Analyzing infectious disease data presents unique challenges for epidemiologists and biostatisticians. Unlike chronic diseases, for which a person’s risk depends only on their personal exposures and risk factors, the risk that someone will acquire an infection is inherently dependent on whether others in the population are infected. As such, traditional statistical methods that assume outcomes are independent cannot always be applied to infectious disease data, and novel statistical methods are often needed. Furthermore, to understand the potential impact of interventions, one must account for the non-linear feedbacks that give rise to population patterns of infection and disease. This requires blending epidemiologic methods with ecologic principles to develop models for transmission dynamics, models that play an integral role in our understanding of infectious disease epidemiology (Figure).
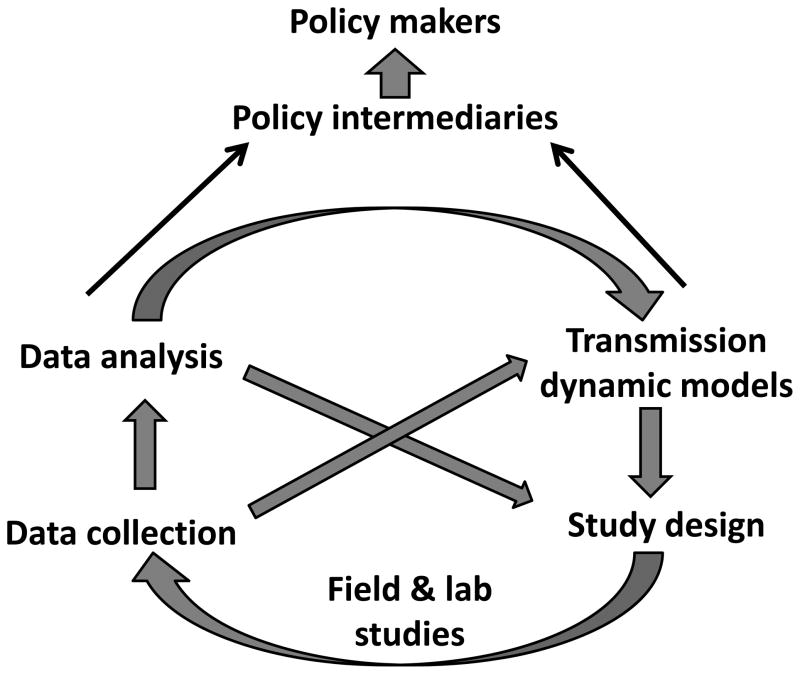
Figure.
Schematic of how empirical and theoretical infectious disease epidemiology research can interact to ultimately inform the policy-making process.
An excellent illustration of the synthesis of data collection, analysis, and transmission dynamic models is provided by Lipsitch and colleagues.1 The authors analyze longitudinal data of Streptococcus pneumoniae nasopharyngeal carriage among children from Kilifi, Kenya. The study design and data collection themselves represent a remarkable feat, with baseline assessment of the carrier status of 2840 children and multiple follow-up assessments of 1868 children who were positive at baseline.1–3 Analyzing these data to obtain unbiased estimates of the serotype-specific rates of acquisition, clearance, and resistance to competition for 27 unique pneumococcal serotypes presents a considerable added challenge. In addition to individual risk factors such as age, the children’s ability to clear colonization could depend on the resident serotype, how often they are exposed to someone shedding a different serotype, how resistant the resident serotype is to competition from other serotypes, and how good those serotypes are at competing. Thus, all the rates of interest are dependent on one another.
To address this challenge, Lipsitch et al. apply a Markov transition model.1 This approach allows them to estimate the competing risks of clearance of the baseline serotype and switching to each of the other serotypes. It also allows for estimation of the rate of acquisition for each of the 27 serotypes from the baseline prevalence distribution. This approach has considerable advantages over previous attempts to estimate serotype-specific parameters for S. pneumoniae, which were limited because they grouped vaccine serotypes and non-vaccine serotypes, estimated parameters for only a limited number of serotypes, assumed certain parameters were the same for all serotypes, or defined separate models for each serotype.3–9 The large, longitudinal cohort study conducted in Kilifi presents a unique opportunity to estimate these parameters, providing data that are essential for understanding competition and coexistence among pneumococcal serotypes.
Complexities underlying pneumococcal epidemiology
A key question for pneumococcal epidemiology is how vaccinations that protect against only certain serotypes will affect the overall diversity of serotypes in the population. Serotype replacement following the introduction of heptavalent pneumococcal conjugate vaccine (PCV7) into national immunization schedules has become a concern for prospects of controlling pneumococcal disease.10 Underlying this is the question of why so many serotypes of S. pneumoniae are able to coexist in the first place.
Transmission dynamic models can aid our understanding of the complex biologic processes that drive epidemiologic patterns such as the coexistence of pneumococcal serotypes, and they can provide an environment for testing hypotheses by simulating counterfactual scenarios. Models for the transmission dynamics are also increasingly used to inform policy decisions, such as whether to introduce a new vaccine into the national immunization schedule. However, to generate meaningful insight with real-world applications, such models must accurately portray the natural history of infection. Too often, transmission dynamic models are structured and parameterized based on little or no data, or on assumptions made by others, and then used to generate “predictions” about future trends in incidence or the impact of interventions. When this occurs, there is often a blurring of the distinction between what is an assumption and what is a prediction of the model. Similarly, models that rely too heavily on parameterizations obtained through fitting to observed incidence data as a substitute for a priori knowledge of the natural history are at best difficult to extrapolate to other settings, and at worst may be misleading. The study by Lipsitch et al.1 plays a crucial role in elucidating the natural history of various serotypes of S. pneumoniae, and provides a basis for modeling the transmission dynamics. Consequently, their findings helped inform the parameter assumptions for an individual-based model for the transmission dynamics of the various pneumococcal serotypes.11
The coexistence of so many different pneumococcal serotypes is surprising, particularly given the evidence of Lipsitch et al.1 that the fittest serotypes tend to be the best across all dimensions of fitness, i.e. tend to have longer durations of infection and higher rates of acquisition, as well as to be more resistant to competition. By modeling the transmission dynamics and interactions of 25 pneumococcal serotypes, Cobey et al.11 provide an explanation for this apparent paradox.
They found that both weak levels of serotype-specific immunity generated by previous infection and a non-specific decrease in the duration of colonization with each subsequent infection regardless of serotype (as observed by Lipsitch et al.1) were necessary to generate patterns of diversity similar to those observed.11 The serotype-specific immunity helps the less fit serotypes gain a competitive advantage among persons previously infected by the more fit types, while the non-specific immunity helps to equalize the fitness differences among serotypes by disproportionately penalizing the more fit types. Without the empirically derived characteristics of pneumococcal natural history obtained by Lipsitch et al.,1 such a well-grounded understanding of the dynamics of co-existence would not have been possible.
Next steps: Modeling serotype replacement
Transmission dynamic models are most useful when they are able to shed light on how incidence patterns depend on factors such as the transmission rate, which may be changing through time or as a result of interventions. The next step for pneumococcal epidemiology is to understand the patterns of serotype replacement that have emerged since the introduction of PCV7 vaccines in the United States and other countries, and to predict the potential impact of the new 13-valent pneumococcal conjugate vaccine (PCV13). While a few modeling studies have attempted to predict patterns of serotype replacement following vaccination,11–19 all of these have had shortcomings. Static models have attempted to predict which serotypes are expected to become more prevalent following vaccination based on their structural properties19 or observed changes in carriage and disease following PCV7 introduction (and assuming similar effects for PCV13),16,17 but these models do not explain why this replacement occurs or how the level of replacement is expected to scale with vaccination coverage. Dynamic models have been largely theoretical13,14 or have considered only vaccine-type versus non-vaccine serotypes, grouping together bacterial strains that may in fact be described by quite different natural histories.12,15 Although Cobey et al.11 make some predictions about the potential impact of vaccination on the distribution and prevalence of serotypes associated with carriage, they do not attempt to extrapolate their findings to the incidence of invasive and non-invasive disease. Furthermore, they note that the precise impact of vaccination will depend on the strength of natural and vaccine-induced cross-immunity,11 which is currently not well understood. Additional epidemiologic studies are needed to assess how the correlates of fitness that determine the prevalence of carriage relate to those that determine a serotype’s invasiveness.
Collecting, analyzing, and utilizing data to inform models for the transmission dynamics and ultimately to provide guidance for policymakers is an iterative process (Figure). Transmission dynamic models are essential for understanding the expected indirect (i.e. “herd immunity”) effects of vaccination, which can make a difference in whether or not a vaccine is deemed cost-effective. Public health officials also rely on dynamic models to interpret patterns such as changes in the age distribution of cases following vaccine introduction. Acknowledging the limitations of existing transmission dynamic models helps to identify key gaps in our understanding and highlight which data should be collected in future epidemiologic and laboratory studies. Once the data are collected, analysis using statistical methods to appropriately deal with the complexities of infectious disease data can lead to better parameter estimates. These parameters can then be used to refine dynamic models and further our understanding of disease transmission.
One of the overarching challenges in infectious disease epidemiology is to understand the comparative multi-strain dynamics of various pathogens and how each is affected by vaccination. Serotype replacement is a concern not just for pneumococcal vaccination, but is also an issue, or potential issue, for other newly introduced vaccines (e.g. rotavirus, human papilloma virus, and meningococcal), vaccines in development (e.g. malaria, HIV, and dengue), and of course existing vaccines (e.g. influenza). Integrating empirical data analysis with transmission dynamic models will enable us to understand the differences and similarities that drive the patterns of strain replacement occurring naturally and under the selective pressures of vaccination. Ultimately, this will help us to design better vaccines and more effective vaccination strategies for staying one step ahead of the pathogens, rather than always struggling to catch up.
Acknowledgments
Conflicts of Interest and Source of Funding: Supported by the RAPIDD program of the Science & Technology Directorate, Department of Homeland Security and the Fogarty International Center, National Institutes of Health (V.E.P. and N.E.B.), the Bill and Melinda Gates Foundation (V.E.P.), and the National Institutes of Health grants 2R37AI032042 (N.E.B.), 7DP5OD009162 (N.E.B.), and R03AI092121 (N.E.B.).
We thank Bryan Grenfell for helpful comments.
Biographies
VIRGINIA PITZER is a Postdoctoral Research Associate in the Department of Ecology and Evolutionary Biology at Princeton University, and will start as an Assistant Professor at Yale School of Public Health in July. Her research focuses on the transmission dynamics of imperfectly immunizing infections, particularly rotavirus.
NICOLE E. BASTA is an Associate Research Scholar in Ecology and Evolutionary Biology at Princeton University and an Affiliate Investigator at the Hutchinson Cancer Research Center. Her research focuses on understanding vaccine effects, particularly for influenza and meningitis.
References
- 1.Lipsitch M, Abdullahi O, D'Amour A, Xie W, Weinberger DM, Tchetgen ET, Scott JA. Estimating rates of carriage acquisition and clearance and competitive ability for pneumococcal serotypes in Kenya with a Markov transition model. Epidemiology. 2012 doi: 10.1097/EDE.0b013e31824f2f32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abdhullahi O, Karani A, Tigoi CC, et al. J Infect Dis. The rates of acquisition and clearance of pneumococcal serotypes in the nasopharynges of children in Kilifi district, Kenya. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abdullahi O, Karani A, Tigoi CC, et al. The prevalence and risk factors for pneumococcal colonization of the nasopharynx among children in Kilifi district, Kenya. PLoS ONE. 2012;7:e30787. doi: 10.1371/journal.pone.0030787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Auranen K, Mehtala J, Tanskanen A, MSK Between-strain competition in acquisition and clearance of pneumococcal carriage--epidemiologic evidence from a longitudinal study of day-care children. Am J Epidemiol. 2010;171(2):169–76. doi: 10.1093/aje/kwp351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cauchemez S, Temime L, Valleron AJ, Varon E, Thomas G, Guillemot D, Boelle PY. S. pneumoniae transmission according to inclusion in conjugate vaccines: Bayesian analysis of a longitudinal follow-up in schools. BMC Infect Dis. 2006;6:14. doi: 10.1186/1471-2334-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Domenech de Celles M, Opatowski L, Salomon J, Varon E, Carbon C, Boelle PY, Guillemot D. Intrinsic epidemicity of Streptococcus pneumoniae depends on strain serotype and antibiotic susceptibility pattern. Antimicrob Agents Chemother. 2011;55(11):5255–61. doi: 10.1128/AAC.00249-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Erasto P, Hoti F, Granat SM, Mia Z, Makela PH, Auranen K. Modelling multi-type transmission of pneumococcal carriage in Bangladeshi families. Epidemiol Infect. 2010;138(6):861–72. doi: 10.1017/S0950268809991415. [DOI] [PubMed] [Google Scholar]
- 8.Melegaro A, Choi Y, Pebody R, Gay N. Pneumococcal carriage in United Kingdom families: estimating serotype-specific transmission parameters from longitudinal data. Am J Epidemiol. 2007;166(2):228–35. doi: 10.1093/aje/kwm076. [DOI] [PubMed] [Google Scholar]
- 9.Melegaro A, Gay NJ, Medley GF. Estimating the transmission parameters of pneumococcal carriage in households. Epidemiol Infect. 2004;132(3):433–41. doi: 10.1017/s0950268804001980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weinberger DM, Malley R, Lipsitch M. Serotype replacement in disease after pneumococcal vaccination. Lancet. 2011;378(9807):1962–73. doi: 10.1016/S0140-6736(10)62225-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cobey S, Lipsitch M. Niche and Neutral Effects of Acquired Immunity Permit Coexistence of Pneumococcal Serotypes. Science. 2012 doi: 10.1126/science.1215947. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choi YH, Jit M, Gay N, Andrews N, Waight PA, Melegaro A, George R, Miller E. 7-Valent pneumococcal conjugate vaccination in England and Wales: is it still beneficial despite high levels of serotype replacement? PLoS One. 2011;6(10):e26190. doi: 10.1371/journal.pone.0026190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lipsitch M. Vaccination against colonizing bacteria with multiple serotypes. Proc Natl Acad Sci U S A. 1997;94(12):6571–6. doi: 10.1073/pnas.94.12.6571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lipsitch M. Bacterial vaccines and serotype replacement: lessons from Haemophilus influenzae and prospects for Streptococcus pneumoniae. Emerg Infect Dis. 1999;5(3):336–45. doi: 10.3201/eid0503.990304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Melegaro A, Choi YH, George R, Edmunds WJ, Miller E, Gay NJ. Dynamic models of pneumococcal carriage and the impact of the Heptavalent Pneumococcal Conjugate Vaccine on invasive pneumococcal disease. BMC Infect Dis. 2010;10:90. doi: 10.1186/1471-2334-10-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rubin JL, McGarry LJ, Strutton DR, Klugman KP, Pelton SI, Gilmore KE, Weinstein MC. Public health and economic impact of the 13-valent pneumococcal conjugate vaccine (PCV13) in the United States. Vaccine. 2010;28(48):7634–43. doi: 10.1016/j.vaccine.2010.09.049. [DOI] [PubMed] [Google Scholar]
- 17.Shea KM, Weycker D, Stevenson AE, Strutton DR, Pelton SI. Modeling the decline in pneumococcal acute otitis media following the introduction of pneumococcal conjugate vaccines in the US. Vaccine. 2011;29(45):8042–8. doi: 10.1016/j.vaccine.2011.08.057. [DOI] [PubMed] [Google Scholar]
- 18.Van Effelterre T, Moore MR, Fierens F, Whitney CG, White L, Pelton SI, Hausdorff WP. A dynamic model of pneumococcal infection in the United States: implications for prevention through vaccination. Vaccine. 2010;28(21):3650–60. doi: 10.1016/j.vaccine.2010.03.030. [DOI] [PubMed] [Google Scholar]
- 19.Weinberger DM, Trzcinski K, Lu YJ, Bogaert D, Brandes A, Galagan J, Anderson PW, Malley R, Lipsitch M. Pneumococcal capsular polysaccharide structure predicts serotype prevalence. PLoS Pathog. 2009;5(6):e1000476. doi: 10.1371/journal.ppat.1000476. [DOI] [PMC free article] [PubMed] [Google Scholar]