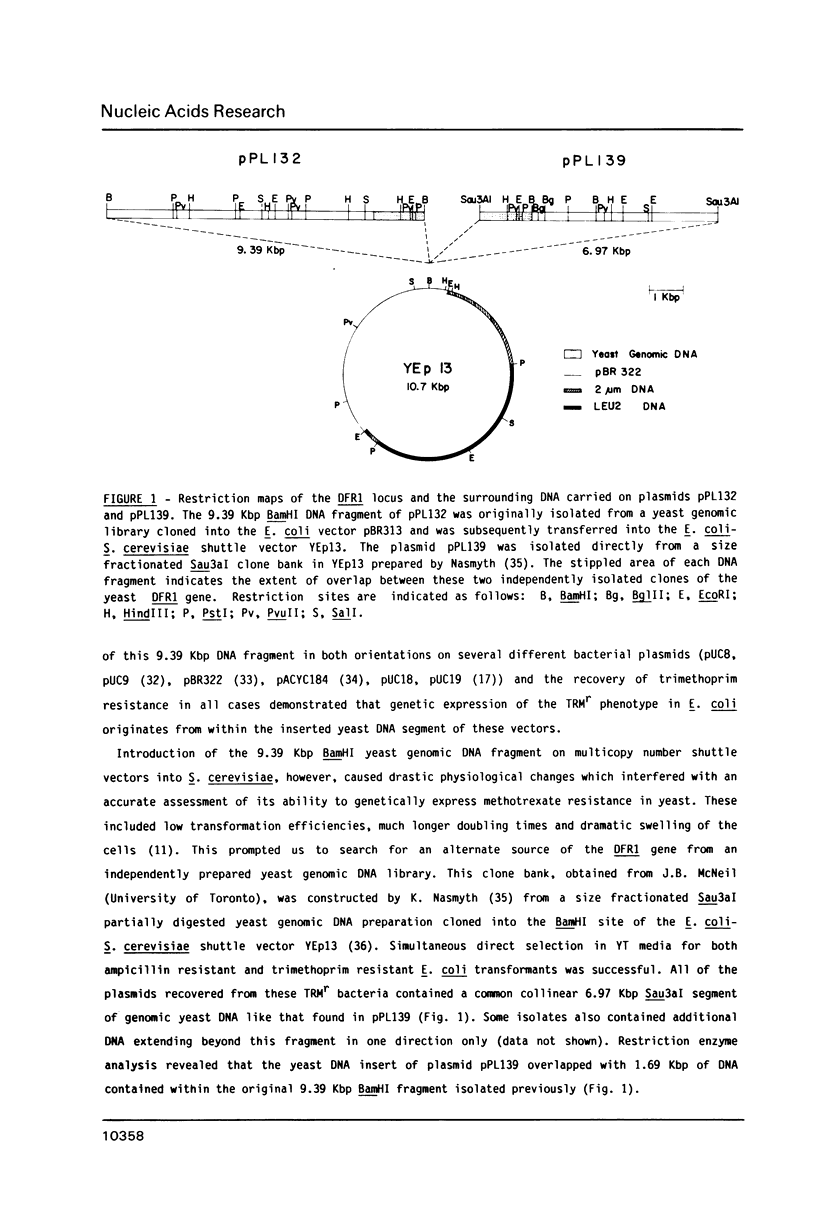
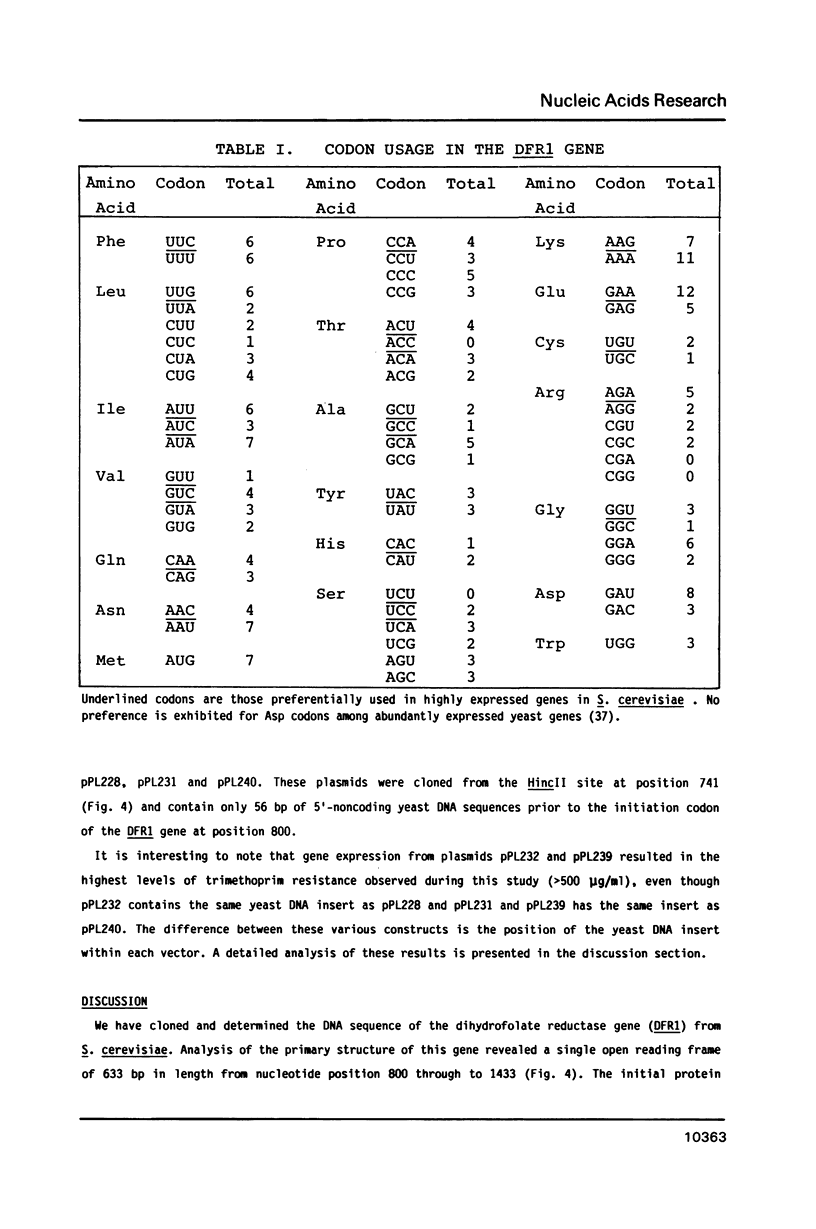
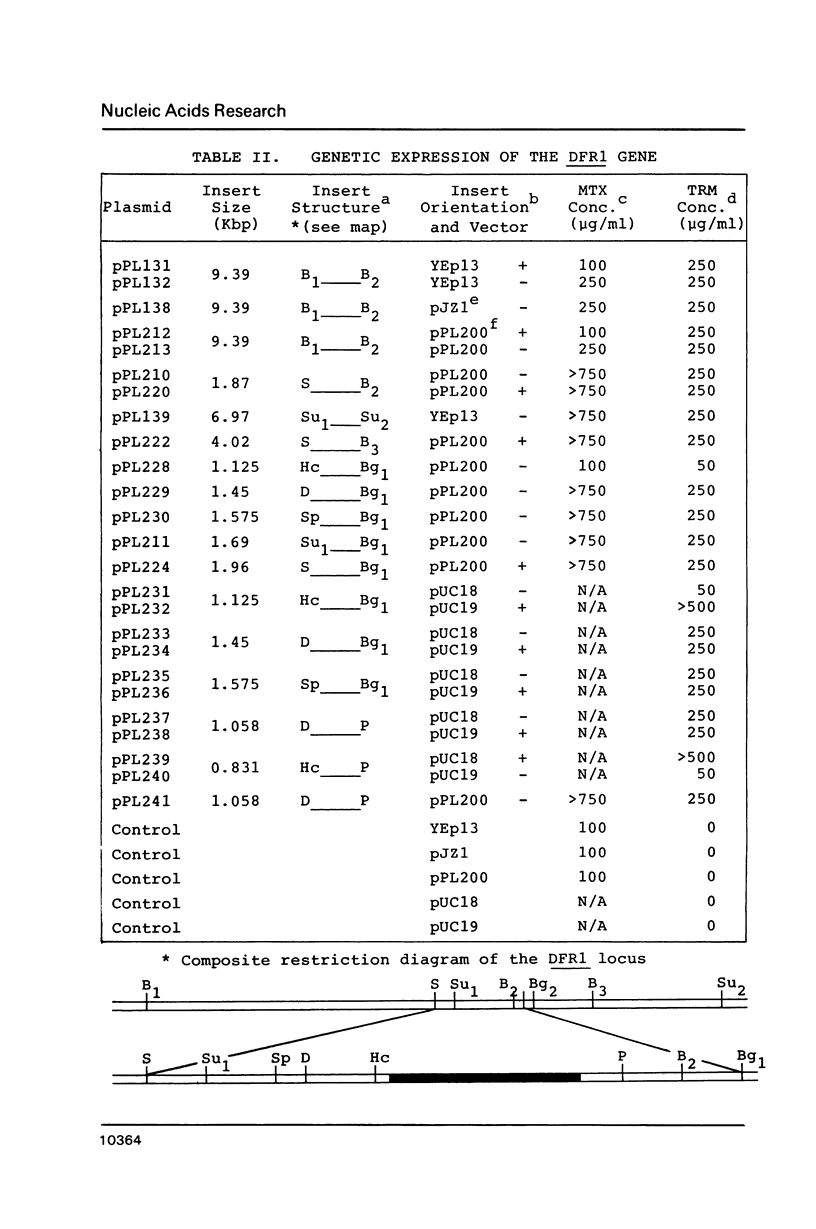
Abstract
The complete nucleotide sequence of a 1957 bp DNA fragment containing the dihydrofolate reductase gene (DFR1) of the yeast Saccharomyces cerevisiae is presented. Within this region a single open reading frame of 633 base pairs was found which is capable of encoding a 211 amino acid residue protein with a calculated Mr of 24,233. The amino acid sequence derived from the yeast DFR1 gene shows limited homology with sequences from both eukaryotic and non-eukaryotic DHFR enzymes. Northern blot hybridization reveals that the mRNA from this gene is a 900 base polyadenylated transcript. Yeast strains containing the cloned DFR1 gene on multicopy number shuttle vector plasmids show dramatically enhanced methotrexate resistance. Consensus DNA sequences responsible for RNA polymerase II interaction and general amino acid control in S. cerevisiae are located within the 5'-noncoding region with respect to the open reading frame. The DNA fragment containing these sequences has been shown to be necessary for DFR1 gene expression in both S. cerevisiae and E. coli.
Full text
PDF












Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bennetzen J. L., Hall B. D. Codon selection in yeast. J Biol Chem. 1982 Mar 25;257(6):3026–3031. [PubMed] [Google Scholar]
- Bennetzen J. L., Hall B. D. The primary structure of the Saccharomyces cerevisiae gene for alcohol dehydrogenase. J Biol Chem. 1982 Mar 25;257(6):3018–3025. [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Betlach M. C., Boyer H. W. Construction and characterization of new cloning vehicles. I. Ampicillin-resistant derivatives of the plasmid pMB9. Gene. 1977;2(2):75–93. doi: 10.1016/0378-1119(77)90074-9. [DOI] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Broach J. R., Strathern J. N., Hicks J. B. Transformation in yeast: development of a hybrid cloning vector and isolation of the CAN1 gene. Gene. 1979 Dec;8(1):121–133. doi: 10.1016/0378-1119(79)90012-x. [DOI] [PubMed] [Google Scholar]
- Burke R. L., Tekamp-Olson P., Najarian R. The isolation, characterization, and sequence of the pyruvate kinase gene of Saccharomyces cerevisiae. J Biol Chem. 1983 Feb 25;258(4):2193–2201. [PubMed] [Google Scholar]
- Caras I. W., Levinson B. B., Fabry M., Williams S. R., Martin D. W., Jr Cloned mouse ribonucleotide reductase subunit M1 cDNA reveals amino acid sequence homology with Escherichia coli and herpesvirus ribonucleotide reductases. J Biol Chem. 1985 Jun 10;260(11):7015–7022. [PubMed] [Google Scholar]
- Casadaban M. J., Cohen S. N. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1980 Apr;138(2):179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- Chang A. C., Cohen S. N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978 Jun;134(3):1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M. J., Shimada T., Moulton A. D., Cline A., Humphries R. K., Maizel J., Nienhuis A. W. The functional human dihydrofolate reductase gene. J Biol Chem. 1984 Mar 25;259(6):3933–3943. [PubMed] [Google Scholar]
- Dobson M. J., Tuite M. F., Roberts N. A., Kingsman A. J., Kingsman S. M., Perkins R. E., Conroy S. C., Fothergill L. A. Conservation of high efficiency promoter sequences in Saccharomyces cerevisiae. Nucleic Acids Res. 1982 Apr 24;10(8):2625–2637. doi: 10.1093/nar/10.8.2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donahue T. F., Daves R. S., Lucchini G., Fink G. R. A short nucleotide sequence required for regulation of HIS4 by the general control system of yeast. Cell. 1983 Jan;32(1):89–98. doi: 10.1016/0092-8674(83)90499-3. [DOI] [PubMed] [Google Scholar]
- Fling M. E., Richards C. The nucleotide sequence of the trimethoprim-resistant dihydrofolate reductase gene harbored by Tn7. Nucleic Acids Res. 1983 Aug 11;11(15):5147–5158. doi: 10.1093/nar/11.15.5147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Game J. C., Little J. G., Haynes R. H. Yeast mutants sensitive to trimethoprim. Mutat Res. 1975 May;28(2):175–182. doi: 10.1016/0027-5107(75)90094-9. [DOI] [PubMed] [Google Scholar]
- Henikoff S., Kelly J. D., Cohen E. H. Transcription terminates in yeast distal to a control sequence. Cell. 1983 Jun;33(2):607–614. doi: 10.1016/0092-8674(83)90441-5. [DOI] [PubMed] [Google Scholar]
- Hope I. A., Struhl K. GCN4 protein, synthesized in vitro, binds HIS3 regulatory sequences: implications for general control of amino acid biosynthetic genes in yeast. Cell. 1985 Nov;43(1):177–188. doi: 10.1016/0092-8674(85)90022-4. [DOI] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz B. A., Barclay B. J., Game J. C., Little J. G., Haynes R. H. Induction of mitotic recombination in yeast by starvation for thymine nucleotides. Proc Natl Acad Sci U S A. 1980 Oct;77(10):6057–6061. doi: 10.1073/pnas.77.10.6057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz B. A. Genetic effects of deoxyribonucleotide pool imbalances. Environ Mutagen. 1982;4(6):695–725. doi: 10.1002/em.2860040609. [DOI] [PubMed] [Google Scholar]
- Kunz B. A., Taylor G. R., Haynes R. H. Induction of intrachromosomal recombination in yeast by inhibition of thymidylate biosynthesis. Genetics. 1986 Oct;114(2):375–392. doi: 10.1093/genetics/114.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagrimini L. M., Brentano S. T., Donelson J. E. A DNA sequence analysis package for the IBM personal computer. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):605–614. doi: 10.1093/nar/12.1part2.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langford C. J., Klinz F. J., Donath C., Gallwitz D. Point mutations identify the conserved, intron-contained TACTAAC box as an essential splicing signal sequence in yeast. Cell. 1984 Mar;36(3):645–653. doi: 10.1016/0092-8674(84)90344-1. [DOI] [PubMed] [Google Scholar]
- Little J. G., Haynes R. H. Isolation and characterization of yeast mutants auxotrophic for 2'-deoxythymidine 5'-monophosphate. Mol Gen Genet. 1979 Jan 10;168(2):141–151. doi: 10.1007/BF00431440. [DOI] [PubMed] [Google Scholar]
- Luzzati M. Isolation and properties of a thymidylate-less mutant in Saccharomyces cerevisiae. Eur J Biochem. 1975 Aug 15;56(2):533–538. doi: 10.1111/j.1432-1033.1975.tb02259.x. [DOI] [PubMed] [Google Scholar]
- Mandel M., Higa A. Calcium-dependent bacteriophage DNA infection. J Mol Biol. 1970 Oct 14;53(1):159–162. doi: 10.1016/0022-2836(70)90051-3. [DOI] [PubMed] [Google Scholar]
- McIntosh E. M., Gadsden M. H., Haynes R. H. Transcription of genes encoding enzymes involved in DNA synthesis during the cell cycle of Saccharomyces cerevisiae. Mol Gen Genet. 1986 Sep;204(3):363–366. doi: 10.1007/BF00331011. [DOI] [PubMed] [Google Scholar]
- McMaster G. K., Carmichael G. G. Analysis of single- and double-stranded nucleic acids on polyacrylamide and agarose gels by using glyoxal and acridine orange. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4835–4838. doi: 10.1073/pnas.74.11.4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K. A., Tatchell K. The structure of transposable yeast mating type loci. Cell. 1980 Mar;19(3):753–764. doi: 10.1016/s0092-8674(80)80051-1. [DOI] [PubMed] [Google Scholar]
- Norrander J., Kempe T., Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983 Dec;26(1):101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- Purohit S., Mathews C. K. Nucleotide sequence reveals overlap between T4 phage genes encoding dihydrofolate reductase and thymidylate synthase. J Biol Chem. 1984 May 25;259(10):6261–6266. [PubMed] [Google Scholar]
- Russell P., Nurse P. Schizosaccharomyces pombe and Saccharomyces cerevisiae: a look at yeasts divided. Cell. 1986 Jun 20;45(6):781–782. doi: 10.1016/0092-8674(86)90550-7. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz L. D., Friesen J. D. Nucleotide sequence of the tcml gene (ribosomal protein L3) of Saccharomyces cerevisiae. J Bacteriol. 1983 Jul;155(1):8–14. doi: 10.1128/jb.155.1.8-14.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D. R., Calvo J. M. Nucleotide sequence of the E coli gene coding for dihydrofolate reductase. Nucleic Acids Res. 1980 May 24;8(10):2255–2274. doi: 10.1093/nar/8.10.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storms R. K., McNeil J. B., Khandekar P. S., An G., Parker J., Friesen J. D. Chimeric plasmids for cloning of deoxyribonucleic acid sequences in Saccharomyces cerevisiae. J Bacteriol. 1979 Oct;140(1):73–82. doi: 10.1128/jb.140.1.73-82.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storms R. K., Ord R. W., Greenwood M. T., Mirdamadi B., Chu F. K., Belfort M. Cell cycle-dependent expression of thymidylate synthase in Saccharomyces cerevisiae. Mol Cell Biol. 1984 Dec;4(12):2858–2864. doi: 10.1128/mcb.4.12.2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talmadge K., Gilberg W. Construction of plasmid vectors with unique PstI cloning sites in a signal sequence coding region. Gene. 1980 Dec;12(3-4):235–241. doi: 10.1016/0378-1119(80)90105-5. [DOI] [PubMed] [Google Scholar]
- Taylor G. R., Barclay B. J., Storms R. K., Friesen J. D., Haynes R. H. Isolation of the thymidylate synthetase gene (TMP1) by complementation in Saccharomyces cerevisiae. Mol Cell Biol. 1982 Apr;2(4):437–442. doi: 10.1128/mcb.2.4.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor G. R., Lagosky P. A., Storms R. K., Haynes R. H. Molecular characterization of the cell cycle-regulated thymidylate synthase gene of Saccharomyces cerevisiae. J Biol Chem. 1987 Apr 15;262(11):5298–5307. [PubMed] [Google Scholar]
- Tsao S. G., Brunk C. F., Pearlman R. E. Hybridization of nucleic acids directly in agarose gels. Anal Biochem. 1983 Jun;131(2):365–372. doi: 10.1016/0003-2697(83)90185-9. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Zaret K. S., Sherman F. DNA sequence required for efficient transcription termination in yeast. Cell. 1982 Mar;28(3):563–573. doi: 10.1016/0092-8674(82)90211-2. [DOI] [PubMed] [Google Scholar]
- Zelikson R., Luzzati M. Mitochondrial and cytoplasmic distribution in Saccharmoyces cerevisiae of enzymes involved in folate-coenzyme-mediated one-carbon-group transfer. A genetic and biochemical study of the enzyme deficiencies in mutants tmp3 and ade3. Eur J Biochem. 1977 Sep 15;79(1):285–292. doi: 10.1111/j.1432-1033.1977.tb11808.x. [DOI] [PubMed] [Google Scholar]
- Zelikson R., Luzzati M. Two forms of serine transhydroxymethylase, one absent in a thymidylate-less mutant in Saccharomyces cerevisiae. Eur J Biochem. 1976 Apr 15;64(1):7–13. doi: 10.1111/j.1432-1033.1976.tb10269.x. [DOI] [PubMed] [Google Scholar]