Abstract
It has been long known that DNA molecules encode information at various levels. The most basic level comprises the base sequence itself and is primarily important for the encoding of proteins and direct base recognition by DNA-binding proteins. A more elusive level consists of the local structural properties of the DNA molecule wherein the DNA sequence only plays an indirect supportive role. These properties are nevertheless an important factor in a large number of biomolecular processes and can be considered as informative signals for the presence of a variety of genomic features. Several recent studies have unequivocally shown the benefit of relying on such DNA properties for modeling and predicting genomic features as diverse as transcription start sites, transcription factor binding sites, or nucleosome occupancy. This review is meant to provide an overview of the key aspects of these DNA conformational and physicochemical properties. To illustrate their potential added value compared to relying solely on the nucleotide sequence in genomics studies, we discuss their application in research on transcription regulation mechanisms as representative cases.
Keywords: DNA structure, structural scales, transcription, functional genomics
Introduction
It is well understood that DNA in living cells is not a uniform linear macromolecule but displays local structural variations that depend on the base sequence. This intrinsic variability in the DNA structure has been found to play a key role in several biological processes. The structure of a DNA molecule is primarily determined by its nucleotide sequence, so that similar DNA sequences have similar DNA structures. The reverse is not always true however: DNA molecules with similar structural properties can arise from different sequences. This redundancy is the reason that the DNA molecule is described as having at least two levels of information.1–3 The first level consists of the basic nucleotide sequence string, which is primarily used as the ‘genetic code’ central to gene coding. A second information level is present in the different properties of the intrinsic DNA structure, where the DNA sequence itself only plays a supporting role. These DNA structural properties are various characteristics of the molecular structure that can be assigned a numeric value based on theoretical simulations or experimental measurements. As the DNA molecule is highly variable at many different levels, from the local stability of the helical duplex to the global conformation of the molecule, so are there many possible DNA structural properties which can be defined. It is not surprising that these structural properties can have a large impact, as most genomic processes involve some sort of change in the DNA structure such as the denaturation required prior to the start of duplication and transcription, protein- induced deformation at DNA-protein complex formation, or the extensive nucleosome packaging of an entire genome. Several studies have shown that to create an accurate model to predict or describe these processes, one must account for the presence of these local structural properties of the DNA molecule. The use of intrinsic DNA structural properties has therefore seen a broad range of applications in genomics in the past decade. In this review we aim to give an overview of the terminology and key aspects of these DNA structural properties, and illustrate their potential added value compared to relying solely on the nucleotide sequence in genomics studies.
Box 1. Definitions as used in this review.
Structural property: specific characteristic of the DNA molecular structure, such as stability, rigidity (or the converse flexibility), or curvature.
Conformational property: structural property relating to the static DNA structure, sometimes termed geometrical property, ground-state structure or structural property in the literature.
Physicochemical property: structural property relating to the dynamic DNA structure implying its potential to change conformation, sometimes termed chemical property or mechanical property in the literature.
Structural scale: Look-up table enumerating all oligonucleotides of a given length and their corresponding values for a certain structural property.
Structural profile: vector of values for a given structural property for every position in a DNA sequence, typically derived from a structural scale.
Higher-order model: A mathematical model which explicitly includes terms for interactions between various observations. Eg, a higher-order dinucleotide sequence model is able to account for the dependency between two sequential base pairs.
Functional genomic element: discrete nucleotide sequence present in the genome with a specific biological role.
We first discuss the basic principles of modeling the properties of the DNA structure from the nucleotide sequence. Epigenetic modifications and specific spatial DNA structures, such as G-quadruplexes, are beyond the scope of this review because their characterization substantially differs from the DNA structural properties as discussed here.4–8 The later sections then discuss two well-studied cases to illustrate the use of DNA structural properties in the classification of genomic elements. Because of the wide array of applications, we have limited ourselves to research on transcription regulation mechanisms as representative cases. The identification of small elements at a position- specific level will be evaluated in the scope of locating potential sites of protein-DNA complex formation in the genome, eg, transcription factor binding sites. The characterization at a coarser level of large genomic elements will be illustrated by gene promoter prediction.
Structural Properties, Scales and Profiles
The structural properties of the DNA molecule can be roughly divided into two categories, the conformational and the physicochemical properties, although these terms are not strictly defined and there is some conflicting terminology in literature.9–12 In this review, we adhere to the most typically used definitions: The conformational properties refer to details of the static DNA structure and how this is influenced by base pair sequences, resulting in translational (eg, slide, rise and shift) or rotational (eg, roll, twist and tilt) variation between successive base pairs and variations in the width and depth of both the major and minor groove (important in several biological processes). At a coarser level, one can also consider the local bends present in the sequence or the curvature of the DNA molecule across large distances or more global properties such as the shape/form in which the DNA molecule is present; in most living cells this is limited to the A-form, B-form or Z-form. The physicochemical properties on the other hand, refer to the dynamic potential of the DNA structure or the free energy stored within different conformations. As the DNA molecule is anisotropically deformable along any axis, several properties can be defined which capture the extent of resistance displayed by the DNA molecule to various changes. The denaturation temperature is also known to vary depending on the molecular structure of the double stranded DNA molecule. This intrinsic variability in denaturation potential can be described by for instance the stacking energy between base pairs or the global free energy of the DNA duplex.
As mentioned before, the structural properties of a stretch of DNA are determined by its nucleotide sequence. A DNA molecule with the same nucleotide sequence will have the same structural properties. It is therefore theoretically possible to predict the entire DNA structure and all of its properties if one is given the DNA sequence. This is currently being done with great accuracy using molecular simulations.13,14 However if one is only interested in a specific set of properties of the DNA structure, there are many models available for DNA structural property prediction derived from experimental or theoretical data. It has been demonstrated that most structural properties are very local features and primarily depend on the neighboring nucleotides of a certain position. Often one can achieve reasonable predictions of the structural properties by simply accounting for the contribution of every di- or trinucleotide to the structural property. Such oligonucleotide contributions are usually represented in a structural scale, a look-up table listing every possible oligonucleotide and a corresponding value which represents the contribution to a given structural property. The length of this oligonucleotide is referred to as the order of the structural scale, eg, a dinucleotide scale is of the second order. Higher order structural scales will always be more informative yet require exponentially more data to enumerate. Most structural scales exist for dinucleotides as they are typically considered the best trade-off between accuracy and complexity. A number of the most frequently used structural properties and their scales are listed in Table 1.
Table 1.
Examples of structural properties.
| Structural property | Description | Category |
|---|---|---|
| Slide-rise-tilt-roll-twist-shift87,88 | The rotational and translational deviations present in DNA base pair steps. | Conformational |
| Curvature89,90 | The large scale curves made by the DNA molecule. They are often derived from the base pair step deviations as per the wedge model. The value of these scales typically corresponds to the intensity of the DNA curvature. | Conformational |
| Minor/major groove depth/width27 | The size of the minor and/or major groove, with larger grooves usually allowing easier access to the bases within the helix. | Conformational |
| A/Z-philicity91,92 | The propensity of the DNA molecule to adopt the A-form or Z-form. Often estimated based on the difference in free energy of these forms. | Conformational |
| Propeller twist93 | Though intrinsically a conformation property, there is a direct link between the twist of the DNA base pairs and it’s rigidity towards deformations. | Conformational |
| Persistence length86 | The molecular distance that the DNA molecule is expected to keep directionality. Also referred to as the DNA bending stiffness. | Physicochemical |
| DNA stability94–96 | Several measures for the DNA helical stability exists, often enumerating the energy theoretically needed per base pair step to disrupt or create the DNA helix. | Physicochemical |
| Stress-induced duplex stability97 | The stability of the DNA helix which accounts for the torsional stress resulting from the superhelical winding. | Physicochemical |
| Base stacking energy98 | The stacking energy of sequential bases that contributes to the overall stability of the DNA helix. | Physicochemical |
| Deformability87 | Deviations accepted by the DNA molecule in response to protein binding. The inverse of these scales is the rigidity, ie, the resistance towards these deviations. | Physicochemical |
| Bendability99 | Usually refers to the propensity of the DNA molecule to bend or be bent in a specific direction. For example, the Brukner scale enumerates the bendability towards the major groove. | Physicochemical |
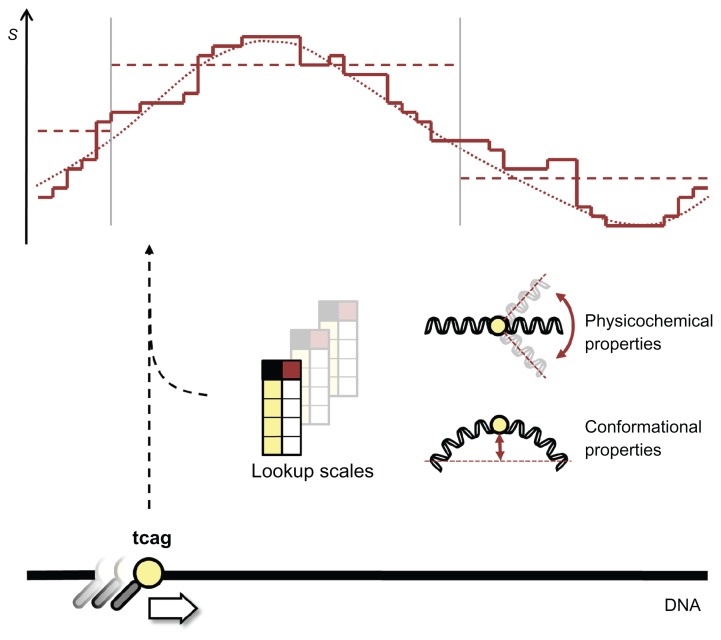
The structural profile represents the variability of a structural property along a given sequence of DNA. It is constructed by looking up the corresponding structural scale values for every successive oligonucleotide in the sequence (Fig. 1). This profile will then correspond to the variation that exists for the given structural property over the given sequence. Note that the vector of this structural profile always has a length equal to the length of the sequence subtracted by one less than the order of the structural scale used, eg, converting a sequence with a dinucleotide scale results in a vector with a length equal to that of the sequence minus one. Three types of features are typically derived from the structural profile that aid in the computational analysis of different genomic elements. The raw profile, or the unmodified structural vector as derived from the structural scales, is often used for position specific effects, eg, modeling the induction of a kink into a single dinucleotide. An average structural profile is the mean of the structural values calculated over all positions in a predefined region. This average profile is usually calculated for broader genetic elements, such as promoter regions. Another common procedure after calculation of the structural profile is the smoothing of the values. This is a rescaling of every value according to a smoothing function that takes into account the values of the neighboring positions, typically by using a short sliding window. The smoothing function could be as simple as averaging the values in the window and results in a smoothed structural profile. As the size of the sliding window is increased (ie, more neighboring positions affect the rescaling), the general patterns in the structural profile will become more pronounced. However if the smoothing range is chosen too high, the information loss will be too great and any important patterns in the structural profile might no longer be visible.2
Figure 1.
Modeling structural properties of the DNA.
Notes: Along the length of the DNA all oligonucleotides of a certain order (usually di- or trinucleotides) are looked up in a table (called a structural scale) which contains corresponding values measuring a certain structural property. These structural scales can represent conformational or physicochemical structural properties, and when viewed along the length of the DNA form a structural profile. Due to the discrete nature of the scale values, a structural profile usually has a staircase-like appearance (full line; the S axis represents the structural property values obtained from the lookup scales). Often these profiles are further smoothed (dotted line), or one might take the average value over a stretch of DNA of a given length (horizontal dashed lines) before they are put to use.
Several databases exist where one can look up or apply a structural scale of interest. The PROPERTY database is one of the earliest collections of structural scales and is at the time of writing still available and listing 35 different structural properties.9 In addition the SITECON web tool can calculate structural profiles for 38 different properties, and if provided with a training set can identify informative features therein.11 The more recent DiProDB contains a list of 125 structural scales.15 A downloadable tool DiProGB allows for calculation of the structural profiles using these scales, or with any user-provided dinucleotide scale.16 Unfortunately both databases are limited to dinucleotides scales, which may be insufficient for some structural properties.17 While higher-order scale collections do exist, they have seemingly never been made available in a straightforward manner.18
Determining Sites of Protein-DNA Complex Formation
DNA structural properties can aid the targeting and functionality of DNA-binding proteins in a wide variety of manners. The common hypothesis is that proteins will find their binding sites by random hopping and sliding along the DNA structure.19 Several studies have shown that rigid and curved DNA molecules will aid in this process.20 Furthermore the specific DNA binding sites of a protein will typically carry recognition features in their structural properties which can be accessed by the protein through ‘indirect’ readout.21 This is distinguished from ‘direct’ readout, were specific bases in the DNA are recognized by the protein binding domain; both direct and indirect readout can contribute to protein-DNA binding.1,3,21,22 The importance of the DNA structure for protein-DNA complex formation lies in the fact that most proteins require the DNA molecule to be present in a specific conformation during complex formation, typically necessitating the deformation of the DNA binding site. It has been shown that proteins will prefer to bind to DNA molecules which easier accept the needed conformation, either because they naturally exist in this state or because they offer little resistance to take on this new state. The energy required for any such deformation can be compensated by favorable contacts made within the complex.23 Many reviews on protein-DNA interactions are available, such as Rohs et al24 The functionality after DNA-binding can also be influenced by the DNA structural properties as many biological processes require the DNA molecule to adapt to a specific conformation, for example a DNA-loop which can facilitate protein-protein interactions at long distances.25
Consensus structural profiles
The most common approach for characterizing protein-DNA binding sites, is through a consensus profile approach. This is especially widely used for transcription factors (TFs). Transcription factors are an important class of DNA binding proteins that will, upon binding the DNA molecule, either activate or repress the transcription of the neighboring gene. TFs typically recognize a specific motif present in the nucleotide sequence and most cause significant distortion of the DNA molecule upon binding. There is great variation between different TFs and no unique nucleotide or structural motif can be attributed to the ensemble of binding sites, and they therefore present an ideal case how DNA structural properties can be beneficial in identifying their binding sites. In a consensus profile framework, the recognition characteristics of a protein are estimated based on an analysis of its known binding sites typically by searching for features (in the DNA sequence and/or the DNA structural properties) which distinguish this set of sites from the genomic background. The direct read-out preferences of a given protein can be represented by a consensus sequence, often represented as a ‘motif logo’.26 In a similar manner the indirect readout can be partially represented by a set of consensus structural profiles.27 This type of consensus profile can be constructed from a set of known binding sites for a certain structural property by many different methods, such as calculating the average profile, averaging specific regions, Fourier analysis or error minimization.9,11,27–29 The structural profiles of unknown sites can be compared to the consensus profile and scored accordingly (Fig. 2, left hand panel) and the contributions of different structural properties can be estimated by simple linear regression or linear discriminant analysis.9,28,30 However, relying on consensus profiles alone provides poor classification performance for the most simple methodologies, likely due to the large amount of possible structural properties and the importance of the direct recognition mechanism in the specificity of many TF proteins.30 Most recent binding site classification methods now use a combination of both sequence data and structural properties and as such require more advanced classification methodologies such as Support Vector Machines, Bayesian Networks, Neural Networks, Hidden Markov Models or Conditional Random Fields.10,12,31–35 These methods have the added advantage of inherently selecting or upweighting the most informative structural profiles and, depending on the model design, the most informative positions. Integration of structural properties with the simple base sequence has been shown to improve classification performance compared to existing methods using only sequence information.32,35 A brief overview of methodologies using the structural consensus approach can be found in Table 2. Unfortunately, as of yet there has been no large scale comparison of different methodologies for representing binding sites by incorporating structural profiles. Such a comparative survey is complicated due to the incompatibility of these methods in two ways. First, the structural profiles that are employed cannot always be separated from the methodology itself. Different structural scales capture different aspects of the DNA structure, and this could partly explain any variation in performance. Secondly, these methodologies generally have a specific application focus and often integrate a wide range of independent data into the model, eg, evolutionary conservation scores or cis-regulatory module information.34 This creates an intrinsic bias in any comparison of different methods and due to the variable nature of TFs and the different organisms in which they reside, no methodology will likely outperform all others in any potential application.
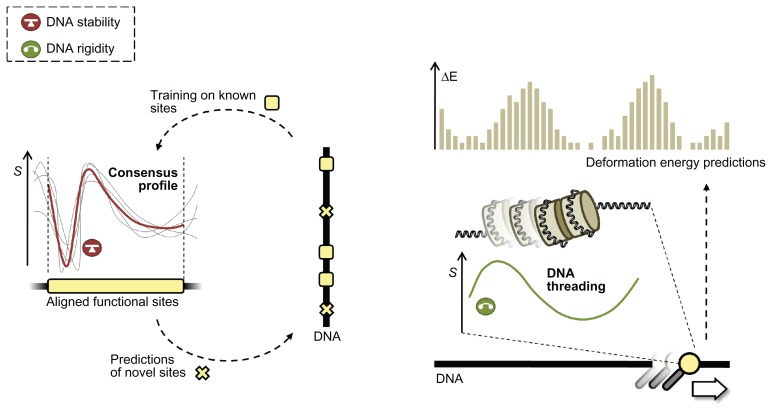
Figure 2.
Predicting DNA binding from DNA structural properties.
Notes: In the case of the consensus approach (left hand panel), all known binding sites are used to generate a consensus profile (red line, representing the structural property stability in this example), which in turn can be used to predict novel binding sites. The consensus profile is an average of structural profiles of aligned known binding sites (grey lines). In DNA threading (right hand panel), a sliding window moves along the DNA calculating the energy required (ΔE axis) for the given stretch of DNA to adapt the required conformation based on structural profiles, in this figure represented by the deformability (green line).
Table 2.
Summary of DNA-binding protein consensus approaches.
| Name | Statistical model | # Properties | Additional information |
|---|---|---|---|
| Karas et al27 | Absolute distance between profiles | 4 conformational + 1 physicochemical properties | |
| ACTIVITY/B–DNA–VIDEO9 | Gaussian distribution | 35 structural properties | |
| Liu et al30 | Gaussian distribution | 5 conformational properties | |
| Gunewardena et al28 | Linear discrimination model | 35 structural properties | Linear steady-state structural templates |
| SITECON11 | Chi-squared test | 35 structural properties | |
| Gardiner et al29 | Fourier transformation | 5 conformational + 18 physicochemical properties | |
| ICSF100 | Moses rank-like test | 6 conformational properties | |
| MDS-HMM12 | Hidden Markov model | 2 conformational properties | |
| ProMapper10/BioBayesNet101 | Bayesian network | 35 structural properties | Additional sequence features, eg, base composition |
| Holloway et al31 | Support vector machine | 1 conformational + 4 physicochemical properties | Additional sequence features, eg, phylogeny |
| DISCOVER34 | Conditional random fields | 1 physicochemical property (DNA stability) | Additional sequence features, eg, phylogeny |
| GANN33 | Neural network + Genetic algorithm | Unspecified | |
| CRoSSeD35 | Conditional random fields | 5 conformational + 7 physicochemical properties | |
| SiteSleuth32 | Support vector machine | 12 conformational + 62 physicochemical properties | Physicochemical properties reduced to 8 eigen vectors |
Protein-DNA threading
In some cases, instead of possessing only a set of binding site locations for a given DNA-interacting protein, a detailed 3D structure of the protein-DNA complex is known. With this information, one could use a protein-DNA threading approach as well (Fig. 2). These screen for target sites of DNA- binding proteins by modeling the complex formation. From solved complexes, the bound conformation of the protein and the DNA molecule can be derived, as well as their intermolecular interactions. A common approach to evaluate other target sites is to then ‘thread’ novel DNA sequences (eg, along a genomic region) into the required conformation and calculate the formation energy, or an equivalent approximation, of the resulting complex. Part of the formation energy can be the result of the deformation needed to change the intrinsic DNA superstructure to the required bound conformation (Fig. 2, right hand panel).36,37 Use of the aforementioned deformability scales are one possible way to calculate the energy that is required for this DNA conformation change.38–41 This deformation energy can then be integrated into the energy function of the protein-DNA complex.42,43 The resulting energy can then be used to assess the viability of the formation of the protein-DNA complex at a given genomic site.
While the actual biomolecular mechanism of protein-DNA binding are more firmly rooted in these threading approaches than relying on consensus profiles, there are drawbacks as well. For instance, there is no guarantee that different DNA sequences will display the same conformation when bound by the protein than the sequence that was used to construct the 3D model.1,44,45 The major limitation for the widespread application of protein-DNA threading for binding site prediction however, is not due to its methodological principals but rather due to a lack of ‘solved’ protein-DNA complexes. Threading approaches require the bound protein-DNA complex or that of a related protein to be known, which is not very common for many DNA-binding proteins such as TFs.46 More often these approaches are used for well characterized protein-DNA complexes, such as the nucleosomes responsible for the higher-order packing of the DNA molecule into chromatin in eukaryotes. Because it affects the genomic accessibility, this DNA packing has a critical role in various cellular processes, among which transcription regulation. Given this interest, predicting nucleosome positioning currently forms the bulk of protein-DNA threading that relies on DNA structural properties.39,41,44,45 While these methodologies do successfully increase prediction, it is well known that there are many other factors that drive the positioning of nucleosomes and that can confound the contribution of certain DNA characteristics in nucleosome formation.47–49 In that respect, there has also been some discussion based on recent evidence suggesting that the main experimental technique for determining nucleosome occupancy may generate biased results, which, if true, would also affect the perceived dependence of nucleosome occupancy on DNA sequence and/or structure.50,51
Identification of Gene Promoter Regions
Different genomic regions are known to display unique structural characteristics.52 Many statistical models have therefore been built to identify the presence of a given genomic region based on its structural properties. The most widely used among these applications are those that attempt to predict the promoter region. Promoters are the regions upstream from genes, where the RNA polymerase is recruited and transcription is started. Promoters seem to share similar structural profiles that can be related to their function. Exploiting the common patterns in these profiles has been known to increase the performance of promoter prediction algorithms. The structural profiles for individual promoters are however very noisy, and identifying these common patterns require complex modeling methods. Even then predictions remain coarse and cannot identify the exact transcription start site (TSS), but only the general promoter region. Furthermore, the structural profiles of promoters differ greatly between eukaryotes and prokaryotes as the transcription complexes are radically different. Most promoter prediction methods are therefore tuned to a single taxonomical domain or even a single species, with few exceptions that typically require retraining for novel organisms. A brief summary of structure-based promoter prediction methods can be found in Table 3.
Table 3.
Summary of structure-based promoter prediction methods.
| Name | Statistical model | Structural property | Organism(s) |
|---|---|---|---|
| EP353 | Average profile | Base stacking energy | Animals, fungi, algae, higher plants and protists |
| ProSOM62 | Unsupervised self organizing map | Base stacking energy | Human |
| Florquin et al2 | Adaptive quality-based clustering | 5 conformational + 8 physicochemical properties | Human, mouse and plant |
| PNNP59 | Pattern-based nearest neighbor search | DNA stability | Human, mouse, Caenorhabditis elegans and plant |
| PromPredict70,102 | Absolute and relative difference | DNA stability | Plants and prokaryotes |
| McPromoter61 | Stochastic segment model | DNA twist and persistence length | Drosophila |
| Prostar63 | Mahalanobis distance | DNA deformability | Human |
| Profisi60 | Average profile | DNA stability | Human |
| ARTS103 | Support vector machine | DNA twist and base stacking energy | Human |
| Gardiner et al104 | Ward’s clustering algorithm | 5 conformational + 18 physicochemical properties | Human |
| Wang et al74 | Linear discrimination model | Stress-induced duplex destability | Escherichia coli |
| N472 | Neural network | DNA stability | Escherichia coli |
| Conilione et al64 | Neural network | Base stacking energy | Escherichia coli |
| Parbhane et al75 | Neural network | DNA wedge and twist | Escherichia coli |
| Mallios et al105 | Stepwise binary logistic regression | 2 conformational + 2 physicochemical properties | Chlamydia trachomatis |
Predicting eukaryote promoters
In eukaryotes the promoter region can be divided into three parts: the core promoter where the basal transcription complex binds, the proximal promoter where most transcription factor binding sites are located and the distal promoter that can contain enhancer elements. The promoters themselves can be grouped according to the RNA polymerase that binds to them. The targets of the different RNA polymerase have different recognition elements and therefore different structural profiles.53 Most promoter prediction methods will focus on RNA polymerase II which transcribes protein-coding genes and most microRNAs. On average, these promoters are described as being more rigid than the remainder of the genome, but the actual structural profile of promoter flexibility is much more complex.54,55 The global rigidity is most likely necessary to exclude nucleosomes from the promoter region, as they will compete with the binding of transcription factors and the basal transcription complex. The proximal promoter is usually characterized by a decrease in the rigidity of the promoter.54 The hypothesis is that the binding sites of transcription factors need to be flexible to allow complex formation. There is likely a careful trade-off between rigid DNA stretches blocking nucleosomes and small flexible regions attracting transcription factors.54,56 Indeed the promoter activity seems to correlate with the proportion of flexible regions in the whole fragment.57 The core promoter is typically characterized by a gradual decrease in rigidity from upstream of the TSS to downstream.2,54,57 Extreme rigidity values embedded in this region match with known promoter elements. For example, the TATA-box corresponds to a very rigid region in the promoter. This rigid peak at the −30 position can still be observed even if no explicit TATA motif is present, which lead to the hypothesis that this rigidity feature is more important than the actual sequence motif and could partially explain why many promoters lack a clear TATA motif.2,58 Eukaryotic promoters are also typically more stable than the genomic average with peaks of heavy instability at the promoter elements, such as the TATA-box and the TSS. Likely this contrast helps direct the transcription complex to the correct transcription start site.59 A conceptual overview of the main structural features of eukaryotic promoters is given in Figure 3 (left hand panel).
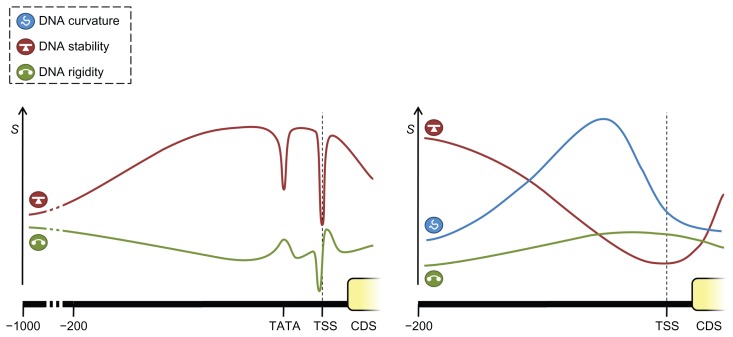
Figure 3.
Conceptual representation of structural features of eukaryotic and prokaryotic promoters.
Notes: Eukaryotic proximal promoters (left hand panel) are generally characterized by an increased stability (red line; the S axis represents the structural property values) and a decreasing rigidity (green line) compared to surrounding regions (although eukaryotic promoters on average still have a higher rigidity than the rest of the genome), with strong peaks or valleys at functional sites such as the TSS or TATA box. In contrast, prokaryotic promoters on average have a decreased stability (red line) and increased rigidity (green line) and have been observed to show a broad curvature peak (blue line) upstream of the TSS.
A number of methods have been proposed to translate the complex structural profiles of eukaryotic promoters into features which can be used in promoter prediction with varying success. The most straightforward approaches classify promoters by comparing an averaged structural profile for a stretch of sequence to a set threshold. Such methods have relied for instance on the base stacking energy as a representation of the stability, or the DNA melting temperature as defined by an extensive calculation of the genome-wide DNA duplex stability.53,60 Averaging out the structural properties will unfortunately ignore the typical structural patterns observed for promoters. Other methods try to directly use the pattern contained in the structural profiles. The McPromoter method does this by dividing the promoter into smaller regions and models the average of the structural profiles in every segment as a single observation from a Hidden Markov Model.61 Out of all the tested structural properties, the DNA twist, the persistence length and the propeller twist were found most informative for predicting Drosophila melanogaster promoters. The PNNP method uses a pattern-based distance nearest neighbor search where promoters are classified if the maximum deviation from the relative profile is smaller than a threshold.59 In this manner PNNP is able to model promoters which have a similar pattern in the structural profile but at a different absolute level, as observed for promoters containing CpG islands and those that don’t. PNNP predictions based on the DNA duplex free energy gave the best performance. ProSOM uses an unsupervised self-organizing map to cluster the base stacking energy profile of various sequences into subgroups.62 Classification of promoters in this case consists of attempting to cluster the structural profile of the unknown sequence with a known set of promoters. This approach has the advantage of accounting for different groups of promoters. The general conclusion for most of these methods using only a single or few structural properties, is that they are able to get a classification performance equal to or often greater than methods using a large number of sequence signals in complex statistical frameworks, despite the relative simplicity of their approaches.53,59,60,63
Predicting prokaryote promoters
Prokaryotic genomes have a very high gene density and therefore promoters are typically much shorter than their eukaryotic counterparts, sometimes spanning less than a hundred base pairs. Because of this short intergenic region between coding regions, divergent promoters have been known to overlap. Many sequential genes are typically transcribed in a single run in so-called operons. On average, promoters are less stable, more rigid and have more extreme curvature than other genomic regions in most prokaryotes (Fig. 3, right hand panel).64,65 The role of the DNA curvature in prokaryotic promoters seems to vary greatly, as does the type of curvature pattern found in these promoters.66 Mostly extreme DNA curvature has been associated with the presence of strong transcription factor binding motifs and it has been postulated to act as a thermosensor under some conditions.65,67,68 The low stability is likely to facilitate helix denaturation prior to the transcription event and is indeed centered around the TSS with the upstream region being less stable than the downstream region.69 This fact is often exploited for the prediction of promoter regions. Indeed the first time it was observed, the absolute value and the difference in values of the DNA duplex stability immediately upstream of the TSS (<100 bp) were shown to be informative in a simple framework where a sequence was classified as a promoter if it exceeded a certain threshold for both measures.70,71 These measures for DNA duplex stability have been frequently used, eg, in the N4 promoter prediction algorithm where they are integrated in a neural network.72 The PromPredict method also uses the difference between upstream and downstream DNA duplex stability but groups the sequences according to GC content prior to classification.69 This is based on the fact that the difference in DNA duplex stability between the TSS and the region downstream varies according to the GC content of the genomic region containing the promoter. These findings have allowed promoter prediction across a wide range of prokaryotes.73 Related is the SIDD method, which uses the stress-induced duplex destability profile for the classification of promoters with a linear discriminatory model.74 The stress-induced duplex stability differs from the standard stability calculations because it accounts for torsional stresses present in the DNA molecule as a result of the genomic negative superhelicity. In this framework, the stress- induced stability was found to be more informative for promoter prediction than other stability profiles, the rigidity profile or the curvature profile.
Discussion
The idea of using DNA structural properties to model and describe genomic elements has been around for some time. Originally they were mostly limited to focused, small-scale studies. Only in the past decade have these types of studies moved into the realm of genome-wide applications. As more and more sequence data became publically available and the quality of annotated genomes steadily increased, so did the number of studies that tried to identify specific patterns of DNA structural properties for various types of genomic elements. In contrast, ‘raw’ nucleotide sequence information has been the standard representation of DNA in computational biology for much longer, and techniques such as consensus sequences and position weight matrices have become the default workhorses for the identification of many genomic elements. Highly advanced sequence-based methods have also been developed over the years and many have proven to be successful approaches for functional genomics applications such as those discussed in this review. Sequence-based methods can sometimes even capture part of the local DNA structure, as the DNA structural properties are generally dependent on interactions between neighboring base pairs. This is illustrated by the fact that they are often calculated from higher-order (mostly di- or trinucleotide) lookup scales. The advantage of using DNA structural properties is that they explicitly assign actual ‘measurement’ values (ie, the structural scales) to a given sequence of DNA. These values represent conformational and physicochemical characteristics, and can thus reveal structural patterns that would remain hidden when only relying on the corresponding categorical higher-order nucleotides.75 Thus there is much to be gained by employing the DNA structure. Grouping different sequences with similar structural properties for example, will always be more difficult for sequence-based methods, as was shown for predicting prokaryotic TF binding sites.35 Correctly identifying the contribution of the structural properties can also generate more powerful models, which has lead for instance to better predictions for nucleosome formation energy.44 Nevertheless, a strong conservation of nucleotide sequence will always correspond to a strong conservation of the DNA structural signal. Even if complex structural mechanics play an essential functional role in such a case, calculated structural properties will not provide any complementary information. Only experimental assessment of the underlying mechanism can then quantify the relative contributions of the DNA structure.
The purpose of this review is not to provide an exhaustive list of structure-based methodologies. There are many other genomic elements for which common patterns of DNA structural properties have been described or even integrated into a classification framework. Examples that were not discussed in detail here include splice sites, replication start sites, transposon insertion sites, methylation events, functional SNPs, plasmid conjugation factor binding and gene prediction.20,76–84 Instead, the goal of this review is to provide a bridge between different application domains and to further promote the added value that these structural properties of DNA could have in functional genomics studies. Indeed, it is becoming more and more clear that DNA structural properties play an important role in a great many biomolecular processes and that their characterization for different genomic elements will be essential to generate a complete understanding. As we have presented here, different genomic elements require different representation methods to capture potential defining structural patterns. This does not imply that these methodologies cannot learn from one and other as there are functional and/or biomolecular relationships between many of these elements, the characterization of which might benefit from more comprehensive approaches. For example, the inherent flexibility of the DNA molecule is often found to be an informative feature in a large number of genetic elements.35,52,63,82,83,85,86 This can be problematic as different genomic elements sharing a number of structural similarities can result in false positive predictions during classification. Proper structural characterization can thus also be important to help understand such intricate relationships between different elements, as was shown to be the case for promoter regions and splice sites.20 Regional relationship between various elements can also occur, eg, TF binding sites are commonly located in the promoter region of genes, and thus identification of TF binding sites could benefit from knowledge the promoter region and vice versa. In the end there is still much to be learned about what DNA structural properties play a role where and, perhaps more critically, how they can contribute to revealing the underlying biomolecular mechanisms.
Footnotes
Author Contributions
Conceived and designed the experiments: PM, KM KE. Analysed the data: PM, KM, KE. Wrote the first draft of the manuscript: PM, KM, KE. Contributed to the writing of the manuscript: PM, KM, KE. Agree with manuscript results and conclusions: PM, KM, KE. Jointly developed the structure and arguments for the paper: PM, KM, KE. Made critical revisions and approved final version: PM, KM, KE. All authors reviewed and approved of the final manuscript.
Funding
This work was supported by the KULeuven Research Council [GOA/08/011, CoE EF/05/007—SymBioSys, CREA/08/023, OT 05-33, OT09/022]; the agency for Innovation by Science and Technology [SBO-BioFrame, SB-81297]; Interuniversity Attraction Poles [P6/25—BioMaGNet]; Research Foundation— Flanders [IOK-B9725-G.0329.09]; and the Human Frontier Science Program [RGY0079/2007C].
Competing Interests
Author(s) disclose no potential conflicts of interest.
Disclosures and Ethics
As a requirement of publication author(s) have provided to the publisher signed confirmation of compliance with legal and ethical obligations including but not limited to the following: authorship and contributorship, conflicts of interest, privacy and confidentiality and (where applicable) protection of human and animal research subjects. The authors have read and confirmed their agreement with the ICMJE authorship and conflict of interest criteria. The authors have also confirmed that this article is unique and not under consideration or published in any other publication, and that they have permission from rights holders to reproduce any copyrighted material. Any disclosures are made in this section. The external blind peer reviewers report no conflicts of interest.
References
- 1.Bareket-Samish A, Cohen I, Haran TE. Direct versus indirect readout in the interaction of the trp repressor with non-canonical binding sites. Journal of Molecular Biology. 1998;277(5):1071–80. doi: 10.1006/jmbi.1998.1638. [DOI] [PubMed] [Google Scholar]
- 2.Florquin K, Saeys Y, Degroeve S, Rouzé P, Van de Peer Y. Large-scale structural analysis of the core promoter in mammalian and plant genomes. Nucleic Acids Research. 2005;33(13):4255–64. doi: 10.1093/nar/gki737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Michael Gromiha M, Siebers JG, Selvaraj S, Kono H, Sarai A. Intermolecular and intramolecular readout mechanisms in protein-DNA recognition. Journal of Molecular Biology. 2004;337(2):285–94. doi: 10.1016/j.jmb.2004.01.033. [DOI] [PubMed] [Google Scholar]
- 4.Lipps HJ, Rhodes D. G-quadruplex structures: in vivo evidence and function. Trends in Cell Biology. 2009;19(8):414–22. doi: 10.1016/j.tcb.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 5.Johnson JE, Smith JS, Kozak ML, Johnson FB. In vivo veritas: using yeast to probe the biological functions of G-quadruplexes. Biochimie. 2008;90(8):1250–63. doi: 10.1016/j.biochi.2008.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huppert JL. Hunting G-quadruplexes. Biochimie. 2008;90(8):1140–8. doi: 10.1016/j.biochi.2008.01.014. [DOI] [PubMed] [Google Scholar]
- 7.Choi JK, Kim YJ. Epigenetic regulation and the variability of gene expression. Nature Genetics. 2008;40(2):141–7. doi: 10.1038/ng.2007.58. [DOI] [PubMed] [Google Scholar]
- 8.Lim SJ, Tan TW, Tong JC. Computational Epigenetics: the new scientific paradigm. Bioinformation. 2010;4(7):331–7. doi: 10.6026/97320630004331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ponomarenko JV, Ponomarenko MP, Frolova S, et al. Conformational and physicochemical DNA features specific for transcription factor binding sites. Bioinformatics (Oxford, England) 1999;15(7–8):654–8. doi: 10.1093/bioinformatics/15.7.654. [DOI] [PubMed] [Google Scholar]
- 10.Pudimat R, Schukat-Talamazzini E-G, Backofen R. A multiple-feature framework for modelling and predicting transcription factor binding sites. Bioinformatics (Oxford, England) 2005;21(14):3082–8. doi: 10.1093/bioinformatics/bti477. [DOI] [PubMed] [Google Scholar]
- 11.Oshchepkov DY, Vityaev EE, Grigorovich DA, Ignatieva EV, Khlebodarova TM. SITECON: a tool for detecting conservative conformational and physicochemical properties in transcription factor binding site alignments and for site recognition. Nucleic Acids Research. 2004;32(Web Server issue):W208–12. doi: 10.1093/nar/gkh474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thayer KM, Beveridge DL. Hidden Markov models from molecular dynamics simulations on DNA. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(13):8642–7. doi: 10.1073/pnas.132148699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheatham TE. Simulation and modeling of nucleic acid structure, dynamics and interactions. Current Opinion in Structural Biology. 2004;14(3):360–7. doi: 10.1016/j.sbi.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 14.Fujii S, Kono H, Takenaka S, Go N, Sarai A. Sequence-dependent DNA deformability studied using molecular dynamics simulations. Nucleic Acids Research. 2007;35(18):6063–74. doi: 10.1093/nar/gkm627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friedel M, Nikolajewa S, Sühnel J, Wilhelm T. DiProDB: a database for dinucleotide properties. Nucleic Acids Research. 2009;37(Database issue):D37–40. doi: 10.1093/nar/gkn597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Friedel M, Nikolajewa S, Sühnel J, Wilhelm T. DiProGB: the dinucleotide properties genome browser. Bioinformatics (Oxford, England) 2009;25(19):2603–4. doi: 10.1093/bioinformatics/btp436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Packer MJ, Dauncey MP, Hunter CA. Sequence-dependent DNA structure: tetranucleotide conformational maps. Journal of Molecular Biology. 2000;295(1):85–103. doi: 10.1006/jmbi.1999.3237. [DOI] [PubMed] [Google Scholar]
- 18.Gardiner E. Sequence-dependent DNA Structure: A Database of Octamer Structural Parameters. Journal of Molecular Biology. 2003;332(5):1025–35. doi: 10.1016/j.jmb.2003.08.006. [DOI] [PubMed] [Google Scholar]
- 19.von Hippel PH, Berg OG. Facilitated target location in biological systems. The Journal of Biological Chemistry. 1989;264(2):675–8. [PubMed] [Google Scholar]
- 20.Cao X-Q, Zeng J, Yan H. Physical signals for protein-DNA recognition. Physical Biology. 2009;6(3):036012. doi: 10.1088/1478-3975/6/3/036012. [DOI] [PubMed] [Google Scholar]
- 21.von Hippel P. Protein-DNA recognition: new perspectives and underlying themes. Science. 1994;263(5148):769–70. doi: 10.1126/science.8303292. [DOI] [PubMed] [Google Scholar]
- 22.Kitayner M, Rozenberg H, Kessler N, et al. Structural basis of DNA recognition by p53 tetramers. Molecular Cell. 2006;22(6):741–53. doi: 10.1016/j.molcel.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 23.Kalodimos CG, Boelens R, Kaptein R. Toward an integrated model of protein-DNA recognition as inferred from NMR studies on the Lac repressor system. Chemical Reviews. 2004;104(8):3567–86. doi: 10.1021/cr0304065. [DOI] [PubMed] [Google Scholar]
- 24.Rohs R, West SM, Sosinsky A, et al. The role of DNA shape in protein-DNA recognition. Nature. 2009;461(7268):1248–53. doi: 10.1038/nature08473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheema AK, Choudhury NR, Das HK. A- and T-Tract-Mediated Intrinsic Curvature in Native DNA between the Binding Site of the Upstream Activator NtrC and the nifLA Promoter of Klebsiella pneumoniae Facilitates Transcription. J Bacteriol. 1999;181(17):5296–302. doi: 10.1128/jb.181.17.5296-5302.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stormo GD. DNA binding sites: representation and discovery. Bioinformatics (Oxford, England) 2000;16(1):16–23. doi: 10.1093/bioinformatics/16.1.16. [DOI] [PubMed] [Google Scholar]
- 27.Karas H, Knüppel R, Schulz W, Sklenar H, Wingender E. Combining structural analysis of DNA with search routines for the detection of transcription regulatory elements. Computer Applications in the Biosciences: CABIOS. 1996;12(5):441–6. doi: 10.1093/bioinformatics/12.5.441. [DOI] [PubMed] [Google Scholar]
- 28.Gunewardena S, Jeavons P, Zhang Z. Enhancing the prediction of transcription factor binding sites by incorporating structural properties and nucleotide covariations. Journal of Computational Biology. 2006;13(4):929–45. doi: 10.1089/cmb.2006.13.929. [DOI] [PubMed] [Google Scholar]
- 29.Gardiner EJJ, Hunter Ca, Willett P. Structural Fingerprints of Transcription Factor Binding Site Regions. Algorithms. 2009;2(1):448–69. [Google Scholar]
- 30.Liu R, Blackwell TW, States DJ. recognition by the E. coli MetJ transcription factor. Bioinformatics. 2001;17(7):622–33. doi: 10.1093/bioinformatics/17.7.622. [DOI] [PubMed] [Google Scholar]
- 31.Holloway DT, Kon M, Delisi C. Machine learning for regulatory analysis and transcription factor target prediction in yeast. Systems and Synthetic Biology. 2007;1(1):25–46. doi: 10.1007/s11693-006-9003-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bauer AL, Hlavacek WS, Unkefer PJ, Mu F. Using sequence-specific chemical and structural properties of DNA to predict transcription factor binding sites. PLoS Computational Biology. 2010;6(11):e1001007. doi: 10.1371/journal.pcbi.1001007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beiko RG, Charlebois RL. GANN: genetic algorithm neural networks for the detection of conserved combinations of features in DNA. BMC Bioinformatics. 2005;6:36. doi: 10.1186/1471-2105-6-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fu W, Ray P, Xing EP. DISCOVER: a feature-based discriminative method for motif search in complex genomes. Bioinformatics (Oxford, England) 2009;25(12):i321–9. doi: 10.1093/bioinformatics/btp230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meysman P, Dang TH, Laukens K, et al. Use of structural DNA properties for the prediction of transcription-factor binding sites in Escherichia coli. Nucleic Acids Research. 2010;39(2):e6. doi: 10.1093/nar/gkq1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sarai A, Kono H. Protein-DNA recognition patterns and predictions. Annual Review of Biophysics and Biomolecular Structure. 2005;34:379–98. doi: 10.1146/annurev.biophys.34.040204.144537. [DOI] [PubMed] [Google Scholar]
- 37.Kono H, Sarai A. Structure-based prediction of DNA target sites by regulatory proteins. Proteins. 1999;35(1):114–31. [PubMed] [Google Scholar]
- 38.Anselmi C, Bocchinfuso G, De Santis P, Savino M, Scipioni A. Dual role of DNA intrinsic curvature and flexibility in determining nucleosome stability. Journal of Molecular Biology. 1999;286(5):1293–301. doi: 10.1006/jmbi.1998.2575. [DOI] [PubMed] [Google Scholar]
- 39.Miele V, Vaillant C, d’Aubenton-Carafa Y, Thermes C, Grange T. DNA physical properties determine nucleosome occupancy from yeast to fly. Nucleic Acids Research. 2008;36(11):3746–56. doi: 10.1093/nar/gkn262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tolstorukov MY, Choudhary V, Olson WK, Zhurkin VB, Park PJ. nuScore: a web-interface for nucleosome positioning predictions. Bioinformatics (Oxford, England) 2008;24(12):1456–8. doi: 10.1093/bioinformatics/btn212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu F, Olson WK. DNA architecture, deformability, and nucleosome positioning. Journal of Biomolecular Structure and Dynamics. 2010;27(6):725–39. doi: 10.1080/073911010010524943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee W, Tillo D, Bray N, et al. A high-resolution atlas of nucleosome occupancy in yeast. Nature Genetics. 2007;39(10):1235–44. doi: 10.1038/ng2117. [DOI] [PubMed] [Google Scholar]
- 43.Becker NB, Wolff L, Everaers R. Indirect readout: detection of optimized subsequences and calculation of relative binding affinities using different DNA elastic potentials. Nucleic Acids Research. 2006;34(19):5638–49. doi: 10.1093/nar/gkl683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morozov AV, Fortney K, Gaykalova DA, et al. Using DNA mechanics to predict in vitro nucleosome positions and formation energies. Nucleic Acids Research. 2009;37(14):4707–22. doi: 10.1093/nar/gkp475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Balasubramanian S, Xu F, Olson WK. DNA sequence-directed organization of chromatin: structure-based computational analysis of nucleosome-binding sequences. Biophysical Journal. 2009;96(6):2245–60. doi: 10.1016/j.bpj.2008.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim R, Corona RI, Hong B, Guo J. Benchmarks for flexible and rigid transcription factor-DNA docking. BMC Structural Biology. 2011;11(1):45. doi: 10.1186/1472-6807-11-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Arya G, Maitra A, Grigoryev SA. A structural perspective on the where, how, why, and what of nucleosome positioning. Journal of Biomolecular Structure and Dynamics. 2010;27(6):803–20. doi: 10.1080/07391102.2010.10508585. [DOI] [PubMed] [Google Scholar]
- 48.Iyer VR. Nucleosome positioning: bringing order to the eukaryotic genome. Trends in Cell Biology. 2012 doi: 10.1016/j.tcb.2012.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Trifonov EN. Cracking the chromatin code: precise rule of nucleosome positioning. Physics of Life Reviews. 2011;8(1):39–50. doi: 10.1016/j.plrev.2011.01.004. [DOI] [PubMed] [Google Scholar]
- 50.Chung H-R, Dunkel I, Heise F, et al. The effect of micrococcal nuclease digestion on nucleosome positioning data. PloS One. 2010;5(12):e15754. doi: 10.1371/journal.pone.0015754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Allan J, Fraser RM, Owen-Hughes T, Keszenman-Pereyra D. Micrococcal nuclease does not substantially bias nucleosome mapping. Journal of Molecular Biology. 2012;417(3):152–64. doi: 10.1016/j.jmb.2012.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pedersen AG, Jensen LJ, Brunak S, Staerfeldt HH, Ussery DW. A DNA structural atlas for Escherichia coli. Journal of Molecular Biology. 2000;299(4):907–30. doi: 10.1006/jmbi.2000.3787. [DOI] [PubMed] [Google Scholar]
- 53.Abeel T, Saeys Y, Bonnet E, Rouzé P, Van de Peer Y. Generic eukaryotic core promoter prediction using structural features of DNA. Genome Research. 2008;18(2):310–23. doi: 10.1101/gr.6991408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cao X-Q, Zeng J, Yan H. Structural property of regulatory elements in human promoters. Physical Review E. 2008;77(4):1–7. doi: 10.1103/PhysRevE.77.041908. [DOI] [PubMed] [Google Scholar]
- 55.Akan P, Deloukas P. DNA sequence and structural properties as predictors of human and mouse promoters. Gene. 2008;410(1):165–76. doi: 10.1016/j.gene.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tirosh I, Berman J, Barkai N. The pattern and evolution of yeast promoter bendability. Trends in Genetics: TIG. 2007;23(7):318–21. doi: 10.1016/j.tig.2007.03.015. [DOI] [PubMed] [Google Scholar]
- 57.Fukue Y, Sumida N, Tanase J-I, Ohyama T. A highly distinctive mechanical property found in the majority of human promoters and its transcriptional relevance. Nucleic Acids Research. 2005;33(12):3821–7. doi: 10.1093/nar/gki700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zeng J, Cao X-Q, Zhao H, Yan H. Finding human promoter groups based on DNA physical properties. Physical Review E. 2009;80(4) doi: 10.1103/PhysRevE.80.041917. [DOI] [PubMed] [Google Scholar]
- 59.Gan Y, Guan J, Zhou S. A pattern-based nearest neighbor search approach for promoter prediction using DNA structural profiles. Bioinformatics (Oxford, England) 2009;25(16):2006–12. doi: 10.1093/bioinformatics/btp359. [DOI] [PubMed] [Google Scholar]
- 60.Dineen DG, Wilm A, Cunningham P, Higgins DG. High DNA melting temperature predicts transcription start site location in human and mouse. Nucleic Acids Research. 2009;37(22):7360. doi: 10.1093/nar/gkp821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ohler U, Niemann H, Liao GC, Rubin GM. Joint modeling of DNA sequence and physical properties to improve eukaryotic promoter recognition. Bioinformatics (Oxford, England) 2001;17(Suppl 1):S199–206. doi: 10.1093/bioinformatics/17.suppl_1.s199. [DOI] [PubMed] [Google Scholar]
- 62.Abeel T, Saeys Y, Rouzé P, Van de Peer Y. ProSOM: core promoter prediction based on unsupervised clustering of DNA physical profiles. Bioinformatics (Oxford, England) 2008;24(13):i24–31. doi: 10.1093/bioinformatics/btn172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Goni JR, Pérez A, Torrents D, Orozco M. Determining promoter location based on DNA structure first-principles calculations. Genome Biology. 2007;8(12):R263. doi: 10.1186/gb-2007-8-12-r263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Conilione P, Wang D, Abraham A, et al. Neural Classification of E. coli Promoters Using Selected DNA profiles. In: Abraham A, Dote Y, Furuhashi T, et al., editors. Soft Computing as Transdisciplinary Science and Technology. Berlin, Heidelberg: Springer Berlin Heidelberg; 2005. pp. 51–60. [Google Scholar]
- 65.Nov Klaiman T, Hosid S, Bolshoy A. Upstream curved sequences in E. coli are related to the regulation of transcription initiation. Computational Biology and Chemistry. 2009;33(4):275–82. doi: 10.1016/j.compbiolchem.2009.06.007. [DOI] [PubMed] [Google Scholar]
- 66.Kozobay-Avraham L, Hosid S, Volkovich Z, Bolshoy A. Prokaryote clustering based on DNA curvature distributions. Discrete Applied Mathematics. 2009;157(10):2378–87. [Google Scholar]
- 67.Prosseda G, Falconi M, Giangrossi M, et al. The virF promoter in Shigella: more than just a curved DNA stretch. Molecular Microbiology. 2004;51:523–37. doi: 10.1046/j.1365-2958.2003.03848.x. [DOI] [PubMed] [Google Scholar]
- 68.Olivares-Zavaleta N, Jáuregui R, Merino E. Genome analysis of Escherichia coli promoter sequences evidences that DNA static curvature plays a more important role in gene transcription than has previously been anticipated. Gene. 2006;87(3):329–37. doi: 10.1016/j.ygeno.2005.11.023. [DOI] [PubMed] [Google Scholar]
- 69.Rangannan V, Bansal M. Relative stability of DNA as a generic criterion for promoter prediction: whole genome annotation of microbial genomes with varying nucleotide base composition. Molecular Bio Systems. 2009;5(12):1758–69. doi: 10.1039/B906535K. [DOI] [PubMed] [Google Scholar]
- 70.Kanhere A, Bansal M. A novel method for prokaryotic promoter prediction based on DNA stability. BMC Bioinformatics. 2005;6:1. doi: 10.1186/1471-2105-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.SantaLucia J. A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(4):1460–5. doi: 10.1073/pnas.95.4.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Askary A, Masoudi-Nejad A, Sharafi R, et al. N4: a precise and highly sensitive promoter predictor using neural network fed by nearest neighbors. Genes and Genetic Systems. 2009;84(6):425–30. doi: 10.1266/ggs.84.425. [DOI] [PubMed] [Google Scholar]
- 73.Rangannan V, Bansal M. High-quality annotation of promoter regions for 913 bacterial genomes. Bioinformatics (Oxford, England) 2010;26(24):3043–50. doi: 10.1093/bioinformatics/btq577. [DOI] [PubMed] [Google Scholar]
- 74.Wang H, Benham CJ. Promoter prediction and annotation of microbial genomes based on DNA sequence and structural responses to superhelical stress. BMC Bioinformatics. 2006;7:248. doi: 10.1186/1471-2105-7-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Parbhane RV, Tambe SS, Kulkarni BD. ANN modeling of DNA sequences: new strategies using DNA shape code. Computers and Chemistry. 2000;24(6):699–711. doi: 10.1016/s0097-8485(00)00072-3. [DOI] [PubMed] [Google Scholar]
- 76.Cayrou C, Coulombe P, Vigneron A, et al. Genome-scale analysis of metazoan replication origins reveals their organization in specific but flexible sites defined by conserved features. Genome Research. 2011 doi: 10.1101/gr.121830.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cao X-Q, Zeng J, Yan H. Structural properties of replication origins in yeast DNA sequences. Physical Biology. 2008;5(3):036012. doi: 10.1088/1478-3975/5/3/036012. [DOI] [PubMed] [Google Scholar]
- 78.Bock C, Paulsen M, Tierling S, et al. CpG island methylation in human lymphocytes is highly correlated with DNA sequence, repeats, and predicted DNA structure. PLoS Genetics. 2006;2(3):e26. doi: 10.1371/journal.pgen.0020026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Parker SCJ, Hansen L, Abaan HO, Tullius TD, Margulies EH. Local DNA topography correlates with functional noncoding regions of the human genome. Science. 2009;324(5925):389–92. doi: 10.1126/science.1169050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wong JJW, Lu J, Edwards RA, Frost LS, Glover JNM. Structural basis of cooperative DNA recognition by the plasmid conjugation factor, TraM. Nucleic Acids Research. 2011;39(15):6775–88. doi: 10.1093/nar/gkr296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Akhtar M, Epps J, Ambikairajah E. Signal Processing in Sequence Analysis: Advances in Eukaryotic Gene Prediction. IEEE Journal of Selected Topics in Signal Processing. 2008;2(3):310–21. [Google Scholar]
- 82.Liao GC, Rehm EJ, Rubin GM. Insertion site preferences of the P transposable element in Drosophila melanogaster. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(7):3347–51. doi: 10.1073/pnas.050017397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rawal K, Ramaswamy R. Genome-wide analysis of mobile genetic element insertion sites. Nucleic Acids Research. 2011:1–15. doi: 10.1093/nar/gkr337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Geurts AM, Hackett CS, Bell JB, et al. Structure-based prediction of insertion-site preferences of transposons into chromosomes. Nucleic Acids Research. 2006;34(9):2803–11. doi: 10.1093/nar/gkl301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhang J, Guo D, Chang Y, et al. Non-random distribution of T-DNA insertions at various levels of the genome hierarchy as revealed by analyzing 13 804 T-DNA flanking sequences from an enhancer-trap mutant library. The Plant Journal: for Cell and Molecular Biology. 2007;49(5):947–59. doi: 10.1111/j.1365-313X.2006.03001.x. [DOI] [PubMed] [Google Scholar]
- 86.Sivolob AV, Khrapunov SN. Translational positioning of nucleosomes on DNA: the role of sequence-dependent isotropic DNA bending stiffness. Journal of Molecular Biology. 1995;247(5):918–31. doi: 10.1006/jmbi.1994.0190. [DOI] [PubMed] [Google Scholar]
- 87.Olson WK, Gorin AA, Lu XJ, Hock LM, Zhurkin VB. DNA sequence-dependent deformability deduced from protein-DNA crystal complexes. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(19):11163–8. doi: 10.1073/pnas.95.19.11163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pérez A, Noy A, Lankas F, Luque FJ, Orozco M. The relative flexibility of B-DNA and A-RNA duplexes: database analysis. Nucleic Acids Research. 2004;32(20):6144–51. doi: 10.1093/nar/gkh954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shpigelman ES, Trifonov EN, Bolshoy A. CURVATURE: software for the analysis of curved DNA. Bioinformatics. 1993;9(4):435–40. doi: 10.1093/bioinformatics/9.4.435. [DOI] [PubMed] [Google Scholar]
- 90.Goodsell DS, Dickerson RE. Bending and curvature calculations in B-DNA. Nucleic Acids Research. 1994;22(24):5497–03. doi: 10.1093/nar/22.24.5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ivanov VI, Minchenkova LE. The A-form of DNA: in search of the biological role. Molekuliarnaia Biologiia. 28(6):1258–71. [PubMed] [Google Scholar]
- 92.Ho PS, Ellison MJ, Quigley GJ, Rich A. A computer aided thermodynamic approach for predicting the formation of Z-DNA in naturally occurring sequences. The EMBO Journal. 1986;5(10):2737–44. doi: 10.1002/j.1460-2075.1986.tb04558.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.el Hassan MA, Calladine CR. Propeller-twisting of base-pairs and the conformational mobility of dinucleotide steps in DNA. Journal of Molecular Biology. 1996;259(1):95–103. doi: 10.1006/jmbi.1996.0304. [DOI] [PubMed] [Google Scholar]
- 94.Sugimoto N. Improved thermodynamic parameters and helix initiation factor to predict stability of DNA duplexes. Nucleic Acids Research. 1996;24(22):4501–5. doi: 10.1093/nar/24.22.4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Blake R. Thermal stability of DNA. Nucleic Acids Research. 1998;26(14):3323–32. doi: 10.1093/nar/26.14.3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Breslauer KJ. Predicting DNA Duplex Stability from the Base Sequence. Proceedings of the National Academy of Sciences. 1986;83(11):3746–50. doi: 10.1073/pnas.83.11.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Benham CJ. Sites of predicted stress-induced DNA duplex destabilization occur preferentially at regulatory loci. Proceedings of the National Academy of Sciences. 1993;90(7):2999–3003. doi: 10.1073/pnas.90.7.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ornstein RL, Rein R, Breen DL, Macelroy RD. An optimized potential function for the calculation of nucleic acid interaction energies I. Base stacking. Biopolymers. 1978;17(10):2341–60. doi: 10.1002/bip.1978.360171005. [DOI] [PubMed] [Google Scholar]
- 99.Brukner I, Sánchez R, Suck D, Pongor S. Sequence-dependent bending propensity of DNA as revealed by DNase I: parameters for trinucleotides. The EMBO journal. 1995;14(8):1812–8. doi: 10.1002/j.1460-2075.1995.tb07169.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Burden HE, Weng Z. Identification of conserved structural features at sequentially degenerate locations in transcription factor binding sites. Genome Informatics International Conference on Genome Informatics. 2005;16(1):49–58. [PubMed] [Google Scholar]
- 101.Nikolajewa S, Pudimat R, Hiller M, Platzer M, Backofen R. BioBayes- Net: a web server for feature extraction and Bayesian network modeling of biological sequence data. Nucleic Acids Research. 2007;35(Web Server issue):W688–93. doi: 10.1093/nar/gkm292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Morey C, Mookherjee S, Rajasekaran G, Bansal M. DNA free energy based promoter prediction and comparative analysis of Arabidopsis and rice genomes. Plant Physiology. 2011 doi: 10.1104/pp.110.167809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sonnenburg S, Zien A, Rätsch G. ARTS: accurate recognition of transcription starts in human. Bioinformatics (Oxford, England) 2006;22(14):e472–80. doi: 10.1093/bioinformatics/btl250. [DOI] [PubMed] [Google Scholar]
- 104.Gardiner EJ, Hunter CA, Lu X-J, Willett P. A structural similarity analysis of double-helical DNA. Journal of Molecular Biology. 2004;343(4):879–89. doi: 10.1016/j.jmb.2004.08.092. [DOI] [PubMed] [Google Scholar]
- 105.Mallios RR, Ojcius DM, Ardell DH. An iterative strategy combining biophysical criteria and duration hidden Markov models for structural predictions of Chlamydia trachomatis sigma66 promoters. BMC Bioinformatics. 2009;10:271. doi: 10.1186/1471-2105-10-271. [DOI] [PMC free article] [PubMed] [Google Scholar]