(lactic-co-glycolic acid) carriers on the humoral immune response to co-delivered antigen. Biomaterials. 2005;26:2991–2999. doi: 10.1016/j.biomaterials.2004.08.023. [DOI] [PubMed] [Google Scholar]
- 14.Norton LW, Park J, Babensee JE. Biomaterial adjuvant effect is attenuated by anti-inflammatory drug delivery or material selection. J Control Release. 2010;146:341–348. doi: 10.1016/j.jconrel.2010.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kohn J. New approaches to biomaterials design. Nat Mater. 2004;3:745–747. doi: 10.1038/nmat1249. [DOI] [PubMed] [Google Scholar]
- 16.Kohn J, Welsh WJ, Knight D. A new approach to the rationale discovery of polymeric biomaterials. Biomaterials. 2007;28:4171–4177. doi: 10.1016/j.biomaterials.2007.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Livingstone DJ, Manallack DT. Neural Networks in 3D QSAR. QSAR CombSci. 2003;22:510–518. [Google Scholar]
- 18.Smith JR, Seyda A, Weber N, Knight D, Abramson S, Kohn J. Integration of combinatorial synthesis, rapid screening, and computational modeling in biomaterials development. Macromol Rapid Comm. 2004;25:127–140. [Google Scholar]
- 19.Smith JR, Knight D, Kohn J, Rasheed K, Weber N, Kholodovych V, et al. Using surrogate modeling in the prediction of fibrinogen adsorption onto polymer surfaces. J Chem Inf Comp Sci. 2004;44:1088–1097. doi: 10.1021/ci0499774. [DOI] [PubMed] [Google Scholar]
- 20.Kholodovych V, Smith JR, Knight D, Abramson S, Kohn J, Welsh WJ. Accurate predictions of cellular response using QSPR: a feasibility test of rational design of polymeric biomaterials. Polymer. 2004;45:7367–7379. [Google Scholar]
- 21.Smith JR, Kholodovych V, Knight D, Welsh WJ, Kohn J. QSAR models for the analysis of bioresponse data from combinatorial libraries of biomaterials. QSAR Comb Sci. 2005;24:99–113. [Google Scholar]
- 22.Kholodovych V, Gubskaya AV, Bohrer M, Harris N, Knight D, Kohn J, et al. Prediction of biological response for large combinatorial libraries of biodegradable polymers: Polymethacrylates as a test case. Polymer. 2008;49:2435–2439. [Google Scholar]
- 23.Chang D, Saidel G, Anderson J. Dynamic systems model for lymphocyte interactions with macrophages at biomaterial surfaces. Cell Mol Bioeng. 2009;2:573–590. [Google Scholar]
- 24.Rojas R, Harris NK, Piotrowska K, Kohn J. Evaluation of automated synthesis for chain and step-growth polymerizations: Can robots replace the chemists? J Polym Sci A1. 2009;47:49–58. [Google Scholar]
- 25.Joy A, Cohen DM, Luk A, Anim-Danso E, Chen C, Kohn J. Control of surface chemistry, substrate stiffness, and cell function in a novel terpolymer methacrylate Library. Langmuir. 2011;27:1891–1899. doi: 10.1021/la103722m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Romani N, Gruner S, Brang D, Kämpgen E, Lenz A, Trockenbacher B, et al. Proliferating dendritic cell progenitors in human blood. J Exp Med. 1994;180:83–93. doi: 10.1084/jem.180.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kou PM, Babensee JE. Validation of a high-throughput methodology to assess the effects of biomaterials on dendritic cell phenotype. Acta Biomater. 2010;6:2621–2630. doi: 10.1016/j.actbio.2010.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kou PM, Schwartz Z, Boyan BD, Babensee JE. Dendritic cell responses to surface properties of clinical titanium surfaces. Acta Biomater. 2011;7:1354–1363. doi: 10.1016/j.actbio.2010.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Janes KA, Yaffe MB. Data-driven modelling of signal-transduction networks. Nat Rev Mol Cell Biol. 2006;7:820–828. doi: 10.1038/nrm2041. [DOI] [PubMed] [Google Scholar]
- 30.Janes KA, Kelly JR, Gaudet S, Albeck JG, Sorger PK, Lauffenburger DA. Cue-signal-response analysis of TNF-induced apoptosis by partial least squares regression of dynamic multivariate data. J Comput Biol. 2004;11:544–561. doi: 10.1089/cmb.2004.11.544. [DOI] [PubMed] [Google Scholar]
- 31.Albeck JG, MacBeath G, White FM, Sorger PK, Lauffenburger DA, Gaudet S. Collecting and organizing systematic sets of protein data. Nat Rev Mol Cell Biol. 2006;7:803–812. doi: 10.1038/nrm2042. [DOI] [PubMed] [Google Scholar]
- 32.Eriksson L, Johansson E, Kettaneh-Wold N, Trygg J, Wikstrom C, Wold S. Multi- and megavariate data analysis. 2nd ed. Umetrics AB; Umea, Sweden: 2006. [Google Scholar]
- 33.Wold S, Sjöström M, Eriksson L. PLS-regression: a basic tool of chemometrics. Chemometr Intell Lab. 2001;58:109–130. [Google Scholar]
- 34.Martens H, Martens M. Multivariate analysis of quality An introduction. John Wiley & Sons; [Google Scholar]
- 35.Eriksson L, Trygg J, Wold S. CV-ANOVA for significance testing of PLS and OPLS models. J Chemometr. 2008;22:594–600. [Google Scholar]
- 36.Chi H, Barry SP, Roth RJ, Wu JJ, Jones EA, Bennett AM, et al. Dynamic regulation of pro- and anti-inflammatory cytokines by MAPK phosphatase 1 (MKP-1) in innate immune responses. P Natl Acad Sci USA. 2006;103:2274–2279. doi: 10.1073/pnas.0510965103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thacker RI, Retzinger GS. Adsorbed fibrinogen regulates the behavior of human dendritic cells in a CD18-dependent manner. Exp Mol Pathol. 2008;84:122–130. doi: 10.1016/j.yexmp.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 38.Wilson CJ, Clegg RE, Leavesley DI, Pearcy MJ. Mediation of biomaterial–cell interactions by adsorbed proteins: A Review. Tissue Eng. 2005;11:1–18. doi: 10.1089/ten.2005.11.1. [DOI] [PubMed] [Google Scholar]
- 39.Kwon OH, Kikuchi A, Yamato M, Sakurai Y, Okano T. Rapid cell sheet detachment from poly(N-isopropylacrylamide)-grafted porous cell culture membranes. J Biomed Mater Res. 2000;50:82–89. doi: 10.1002/(sici)1097-4636(200004)50:1<82::aid-jbm12>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 40.Shankar SP, Chen II, Keselowsky BG, García AJ, Babensee JE. Profiles of carbohydrate ligands associated with adsorbed proteins on self-assembled monolayers of defined chemistries. J Biomed Mater Res A. 2010;92A:1329–1342. doi: 10.1002/jbm.a.32457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kou PM, Babensee JE. Macrophage and dendritic cell phenotypic diversity in the context of biomaterials. J Biomed Mater Res A. 2011;96A:239–260. doi: 10.1002/jbm.a.32971. [DOI] [PubMed] [Google Scholar]
- 42.Tang L, Wu Y, Timmons RB. Fibrinogen adsorption and host tissue responses to plasma functionalized surfaces. J Biomed Mater Res. 1998;42:156–163. doi: 10.1002/(sici)1097-4636(199810)42:1<156::aid-jbm19>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 43.Shen MC, Martinson L, Wagner MS, Castner DG, Ratner BD, Horbett TA. PEO-like plasma polymerized tetraglyme surface interactions with leukocytes and proteins: in vitro and in vivo studies. J Biomater Sci-Polym E. 2002;13:367–390. doi: 10.1163/156856202320253910. [DOI] [PubMed] [Google Scholar]
- 44.Thevenot P, Hu WJ, Tang LP. Surface chemistry influences implant biocompatibility. Curr Top Med Chem. 2008;8:270–280. doi: 10.2174/156802608783790901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kamath S, Bhattacharyya D, Padukudru C, Timmons RB, Tang L. Surface chemistry influences implant-mediated host tissue responses. J Biomed Mater Res A. 2008;86A:617–626. doi: 10.1002/jbm.a.31649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ghosh J, Lewitus DY, Chandra P, Joy A, Bushman J, Knight D, et al. Computational modeling of in vitro biological responses on polymethacrylate surfaces. Polymer. 2011;52:2650–2660. doi: 10.1016/j.polymer.2011.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Anderson P. Post-transcriptional control of cytokine production. Nat Immunol. 2008;9:353–359. doi: 10.1038/ni1584. [DOI] [PubMed] [Google Scholar]
- 48.Moser M, Murphy KM. Dendritic cell regulation of TH1-TH2 development. Nat Immunol. 2000;1:199–205. doi: 10.1038/79734. [DOI] [PubMed] [Google Scholar]
- 49.Stockinger B, Veldhoen M. Differentiation and function of Th17 T cells. Curr Opin Immunol. 2007;19:281–286. doi: 10.1016/j.coi.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 50.Sakaguchi S. Naturally arising CD4(+) regulatory T cells for immunologic self-tolerance and negative control of immune responses. Annu Rev Immunol. 2004;22:531–562. doi: 10.1146/annurev.immunol.21.120601.141122. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Prediction results using PLSR models similar to first PLSR model built with surface material properties but with high resolution experimental surface chemical composition fitted (A) at GT (no beta carbons and C=O) removed from the X-block, and (B) at UT (with beta carbons and C=O) removed from the X-block. After pruning steps, prediction of MF induced by the prediction set terpolymers in six independent experiments with different donors was determined. The error bars indicate the standard errors of the experimental values (n = 6).
Fgn adsorption (A) and fibroblast proliferation (B) on the pMAs. The Fgn adsorption on each pMA was normalized by that on the control polypropylene surface (Mean ± SD; n = 14 wells). Fibroblast proliferation on each pMA was normalized by that on the control TCPS surface and is represented in terms of relative proliferation index. Detailed methods are published in [46]. The corresponding CD86/DC-SIGN ratios (solid squares, Mean ± SEM) are also shown for comparison. Brackets: p < 0.05 different between treatments for fibrinogen adsorption (A) or relative proliferation index (B).