Abstract
NK cell function is important in the immune response to HIV infection. NKG2C and NKG2A are activating and inhibitory NK cell receptors, respectively, and their only known ligand, HLA-E, demonstrates increased expression in HIV infection and presents at least one HIV-derived peptide. A variation in chromosome 12 exists in which the 16-kb section of DNA encompassing the nkg2c gene is completely absent. DNA samples of 433 HIV-1-infected patients and 280 controls were genotyped by PCR, and revealed an association of the absence variation with a higher risk of HIV infection, as well as faster progression and higher pretreatment viral loads (p<0.05, respectively). Surface NKG2C expression, analyzed by FACS, on the freshly isolated lymphocytes of 20 control and 19 HIV-infected donors revealed that NKG2C expression is genotype dependent in both populations: no NKG2C expression in the −/− groups, intermediate expression in the +/− groups, and highest expression in the +/+ groups. The comparison of NKG2C and NKG2A expression in HIV and control groups (+/− and +/+ included) indicates an increased NKG2C expression on HIV patient NK cells (p<0.05) and decreased inhibitory NKG2A expression on CD8 T cells (p<0.001), and both these effects are more striking in the +/+ genotype (p<0.005). Furthermore, a positive correlation was found between HIV viral load and the proportion of NKG2C+ NK cells. The increased expression of NKG2C in HIV patients, in combination with the genetic association of the absence variation with an increased susceptibility to HIV infection, higher HIV viral set point, and a faster progression, indicate that NKG2C is important in the defense against HIV infection and progression.
Introduction
Natural killer (NK) cells are important effectors of the innate immune response to HIV infection and their activity, be it cytotoxicity or cytokine release, is controlled by a balance of inhibitory and activating NK cell receptor signaling.1 The relative expression of these receptors, and thus the outcome of signaling, is determined by a combination of genetic and stress factors.2 Impaired NK cell functions have been demonstrated in HIV patients, including reduced cytotoxicity, correlated with decreased expression of various activating NK cell receptors,3 and deficient production of tumor necrosis factor (TNF)-α and interferon (IFN)-γ.4 Although HIV downregulates the expression of activating receptor-ligands, it has been shown to upregulate the expression of HLA-E,5 a nonclassical HLA molecule that presents both self and viral nonamer peptides, including an HIV p24-derived peptide.6 HLA-E is the only known ligand of both the inhibitory NKG2A/CD94 and the activating NKG2C/CD94 complexes, which are expressed on NK cells and some T cell subsets.6 A variation in the 12p13 region of chromosome 12 was found in which the 16-kb section of DNA encompassing the NKG2C gene is completely absent.7 Both the specific peptide presented by HLA-E, which affects receptor/ligand affinity, and the relative surface expression of the NKG2A- and NKG2C/CD94 receptor complexes will together determine the outcome of HLA-E binding.8 The aim of this study was to determine any genetic association of this NKG2C absence (“−”) or presence (“+”) variation with HIV. We also investigated the relative expression of NKG2C/CD94 according to genotype in both controls and HIV patients.
Materials and Methods
Study population
DNA samples
DNA was isolated from whole blood samples using the Qiagen Midi kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. DNA samples of all available 433 HIV-1-infected patients, unrelated German whites, were selected from the DNA bank of the Immunological Outpatient Clinic at Hannover Medical University, Germany. The demographic and clinical parameters of the HIV cohort used are listed in Table 1. In addition, 280 DNA samples of German white control donors (HIV-1, HCV, and HBV seronegative) were also obtained from the blood bank at Hannover Medical University, Germany. All donors provided informed consent, which has been approved by the local ethical committee (ethical volume number 2384).
Table 1.
Demographics of HIV Patients
| Characteristic | HIV-Infected patients, n (%) | |
|---|---|---|
| Number | 433 | |
| Male | 351 | (81.1%) |
| Female | 82 | (18.9%) |
| Age at diagnosis | 33.7 years | (10.5–67.3) |
| HIV infection risk category | ||
| Men-who-have-sex-with-men (MSM) | 175 | (37.9%) |
| Unknown/undeclared | 146 | (31.6%) |
| Intravenous drug abuse (IVDA) | 93 | (20.1%) |
| Heterosexual contact | 29 | (6.3%) |
| Hemophilia | 13 | (2.8%) |
| Blood transfusion | 5 | (1.1%) |
| Workplace exposure | 1 | (0.2%) |
| Progression categories | ||
| Long term nonprogressors | 78 | (17.9%) |
| Short term progressors | 113 | (25.9%) |
| Normal progressors | 121 | (27.8%) |
| Others | 123 | (28.3%) |
| HAART treatment | ||
| On treatment | 5 | (26.3%) |
| Treatment naíve | 14 | (73.7%) |
Risk category definitions
The cohort of HIV-1-infected patients was subgrouped according to their risk categories, as seen in Table 1. The two categories later compared for genotype distributions were the high risk intravenous drug abusers (IVDA) and the lower risk men who have sex with men (MSM). IVDA are considered as high risk as a higher viral load is injected directly into the blood, with a greater chance of overwhelming the immune response and establishing infection. MSM are considered lower risk as the viral input is lower and the virus must both escape the mucosal immune system and pass the endothelial barrier in order to enter the blood and establish infection.
Progression category definitions
The cohort of HIV-1-infected patients was subdivided into four categories, according to the speed of their disease progression (Table 1): short-term progressors, long-term nonprogressors, normal progressors, and other. These categories were defined based on the time of diagnosis until death, initiation of antiretroviral therapy (ART) treatment, and/or absolute CD4+T cell counts falling below 200/μl blood. A short-term progressor meets any of these criteria within 2 years (730 days), a normal progressor between 2 and 10 years (730 to 3652 days), and a long-term nonprogressor meets none of these criteria during a minimum of 10 years (3652 days) of follow-up. The category “other” includes patients who do not meet any of these criteria, but have less than 10 years of follow-up or insufficient regularity of monitoring in their patient records.
Lymphocytes
Lymphocytes were isolated by density gradient centrifugation (Biocoll; Biochrom AG, Berlin, Germany) from fresh blood samples collected in heparin bead tubes from 19 HIV patients from the Immunological Outpatient Clinic at Hannover Medical University, as well as 20 control subjects from the blood bank at Hannover Medical University, Germany (see Table 2 for sample characteristics). Following separation, peripheral blood mononuclear cells (PBMCs) were washed twice with phosphate-buffered saline (PBS). A third washing in PBS with 3% fetal calf serum (FCS) was used for lymphocytes destined for immediate antibody staining, or in R10 medium for cell culture or functional assays. All donors provided informed consent.
Table 2.
Genotypes of the Control and HIV Subjects Included in Surface Expression Analysis
| |
NKG2C genotype |
|||
|---|---|---|---|---|
| Subject group | n | −/− | +/− | +/+ |
| HIV patients | 19 | 3 | 5 | 11 |
| Controls | 20 | 2 | 4 | 14 |
Viral load
Viral setpoint (pretreatment)
The pretreatment viral loads were collected from HIV patient files retrospectively, and the viral set point calculated by averaging two or more pretreatment counts.
Viral load
The viral load of HIV patients, corresponding to the time their samples were collected, was retrieved from their patient files. Only patients of the +/+ genotype were included in the analysis.
NKG2C genotyping
PCR was performed on 280 control and 433 HIV DNA samples in order to determine the NKG2C genotype according to the 16 kb absence, as previously described.7 Briefly, two separate PCRs were performed: an “absence” PCR targeting a specific sequence only present in the recombined absence variation and a “presence” PCR targeting a sequence within the NKG2C gene. Successful amplification was analyzed by electrophoresis on a 1.5% agarose gel and resulted in a 411-bp and a 363-bp fragment for the absence and presence PCRs, respectively.
FACS analysis
Antibodies
The following antibodies were used throughout the study: NKG2C-PE (R&D Systems, USA); NKG2A-PE (Beckman Coulter, USA); HLA-E-PE (eBioscience, USA); CD4-PerCP, CD8-PerCP, isotype control IgG1-PerCP (BD, Germany); CD3-FITC, CD56-FITC, CD14-APC, CD3-APC, and Isotypes IgG1-PE, IgG2b-PE, IgG2a-FITC, IgG1-FITC, IgG1-APC, and IgG2a-APC (Immunotools, Germany). All of the antibodies used were directly labeled mouse-antihuman monoclonal antibodies.
Lymphocyte staining
Freshly isolated lymphocytes, at a concentration of 3.0×106 cells per ml, were stained with titered concentrations of monoclonal antibodies for 25 min at 4°C and washed three times in PBS with 3% FCS. Following a 15 min fixation at 4°C in 4% paraformaldehyde, cells were washed three times in PBS with 3% FCS, and later acquired by four color flow cytometry using a Becton Dickinson FACS Calibur. Acquired samples were analyzed with Summit v4.3 software (Dako Colorado, Inc., USA).
Statistical analysis
The statistical analysis was carried out using GraphPad Prism software (GraphPad Software Inc., USA). The frequency of homozygous presence variation genotype was compared between HIV patients and controls using Fisher's exact test (two-tailed). Statistical analysis of surface expression patterns was carried out by Mann–Whitney test (one-tailed). A value of p<0.05 was considered significant. The correlation of the percentage of NKG2A+ or NKG2C+ NK cells and the HIV viral load was calculated with Spearman`s test.
Results
Genetic association of NKG2C absence with initial HIV infection and disease progression
The genotypic distribution of NKG2C in HIV patient and control groups is summarized in Table 3. A significantly higher proportion of controls had two copies of the NKG2C gene (genotype +/+) as compared to HIV patients (p<0.05). Conversely stated, the absence variation (including homozygous −/− and heterozygous +/−) is significantly associated with HIV. Furthermore, a significantly higher proportion of MSM carries the NKG2C absence variation than IVDA (p<0.05). The HIV patients were also subdivided according to progression categories and the comparison of genotype frequencies shows that the +/+ genotype was more frequent in the long-term nonprogressors (LTNP) group than in the STP and NP groups, significantly so in the latter case (p<0.05). Additionally, the division of HIV patients according to pretreatment viral set points reveals that a significantly higher proportion of those with low viral load (<30,000 copies/ml) carry the +/+ genotype (p<0.05).
Table 3.
Genotype Distributions and Statistical Comparisons of NKG2C Genotypes in Control and HIV Patients
|
Genotype distributions | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| |
|
|
HIV-infected patient |
|||||||||
| |
|
|
|
|
Risk category |
Pretreatment virus load (copies/ml) |
||||||
| Control | All | IVDA | MSM | <30,000 | >30,000 | |||||||
| −/− | 13 | 5% | 33 | 8% | 6 | 7% | 10 | 6% | 5 | 11% | 8 | 13% |
| +/− | 71 | 25% | 130 | 30% | 19 | 20% | 60 | 34% | 8 | 17% | 25 | 39% |
| +/+ | 196 | 70% | 270 | 62% | 68 | 73% | 105 | 60% | 34 | 72% | 31 | 48% |
| |
|
|
|
Progression category |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LTNP | NP | STP | Other | |||||||||
| −/− | 5 | 9% | 9 | 7% | 7 | 6% | 11 | 9% | ||||
| +/− | 11 | 19% | 44 | 36% | 33 | 29% | 37 | 30% | ||||
| +/+ | 42 | 72% | 68 | 56% | 74 | 65% | 75 | 81% | ||||
|
Statistical comparisons | |||||||
|---|---|---|---|---|---|---|---|
| Comparison | Genotype | Significance | Odds ratio | 95% CI | |||
| Control vs. HIV | −/− | vs. | −/+ and +/+ | NS | 0.59 | 0.31 to 1.14 | |
| −/− and −/+ | vs. | +/+ | * | p=0.037 | 0.71 | 0.52 to 0.98 | |
| IVDA vs. MSM | −/− | vs. | −/+ and +/+ | NS | 1.34 | 0.40 to 3.24 | |
| −/− and −/+ | vs. | +/+ | * | p=0.044 | 0.55 | 0.32 to 0.96 | |
| LTNP vs. NP | −/− | vs. | −/+ and +/+ | NS | 1.17 | 0.38 to 3.68 | |
| −/− and −/+ | vs. | +/+ | * | p=0.049 | 0.49 | 0.25 to 0.96 | |
| LTNP vs. STP+NP | −/− | vs. | −/+ and +/+ | NS | 1.29 | 0.45 to 3.68 | |
| −/− and −/+ | vs. | +/+ | - | p=0.097 | 0.58 | 0.31 to 1.10 | |
| LTNP vs. rest | −/− | vs. | −/+ and +/+ | NS | 1.16 | 0.43 to 3.14 | |
| −/− and −/+ | vs. | +/+ | - | p=0.108 | 0.59 | 0.32 to 1.08 | |
| <30,000 vs. >30,000 (copies/ml) | −/− | vs. | −/+ and +/+ | NS | 0.83 | 0.25 to 2.73 | |
| −/− and −/+ | vs. | +/+ | * | p=0.019 | 2.78 | 1.24 to 6.23 | |
IVDA, intravenous drug abuse; MSM, men who have sex with men; LTNP, long-term nonprogressor; NP, normal progressor; STP, short-term progressor; OR, odds ratio; CI, confidence interval.
NKG2C expression is genotype dependent
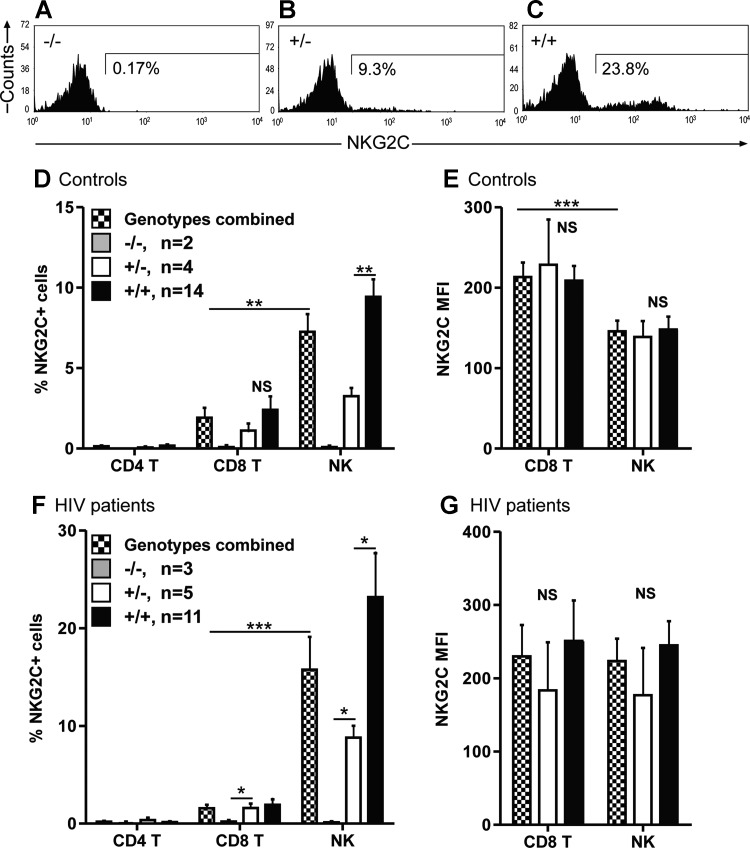
The effect of the genetic absence on surface protein expression of NKG2C was examined on freshly isolated lymphocytes of 20 control and 19 HIV samples by four color flow cytometry. NKG2C expression is determined both by cell type and by genotype. No NKG2C was detected on the surface of CD4 T cells in either controls or HIV patients, while the proportion of NKG2C+ NK cells is significantly higher than NKG2C+ CD8 T cells. The expression of NKG2C is genotype dependent in both controls and HIV patients, as no NKG2C+ CD8 T cells or NK cells were detected in the −/− groups, an intermediate expression was detected in the +/− groups, and the highest expression in the +/+ groups (see Fig. 1A, B, and C for representative histograms, and Fig. 1D and F for a summary). Although the MFIs did not significantly differ between the genotypes, control subjects demonstrated a significantly higher MFI on CD8 T cells as compared to NK cells (this difference was not seen in HIV patients) (Fig. 1E and G).
FIG. 1.
Expression of NKG2C on peripheral blood mononuclear cells (PBMCs). The proportion of NKG2C+ cells is stratified by genotype. Fresh blood samples of 20 control donors and 19 HIV patients were evaluated by four-color FACS for the expression of NKG2C on CD4 T cells (CD3+CD4+), CD8 T cells (CD3+CD8+), and NK cells (CD3-CD56+). The samples were subdivided according to NKG2C genotype: homozygous absence (−/−; dark gray bars), homozygous presence (+/+; black bars), heterozygous (+/−; white bars), and combined heterozygous and homozygous presence (+/− and +/+ ; checked bars). A, B, and C show the genotype-dependent nature of NKG2C expression on the NK cells of three representative HIV patients. The percentages of CD4 T cells, CD8 T cells, and NK cells that express NKG2C are depicted in D and F, and the mean fluorescence intensities in E and G. *p<0.05, **p<0.01, ***p<0.001.
Modulation of activating NKG2C receptor expression in HIV patients
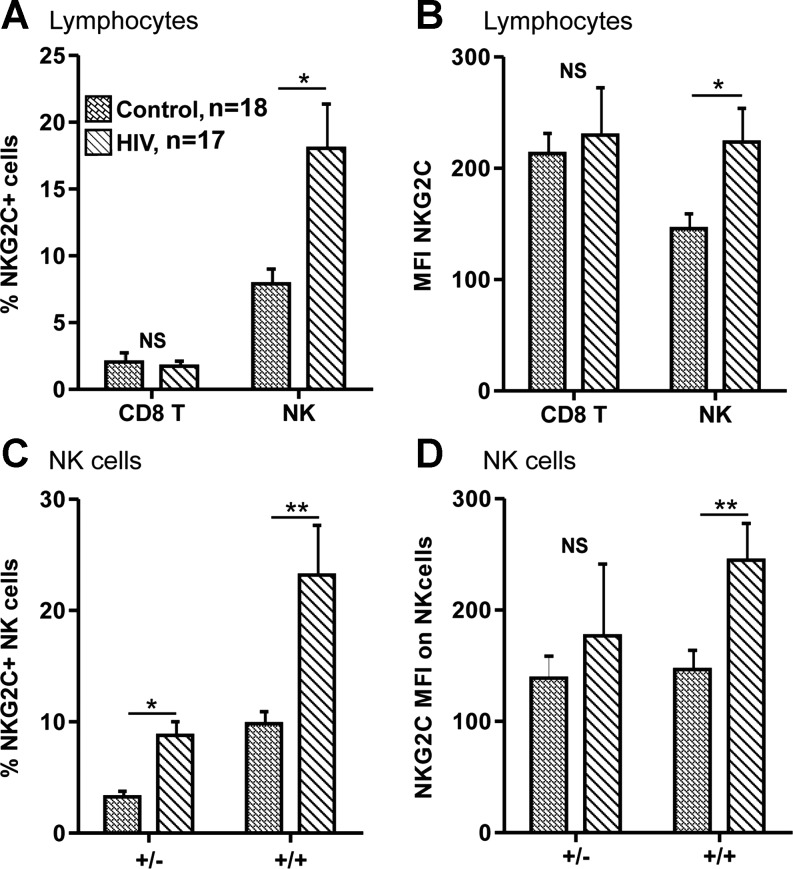
The overall expression of NKG2C was compared between controls and HIV patients by results from above for the genotypes +/− and +/+. These results are depicted graphically in Fig. 2A and B. Both the proportion and MFI of NKG2C on the NK cells patients were significantly higher in HIV patients than in controls. NKG2C expression on CD8 T cells, however, did not show any significant differences between patient and control groups.
FIG. 2.
Comparison of NKG2C expression in control and HIV patients. HIV patients show increased NKG2C expression on NK cells, and this increase is proportional to genotype. CD8 T cells (CD3+CD8+) and NK cells (CD3-CD56+) freshly isolated from 18 control (hatched bars; +/− n=4, +/+ n=14) and 16 HIV patients (diagonally striped bars; +/− n=5, +/+ n=11) were evaluated for NKG2C expression. Only individuals carrying at least one copy of NKG2C (+/− and +/+) were included. The percentage of NKG2C positive cells and the MFI are depicted in A and B, respectively. The HIV patients and controls were also subdivided according to genotype, and NKG2C expression on NK cells is presented in C and D. *p<0.05, **p<0.01, ***p<0.001.
Figure 2C and D breaks down this comparison between controls and HIV into their genotype subgroups in order to evaluate the relative contributions of genotype to the increased NKG2C expression on NK cells. In both +/− and +/+ genotypes the mean percents of NKG2C expressing NK cells were more than twice as high in HIV than in controls (see Fig. 2C). Though both genotype groups show an increased MFI, this difference is significant only in the +/+ group (Fig. 2D). The higher expression levels in the +/+ group indicate that the increased NKG2C expression in HIV patients could be predominantly due to the significant increase on NK cells of +/+ HIV patients.
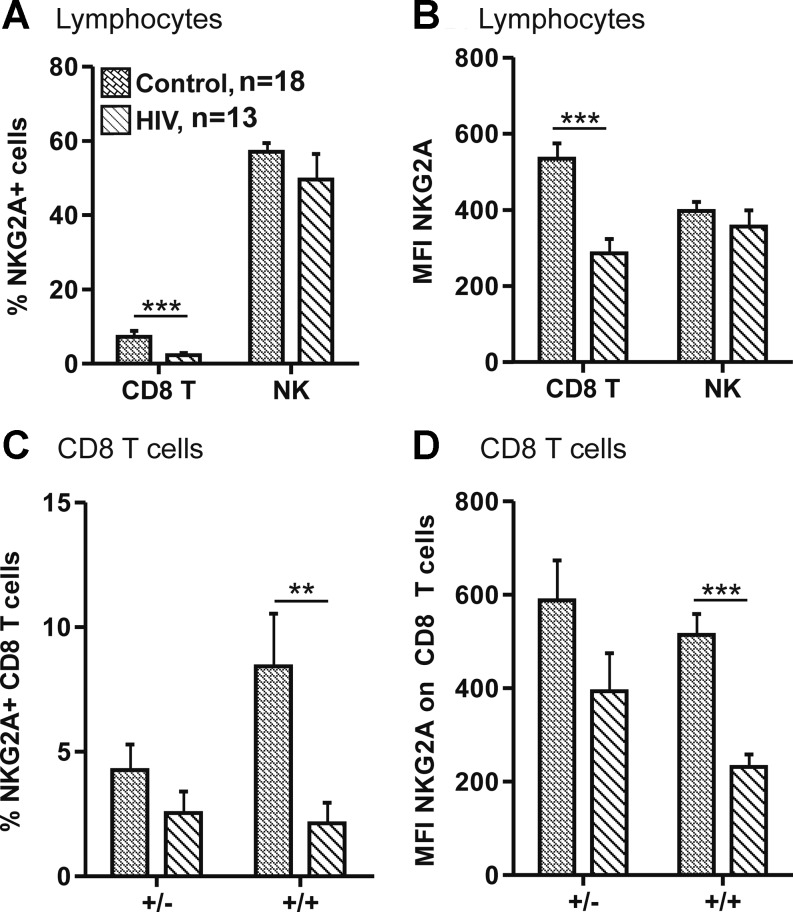
Modulation of inhibitory NKG2A receptor expression in HIV patients
NKG2A is an inhibitory receptor that also recognizes HLA-E as its ligand, and so the surface expression patterns of NKG2A on the PBMCs of 18 controls and 13 HIV patients was compared (Fig. 3A and B). A significantly lower expression of NKG2A was found on the CD8 T cells of HIV patients as compared to controls, both in terms of the proportion of NKG2A+ cells and the MFI. Neither the proportion of NKG2A+ NK nor the MFIs were significantly different between HIV patients and controls. This data were reevaluated by segregating controls and HIV patients by genotype, focusing on CD8 T cells (Fig. 3C and D). Although a decrease in NKG2A expression occurs in both +/− and +/+ genotypes, the reduction is more striking (and highly significant) in +/+ CD8 T cells.
FIG. 3.
Comparison of NKG2A expression in control and HIV patients. HIV patients show decreased NKG2A expression on CD8 T cells, and this increase is proportional to genotype. CD8 T cells (CD3+CD8+) and NK cells (CD3-CD56+) freshly isolated from 18 control (hatched bars; +/− n=5, +/+ n=13) and 12 HIV patients (diagonally striped bars; +/− n=4, +/+ n=8) were evaluated for NKG2A expression. Only individuals carrying at least one copy of NKG2C (+/− and +/+) were included. The percentages of NKG2A positive cells and the MFI are depicted in A and B, respectively. The HIV patients and controls were also subdivided according to genotype, and NKG2A expression on CD8 T cells is presented in C and D. *p<0.05, **p<0.01, ***p<0.001.
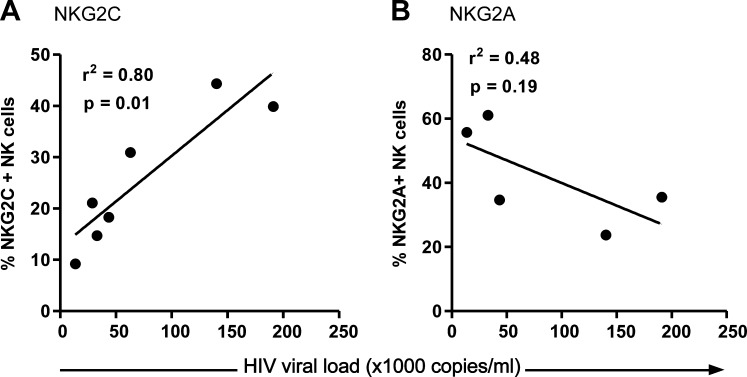
Receptor expression correlated to HIV viral load
The observation that NKG2C expression is upregulated and NKG2A downregulated in HIV-infected patients leads to questions of whether a higher viral load influences receptor expression. Figure 4A shows a positive correlation of HIV viral load and the proportion of NKG2C+ NK cells, while a trend toward a negative correlation is seen in Fig. 4B in terms of NKG2A+ NK cells.
FIG. 4.
The expression of NKG2C and NKG2A in relation to HIV viral load. The percentages of freshly isolated NK cells (CD3-CD56+) of HIV patients expressing NKG2C (A) and NKG2A (B) were plotted against the viral load of the patient corresponding to the time the sample was taken. All subjects included are of the +/+ genotype.
Discussion
The importance of cytotoxic cells in the control of HIV is well known, and their functions can be mediated through both antigen-specific (e.g., T cell receptor) and antigen-independent mechanisms (e.g., NK cell receptors). An increased recognition and signaling by activating NK cell receptors shift the balance toward activation and effector function. In fact, the epistatic association of the activating KIR allele (KIR3DS1) and its ligand HLA-BW4 with a slower progression to AIDS indicates that the transduction of activating signals via NK cell receptors facilitates the clearance of HIV-infected cells and retards progression.9
NKG2C is an activating receptor expressed on γδ T cells, CD8 αβ T cells, and to a greater extent on NK cells. Ligation of NKG2C results in the transduction of an activating signal via its associated DAP12 molecule and ultimately leads to cytokine production or cytotoxicity (in a TCR-independent manner in CD8 T cells).10 The importance of NKG2C in the response to HIV infection has been indicated by the expansion of NKG2C+ NK cells and γδ T cells in HIV-infected patients.11,12 Our results similarly revealed an increase in the proportion of NKG2C+ NK cells and further demonstrate a concurrent increase in cell surface density (MFI) on NK cells of HIV patients, as compared to controls. This increase supports the hypothesis that NKG2C is upregulated in response to HIV infection.
A variation in chromosome 12 was identified, in which a 16-kb segment of DNA encompassing the NKG2C gene is either present (+) or entirely absent (−).7 The importance of NKG2C in HIV, as indicated by its increased expression during infection, suggests that the genetic presence/absence variation could affect an individual's immune responses to HIV. As a genome-wide association study is unlikely to detect the impact of a large genetic deletion (16 kb) that has a high degree of homology with other receptors (e.g., NKG2A), a candidate gene approach was employed to investigate any association with HIV infection and/or disease progression. The genotype distributions of 280 control subjects and 433 HIV patients were compared and it was found that a significantly higher proportion of controls had two copies of the NKG2C gene (+/+) as compared to HIV patients (70% vs. 62%, p<0.05). Conversely stated, the deletion variations (−/− and +/−) are significantly associated with HIV.
This genetic association with HIV infection led to the question of whether the NKG2C deletion is associated with initial HIV infection or disease progression. The HIV patients were subgrouped according to their risk categories and the genotype distributions were compared between the high-risk IVDA and lower risk MSM group. MSM had a significantly higher proportion of the deletion variations (−/− and +/−) than IVDA (40% vs. 27%; p<0.05), indicating that the genetic absence of NKG2C increases susceptibility to the initial HIV infection.
The only known ligand of NKG2C is the nonclassical MHC class I molecule HLA-E, which presents nonamer peptides such as those derived from the leader sequences of other HLA molecules, as well as some viral peptides including HCV coreaa35–44,13 and HIV p24aa4–22,5, both of which stabilize HLA-E surface expression. Furthermore, a particular variant of HLA-E known as HLA-EG has both a higher peptide-binding affinity and surface expression than the HLA-ER wild-type variation14 and is associated with a decreased susceptibility to HIV infection.15 Together, these data suggest that the combination of a higher “baseline” expression of NKG2C and HLA-E could result in a more immediate recognition and destruction of HIV-infected cells. In fact, preliminary HLA-E genotyping data collected by J. Pollak in our laboratory, in combination with the NKG2C genotype results mentioned above, indicate that together NKG2C+/+/HLA-EG/G may provide additional protection against initial HIV infection, as this combination is less commonly found in MSM risk patients than IVDA or controls (8% vs. 13% and 12%, respectively) (unpublished data). This relationship between NKG2C and HLA-EG also decreases the chance that the NKG2C association is an artifact of linkage disequilibrium. An efficient first line response to HIV infection would curb viral replication and budding, thereby decreasing the chance of disseminated infection and thus increasing the likelihood of viral clearance.
The transmission of HIV by a high dose direct injection into the blood is more likely to overwhelm innate immune responses and establish a chronic infection than a lower dose sexual transmission in which HIV must contend with both endothelial barriers and the innate mucosal immune system. If, as we expect, NKG2C receptor expression on mucosal γδ T cells in the lining of the rectum follows the same genotype-specific pattern as on PBMCs, a higher NKG2C expression could more readily and successfully respond to the initial HIV input and explain the decreased susceptibility associated with the +/+ genotype in MSM.
A role for the NKG2C receptor in the immune response to chronic HIV infection, as well as HBV and HCMV, is evidenced by increased proportions of NKG2C+ NK cells.12,16–18 Although our findings revealed no increase in NKG2C expression on CD8 T cells, an approximately 2-fold higher expression on the NK cells of HIV patients as compared to controls further supports such a role. Interestingly, this increase was also observed when the comparison was completed according to genotype, revealing that both +/− and +/+ groups undergo a similar scale of expansion of NKG2C+ NK cells during chronic HIV infection. However, although the same 2:1 ratio of increase was seen in both genotypes, the actual mean percentages of positive staining cells in HIV patients were much higher in the +/+ than the +/− genotype group (23% vs. 9%, p<0.05). The increase in MFI was also significantly higher in +/+ HIV patients (p=0.005). In contrast, the expression of NKG2A was found to be significantly downregulated on the CD8 T cells of HIV patients as compared to healthy controls, which is in agreement with the findings of Zeddou et al.19 Surprisingly, this downregulation was also found to be genotype dependent (i.e., a greater degree of downregulation was seen in the +/+ genotype).
An increased NKG2C/NKG2A ratio has been reported in HIV-exposed uninfected individuals,20 indicating that a quicker activating response may be beneficial in terms of controlling or clearing infection with HIV.
The observation that the extent of NKG2C and NKG2A modulation in HIV-infected patients is genotype determined raised the question as to whether a greater “inducible” modulation of NKG2C and NKG2A is beneficial or detrimental in terms of the progression to AIDS. To determine this relationship, HIV patients were subdivided according to their speed of progression and their genotype frequencies were compared.
The prevalence of the +/+ genotype in LTNP was significantly higher as compared to normal progressors (72% vs. 56%, p<0.05), and tended to be higher than all other progression categories combined (72% vs. 61%, p=0.09). The significantly different distribution of genotypes between HIV patient groups segregated by pretreatment viral load also reveals a higher prevalence of the +/+ genotype in those with low viral set points. Although greater numbers of patients would very likely improve the power of these statistics, even at these low numbers they nonetheless indicate that carrying two copies of nkg2c results in higher expression of NKG2C on NK cells and lower expression of NKG2A on CD8 T cells and that this altered ratio is beneficial in HIV infection in terms of a slower progression to AIDS. In addition, we have found that NKG2C and NKG2A expression is correlated to HIV viral load, similar to the findings of Ballan et al. and Zeddou et al., respectively.19,21 As only +/+ individuals were used in our comparison, it can be suggested that HIV infection itself impacts the level of receptor expression. Therefore, the level of NKG2C and NKG2A expression will depend not only on a patient's genotype, but also on the extent of viremia.
Other genetic associations with delayed progression, such as the epistatic genetic association of HLA-BW4 with either the activating KIR2DS1 or the nonsurface expressed inhibitory KIR3DL1*044,22 also demonstrate that NK cells effector functions are crucial. Furthermore, the combination of preliminary HLA-E genotyping data, collected by J. Pollak in our laboratory, with the NKG2C genotype results mentioned further indicates that the NKG2C+/+/HLA-EG/G genotype is protective against HIV progression, as this combination is more frequently found in LTNP as compared to the other progression categories (17% vs. 8%) (unpublished data). These findings once again indicate that higher NKG2C expression leads to a more rapid recognition and killing of HIV replicating cells, thereby reducing the viral load and the infection of neighboring cells and hence slowing the depletion of CD4 T cells. Although decreased NK cell and CD4 T cell populations are correlated with a faster disease progression, no significant differences in the relative proportions of lymphocytes in HIV patients were seen according to genotype (data not shown). It is possible that such differences may be observable in terms of the absolute numbers of NK and CD4 T cells and/or may be observable only after a certain period of infection. Conversely, the degree of NKG2C expression may not obviously affect the size of lymphocyte populations.
The genetic associations of the NKG2C deletion variation with HIV infection, particularly the initial infection, imply that NKG2C+ cytotoxic lymphocytes are able to quickly recognize and kill HIV-infected cells. Work with Vδ1 T cells has shown that NKG2C is involved in the lysis of HLA-E+ target cells and that activated NKG2C+ Vδ1 T cells of HIV-infected patients lyse autologous HIV-infected CD4 T cells.11 As γδ T cells are an integral component of the mucosal immune system, rapid recognition and killing of HIV-infected cells via NKG2C signaling could represent a first line defense that facilitates early clearance of the virus. Furthermore, the association of the homozygous presence variation (+/+) with LTNPs indicates that NKG2C-mediated responses also play an important role in the control of virus in circulation once chronic HIV infection has been established. We believe that this improved prognosis is partly due to NKG2C's contribution to the rapid killing of HIV-infected cells in which active viral replication is taking place, thereby reducing viral load and the likelihood of bystander cells becoming infected.
Conclusions
The decreased susceptibility to initial infection and a slower disease progression associated with the homozygous presence (+/+) genotype indicate that NKG2C is protective in HIV infection, a conclusion further supported by the skewed NKG2C+/+/HLA-EG/G frequencies. This NKG2C+/+/HLA-EG/G combination would result in higher “baseline” expressions of both the activating NKG2C receptor and its HLA-E ligand and a lower expression of the inhibitory NKG2A receptor, thereby facilitating a faster and more effective recognition and destruction of HIV-infected cells and thus curbing viral replication and budding. Indeed, this haplotype is found less frequently in MSM HIV-infected patients than in controls and more frequently in LTNP, both of which indicate that such an efficient response is beneficial in terms of clearing the initial infection as well as retarding the infection of additional CD4+ cells. In contrast to other NK cell receptors involved in HIV defense, NKG2C is not downregulated but rather the expression of the NKG2C receptor as well as its ligand are upregulated during HIV infection. This may indicate the importance of NKG2C not only in the response to initial infection, but also in slowing the progression of HIV infection.
Acknowledgments
We thank all the patients and controls who participated in this study and the staff of the immunologic outpatient clinic of Hannover Medical School. R. Thomas is a member of the Ph.D. Program Infection Biology, Hannover Biomedical Research School, and is supported by a fellowship from the Georg Christoph Lichtenberg-Foundation of Lower Saxony. This work was also supported by the BMBF Kompetenznetz Rheuma C2.12 and by DFG KFO 250, Grant WI 1031/6-1.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Lanier LL. Up on the tightrope: Natural killer cell activation and inhibition. Nat Immunol. 2008;9:495–502. doi: 10.1038/ni1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jie HB. Sarvetnick N. The role of NK cells and NK cell receptors in autoimmune disease. Autoimmunity. 2004;37:147–153. doi: 10.1080/0891693042000196174. [DOI] [PubMed] [Google Scholar]
- 3.De Maria A. Fogli M. Costa P, et al. The impaired NK cell cytolytic function in viremic HIV-1 infection is associated with a reduced surface expression of natural cytotoxicity receptors (NKp46, NKp30 and NKp44) Eur J Immunol. 2003;33:2410–2418. doi: 10.1002/eji.200324141. [DOI] [PubMed] [Google Scholar]
- 4.Jacobs R. Heiken H. Schmidt RE. Mutual interference of HIV and natural killer cell-mediated immune response. Mol Immunol. 2005;42:239–249. doi: 10.1016/j.molimm.2004.06.021. [DOI] [PubMed] [Google Scholar]
- 5.Nattermann J. Nischalke HD. Hofmeister V, et al. HIV-1 infection leads to increased HLA-E expression resulting in impaired function of natural killer cells. Antivir Ther. 2005;10:95–107. doi: 10.1177/135965350501000107. [DOI] [PubMed] [Google Scholar]
- 6.Tripathi P. Agrawal S. The role of human leukocyte antigen E and G in HIV infection. AIDS. 2007;21:1395–1404. doi: 10.1097/QAD.0b013e32810c8bbc. [DOI] [PubMed] [Google Scholar]
- 7.Miyashita R. Tsuchiya N. Hikami K, et al. Molecular genetic analyses of human NKG2C (KLRC2) gene deletion. Int Immunol. 2004;16:163–168. doi: 10.1093/intimm/dxh013. [DOI] [PubMed] [Google Scholar]
- 8.Kaiser BK. Barahmand-Pour F. Paulsene W. Medley S. Geraghty DE. Strong RK. Interactions between NKG2x immunoreceptors and HLA-E ligands display overlapping affinities and thermodynamics. J Immunol. 2005;174:2878–2884. doi: 10.4049/jimmunol.174.5.2878. [DOI] [PubMed] [Google Scholar]
- 9.Martin MP. Gao X. Lee JH, et al. Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat Genet. 2002;31:429–434. doi: 10.1038/ng934. [DOI] [PubMed] [Google Scholar]
- 10.Guma M. Busch LK. Salazar-Fontana LI, et al. The CD94/NKG2C killer lectin-like receptor constitutes an alternative activation pathway for a subset of CD8+ T cells. Eur J Immunol. 2005;35:2071–2080. doi: 10.1002/eji.200425843. [DOI] [PubMed] [Google Scholar]
- 11.Fausther-Bovendo H. Wauquier N. Cherfils-Vicini J. Cremer I. Debre P. Vieillard V. NKG2C is a major triggering receptor involved in the V[delta]1 T cell-mediated cytotoxicity against HIV-infected CD4 T cells. AIDS. 2008;22:217–226. doi: 10.1097/QAD.0b013e3282f46e7c. [DOI] [PubMed] [Google Scholar]
- 12.Mela CM. Burton CT. Imami N, et al. Switch from inhibitory to activating NKG2 receptor expression in HIV-1 infection: Lack of reversion with highly active antiretroviral therapy. AIDS. 2005;19:1761–1769. doi: 10.1097/01.aids.0000183632.12418.33. [DOI] [PubMed] [Google Scholar]
- 13.Schulte D. Vogel M. Langhans B, et al. The HLA-E(R)/HLA-E(R) genotype affects the natural course of hepatitis C virus (HCV) infection and is associated with HLA-E-restricted recognition of an HCV-derived peptide by interferon-gamma-secreting human CD8(+) T cells. J Infect Dis. 2009;200:1397–1401. doi: 10.1086/605889. [DOI] [PubMed] [Google Scholar]
- 14.Strong RK. Holmes MA. Li P. Braun L. Lee N. Geraghty DE. HLA-E allelic variants. Correlating differential expression, peptide affinities, crystal structures, and thermal stabilities. J Biol Chem. 2003;278:5082–5090. doi: 10.1074/jbc.M208268200. [DOI] [PubMed] [Google Scholar]
- 15.Lajoie J. Hargrove J. Zijenah LS. Humphrey JH. Ward BJ. Roger M. Genetic variants in nonclassical major histocompatibility complex class I human leukocyte antigen (HLA)-E and HLA-G molecules are associated with susceptibility to heterosexual acquisition of HIV-1. J Infect Dis. 2006;193:298–301. doi: 10.1086/498877. [DOI] [PubMed] [Google Scholar]
- 16.Hadaya K. de Rham C. Bandelier C, et al. Natural killer cell receptor repertoire and their ligands, and the risk of CMV infection after kidney transplantation. Am J Transplant. 2008;8:2674–2683. doi: 10.1111/j.1600-6143.2008.02431.x. [DOI] [PubMed] [Google Scholar]
- 17.Guma M. Budt M. Saez A, et al. Expansion of CD94/NKG2C+ NK cells in response to human cytomegalovirus-infected fibroblasts. Blood. 2006;107:3624–3631. doi: 10.1182/blood-2005-09-3682. [DOI] [PubMed] [Google Scholar]
- 18.Oliviero B. Varchetta S. Paudice E, et al. Natural killer cell functional dichotomy in chronic hepatitis B and chronic hepatitis C virus infections. Gastroenterology. 2009;137:1151–1160. doi: 10.1053/j.gastro.2009.05.047. [DOI] [PubMed] [Google Scholar]
- 19.Zeddou M. Rahmouni S. Vandamme A, et al. Downregulation of CD94/NKG2A inhibitory receptors on CD8+ T cells in HIV infection is more pronounced in subjects with detected viral load than in their aviraemic counterparts. Retrovirology. 2007;4:72. doi: 10.1186/1742-4690-4-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ravet S. Scott-Algara D. Bonnet E, et al. Distinctive NK-cell receptor repertoires sustain high-level constitutive NK-cell activation in HIV-exposed uninfected individuals. Blood. 2007;109:4296–4305. doi: 10.1182/blood-2006-08-040238. [DOI] [PubMed] [Google Scholar]
- 21.Ballan WM. Vu BA. Long BR, et al. Natural killer cells in perinatally HIV-1-infected children exhibit less degranulation compared to HIV-1-exposed uninfected children and their expression of KIR2DL3, NKG2C, and NKp46 correlates with disease severity. J Immunol. 2007;179:3362–3370. doi: 10.4049/jimmunol.179.5.3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lanier LL. Evolutionary struggles between NK cells and viruses. Nat Rev Immunol. 2008;8:259–268. doi: 10.1038/nri2276. [DOI] [PMC free article] [PubMed] [Google Scholar]