Abstract
Access to antiretroviral therapy is increasing globally and drug resistance evolution is anticipated. Currently, protease (PR) and reverse transcriptase (RT) sequence generation is increasing, including the use of in-house sequencing assays, and quality assessment prior to sequence analysis is essential. We created a computational HIV PR/RT Sequence Quality Analysis Tool (SQUAT) that runs in the R statistical environment. Sequence quality thresholds are calculated from a large dataset (46,802 PR and 44,432 RT sequences) from the published literature (http://hivdb.Stanford.edu). Nucleic acid sequences are read into SQUAT, identified, aligned, and translated. Nucleic acid sequences are flagged if with >five 1–2-base insertions; >one 3-base insertion; >one deletion; >six PR or >18 RT ambiguous bases; >three consecutive PR or >four RT nucleic acid mutations; >zero stop codons; >three PR or >six RT ambiguous amino acids; >three consecutive PR or >four RT amino acid mutations; >zero unique amino acids; or <0.5% or >15% genetic distance from another submitted sequence. Thresholds are user modifiable. SQUAT output includes a summary report with detailed comments for troubleshooting of flagged sequences, histograms of pairwise genetic distances, neighbor joining phylogenetic trees, and aligned nucleic and amino acid sequences. SQUAT is a stand-alone, free, web-independent tool to ensure use of high-quality HIV PR/RT sequences in interpretation and reporting of drug resistance, while increasing awareness and expertise and facilitating troubleshooting of potentially problematic sequences.
Introduction
The global impact of HIV-1 on public health is immense.1 Treatment regimens typically consisting of three or more antiretroviral (ARV) agents, referred to as highly active antiretroviral therapy (HAART) or combination antiretroviral therapy (cART), have significantly decreased morbidity and mortality in infected individuals since 1996.2 Currently, the mainstay of cART in resource-limited countries consists of nonnucleoside reverse transcriptase inhibitors (NNRTIs) or protease inhibitors (PIs), in combination with nucleoside or nucleotide reverse transcriptase inhibitors (NRTIs), which effectively target the HIV reverse transcriptase (RT) and protease (PR) to inhibit HIV replication. The delivery of optimal care for patients on cART is challenging and complex, largely due to the evolution of viral resistance, which is a consequence of the genetic diversity of HIV and its ability to mutate rapidly in vivo under drug selection pressure. Current guidelines in resource-rich settings recommend incorporation of resistance testing upon HIV diagnosis, before ARV initiation and upon suspected treatment failure.3,4 Resistance testing provides valuable information about drug regimen choices, which can improve clinical outcomes.5,6
The majority of research on the evolution of drug resistance has been conducted on HIV-1 subtype B virus, which is most common in North America, Europe, and Australia, yet comprises only 10% of global HIV cases.7,8 Lessons of ARV development, practice, and research in resource-rich settings currently inform guidelines in resource-limited settings. The ongoing scaling up of ARV therapy in resource-limited settings poses daunting challenges, particularly as in those settings non-B HIV-1 subtypes predominate and patient monitoring resources are limited.9–13
Genotypic testing for drug resistance is a multistep process. Viral RNA is extracted from a patient's blood, reverse transcribed to DNA, and amplified to generate multiple copies. The DNA is then subjected to sequencing reactions to derive the exact nucleic acid (NA) sequence that is identical (but reverse complemented) to the original viral RT and PR RNA. These processes are challenging and require extensive expertise and careful sample preparations, and errors can occur during any stage of the process.14 Despite two FDA-approved genotypic assays,15 less costly, in-house assays are abundantly used. Although at times comparisons are made between approved and in-house assays14 and external quality assurance evaluations are performed,16 this is usually not the case and thus examination of sequence quality after sequence assembly and prior to resistance interpretation is essential. This is particularly relevant with the increasing popularity of resistance testing in individual laboratories worldwide as global ARV treatment access escalates.
There is no clear definition of a good quality sequence and there are no clear guidelines on its generation. Quality assessment of the resistance testing process has focused primarily on laboratory stages of sequence production17,18 and base calling,19,20 and some on phylogenetic21,22 and sequence23,24 analyses. Abundant tools exist for sequence alignment,25,26 phylogenetics,27,28 and interpretation29,30; however these tools are discrete, may mandate individual sequence submission in diverse formats, require Internet connection and varying levels of expertise, and most are nonspecific. We developed a publicly available, comprehensive, interactive, quantitative and standalone sequence quality analysis tool (SQUAT) to evaluate PR and RT sequence quality for HIV drug resistance analysis using real HIV data-derived thresholds. We believe that widespread use of this tool will enhance expertise and increase awareness of its importance as an integrated part of sequence analysis and data dissemination.31–35
Materials and Methods
To create a tool that could be used globally, including in resource-limited settings with no or limited Internet service, SQUAT was written in R36 and Perl,37 both publicly available programs through the GNU General Public License (http://www.gnu.org/licenses/gpl.html). SQUAT runs on windows-based (Windows XP; Windows Vista; Windows 7), Linux and Macintosh computers in the R statistical environment. Installation can be accomplished by downloading R, Perl, and SQUAT directly from the Internet or by installing these programs from a mobile storage device.
Sequence FASTA files are read into SQUAT, aligned, labeled as RT and/or PR, assessed for quality at the NA level (i.e., ambiguous NA, insertions, deletions, stop codons, and frame shifts), translated to AA, assessed for quality at the AA level (i.e. ambiguous AA, unique AA, and frame shifts), and assessed for extreme genotypic diversity. Sequence diversity is assessed by genetic distances, calculated using publicly available SynScan code.38 Phylogenetic analyses are performed with “Ape.”39 Provided visual and textual output files allow the user to evaluate individual sequence quality and determine which sequences are potentially problematic and should be reevaluated.
Thresholds to flag sequences are determined from an analysis of a large, publicly available PR and RT sequence threshold dataset, which stores sequences that have been published in the literature and in GenBank.31–35 The dataset, downloaded on October 27, 2010 (Table 1), contains 44,432 RT sequences covering the most commonly sequenced AA positions 40–240 (28,222 subtype B, 64%; 2561 A, 6%; 5601 C, 13%; 1040 D, 2%; 649 F, 2%; 1201 G, 3%; 60 H, 0.1%; 28 J, 0.1%; 340 K, 0.8%; 2469 CRF01_AE, 6%; and 2261 CRF02_AG, 5%) and 46,802 PR sequences covering the most commonly sequenced AA positions 10–90 (30,137 subtype B, 64%; 2628 A, 6%; 4902 C, 11%; 984 D, 2%; 1400 F, 3%; 1226 G, 3%; 45 H, 0.1%; 196 J, 0.4%; 390 K, 0.8%; 2182 CRF01_AE, 5%; and 2712 CRF02_AG, 6%).
Table 1.
Sequence Threshold Dataset by Gene, Subtype, and Treatment
| Subtype | Total | Subtype % | Treated | Untreated | Treated % | Untreated % |
|---|---|---|---|---|---|---|
| Protease | ||||||
| A | 2628 | 5.6 | 204 | 2424 | 8 | 92 |
| B | 30137 | 64.4 | 13060 | 17077 | 43 | 57 |
| C | 4902 | 10.5 | 429 | 4473 | 9 | 91 |
| D | 984 | 2.1 | 136 | 848 | 14 | 86 |
| F | 1400 | 3.0 | 623 | 777 | 45 | 56 |
| G | 1226 | 2.6 | 264 | 962 | 22 | 78 |
| H | 45 | 0.1 | 6 | 39 | 13 | 87 |
| J | 196 | 0.4 | 17 | 179 | 9 | 91 |
| K | 390 | 0.8 | 64 | 326 | 16 | 84 |
| CFR01_AE | 2182 | 4.7 | 161 | 2021 | 7 | 93 |
| CRF02_AG | 2712 | 5.8 | 165 | 2547 | 6 | 94 |
| Total | 46,802 | 100 | 15,129 | 31,673 | 32 | 68 |
| Reverse Transcriptase | ||||||
| A | 2561 | 5.8 | 635 | 1926 | 25 | 75 |
| B | 28222 | 63.5 | 15791 | 12431 | 56 | 44 |
| C | 5601 | 12.6 | 1600 | 4001 | 29 | 71 |
| D | 1040 | 2.3 | 454 | 586 | 44 | 56 |
| F | 649 | 1.5 | 248 | 401 | 38 | 62 |
| G | 1201 | 2.7 | 551 | 650 | 46 | 54 |
| H | 60 | 0.1 | 16 | 44 | 27 | 73 |
| J | 28 | 0.1 | 3 | 25 | 11 | 89 |
| K | 340 | 0.8 | 75 | 265 | 22 | 78 |
| CFR01_AE | 2469 | 5.6 | 661 | 1808 | 27 | 73 |
| CRF02_AG | 2261 | 5.1 | 462 | 1799 | 20 | 80 |
| Total | 44,432 | 100 | 20,496 | 23,936 | 46 | 54 |
Results
General description
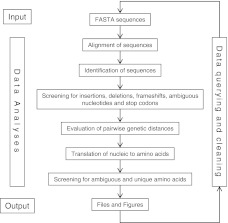
The SQUAT R package, user manual, and sample dataset are available for download at http://www.stat.brown.edu/CFAR/SQUAT; the user manual is also available as Supplementary Data (Fig. S1; Supplementary Data are available online at www.liebertonline.com/aid). SQUAT goes through several steps (Fig. 1) from data input through data analyses, and results in the identification and description of potentially problematic sequences. Based on this process, submitted sequences are either approved or flagged, and the final summary reports include troubleshooting information to assist users in data querying and cleaning.
FIG. 1.
Schematic representation of the SQUAT flow. The figure describes the different steps of a SQUAT session starting from file input through data analyses to creation of output files. The arrows depict the order in which analyses are performed, with the circular loop representing the iterative process of sequence quality analysis. SQUAT, sequence quality analysis tool.
The program defaults to a set of data-driven thresholds calculated using the threshold dataset. The process is interactive, and users may modify thresholds and select which processes are to be included in the analysis. The duration of a SQUAT session depends upon the computer, the number of sequences submitted, and whether sequence alignment and/or phylogenetic analysis are conducted. For example, the time required for a SQUAT session with both alignment and phylogenetic analysis on a 2.4-GHz Intel Core2 Quad CPU with 3.25 GB of RAM running Microsoft Windows XP takes about 1 min for a dataset of 100 PR sequences and less than 5 min for a dataset of 100 RT sequences.
SQUAT's algorithm is made of the following four steps.
1. Sequence input, identification, and alignment
Upon submission of an input file of FASTA sequences40 to SQUAT, alignment is offered. If chosen, sequences are aligned by a Perl program that incorporates the publicly available lap.c program41 to perform a comparison and alignment of each of the NA sequences against reference consensus B PR and RT AA sequences.28 This method aligns an NA sequence to an AA sequence, thus maintaining biological significance as much as possible. Subsequently, sequences are classified as PR, RT, or neither, based upon the strength of the homology between the NA sequence and the reference Consensus B sequence. Classification is performed using “percent identity” (how many of the aligned codons from the submitted NA sequence are identical to the AA in the protein reference sequence) and “coverage” (how many of the aligned codons from the reference sequence are included in the submitted sequence). Empiric “percent identity” thresholds for identifying HIV PR and RT sequences were determined to be above the minimum percent identity that was observed in the threshold dataset. Those values for the PR and RT were 66% and 71%, respectively. “Coverage” thresholds are AA 2 to 98 in PR and 40 to 240 in RT, and are user-modifiable.
2. Screening of nucleic acid sequences
After sequence identification and alignment, SQUAT screens the submitted sequences for insertions, deletions, frame shifts, ambiguous NA, and stop codons. Insertions are encoded as “#” and deletions as “∼” and the aligned sequences are written to output FASTA-formatted NA sequence files. In SQUAT, when an insertion of three or multiple of three NA is identified, this is considered a “true” insertion. If the insertion is not a multiple of three NA long, it is most likely a sequencing alignment or editing error, and biologically insignificant, and is therefore considered a “false” insertion. In both cases the appropriate comment is incorporated in the summary report. A sequence is approved if it has five or fewer “false” insertions or one or fewer “true” insertions. Using the same criteria, appropriate comments are incorporated into the summary report for “false” and “true” deletions. A sequence is approved if it has only one deletion or fewer, either “false” or “true.”
The presence of a frame shift in the NA sequence, as a result of insertions, deletions, or bad sequence quality, can lead to a subsequent string of multiple mutations, defined as differences from the consensus sequence or an NA mixture. To identify potential frame shifts despite prior sequence alignment, we examined the threshold dataset for strings of consecutive mutations and determined thresholds for sequence approval by calculating the percentiles of the maximum length of consecutive mutations per sequence (Table 2). In 95% of the RT and PR sequences, the maximum length of consecutive mutations was four and three, respectively, which are the default thresholds above which sequences are flagged.
Table 2.
Percentiles of Sequences with (A) Consecutive Mutations and (B) Ambiguous Characters per Sequence in the Threshold Dataset
| 0% | 1% | 5% | 25% | 50% | 75% | 95% | 99% | 100% | |
|---|---|---|---|---|---|---|---|---|---|
| A: Maximum length of consecutive mutations | |||||||||
| PR nucleic acids | 0 | 1 | 1 | 1 | 2 | 2 | 3 | 4 | 18 |
| PR amino acids | 0 | 1 | 1 | 1 | 2 | 2 | 3 | 4 | 7 |
| RT nucleic acids | 1 | 1 | 1 | 2 | 2 | 3 | 4 | 4 | 17 |
| RT amino acids | 0 | 1 | 1 | 1 | 2 | 2 | 4 | 4 | 21 |
| B: Number of ambiguous characters | |||||||||
| PR nucleic acids | 0 | 0 | 0 | 0 | 0 | 2 | 6 | 11 | 32 |
| PR amino acids | 0 | 0 | 0 | 0 | 0 | 1 | 3 | 6 | 19 |
| RT nucleic acids | 0 | 0 | 0 | 0 | 2 | 7 | 18 | 29 | 103 |
| RT amino acids | 0 | 0 | 0 | 0 | 1 | 2 | 6 | 10 | 29 |
PR, protease; RT, reverse transcriptase.
High occurrences of ambiguous NA (i.e., an NA at a sequenced position that has a value other than A, C, G, T, e.g. R, that equals A or G) may represent degraded sequence quality. Ninety-five percent of the PR training sequences had fewer than seven (2% of 297 NA) ambiguous NA, and 95% of the RT sequences had fewer than 19 (2% of 750 NA) ambiguous NA (Table 2), which are the default thresholds above which sequences are flagged.
Stop codons (TGA, TAA, TAG) should not exist in biologically functional protein sequences, and those were rare in the threshold dataset, (<0.3% for both RT and PR). By default, if one or more stop codon is found, the sequence is flagged and the NA location is noted in the report.
Genetic distances, calculated using Syn-scan,38 reflect differences between sequences. By default, sequence pairs are flagged if their genetic distance is less than 0.5%, indicating possible contamination or duplication, or greater than 15%, indicating possible sequence quality problems. The thresholds may be incorrect if sequences are from epidemiologically linked individuals, and therefore can be <0.5%, or if sequences are more genetically diverse, such as those that do not belong to group M HIV-1, and therefore can be >15%. Visual outputs in the form of genetic distance histograms and neighbor-joining phylogenetic trees are also created as part of SQUAT output to assist in data inspection and troubleshooting.
3. Screening of amino acid sequences
Prior to conversion to AA the “#”s are removed, and if the insertion is “true,” the AA output file will contain a “#” indicating the presence of an insertion after that position. As with frame shift inspection in NA sequences, SQUAT screens for the presence of frame shifts in the AA sequences (Table 2). The longest string of consecutive AA mutations was three AAs in 95% of the PR and four AAs in 95% of the RT sequences; therefore these values were used as thresholds.
Analogous to NA sequences, a high number of ambiguous AA (i.e., an AA that has a value other than the 20 known AAs, e.g. X, that equals an undetermined AA) may represent degraded sequence quality (Table 2). Overall, 95% of the RT and PR sequences had a maximum of six and three ambiguous AA, respectively, which were used as flagging thresholds.
A unique AA mutation at a certain position is one that does not occur at the same position in the threshold dataset. In the case of an AA mixture, the position would be declared as unique if none of the AAs in the mixture occurs in the threshold dataset at that position. By default, sequences with one or more unique AA are flagged and the AA location is noted in the output summary file.
4. Output files and figures
Sequence files
Sequence output files include aligned PR and RT NA and AA sequences, approved and flagged, and a file containing the input FASTA sequences that were not found to contain a PR or RT sequence.
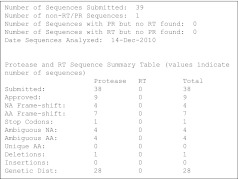
Summary file (Fig. 2)
FIG. 2.
SQUAT summary report (initial section). Shown is the top section of the main output file from a SQUAT session of the sample dataset, which summarizes the submitted sequences and their approval, along with reasons for flagging. This initial section is followed by a detailed description of each flagged sequence, including the location(s) within the sequence at which the potential problem occurred.
A summary report is created, containing a review of approved and flagged sequences with detailed information on the reason for flagging, based on the screening categories. This information includes the specific location within the sequence of each category (insertion, deletion, frame shift, ambiguous nucleotide, stop codon, unique mutation), as well as pairwise genetic distances information.
Phylogenetic information
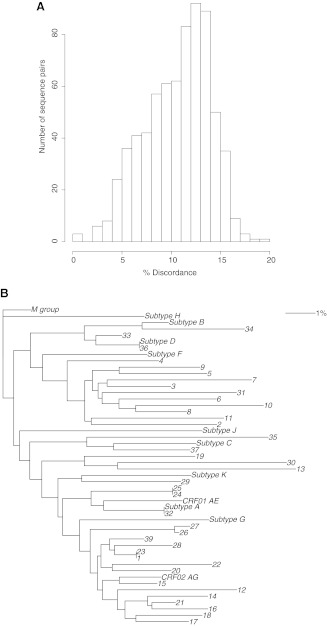
In addition to the summary file, a histogram (Fig. 3A) depicts a summary of all genetic distances to allow for visualization of their spectrum in the sequence input file. A neighbor-joining tree of the submitted sequences is provided, together with reference sequences of HIV group M subtypes, rooted with HIV-1 group M (Fig. 3B).
FIG. 3.
SQUAT-generated output figures. The sequences used are from the sample dataset provided with SQUAT. (A) Histogram of the distribution of pairwise genetic distances between submitted sequences. (B) Neighbor-joining phylogenetic tree of submitted sequences. Numbers (1–39) represent sample dataset sequences. Reference sequences from subtypes A–D, F–H, J–K (GenBank accession numbers M62320, K03455, U46016, M27323, AF005494, AF061642, AF005496, AF082394, AJ249235, CM240) and circulating recombinant forms CRF01AE and CRF02 AG (accession numbers IbNG, AJ006022) are included. The tree is rooted with the group M consensus sequence (www.lanl.gov).
Discussion
Due to large datasets or lack of a comprehensive definition of good sequence quality or expertise, sequence quality inspection is often omitted, leading to naive inclusion of potentially problematic data in analysis. SQUAT is a free, stand-alone, interactive computational tool, which allows examination of the quality of HIV PR and RT sequences after their generation and prior to subsequent analysis and interpretation. In the published literature, assessment of HIV sequences has mostly focused on laboratory stages of producing a sequence text file. Although internet-based programs and open-source computer code exist for certain quality control steps of generated sequences, SQUAT provides a unique method for comprehensive assessment of sequence quality in one streamlined interactive session, with detailed output on potential problems, allowing troubleshooting and sequence reexamination. SQUAT (current version 1.0.0) will be updated periodically to reflect available sequence data. Version numbers (x.y.z; “z”=any change in the package; “y”=change that will possibly affect results; “x”=major change in the package) will be updated as needed.
Ensuring adequate data quality prior to analysis is particularly important with HIV drug resistance sequence data, which involve multiple locations throughout the RT (43 resistance-associated mutations in 30 positions) and PR (69 resistance-associated mutations in 39 positions).42 Missing mutations or identification of mutations that are not “real” and that occur due to sequencing or editing errors would lead to misinterpretation and erroneous conclusions. This is emphasized more and is potentially problematic in small datasets that have high impact, such as those used in the World Health Organization (WHO) resistance surveillance program.43–45 SQUAT was modified for specific WHO needs, and has been made available to the WHO laboratories in the HIV Drug Resistance Network, which currently consists of 24 laboratories in 17 countries (http://www.who.int/hiv/topics/drugresistance/laboratory/en/index1. html). It has been sent out to other candidate laboratories as well. SQUAT flags potentially problematic sequences that can be inspected, resequenced, and corrected after examining electropherograms or additional data, or excluded from the analysis. WHO recommends review of chromatograms of flagged sequences by a WHO virologist and a virologist from the laboratory generating the sequences. Flagged sequences may still be approved, or may require further editing or retesting of the specimen. Positive comments from laboratory users include the fact that the application includes the ability to perform multiple sequence quality checks (including phylogeny) on larger batches of sequences all at once with modifiable thresholds and that an internet connection is not required to run the application.
Additionally SQUAT has been used for sequence quality analysis in the TREAT Asia Quality Assurance Scheme, an external quality assurance program to evaluate HIV sequencing quality in southeast Asia.16 Methodology within SQUAT was also previously used for quality control in analysis of a large and diverse HIV sequence dataset.10
Default thresholds for sequence quality used in SQUAT were derived from a large threshold dataset represented in the Stanford HIV Sequence Database. This is an HIV drug resistance specific database that accrues all available sequences from the published literature, GenBank,46 and the Los Alamos database,28 and thus represents the largest set of PR and RT sequence data available. All thresholds can be modified by the user, allowing personal preferences and relaxing of criteria based on individual results and sequence modification. This interactive nature of SQUAT also provides learning opportunities for the user to better understand the quality control steps incorporated in this tool, and allows more “active” user involvement as opposed to a “passive” process in which only final results are provided. In other words, SQUAT provides information on sequence quality based on which users need to make decisions regarding sequence analysis.
Inhibitors of the PR and RT proteins are currently the mainstay of global ARV HIV therapy; hence SQUAT was developed for examination of those sequences. Additional ARVs are needed and are continuously introduced, including novel agents from known drug classes, as well as novel drug classes.47,48 The methodology incorporated into SQUAT can be expanded to include the examination of additional HIV genes as more HIV-infected patients are exposed to more novel ARVs and respective sequencing for resistance testing is mandated. Additionally, as more microbes are being sequenced and as genomic medicine becomes more widespread, this methodology can also be implemented in non-HIV sequences.
Limitations of SQUAT first include mandated user expertise and understanding of molecular biology and the SQUAT process. It is currently command-line driven and lacks a simple user interface. Second, due to the magnitude of the various steps incorporated into SQUAT, it may take some time to run for large datasets. This will be overcome in time with faster processors. Third, the majority of the threshold dataset is HIV-1 subtype B and is potentially “biased.” However, this is the largest available dataset and future updates will incorporate newly accrued sequence data. This serves as a strong justification for making HIV sequences publicly available through GenBank, the Los Alamos Databases, and the Stanford HIV Sequence database. Lastly, SQUAT is currently in English only, which may be problematic in some parts of the world.
In summary, we describe SQUAT, a new sequence quality analysis tool for HIV PR and RT sequences, to assist laboratories generating them and researchers analyzing them to confirm adequate quality. With the continued global rise in access to antiretroviral medications and drug resistance research in diverse subtypes, it is anticipated that the need to evaluate and analyze HIV sequences will increase, especially in resource-limited settings using in-house assays. SQUAT's profile, being a stand-alone product, not requiring an internet connection, as well as its interactive performance and guiding outputs should address those challenges.
Supplementary Material
Acknowledgments
We acknowledge Drs. Robert Shafer, David Katzenstein, and Matthew Gonzales for early related discussions and collaborations at Stanford University, and Chris Kemp at the Center for Statistical Sciences for website support. This work was supported by the National Institute of Allergy and Infectious Diseases at the National Institutes of Health, grant numbers RO1AI66922 and P30AI042853.
Author Disclosure Statement
No competing financial interests exist. The conclusions and opinions expressed in this article are those of the authors and do not reflect those of their respective organizations, including, the Centers for Disease Control and Prevention and the US Department of Health and Human Services.
References
- 1.Joint United Nations Programme on HIV/AIDS (UNAIDS) and World Health Organization (WHO) AIDS epidemic update. 2007. http://www.unaids.org/en/HIV_data/2007EpiUpdate/default.asp http://www.unaids.org/en/HIV_data/2007EpiUpdate/default.asp
- 2.Palella FJ., Jr Delaney KM. Moorman AC, et al. Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection. HIV Outpatient Study Investigators. N Engl J Med. 1998;338(13):853–860. doi: 10.1056/NEJM199803263381301. [DOI] [PubMed] [Google Scholar]
- 3.Hirsch MS. Gunthard HF. Schapiro JM, et al. Antiretroviral drug resistance testing in adult HIV-1 infection: 2008 recommendations of an International AIDS Society-USA panel. Clin Infect Dis. 2008;47(2):266–285. doi: 10.1086/589297. [DOI] [PubMed] [Google Scholar]
- 4.DHHS Panel on Antiretroviral Guidelines for Adults and Adolescents––A Working Group of the Office of AIDS Research Advisory Council (OARAC) Guidelines for the use of antiretroviral agents in HIV-1 infected adults and adolescents. http://www.aidsinfo.nih.gov/guidelines http://www.aidsinfo.nih.gov/guidelines
- 5.Richman DD. Morton SC. Wrin T, et al. The prevalence of antiretroviral drug resistance in the United States. AIDS. 2004;18(10):1393–1401. doi: 10.1097/01.aids.0000131310.52526.c7. [DOI] [PubMed] [Google Scholar]
- 6.Little SJ. Holte S. Routy JP, et al. Antiretroviral-drug resistance among patients recently infected with HIV. N Engl J Med. 2002;347(6):385–394. doi: 10.1056/NEJMoa013552. [DOI] [PubMed] [Google Scholar]
- 7.McCutchan FE. Global epidemiology of HIV. J Med Virol. 2006;78(Suppl 1):S7–S12. doi: 10.1002/jmv.20599. [DOI] [PubMed] [Google Scholar]
- 8.Hemelaar J. Gouws E. Ghys PD. Osmanov S. Global and regional distribution of HIV-1 genetic subtypes and recombinants in 2004. AIDS. 2006;20(16):W13–23. doi: 10.1097/01.aids.0000247564.73009.bc. [DOI] [PubMed] [Google Scholar]
- 9.Kantor R. Impact of HIV-1 pol diversity on drug resistance and its clinical implications. Curr Opin Infect Dis. 2006;19(6):594–606. doi: 10.1097/QCO.0b013e3280109122. [DOI] [PubMed] [Google Scholar]
- 10.Kantor R. Katzenstein D. Efron B, et al. Impact of HIV-1 subtype and antiretroviral therapy on protease and reverse transcriptase genotypic evolution: Results of a global collaboration. PLOS Med. 2005;2:e112. doi: 10.1371/journal.pmed.0020112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brenner B. Turner D. Oliveira M, et al. A V106M mutation in HIV-1 clade C viruses exposed to efavirenz confers cross-resistance to non-nucleoside reverse transcriptase inhibitors. AIDS. 2003;17(1):F1–5. doi: 10.1097/00002030-200301030-00001. [DOI] [PubMed] [Google Scholar]
- 12.Brenner BG. Oliveira M. Doualla-Bell F, et al. HIV-1 subtype C viruses rapidly develop K65R resistance to tenofovir in cell culture. AIDS. 2006;20(9):F9–F13. doi: 10.1097/01.aids.0000232228.88511.0b. [DOI] [PubMed] [Google Scholar]
- 13.Grossman Z. Sugarman K. Auerbuch D, et al. Reverse transcriptase T69 6-bo insertion and multi-class resistance within a population of nucleoside reverse transcriptase inhibitor-treated subtype C patients. Antivir Ther. 2002;7(Suppl 1):S194. [Google Scholar]
- 14.Saravanan S. Vidya M. Balakrishanan P, et al. Evaluation of two human immunodeficiency virus-1 genotyping systems: ViroSeq 2.0 and an in-house method. J Virol Methods. 2009;159(2):211–216. doi: 10.1016/j.jviromet.2009.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.U.S. Food and Drug Administration. FDA licensed and approved HIV tests. http://www.fda.gov/ohrms/dockets/ac/03/briefing/3982b1_Licensed-Approved%20HIV%20Tests.doc http://www.fda.gov/ohrms/dockets/ac/03/briefing/3982b1_Licensed-Approved%20HIV%20Tests.doc
- 16.Land S. Cunningham P. Zhou J, et al. TREAT Asia Quality Assessment Scheme (TAQAS) to standardize the outcome of HIV genotypic resistance testing in a group of Asian laboratories. J Virol Methods. 2009;159(2):185–193. doi: 10.1016/j.jviromet.2009.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galli RA. Sattha B. Wynhoven B, et al. Sources and magnitude of intralaboratory variability in a sequence-based genotypic assay for human immunodeficiency virus type 1 drug resistance. J Clin Microbiol. 2003;41(7):2900–2907. doi: 10.1128/JCM.41.7.2900-2907.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang DD. Eshleman SH. Brambilla DJ, et al. Evaluation of the editing process in human immunodeficiency virus type 1 genotyping. J Clin Microbiol. 2003;41(7):3265–3272. doi: 10.1128/JCM.41.7.3265-3272.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gene Codes Corporation. DNA Sequence Assembly Software: For Mac and Windows; Sequencher 4.10.1. [Google Scholar]
- 20.Ewing B. Hillier L. Wendl MC. Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8(3):175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 21.Korber B. Myers G. Signature pattern analysis: A method for assessing viral sequence relatedness. AIDS Res Hum Retroviruses. 1992;8:1549–1560. doi: 10.1089/aid.1992.8.1549. [DOI] [PubMed] [Google Scholar]
- 22.Boeri E. Canducci F. Grasso MA, et al. Phylogenetic internal control for HIV-1 genotypic antiretroviral testing. New Microbiol. 2004;27(2 Suppl 1):105–109. [PubMed] [Google Scholar]
- 23.Learn GH., Jr Korber BT. Foley B, et al. Maintaining the integrity of human immunodeficiency virus sequence databases. J Virol. 1996;70:5720–5730. doi: 10.1128/jvi.70.8.5720-5730.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kjaer J. Ledergerber B. HIV cohort collaborations: Proposal for harmonization of data exchange. Antivir Ther. 2004;9(4):631–633. [PubMed] [Google Scholar]
- 25.Larkin MA. Blackshields G. Brown NP, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 26.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Korber B. HIV signature, sequence variation analysis. In: Rodrigo AG, editor; Learn GH, editor. Computational and Evolutionary Analysis of HIV Molecular Sequences. Kluwer Academic Publishers, Dordrecht; The Netherlands: 2000. pp. 55–72. [Google Scholar]
- 28.Los Alamos HIV Database. www.hiv.lanl.gov. http://www.hiv.lanl.gov/content/index www.hiv.lanl.govhttp://www.hiv.lanl.gov/content/index
- 29.Liu TF. Shafer RW. Web resources for HIV type 1 genotypic-resistance test interpretation. Clin Infect Dis. 2006;42(11):1608–1618. doi: 10.1086/503914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sturmer M. Doerr HW. Preiser W. Variety of interpretation systems for human immunodeficiency virus type 1 genotyping: Confirmatory information or additional confusion? Curr Drug Targets Infect Disord. 2003;3(4):373–382. doi: 10.2174/1568005033481006. [DOI] [PubMed] [Google Scholar]
- 31.Kantor R. Machekano R. Gonzales MJ, et al. Human immunodeficiency virus reverse transcriptase and protease sequence database: An expanded model integrating natural language text and sequence analysis. Nucleic Acids Res. 2001;29:296–299. doi: 10.1093/nar/29.1.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rhee SY. Gonzales MJ. Kantor R, et al. Human immunodeficiency virus reverse transcriptase and protease sequence database. Nucleic Acids Res. 2003;31(1):298–303. doi: 10.1093/nar/gkg100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shafer RW. Jung DR. Betts BJ. Human immunodeficiency virus type 1 reverse transcriptase and protease mutation search engine for queries. Nat Med. 2000;6(11):1290–1292. doi: 10.1038/81407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shafer RW. Jung DR. Betts BJ, et al. Human immunodeficiency virus reverse transcriptase and protease sequence database. Nucleic Acids Res. 2000;28:346–348. doi: 10.1093/nar/28.1.346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shafer RW. Stevenson D. Chan B. Human immunodeficiency virus reverse transcriptase and protease sequence database. Nucleic Acids Res. 1999;27:348–352. doi: 10.1093/nar/27.1.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.R: A language and environment for statistical computing [computer program] Vienna, Austria: 2009. Version. [Google Scholar]
- 37.Perl. Programming language. http://www.perl.org/ http://www.perl.org/
- 38.Gonzales MJ. Dugan JM. Shafer RW. Synonymous-non-synonymous mutation rates between sequences containing ambiguous nucleotides (Syn-SCAN) Bioinformatics. 2002;18(6):886–887. doi: 10.1093/bioinformatics/18.6.886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paradis E. Claude J. Strimmer K. APE: Analyses of Phylogenetics and Evolution in R language. Bioinformatics. 2004;20(2):289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- 40.FASTA. Sequence Format. http://www.ncbi.nlm.nih.gov/BLAST/fasta.shtml http://www.ncbi.nlm.nih.gov/BLAST/fasta.shtml
- 41.Huang X. Zhang J. Methods for comparing a DNA sequence with a protein sequence. Comput Appl Biosci. 1996;12(6):497–506. doi: 10.1093/bioinformatics/12.6.497. [DOI] [PubMed] [Google Scholar]
- 42.Johnson VA. Brun-Vezinet F. Clotet B, et al. Update of the drug resistance mutations in HIV-1: December 2009. Top HIV Med. 2009;17(5):138–145. [PubMed] [Google Scholar]
- 43.Bennett DE. Bertagnolio S. Sutherland D. Gilks CF. The World Health Organization's global strategy for prevention and assessment of HIV drug resistance. Antivir Ther. 2008;13(Suppl 2):1–13. [PubMed] [Google Scholar]
- 44.Bennett DE. Myatt M. Bertagnolio S, et al. Recommendations for surveillance of transmitted HIV drug resistance in countries scaling up antiretroviral treatment. Antivir Ther. 2008;13(Suppl 2):25–36. [PubMed] [Google Scholar]
- 45.Jordan MR. Bennett DE. Bertagnolio S, et al. World Health Organization surveys to monitor HIV drug resistance prevention and associated factors in sentinel antiretroviral treatment sites. Antivir Ther. 2008;13(Suppl 2):15–23. [PubMed] [Google Scholar]
- 46.Genbank. http://www.ncbi.nlm.nih.gov/ http://www.ncbi.nlm.nih.gov/
- 47.Hughes CA. Robinson L. Tseng A. MacArthur RD. New antiretroviral drugs: A review of the efficacy, safety, pharmacokinetics, and resistance profile of tipranavir, darunavir, etravirine, rilpivirine, maraviroc, and raltegravir. Expert Opin Pharmacother. 2009;10(15):2445–2466. doi: 10.1517/14656560903176446. [DOI] [PubMed] [Google Scholar]
- 48.Bhattacharya S. Osman H. Novel targets for anti-retroviral therapy. J Infect. 2009;59(6):377–386. doi: 10.1016/j.jinf.2009.09.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.