Abstract
The objective of this study was to compare in vivo the effects of bicarbonate and phosphate buffers on surviving and menadione-induced oxidative stress in yeast cells. The latter were treated with different concentrations of menadione in the presence of these two buffers. If at 25 mM concentration of buffers menadione only slightly reduced yeast surviving, at 50 mM concentration cell killing by menadione was much more pronounced in bicarbonate than in phosphate buffer. Although the content of protein carbonyl groups did not show development of oxidative stress under menadione-induced stress, inactivation of aconitase and decrease in glutathione level mirrored its induction. However, cellular glutathione and aconitase activity decrease did not correlate with yeast survival. In vitro, aconitase was more quickly inactivated in 50 mM carbonate, than in 50 mM phosphate buffer. The possible involvement of the carbonate radical in these processes is discussed.
Keywords: Aconitase, Antioxidant enzymes, Glutathione, Protein carbonyls, Reactive oxygen species, Saccharomyces cerevisiae
1. Introduction
Under normal conditions, cells precisely regulate their redox status. Reactive oxygen species (ROS), namely superoxide anion (O•2 −), hydrogen peroxide (H2O2), and hydroxyl radical (•OH), are continuously generated and decomposed. These processes of generation and degradation are typically well balanced, and usually certain ROS concentrations are maintained. However, intracellular and extracellular changes may cause an imbalance between generation and decomposition of ROS, and increasing their steady state concentrations may result in oxidative stress.1–4
Oxidative stress may be induced by exogenous compounds such as hydrogen peroxide and superoxide or by compounds such as menadione or paraquate which cause an oxygen-dependent redox cycling. Reactive oxygen species can modify most cellular constituents and are believed to cause many degenerative diseases. The budding yeast Saccharomyces cerevisiae is widely used model for studying cellular response to oxidative stress.5–7 Previously, we used this in vivo model to extend in vitro findings of McCord and colleagues, who found a nonlinear effect of superoxide dismutase (SOD) activity on the lipid peroxidation induced by free radicals.8 We also demonstrated a nonlinear dependency of protein carbonyl concentrations, a measure of free radical damage to proteins, on SOD activity in yeast.7,9
Bicarbonate and carbon dioxide form the major physiological buffering system and were thus employed for many years in physiological and biochemical investigations. Because work with a bicarbonate buffer is somewhat inconvenient, it is not often employed in contemporary investigations. However, there are a number of reactions which are dependent on bicarbonate or carbon dioxide, and many of them are of potential physiological or pathological significance. For example, carbon dioxide increases the intensity of peroxidation catalyzed by transition metals10–15 and by Cu,Zn-SOD.16–21 Studies by Liochev and Fridovich of peroxidation catalyzed by Mn(II) demonstrated a synergism between carbon dioxide and bicarbonate.15 It has been proposed that this phenomenon might involve the generation of the carbonate radical as the distal oxidant.11,15,17,18 In contrast, Stadtman and colleagues established that bicarbonate can be protective.11–14 They showed that a manganese, bicarbonate, amino acid complex can scavenge hydrogen peroxide through both catalatic and peroxidatic reactions. The articles cited above and many similar articles used in vitro systems. It is not known whether similar processes take place in vivo.
These considerations led us to investigate oxidative stress induced in S. cerevisiae by menadione in carbonate and phosphate buffers. We found that menadione reduced yeast survival in 50 mM bicarbonate, but not in phosphate buffer.
2. Materials and methods
Chemicals and growth conditions
Guanidine-HCl was obtained from Fluka (Germany). All other chemicals, yeast extract, and peptone were obtained from Sigma-Aldrich Chemicals Company.
The yeast Saccharomyces cerevisiae of the YPH250 strain (MATa trp1-Δ1, his3-Δ200, lys2-801, leu2-Δ1, ade2-101, ura3-52) was kindly provided by Professor Yoshiharu Inoue (Division of Applied Life Sciences, Graduate School of Agriculture, Kyoto, Japan). Cells were grown to stationary phase (72 h) at 28°C in an orbital shaker (175 rpm) in a liquid YPD medium (1% w/v yeast extract, 2% w/v bacto-peptone, and 2% w/v glucose). Cells prepared from an overnight culture grown in YPD medium were used for all experiments. Cultures were inoculated with ~0.3 × 106 cells/ml.
Bicarbonate buffer was prepared from NaHCO3. The salt was dissolved in bi-distilled, degassed water, and the pH was adjusted to 7.50. Yeast cells were resuspended in bicarbonate buffer and treated with menadione. During the incubation, the pH of the yeast suspension remained at 7.50 in phosphate buffer and decreased slightly to 7.46 in bicarbonate buffer. Cell survival was determined by counting the number of colony forming units. Cell suspensions treated with menadione for 1 h were diluted in sterile distilled water; 75 μl of the resulting suspensions were plated onto YPD-agar in Petri dishes (10-cm diameter).
Preparation of cell extracts and assay of enzyme activity
Extracts were prepared by vortexing yeast cells with glass beads (0.5 mm), as described previously.7 They were kept on ice for immediate use. The activities of glutathione reductase, aconitase and catalase were measured as described.7,9 Hydrogen peroxide consumption by catalase was measured at 240 nm using an extinction coefficient for hydrogen peroxide of 39.4 M−1cm−1. The reaction was started by addition of cell-free extracts. One unit of catalase activity decomposes 1 μmol of hydrogen peroxide per minute. All enzyme activities were measured at 25°C and expressed per mg soluble protein in the supernatant. Native electrophoresis was performed, and SOD activity was assayed using N,N,N',N'-tetramethylethylenediamine-riboflavin treatment immediately after staining with nitroblue tetrasodium.22 Gels were scanned at 430 nm with a Specord M40 gel scanner.
Measurement of protein carbonyls and total glutathione
The content of carbonyl groups in proteins was measured by determining the amount of 2,4-dinitrophenylhydrazone formed upon reaction with 2,4-dinitrophenylhydrazine.7,9 Carbonyl content was calculated from the absorbance maximum of 2,4-dinitrophenylhydrazone, measured at 370 nm using an extinction coefficient of 22 mM−1cm−1.23 The results are expressed as nmol carbonyl per mg protein. The level of total glutathione was measured as described.24 Yeast cells were resuspended in 1.3% dinitrosalicilic acid and disrupted by vortexing with glass beads (0.5 mm) for three cycles (1 min of disruption and 3 min of cooling on ice).
Determination of protein concentrations and statistical analysis
Protein concentrations were determined using the Coomassie brilliant blue G-250 dye-binding method25 with bovine serum albumin as the standard. Experimental data are expressed as the mean ± SEM. Statistical testing was performed using ANOVA followed by Dunnett’s test for multiple comparisons.26
3. Results
Killing by menadione is biphasic
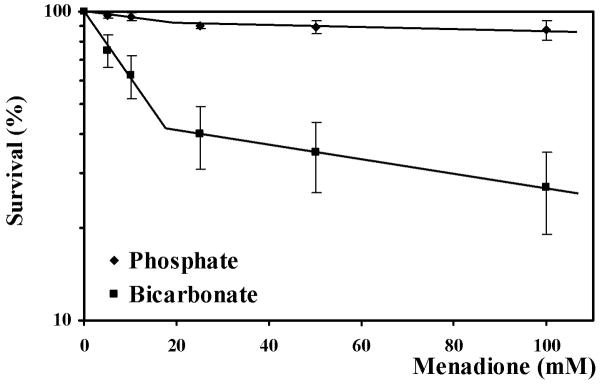
Treatment of yeast with menadione for 60 min with 25 mM phosphate and bicarbonate buffers (data not shown) or 50 mM phosphate buffer only slightly reduced yeast survival (Fig. 1). However, in 50 mM bicarbonate the loss of viability was substantial and biphasic (Fig. 1). Low concentrations of menadione were much more lethal in 50 mM bicarbonate buffer than in 50 mM phosphate one. At higher concentrations of menadione, the slope of concentration dependent killing was similar to that in the other three buffers.
Fig. 1.
Survival of yeast cells treated with menadione (5–100 mM) in 50 mM phosphate or bicarbonate buffers, presented in half-logarithmical coordinates. Data shown are the mean ± SEM (n = 4). *Significantly different from untreated (control) cells, P<0.05. The Y-axis is logarithmic, allowing visualization of the first-order process.
Protein carbonyl and glutathione
Protein carbonylation is a widely used marker of oxidative stress and oxidative damage. Even in the absence of menadione, the protein carbonyl content varied with buffer conditions (Table 1). Protein carbonyl concentration was highest in 25 mM phosphate buffer, and this was the only buffer used in which menadione caused a significant change in protein carbonyl concentrations. The level actually decreased for 43%, most likely due to further oxidation of the carbonyl moieties to carboxylic acids, which are not detected in the protein carbonyl assay.
Table I.
Concentrations of protein carbonyls (nmol mg−1 protein) in cell extracts of yeast treated with different menadione concentrations (5–100 mM) in phosphate or bicarbonate buffers. Data shown are the mean ± SEM (n = 4).
| Buffer | Menadione (mM)
|
|||||
|---|---|---|---|---|---|---|
| 0 | 5 | 10 | 25 | 50 | 100 | |
| 25 mM | ||||||
| Phosphate | 6.2 ± 0.61 | 5.0 ± 0.77 | 4.4 ± 0.29* | 3.4 ± 0.27* | 3.7 ± 0.43* | 3.5 ± 0.28* |
| Bicarbonate | 4.3 ± 0.58 | 4.0 ± 0.21 | 4.2 ± 0.58 | 4.9 ± 0.64 | 3.9± 0.48 | 4.4 ± 0.43 |
|
| ||||||
| 50 mM | ||||||
| Phosphate | 2.7 ± 0.21 | 3.3 ± 0.36 | 2.9 ± 0.36 | 2.5 ± 0.35 | 3.4 ± 0.68 | 3.0 ± 0.29 |
| Bicarbonate | 2.8 ± 0.26 | 3.6 ± 0.68 | 3.0 ± 0.46 | 3.8 ± 0.32 | 3.3 ± 0.15 | 3.4 ± 0.29 |
Significantly different from respective controls (untreated cells), P<0.05.
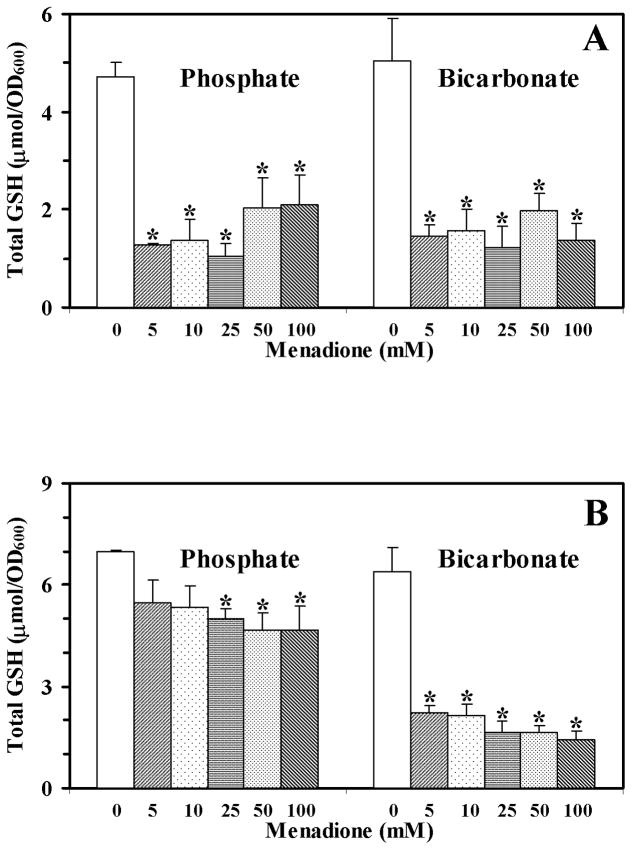
The basal level of total glutathione in all four buffers was similar, and it decreased substantially in the presence of menadione (Fig. 2). However, the decrease in 50 mM phosphate buffer was modest compared to the other three buffer conditions. Comparison of Figs. 1 and 2 shows, that glutathione content does not correlate with yeast survival.
Fig. 2.
Levels of total glutathione in yeast cells treated with menadione (5–100 mM) in 25 mM (A) or 50 mM (B) phosphate or bicarbonate buffers. Data shown are the mean ± SEM (n = 4). *Significantly different from untreated cells, P<0.05.
Enzyme activities
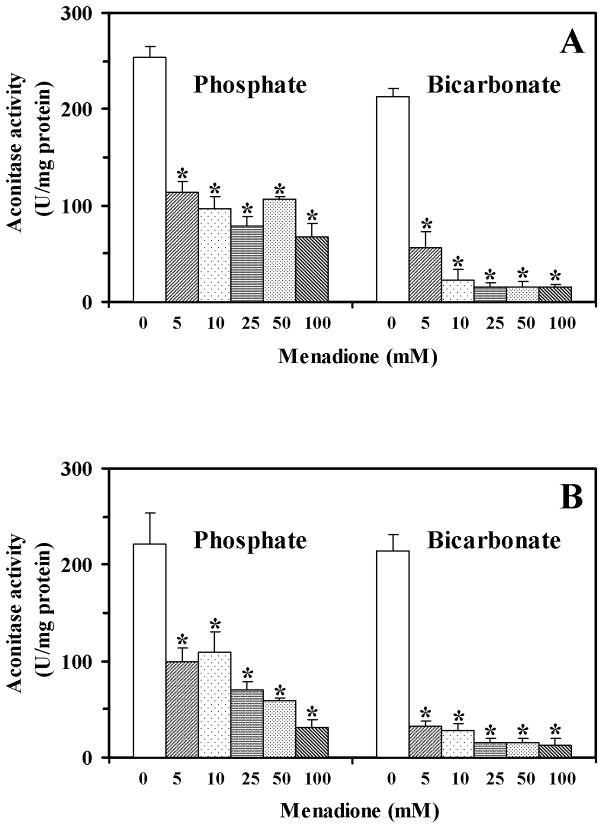
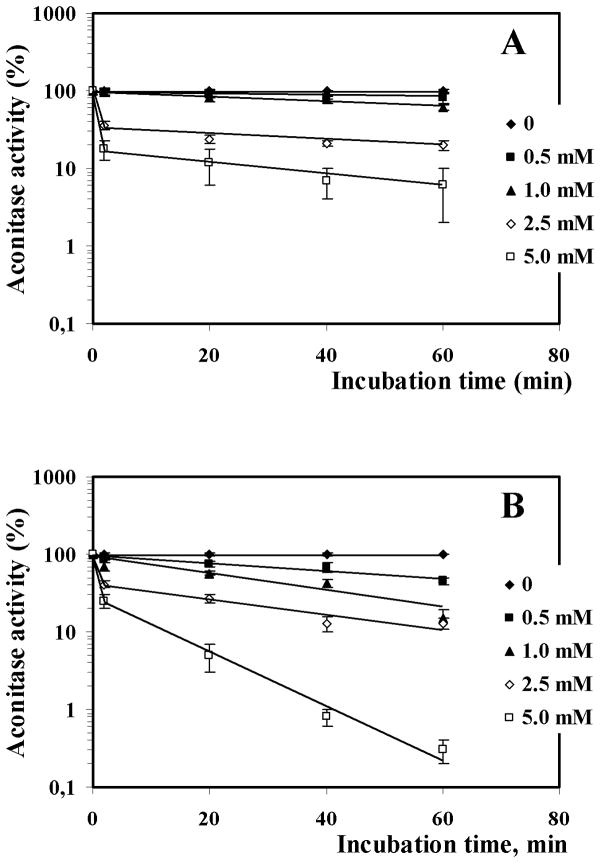
Aconitase, which has a cubic [4Fe-4S] center, may be reversibly inactivated by superoxide and can serve as a marker of oxidative stress.27–30 With intact yeast cells, menadione inactivated aconitase in a concentration-dependent manner with a slightly greater effect in bicarbonate buffer compared to phosphate (Fig. 3). Cell-free extracts also supported a menadione-dependent inactivation of aconitase and allowed measurement of the actual initial rate of the inactivation reaction. The enzyme was inactivated faster in 50 mM bicarbonate buffer, than in 50 mM phosphate buffer (Fig. 4).
Fig. 3.
Activity of aconitase in yeast cells treated with menadione (5–100 mM) in 25 mM (A) or 50 mM (B) phosphate or bicarbonate buffers. Data shown are the mean ± SEM (n = 4). *Significantly different from untreated (control) cells, P<0.05.
Fig. 4.
In vitro inactivation of aconitase by menadione (0.5–5 mM) in 50 mM phosphate (A) and bicarbonate (B) buffers. Activity was determined at 0, 2.5, 20, 40, and 60 min incubation. The data shown are the mean ± SEM (n = 3). The Y-axis is logarithmic, allowing visualization of the first-order process. The 2.5 min point gives the initial inactivation rate.
The activity of catalase was essentially unchanged except for a modest reduction (~28%) in 50 mM phosphate buffer (Table 2), which may reflect a protective role of bicarbonate, as observed by Stadtman and colleagues in vitro.11,12
Table II.
Catalase activity (U mg−1 protein) in cell extracts of yeast treated with different menadione concentrations (5–100 mM) in phosphate or bicarbonate buffers. Data shown are the mean ± SEM (n = 4).
| Buffer | Menadione (mM)
|
|||||
|---|---|---|---|---|---|---|
| 0 | 5 | 10 | 25 | 50 | 100 | |
| 25 mM | ||||||
| Phosphate | 223 ± 33 | 208 ± 28 | 203 ± 27 | 217 ± 25 | 177 ± 20 | 190 ± 17 |
| Bicarbonate | 193 ± 26 | 208 ± 26 | 174 ± 22 | 196 ± 18 | 181 ± 22 | 194 ± 34 |
|
| ||||||
| 50 mM | ||||||
| Phosphate | 209 ± 9 | 177 ± 14 | 180 ± 14 | 174 ± 13 | 176 ± 17 | 151 ± 24* |
| Bicarbonate | 175 ± 7 | 174 ± 5 | 168 ± 7 | 168 ± 6 | 162 ± 6 | 151 ± 5 |
Significantly different from respective controls (untreated cells), P<0.05.
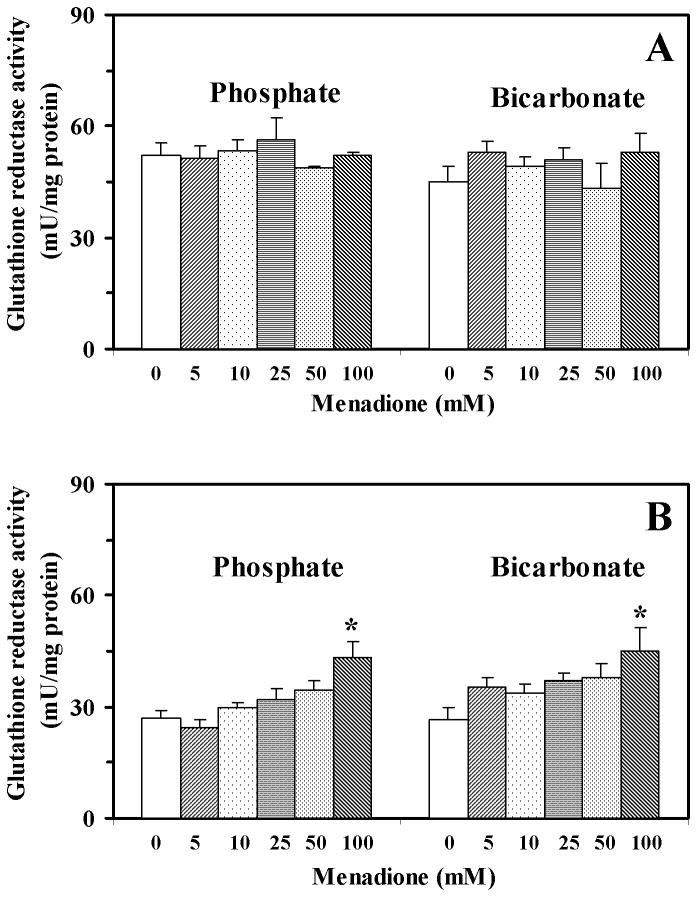
The activity of glutathione reductase was approximately 2-fold higher following incubation in 25 mM buffer (phosphate and bicarbonate) than after incubation in 50 mM buffer (data not shown). Menadione treatment in 25 mM buffer did not affect glutathione reductase activity (data not shown), while treatment with 100 mM menadione in 50 mM phosphate buffer and 50 mM bicarbonate buffer increased the activity by 61% and 71%, respectively (Fig. 5).
Fig. 5.
Activity of glutathione reductase in yeast cells treated with menadione (5–100 mM) in 25 mM (A) or 50 mM (B) phosphate or bicarbonate buffers. Data shown are the mean ± SEM (n = 4). *Significantly different from untreated (control) cells, P<0.05.
Menadione seems interfered with traditional for our laboratory quercetin method for evaluating the activity of superoxide dismutase.7,9 Therefore, we performed a semi-quantitative assay using native gel electrophoresis and an in-gel activity assay. These studies showed that menadione increased SOD activity in both buffers (data not shown). The increase in activities of both, SOD and glutathione reductase, by yeast treatment with menadione confirms previous findings.5,31
4. Discussion
In mammalian tissues bicarbonates present at relatively high concentrations –approximately 25 mM32. They were suggested to interact with free radicals yielding other radicals, such as carbonate radical anion.33 The products of similar reaction(s) could increase the susceptibility of S. cerevisiae to menadione (Fig. 1). Similar results were previously reported for several bacteria, including E. coli.34 Addition of hydroxyl radical scavengers eliminated the increased killing in bicarbonate buffer, which was attributed to carbonate radical generated in the external medium. Hurst et al. extended their previous work34 to yeast and found that sodium bicarbonate increased the sensitivities of both E. coli and S. cerevisiae to γ–irradiation.35 However, the reactivity of the carbonate radical anion is much less than that of the hydroxyl radical. Hurst et al. proposed that the greater toxicity of the carbonate radical anion compared to the hydroxyl radical is a consequence of the greater stability of the carbonate radical anion, allowing greater oxidation of cellular molecules.35 It was also suggested that carbonate radical might be more specific that hydroxyl one, but the issue needs further investigation.
The exponential loss of viability described here and elsewhere35 suggested that microorganisms were killed by reactants randomly reaching vulnerable targets, presumably after depletion of cellular antioxidants. Thus, there must be at least two pathways for generating species which can kill yeast or there are two distinct targets, both of which must be hit to kill the cell. However, when we compared the loss of cellular glutathione with cell survival we did not find a correlation. Glutathione concentrations were decreased in all experiments, regardless of survival. Thus, while glutathione may be involved in protecting cellular macromolecules during oxidative stress, but it is not a key determinant of survival.
The activity of aconitase was another measure we used to identify the reason for yeast killing in carbonate buffer under our experimental conditions. Again, like in the case with glutathione, we disclosed that aconitase was inactivated in all experiments, but in carbonate the process was more pronounced (Fig. 3). The very small difference was also found between 25 and 50 mM carbonate buffers; in the second case inactivation looked stronger. The in vivo experiments were confirmed in vitro, where the enzyme was stronger inactivated in bicarbonate, than in phosphate buffer. There are several mechanisms of aconitase inactivation27, and at least two of known involve oxidative steps. The first and commonly discussed way includes oxidation of Fe2+ localizing in cubic [4Fe-4S] cluster and following iron ion abstraction prolonged or strong stress results in irreversible inactivation and ATP-dependent degradation of the enzyme.36 Under certain conditions upon resolution of the oxidative stress, oxidatively inhibited aconitase can be reactivated. Therefore, it is not clear, if aconitase is capable to participate in redox regulation and/or it is a potential target for oxidative damage. In addition, there is the second oxidative way for aconitase inactivation. L. Szweda and colleagues found that during heart ischemia/reperfusion aconitase is reversibly inactivated and the disclosed mechanism involved reversible oxidation of cysteine residue.36 Theoretically, cysteine residues may be oxidized to sulfenic, sulfinic and sulfonic acids and may form disulfide and mixed disulfide bounds at these modifications, but only oxidation of sulfenic acid and disulfide bonds may be reversed.37 In vitro treatment of oxidized mitochondrial aconitase from myocardial tissue by ditiothrietol resulted in full recovery of the activity.27 It can be concluded that in our case although yeast aconitase is inactivated in both in vivo and in vitro and this inactivation is better pronounced in carbonate than in phosphate buffer that probably, cannot be the only reason for difference between experiments in two buffers.
There were positive correlations between the activity of aconitase and the concentration of glutathione for all four experimental sets (R2=0.81–0.99). These correlative data suggest that under our experimental conditions glutathione could be important for protecting enzymes containing [4Fe-4S] clusters and/or cysteine.
The activation of SOD and GR at yeast treatment by menadione confirms the previous works.5,31 However, we have not found any activation of catalase under these conditions and this may reflect the strain specificity or particular conditions of stress induction. The reduction of catalase activity could be connected with the inactivation of the enzymes by superoxide anion and the listed issues were recently analyzed in one of our works.38
Although both glutathione reductase and catalase are known to be sensitive to inactivation by free radicals,7,9 this did not occur when yeast cells were incubated with menadione in 25 mM buffers. Both were virtually unaffected by incubation with menadione under the conditions used (Table 2 and Fig. 5). However, when the buffer concentration was 50 mM, treatment with 100 mM menadione increased glutathione reductase activity, while catalase activity was decreased in phosphate buffer. This observation again suggests that bicarbonate may have protective effects, as it was proposed by Stadtman and colleagues in their in vitro works.11–14 However, the issue needs further investigation.
Overall, the toxicity of menadione (2-methyl-1,4-naphthoquinone: vitamin K3) as other quinones, in biological systems is related with their possibility to undergo one-electron reduction by such enzymes as microsomal NADPH-cytochrome P-450 reductase or mitochondrial NADH ubiquinone oxidoreductase, yielding the corresponding semiquinone radicals. These radicals under aerobic conditions participate in redox cycling to generate ROS. Other mechanism is related with their electrophile properties being capable to react with thiol groups of proteins and glutathione. In fact, the generation of conjugates with glutathione catalyzed by glutathione tranferase (GST) with depletion of glutathione has been associated with menadione-induced cytotoxicity and oxidative stress.39 These two mechanisms were found to be responsible for the production of conjugates of glutathione and menadione, catalyzed by GST, and could be responsible for yeast cell killing. However, yeasts used were grown to mid log phase culture, while we used the stationary one. Therefore, the comparison of results of both groups is not very correct. If menadione toxicity is really related with GST operation, at least two aspects may be important in order to demonstrate biphasic concentration-dependent killing: depletion of glutathione and inactivation of GST by ROS.40 Although we found the decrease in glutathione content, it did not correlate with cell killing. The possible involvement of GST is another possibility tested39 and was found to be, at least partially, responsible for yeast cell killing. One may suggest that at least two mentioned mechanisms could be responsible, and received results in fact reflect the complex superposition of those possibilities, along with other ones.
In summary, our data show that the incubation of yeast cells with menadione in 50 mM carbonate buffer renders them more susceptible to oxidative stress than at treatment in phosphate buffer of the same concentration. Although the development of oxidative stress was evident, it differed in two buffers. However, the markers of oxidative stress which we measured did not correlate well with yeast susceptibility to menadione, indicating that other mechanisms might be also primarily responsible for the processes culminating in cell death.
Acknowledgments
We are grateful to Professor Y. Inoue for the providing S. cerevisiae strains. OVL, MMB and VIL, were supported partially by the grant of Ministry of Education and Science of Ukraine, #18/051. RLL was supported by the Intramural Research Program of the National Heart, Lung, and Blood Institute, National Institutes of Health.
Abbreviations
- ROS
reactive oxygen species
- SOD
superoxide dismutase
References
- 1.Sies H. Oxidative stress: introduction. In: Sies H, editor. Oxidative stress: Oxidants and antioxidants. Academic Press; San Diego: 1991. pp. 21–48. [Google Scholar]
- 2.Demple B. Regulation of bacterial oxidative stress genes. Annu Rev Genet. 1991;25:315–337. doi: 10.1146/annurev.ge.25.120191.001531. [DOI] [PubMed] [Google Scholar]
- 3.Lushchak VI. Oxidative stress and mechanisms of protection against it in bacteria. Biochemistry (Moscow) 2001;66:476–489. doi: 10.1023/a:1010294415625. [DOI] [PubMed] [Google Scholar]
- 4.Stadtman ER, Van Remmen H, Richardson A, Wehr NB, Levine RL. Methionine oxidation and aging. Biochim Biophys Acta. 2005;1703:135–140. doi: 10.1016/j.bbapap.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 5.Jamieson DJ. Oxidative stress responses of the yeast Saccharomyces cerevisiae. Yeast. 1998;14:1511–1527. doi: 10.1002/(SICI)1097-0061(199812)14:16<1511::AID-YEA356>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 6.Costa VM, Moradas-Ferreira P. Oxidative stress and signal transduction in S. cerevisiae: insights into ageing, apoptosis and diseases. Mol Aspects Med. 2001;22:217–246. doi: 10.1016/s0098-2997(01)00012-7. [DOI] [PubMed] [Google Scholar]
- 7.Lushchak V, Semchyshyn H, Mandryk S, Lushchak O. Possible role of superoxide dismutases in the yeast Saccharomyces cerevisiae under respiratory conditions. Arch Biochem Biophys. 2005;441:35–40. doi: 10.1016/j.abb.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 8.McCord JM. Oxidative Stress, Cancer, AIDS, and Neurodegenerative Diseases. Marcel Dekker; New York: 1997. The importance of oxidant-antioxidant balance; pp. 1–7. [Google Scholar]
- 9.Lushchak V, Semchyshyn H, Lushchak O, Mandryk S. Diethyldithiocarbamate inhibits in vivo Cu,Zn-superoxide dismutase and perturbs free radical processes in yeast Saccharomyces cerevisiae. Biochem Biophys Res Commun. 2005;338:1739–1744. doi: 10.1016/j.bbrc.2005.10.147. [DOI] [PubMed] [Google Scholar]
- 10.Liochev SI, Fridovich I. The role of CO2 in cobalt-catalyzed peroxidations. Arch Biochem Biophys. 2005;439:99–104. doi: 10.1016/j.abb.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 11.Stadtman ER, Berlet BS, Chock PB. Manganese-dependent disproportionation of hydrogen peroxide in bicarbonate buffer. Proc Natl Acad Sci USA. 1990;87:384–388. doi: 10.1073/pnas.87.1.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berlett BS, Chock PB, Yim MB, Stadtman ER. Manganese (II) catalyzes the bicarbonate-dependent oxidation of amino acids by hydrogen peroxide and the amino acid-facilitated dismutation of hydrogen peroxide. Proc Natl Acad Sci USA. 1990;87:389–393. doi: 10.1073/pnas.87.1.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yim MB, Berlett BS, Chock PB, Stadtman ER. Manganese(II)-bicarbonate-mediated catalytic activity for hydrogen peroxide dismutation and amino acid oxidation: Detection of free radical intermediates. Proc Natl Acad Sci USA. 1990;87:394–398. doi: 10.1073/pnas.87.1.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stadtman ER, Berlett BS. Fenton chemistry. Amino acid oxidation. J Biol Chem. 1991;266:17201–17211. [PubMed] [Google Scholar]
- 15.Liochev SI, Fridovich I. Carbon dioxide mediates Mn(II)-catalyzed decomposition of hydrogen peroxide and peroxidation reactions. Proc Natl Acad Sci USA. 2004;101:12485–12490. doi: 10.1073/pnas.0404911101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sankarapandi S, Zweier JL. Bicarbonate is required for the peroxidase function of Cu,Zn-superoxide dismutase at physiological pH. J Biol Chem. 1999;274:1226–1232. doi: 10.1074/jbc.274.3.1226. [DOI] [PubMed] [Google Scholar]
- 17.Ramirez DC, Gomez-Mejiba SE, Corbett JT, Deterding LJ, Tomer KB, Mason RP. Cu,Zn-superoxide dismutase-driven free radical modifications: copper- and carbonate radical anion-initiated protein radical chemistry. Biochem J. 2009;417:341–53. doi: 10.1042/BJ20070722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liochev SI, Fridovich I. CO2, not HCO3−, facilitates oxidations by Cu,Zn superoxide dismutase plus H2O2. Proc Natl Acad Sci USA. 2004;101:743–744. doi: 10.1073/pnas.0307635100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gunther MR, Donahue JA. Bicarbonate and active site zinc modulate the self-peroxidation of bovine copper-zinc superoxide dismutase. Free Radic Res. 2007;41:1005–1016. doi: 10.1080/10715760701516308. [DOI] [PubMed] [Google Scholar]
- 20.Zhang H, Joseph J, Gurney M, Becker D, Kalyanaraman B. Bicarbonate enhances peroxidase activity of Cu,Zn-superoxide dismutase. Role of carbonate anion radical and scavenging of carbonate anion radical by metalloporphyrin antioxidant enzyme mimetics. J Biol Chem. 2002;277:1013–1020. doi: 10.1074/jbc.M108585200. [DOI] [PubMed] [Google Scholar]
- 21.Bonini MG, Fernandes DC, Augusto O. Albumin oxidation to diverse radicals by the peroxidase activity of Cu,Zn-superoxide dismutase in the presence of bicarbonate or nitrite: diffusible radicals produce cysteinyl and solvent-exposed and unexposed tyrosyl radicals. Biochemistry. 2004;43:344–351. doi: 10.1021/bi035606p. [DOI] [PubMed] [Google Scholar]
- 22.Beauchamp C, Fridovich I. Superoxide dismutase: improved assays and an assay applicable to acrylamide gels. Anal Biochem. 1971;44:276–287. doi: 10.1016/0003-2697(71)90370-8. [DOI] [PubMed] [Google Scholar]
- 23.Lenz AG, Costabel U, Shaltiel S, Levine RL. Determination of carbonyl groups in oxidatively modified of proteins by reduction with tritiated sodium borohydride. Anal Biochem. 1989;177:419–425. doi: 10.1016/0003-2697(89)90077-8. [DOI] [PubMed] [Google Scholar]
- 24.Lushchak V, Lushchak L, Mota A, Hermes-Lima M. Oxidative stress and antioxidant defences in goldfish Carassius auratus during anoxia and reoxygenation. Am J Physiol. 2001;280:R100–R107. doi: 10.1152/ajpregu.2001.280.1.R100. [DOI] [PubMed] [Google Scholar]
- 25.Bradford MM. A rapid and sensitive method for the quantification of microgram quantities of protein using the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 26.Brooks SPJ. A simple computer program with statistical tests for the analysis of enzyme kinetics. BioTechniques. 1992;13:906–911. [PubMed] [Google Scholar]
- 27.Bulteau AL, Lundberg KC, Ikeda-Saito M, Isaya G, Szweda LI. Reversible redox-dependent modulation of mitochondrial aconitase and proteolytic activity during in vivo cardiac ischemia/reperfusion. Proc Natl Acad Sci USA. 2005;102:5987–5991. doi: 10.1073/pnas.0501519102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vasquez-Vivar J, Kalyanaraman B, Kennedy MC. Mitochondrial aconitase is a source of hydroxyl radical an electron spin resonance investigation. J Biol Chem. 2000;275:14064–14069. doi: 10.1074/jbc.275.19.14064. [DOI] [PubMed] [Google Scholar]
- 29.Maity P, Bindu S, Dey S, et al. Indomethacin, a non-steroidal anti-inflammatory drug, develops gastropathy by inducing reactive oxygen species-mediated mitochondrial pathology and associated apoptosis in gastric mucosa: a novel role of mitochondrial aconitase oxidation. J Biol Chem. 2009;284:3058–3068. doi: 10.1074/jbc.M805329200. [DOI] [PubMed] [Google Scholar]
- 30.Gardner PR. Aconitase: sensitive target and measure of superoxide. Methods Enzymol. 2002;349:9–23. doi: 10.1016/s0076-6879(02)49317-2. [DOI] [PubMed] [Google Scholar]
- 31.Godon C, Lagniel G, Lee J, et al. The H2O2 stimulon in Saccharomyces cerevisiae. J Biol Chem. 1998;273:22480–22489. doi: 10.1074/jbc.273.35.22480. [DOI] [PubMed] [Google Scholar]
- 32.Medinas DB, Cerchiaro G, Trindade DF, Augusto O. The carbonate radical and related oxidants derived from bicarbonate buffer. IUBMB Life. 2007;59:255–262. doi: 10.1080/15216540701230511. [DOI] [PubMed] [Google Scholar]
- 33.Augusto O, Bonini MG, Amanso AM, et al. Nitrogen dioxide and carbonate radical anion: two emerging radicals in biology. Free Radic Biol Med. 2002;32:841–859. doi: 10.1016/s0891-5849(02)00786-4. [DOI] [PubMed] [Google Scholar]
- 34.Wolcott RG, Franks BS, Hannum DM, Hurst JK. Bactericidal potency of hydroxyl radical in physiological environments. J Biol Chem. 1994;269:9721–9728. [PubMed] [Google Scholar]
- 35.King DA, Sheafor MW, Hurst JK. Comparative toxicities of putative phagocyte-generated oxidizing radicals toward a bacterium (Escherichia coli) and a yeast (Saccharomyces cerevisiae) Free Radic Biol Med. 2006;41:765–774. doi: 10.1016/j.freeradbiomed.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 36.Bulteau A-L, Ikeda-Saito M, Szweda LI. Redox-dependent modulation of aconitase activity in intact mitochondria. Biochemistry. 2003;42:14846–14855. doi: 10.1021/bi0353979. [DOI] [PubMed] [Google Scholar]
- 37.Stadtman ER, Levine RL. Free radical-mediated oxidation of free amino acids and amino acid residues in proteins. Amino Acids. 2003;25:207–218. doi: 10.1007/s00726-003-0011-2. [DOI] [PubMed] [Google Scholar]
- 38.Bayliak M, Semchyshyn H, Lushchak V. Effect of hydrogen peroxide on antioxidant enzyme activities in Saccharomyces cerevisiae is strain specific. Biochemistry (Moscow) 2006;71:1243–252. doi: 10.1134/s0006297906090100. [DOI] [PubMed] [Google Scholar]
- 39.Castro FA, Mariani D, Panek AD, Eleutherio EC, Pereira MD. Cytotoxicity mechanism of two naphthoquinones (menadione and plumbagin) in Saccharomyces cerevisiae. PLoS One. 2008;3(12):e3999. doi: 10.1371/journal.pone.0003999. Epub 2008 Dec 22, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hermes-Lima M, Storey KB. In vitro oxidative inactivation of glutathione S-transferase from a freeze tolerant reptile. Mol Cell Biochem. 1993;124(2):149–158. doi: 10.1007/BF00929207. [DOI] [PubMed] [Google Scholar]