Abstract
We studied drug resistance mutations (DRMs) in 2623 pol sequences. Out of 94,828 amino acid substitutions that were detected, 8749 corresponded to nucleoside reverse transcriptase inhibitor (NRTI), 3765 to nonnucleoside reverse transcriptase inhibitor (NNRTI), and 7141 to protease inhibitor (PI) resistance-associated mutations. The most common DRMs were L10I, I54V, L90M, V82A, A71V, L10V, M46I, M184V, M41L, T215Y, D67N, L210W, K70R, N348I, V118I, K103N, Y181C, G190A, K101E, V108I, L100I, V90I, K101Q, and A98G. As expected, DRMs frequencies depended on viral genotype. The amounts of NRTI and PI resistance mutations among B and BF sequences from children were higher than among sequences from adults. The frequencies of PI and NRTI resistance mutations among B and BF sequences from adult men were higher than among sequences from women. Some of these observations can be explained in light of the available epidemiological information, but some cannot, indicating that further studies are needed to understand the antiretroviral resistance epidemics in Argentina.
Antiretroviral drugs are the only available treatment for preventing the development of acquired immunodeficiency syndrome (AIDS) in persons living with human immunodeficiency virus (HIV). Based on its mechanism of action, these drugs are classified as protease inhibitors (PI), nucleoside and nonnucleoside reverse transcriptase inhibitors (NRTI and NNRTI, respectively), CCR5 inhibitors, and integrase inhibitors. Integrase and CCR5 inhibitors are relatively recent compared to PIs, NNRTIs, and NRTIs, which have been used for many years, first in single-drug treatments and later combined in what is called highly active antiretroviral therapy or HAART. The short generation times and lack of proofreading activity of reverse transcriptase make HIV highly plastic upon selective pressures. As a consequence, virus mutation can drive the emergence of antiretroviral-resistant viruses. The genetic basis of most of the resistance mechanisms are already known and therefore it is possible to monitor the resistance profile of a strain by means of sequence analysis.1
Argentina is a developing country with 88,000 to 140,000 persons living with HIV.2 Starting with AZT in 1987, antiretroviral drugs have been extensively used in Argentina. Herein, sequence data collected at the National Reference Center for AIDS (CNRS, Argentina) through a period of 7 years (2001 to 2007), corresponding to patients with virologic failure, were screened in search for antiretroviral resistance mutations. The sequences (n=2623) studied here encompass codons 1 to 99 of the viral protease and 1 to 400 of the viral reverse transcriptase. The dataset included 2016 sequences newly reported in this article (GenBank accession numbers JN669427–JN671442), 577 sequences previously published by Gomez-Carrillo et al.3 (GenBank accession numbers AY365480.1–AY365987.1 and AY365990.1–AY366058.1), and 30 sequences from Vignoles et al.4 (GenBank accession numbers DQ995522.1–DQ995533.1, DQ995535.1–DQ995550.1, DQ995587.1, and DQ995588.1). Out of 2376 patients for whom we had gender data, 767 were female and 1609 were male. Data on patient age were available for 2445 of the sequences; of these, 671 sequences corresponded to infants and children (patients under 18 years old). Genotyping and recombination analyses were performed by bootscanning.5 Only bootscanning profiles supported by bootstrap values above 70 were considered in the analyses of genotype-related issues. The presence of resistance mutations and the corresponding resistance scores were obtained by the Stanford algorithm.6 All the statistic analyses were performed with the R Statistical Package.7
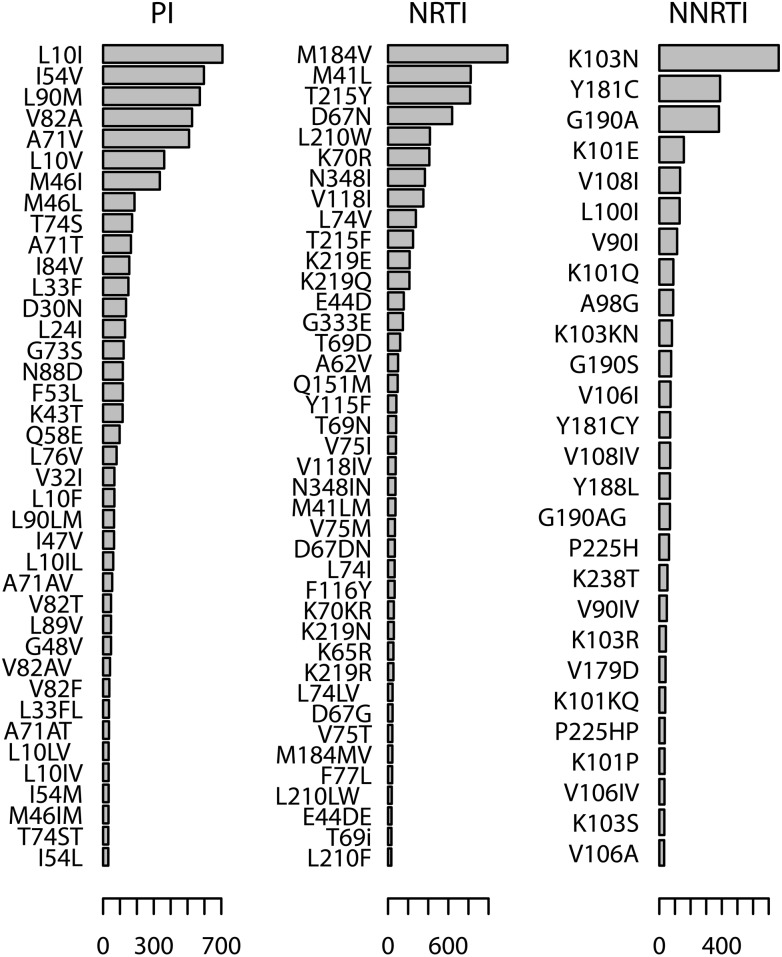
A total of 94,828 amino acid substitutions were detected. Of these, 8749 corresponded to NRTI, 3765 to NNRTI, and 7141 to PI resistance-associated mutations. The most frequent PI resistance mutations were L10I (707 sequences), I54V (597 sequences), L90M (573 sequences), V82A (527 sequences), A71V (509 sequences), L10V (362 sequences), and M46I (336 sequences). The most frequent NRTI resistance mutations were M184V (1118 sequences), M41L (823 sequences), T215Y (817 sequences), D67N (638 sequences), L210W (417 sequences), K70R (411 sequences), N348I (367 sequences), and V118I (352 sequences). The most frequent NNRTI resistance mutations were K103N (764 sequences), Y181C (389 sequences), G190A (382 sequences), K101E (157 sequences), V108I (133 sequences), L100I (130 sequences), V90I (114 sequences), K101Q (90 sequences), and A98G (89 sequences). These data are summarized in Fig. 1.
FIG. 1.
Frequencies of protease inhibitor (PI), nucleoside reverse transcriptase inhibitor (NRTI), and nonnucleoside reverse transcriptase inhibitor (NNRTI) resistance mutations among 2623 pol sequences from Argentina.
Drug resistance mutation (DRM) frequencies varied depending on viral genotype. As expected, most sequences were subtype B or BF recombinants. There were 1148 subtype B, 4 subtype A, 5 subtype C, and 11 subtype F sequences. Eight hundred and eighty-one sequences were BF recombinants, and 574 sequences presented an unsupported (bootstrap <70) bootscanning profile and thus their genotype was recorded as undetermined. Protease inhibitor resistance mutations A71V, M46I, A71T, I84V, G73S, K43T, L10F, V32I, I47V, L89V, and I54M were most frequent among subtype B sequences, whereas mutations I54V, L90M, L10V, and T74S prevailed among BF sequences (Table 1). NRTI resistance mutation L210F was the most frequent DRM among BF sequences, whereas M41L, T215Y, D67N, L210W, V118I, E44D, G333E, A62V, K219N, and K219R were present in higher proportions among B sequences (Table 2). NNRTI resistance mutation K103R was more frequent in B than in BF sequences (Table 3).
Table 1.
Distribution of Protease Inhibitor Resistance Mutations Among Subtype B and BF Recombinant Sequences
| Mutation | Globala | Subtype Bb | BF recombinantsb | p-valuec |
|---|---|---|---|---|
| L10I | 26.95 | 27.26 | 26.90 | 0.9 |
| I54V | 22.76 | 18.38 | 24.52 | 9E-4 |
| L90M | 21.85 | 2.79 | 19.86 | 5E-3 |
| V82A | 20.09 | 17.07 | 21.34 | 0.01 |
| A71V | 19.41 | 24.39 | 16.35 | 1E-5 |
| L10V | 13.80 | 8.01 | 19.64 | 2E-14 |
| M46I | 12.81 | 17.25 | 6.47 | 6E-13 |
| M46L | 7.09 | 7.32 | 5.68 | 0.17 |
| T74S | 6.56 | 2.70 | 9.99 | 8E-12 |
| A71T | 6.29 | 8.54 | 3.52 | 6E-6 |
| I84V | 5.91 | 8.89 | 2.72 | 2E-8 |
| L33F | 5.72 | 6.79 | 4.99 | 0.11 |
| D30N | 5.18 | 5.05 | 5.56 | 0.68 |
| L24I | 4.96 | 4.44 | 4.54 | 0.99 |
| G73S | 4.65 | 6.45 | 2.61 | 9E-5 |
| F53L | 4.46 | 0.61 | 4.09 | 0.93 |
| N88D | 4.46 | 4.09 | 4.54 | 0.7 |
| K43T | 4.42 | 5.57 | 2.72 | 2E-3 |
| Q58E | 3.74 | 4.18 | 2.38 | 0.03 |
| L76V | 3.05 | 3.31 | 1.70 | 0.03 |
| L10F | 2.55 | 4.97 | 0.00 | 4E-11 |
| V32I | 2.55 | 4.44 | 1.02 | 1E-5 |
| I47V | 2.48 | 3.48 | 1.48 | 7E-3 |
| L90LM | 2.48 | 2.79 | 2.27 | 0.56 |
| L10IL | 2.29 | 2.53 | 2.16 | 0.69 |
| A71AV | 2.06 | 1.48 | 2.50 | 0.14 |
| G48V | 1.79 | 1.66 | 1.82 | 0.92 |
| L89V | 1.79 | 2.70 | 0.68 | 1E-3 |
| V82T | 1.79 | 1.57 | 2.38 | 0.24 |
| V82AV | 1.56 | 0.96 | 1.25 | 0.68 |
| V82F | 1.41 | 1.74 | 1.25 | 0.47 |
| A71AT | 1.33 | 1.57 | 1.25 | 0.68 |
| L33FL | 1.33 | 1.13 | 2.04 | 0.14 |
| I54M | 1.26 | 2.26 | 0.45 | 1E-3 |
| L10IV | 1.26 | 0.96 | 1.02 | 0.99 |
| L10LV | 1.26 | 0.52 | 1.59 | 0.02 |
| M46IM | 1.22 | 1.39 | 1.02 | 0.58 |
| I54L | 1.18 | 1.83 | 0.57 | 0.02 |
| T74ST | 1.18 | 0.96 | 1.59 | 0.28 |
| I54IV | 1.14 | 0.78 | 1.36 | 0.29 |
| L33I | 1.14 | 1.39 | 0.00 | 0.01 |
Proportion (%) of sequences displaying the mutation among all the studied sequences (n=2623).
Percentage of sequences displaying the mutation among subtype B (n=1148) or BF (n=881) sequences. Only sequences with confident subtype/CRF assignment were included.
Pearson's chi-squared test with Yate's continuity correction (H0: p1=p2). p-values were adjusted for false discovery rates using the Benjamini–Hochberg method. Significant values (p<0.01) are in bold.
Table 2.
Distribution of Nucleoside Reverse Transcriptase Inhibitor Resistance Mutations Among Subtype B and BF Recombinant Sequences
| Mutation | Globala | Subtype Bb | BF recombinantb | p-valuec |
|---|---|---|---|---|
| M184V | 45.29 | 47.65 | 42.91 | 0.03 |
| M41L | 31.38 | 35.89 | 26.22 | 4E-6 |
| T215Y | 31.15 | 34.49 | 27.24 | 5E-4 |
| D67N | 24.32 | 27.00 | 21.91 | 9E-3 |
| L210W | 15.90 | 23.00 | 6.92 | <1E-16 |
| K70R | 15.67 | 15.42 | 16.35 | 0.61 |
| N348I | 13.99 | 12.89 | 15.32 | 0.13 |
| V118I | 13.42 | 15.51 | 9.65 | 1E-4 |
| L74V | 10.56 | 11.24 | 9.88 | 0.36 |
| T215F | 9.49 | 10.02 | 9.19 | 0.58 |
| K219E | 8.20 | 8.10 | 7.72 | 0.82 |
| K219Q | 8.12 | 7.75 | 9.53 | 0.18 |
| E44D | 5.99 | 7.84 | 2.72 | 1E-6 |
| G333E | 5.64 | 8.19 | 3.18 | 4E-6 |
| T69D | 4.61 | 4.79 | 3.86 | 0.36 |
| A62V | 3.81 | 5.49 | 2.61 | 2E-3 |
| Q151M | 3.66 | 4.79 | 2.95 | 0.04 |
| Y115F | 3.16 | 3.66 | 3.06 | 0.54 |
| T69N | 3.09 | 2.79 | 3.18 | 0.7 |
| V75I | 3.01 | 3.66 | 2.72 | 0.25 |
| V118IV | 2.90 | 3.14 | 2.50 | 0.47 |
| M41LM | 2.82 | 2.79 | 3.06 | 0.81 |
| N348IN | 2.82 | 2.26 | 2.50 | 0.85 |
| V75M | 2.63 | 2.87 | 1.59 | 0.07 |
| D67DN | 2.59 | 3.14 | 1.70 | 0.05 |
| F116Y | 2.52 | 3.48 | 1.59 | 0.01 |
| L74I | 2.52 | 2.61 | 2.50 | 0.98 |
| K219N | 2.21 | 3.31 | 0.91 | 5E-4 |
| K70KR | 2.21 | 2.61 | 2.27 | 0.73 |
| K219R | 2.06 | 2.61 | 0.68 | 1E-3 |
| K65R | 2.06 | 2.87 | 1.25 | 0.01 |
| D67G | 1.72 | 1.92 | 1.14 | 0.22 |
| L74LV | 1.72 | 2.09 | 1.59 | 0.51 |
| M184MV | 1.64 | 1.74 | 1.48 | 0.77 |
| V75T | 1.64 | 1.66 | 1.82 | 0.92 |
| F77L | 1.56 | 2.44 | 1.14 | 0.04 |
| L210LW | 1.41 | 2.09 | 0.68 | 0.01 |
| E44DE | 1.33 | 1.22 | 1.14 | 0.99 |
| T69i | 1.30 | 1.39 | 1.59 | 0.86 |
| L210F | 1.26 | 0.70 | 2.16 | 8E-3 |
| T69NT | 1.14 | 1.05 | 0.91 | 0.93 |
Proportion (%) of sequences displaying the mutation among all the studied sequences (n=2623). Only mutations detected in more than 30 sequences are reported.
Percentage of sequences displaying the mutation among subtype B (n=1148) or BF (n=881) sequences. Only sequences with confident subtype/CRF assignment were included.
Pearson's chi-squared test with Yate's continuity correction (H0: p1=p2). p-values were adjusted for false discovery rates using the Benjamini–Hochberg method. Significant values (p<0.01) are in bold.
Table 3.
Distribution of Nonnucleoside Reverse Transcriptase Inhibitor Resistance Mutations Among Subtype B and BF Recombinant Sequences
| Mutation | Globala | Subtype Bb | BF recombinantb | p-valuec |
|---|---|---|---|---|
| K103N | 29.02 | 30.92 | 27.24 | 0.07 |
| Y181C | 14.77 | 15.94 | 13.05 | 0.07 |
| G190A | 14.51 | 13.85 | 14.42 | 0.77 |
| K101E | 5.96 | 6.18 | 6.02 | 0.95 |
| V108I | 5.05 | 6.45 | 3.97 | 0.01 |
| L100I | 4.94 | 4.53 | 5.56 | 0.34 |
| V90I | 4.33 | 3.83 | 4.31 | 0.67 |
| K101Q | 3.42 | 4.18 | 2.04 | 0.01 |
| A98G | 3.38 | 3.75 | 3.06 | 0.48 |
| K103KN | 3.08 | 2.87 | 3.06 | 0.90 |
| G190S | 2.89 | 2.44 | 3.52 | 0.19 |
| V106I | 2.73 | 2.44 | 2.61 | 0.92 |
| V108IV | 2.66 | 3.05 | 2.38 | 0.44 |
| Y181CY | 2.66 | 2.70 | 2.72 | 0.99 |
| G190AG | 2.58 | 2.96 | 2.61 | 0.73 |
| Y188L | 2.58 | 2.70 | 2.16 | 0.52 |
| P225H | 2.35 | 3.05 | 2.04 | 0.21 |
| K238T | 1.97 | 2.00 | 1.93 | 0.99 |
| V90IV | 1.82 | 1.74 | 2.38 | 0.39 |
| K103R | 1.60 | 2.26 | 0.68 | 7E-3 |
| V179D | 1.52 | 1.92 | 0.91 | 0.09 |
| K101KQ | 1.44 | 2.09 | 0.68 | 0.01 |
| P225HP | 1.33 | 1.66 | 0.91 | 0.21 |
| K101P | 1.29 | 0.87 | 1.59 | 0.20 |
| V106IV | 1.25 | 0.78 | 1.14 | 0.56 |
| K103S | 1.22 | 1.05 | 1.25 | 0.83 |
| V106A | 1.18 | 0.78 | 1.36 | 0.29 |
| F227L | 1.14 | 1.05 | 1.14 | 0.99 |
Proportion (%) of sequences displaying the mutation among all the studied sequences (n=2623). Only mutations detected in more than 30 sequences are reported.
Percentage of sequences displaying the mutation among subtype B (n=1148) or BF recombinant (n=881) sequences. Only sequences with confident subtype/CRF assignment were included.
Pearson's chi-squared test with Yate's continuity correction (H0: p1=p2). p-values were adjusted for false discovery rates using the Benjamini–Hochberg method. Significant values (p<0.01) are in bold.
The majority of sequences harbored multiple DRMs (Table 4). In adults, the most frequent number of NRTI resistance mutations was 1, whereas the most frequent numbers of such mutations in children were 3 in the case of subtype B sequences from females and 5 in the rest of the sequences. In subtype B sequences from males, the most frequent number of PI resistance mutations was 4, whereas strains from adult and child females usually presented 2 and 3 such mutations, respectively. BF recombinant sequences usually harbored 2 PI resistance mutations, with the exception of BF sequences from children, which generally displayed 3 such mutations. The most frequent number of NNRTI resistance mutations per sequence was 2, regardless of viral genotype, patient age, or gender.
Table 4.
Number of Drug Resistance Mutations per Sequence1 According to Gender, Antiretroviral Type, Age, and Sequence Genotype2
| |
|
|
|
Number of resistance mutations |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ARV | Genotype | Gender | Age | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 |
| NRTI | B | Male | Adult | 74 | 72 | 67 | 63 | 73 | 46 | 48 | 30 | 19 | 10 | 6 |
| Children | 11 | 9 | 15 | 14 | 20 | 18 | 12 | 8 | 5 | 2 | — | |||
| Female | Adult | 26 | 21 | 19 | 15 | 23 | 8 | 10 | 3 | 1 | 2 | 1 | ||
| Children | 2 | 4 | 6 | 5 | 3 | 3 | 2 | 3 | 2 | — | — | |||
| BF | Male | Adult | 46 | 44 | 45 | 39 | 38 | 26 | 15 | 10 | 4 | 2 | — | |
| Children | 8 | 10 | 11 | 6 | 13 | 10 | 5 | 5 | 5 | 2 | 2 | |||
| Female | Adult | 38 | 31 | 24 | 29 | 11 | 13 | 4 | 2 | 1 | — | — | ||
| Children | 9 | 6 | 6 | 7 | 11 | 4 | 6 | 5 | 2 | — | — | |||
| PI | B | Male | Adult | 31 | 56 | 50 | 58 | 52 | 16 | 10 | — | — | — | — |
| Children | 10 | 8 | 15 | 19 | 9 | 8 | 4 | 1 | — | — | — | |||
| Female | Adult | 11 | 16 | 10 | 10 | 5 | 2 | 2 | — | — | — | — | ||
| Children | 6 | 3 | 8 | 3 | — | — | — | — | — | — | — | |||
| BF | Male | Adult | 34 | 37 | 31 | 26 | 17 | 7 | 1 | — | — | — | — | |
| Children | 9 | 16 | 16 | 11 | 10 | 4 | 2 | — | — | — | — | |||
| Female | Adult | 16 | 17 | 21 | 5 | 8 | 4 | — | — | — | — | — | ||
| Children | 8 | 10 | 14 | 6 | 3 | 3 | 1 | — | — | — | — | |||
| NNRTI | B | Male | Adult | 107 | 148 | 80 | 34 | 12 | 2 | — | — | — | — | — |
| Children | 29 | 30 | 17 | 9 | 2 | 3 | — | — | — | — | — | |||
| Female | Adult | 27 | 42 | 34 | 6 | 4 | 1 | — | — | — | — | — | ||
| Children | 6 | 8 | 6 | 2 | — | — | — | — | — | — | — | |||
| BF | Male | Adult | 62 | 86 | 50 | 19 | 2 | 1 | 1 | — | — | — | — | |
| Children | 14 | 18 | 15 | 7 | 1 | — | — | — | — | — | — | |||
| Female | Adult | 39 | 54 | 31 | 7 | 2 | 1 | — | — | — | — | — | ||
| Children | 10 | 19 | 13 | 4 | 1 | — | — | — | — | — | — | |||
Modal values are bolded.
Only bootscanning profiles supported by values above 70 were used.
Sequences from men displayed higher predicted resistance to many PIs and NRTIs than sequences from women. Given that the amounts of DRMs were odd among B and BF sequences (Tables 1, 2, and 3), and that these genotypes were heterogeneously distributed among genders (p<2E-16), the frequencies of DRMs among virus from men and women were grouped based on the corresponding viral genotype. Subtype B strains from adult, male patients were more resistant to PIs ATVr, DRVr, FPVr, IDVr, LPVr, SQVr, and TPVr than strains from women (Table 5). Subtype B sequences from adult women usually displayed higher predicted resistance to NNRTIs than sequences from adult men, but only the difference observed for NVP was statistically significant (Table 5). Recombinant (BF) strains from men were more resistant to PIs ATVr, DRVr, FPVr, IDVr, LPVr, NFV, SQVr, TPVr, and NRTIs ABC, AZT, D4T, DDI, and TDF than sequences from women (Table 6). There were no significant differences in the levels of resistance, regarding gender, among sequences from children (not shown).
Table 5.
Comparison of Stanford Resistance Scores of Subtype B Sequences from 154 Adult Female and 594 Adult Male HIV-1 Patients from Argentina
| |
PI |
NRTI |
NNRTI |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ARVa | ATVr | DRVr | FPVr | IDVr | LPVr | NFV | SQVr | TPVr | X3TC | ABC | AZT | D4T | DDI | FTC | TDF | DLV | EFV | ETR | NVP |
| MnMb | 2 | 0 | 2 | 2 | 2 | 2 | 2 | 0 | 60 | 40 | 39 | 37 | 37 | 60 | 17 | 60 | 55 | 10 | 60 |
| MnFc | 2 | 0 | 2 | 2 | 2 | 2 | 2 | 0 | 60 | 28 | 30 | 27 | 28 | 60 | 12 | 60 | 60 | 20 | 70 |
| MdMd | 27 | 9.2 | 26 | 32 | 22 | 53 | 29 | 13 | 41 | 39 | 39 | 39 | 42 | 41 | 20 | 47 | 48 | 19 | 59 |
| MdFe | 18 | 5.8 | 16 | 21 | 14 | 38 | 20 | 8.2 | 38 | 32 | 34 | 32 | 33 | 38 | 16 | 54 | 57 | 22 | 70 |
| Q3Mf | 54 | 16 | 47 | 68 | 47 | 100 | 54 | 26 | 68 | 62 | 75 | 71 | 65 | 68 | 40 | 85 | 90 | 30 | 100 |
| Q3Fg | 34 | 8 | 23 | 37 | 21 | 81 | 32 | 8 | 64 | 54 | 64 | 62 | 57 | 64 | 32 | 90 | 100 | 35 | 120 |
| ph | 2.9E-3 | 3.9E-3 | 1.9E-3 | 1.5E-3 | 9.2E-4 | 0.01 | 4.1E-3 | 1.0E-3 | 0.16 | 0.015 | 0.062 | 0.026 | 0.016 | 0.16 | 0.014 | 0.045 | 0.022 | 0.032 | 9.1E-3 |
Antiretroviral.
Mean score among males.
Mean score among females.
Median score among males.
Median score among females.
Third quartile for males.
Third quartile for females.
P-value. Wilcoxon rank sum test with continuity correction; significant values (p<0.01) are in bold.
Table 6.
Comparison of Stanford Resistance Scores of BF Recombinant Sequences from 289 Adult Female and 424 Adult Male HIV-1 Patients From Argentina
| |
PI |
NRTI |
NNRTI |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ARVa | ATVr | DRVr | FPVr | IDVr | LPVr | NFV | SQVr | TPVr | X3TC | ABC | AZT | D4T | DDI | FTC | TDF | DLV | EFV | ETR | NVP |
| MnMb | 17 | 0 | 2 | 5 | 2 | 38 | 9.5 | 0 | 30 | 32 | 35 | 30 | 30 | 30 | 14 | 52 | 48 | 15 | 60 |
| MnFc | 2 | 0 | 2 | 2 | 2 | 2 | 2 | 0 | 22 | 12 | 0 | 0 | 10 | 22 | 0 | 40 | 55 | 10 | 60 |
| MdMd | 24 | 7 | 20 | 30 | 21 | 51 | 26 | 13 | 36 | 34 | 35 | 35 | 37 | 36 | 17 | 47 | 49 | 20 | 61 |
| MdFe | 17 | 5.4 | 16 | 22 | 16 | 37 | 18 | 9.1 | 33 | 25 | 24 | 23 | 26 | 33 | 11 | 43 | 47 | 18 | 57 |
| Q3Mf | 44 | 11 | 37 | 59 | 43 | 92 | 42 | 23 | 64 | 52 | 67 | 62 | 60 | 64 | 32 | 80 | 90 | 30 | 100 |
| Q3Fg | 31 | 7 | 22 | 44 | 31 | 74 | 30 | 15 | 60 | 44 | 47 | 45 | 43 | 60 | 24 | 75 | 85 | 30 | 100 |
| ph | 1.0E-3 | 4.7E-3 | 6.1E-3 | 1.6E-3 | 4.7E-3 | 2.1E-3 | 1.2E-3 | 2.8E-3 | 0.040 | 6E-5 | 1.1E-4 | 2.8E-5 | 3.6E-5 | 0.040 | 1.2E-5 | 0.42 | 0.57 | 0.39 | 0.49 |
Antiretroviral.
Mean score among males.
Mean score among females.
Median score among males.
Median score among females.
Third quartile for males.
Third quartile for females.
P-value. Wilcoxon rank sum test with continuity correction; significant values (p<0.01) are in bold.
Discussion
The patterns and frequencies of the DRMs described (Fig. 1) are consistent with present treatment strategies. The World Health Organization (WHO) recommends AZT or TDF plus 3TC together with EFV or NVP as a first-line antiretroviral regimen.2 The highly frequent M184V substitution is the first mutation that appears under 3TC or 3TC-containing regimens, resulting in complete resistance to 3TC. Mutations M41L, T215Y, D67N, L210W, and K70R are selected by thymidine analogs such as AZT, which has been heavily used in our country. These mutations belong to a group of substitutions known as thymidine analog mutations or TAMs, which decrease susceptibility to almost all nucleoside and nucleotide analogs. Furthermore, N348I and V118I are accessory mutations that, in combination with TAMs, also reduce susceptibility to most NRTIs.8
Analogously, K103N and Y181C are the mutations most frequently selected by EFV and NVP, respectively. K103N confers high resistance to all first-generation NNRTIs.9 K103N has the potential to persist for years,10 and both K103N and Y181C have been shown to be present as minor variants prior to antiretroviral treatment and are correlated with the risk of virologic failure.9 Furthermore, these two mutations, together with G190A, are known to be common in transmitted drug resistance.11 The WHO also recommends the inclusion of PIs in second-line regimens2. DRMs L10I, I54V, A71V, and M46I are known to accumulate during failure of therapy with most PIs, causing gradual increases of resistance levels.12 Mutation L90M confers resistance to several Pis,8 whereas V82A is selected by ritonavir and produces failure of therapy with most PIs.8 Mutation L10V and other mutations at position 10 compensate for the loss of fitness associated with the major PI resistance mutations.8 Interestingly, this mutation has been observed in antiretroviral-naive patients in three independent studies,13–15 which, together with the data described here, suggests that transmitted drug resistance could be important in our country.
Viral genotype was a good predictor of the frequencies of different DRMs (Tables 1, 2, and 3). These are not unexpected results, as it has been shown that non-B HIV-1 subtypes can present resistance profiles that differ from those observed in subtype B viruses, a fact that is attributed to differences in the number of mutations needed by viruses with different genetic backgrounds to achieve resistance (the so-called genetic barrier), which differs among viral subtypes.16,17 The high frequency of mutation T74S is unexpected and interesting, as it is rare among non-C subtype viruses.8 This mutation was particularly frequent among the BF sequences studied here (Table 1), suggesting that recombination could drive the emergence of otherwise rare resistance mutations.
The presence of multiple DRMs was fairly common among the sequences studied here (Table 4). This condition is related basically to two facts: first, high level resistance and resistance to multiple antiretrovirals necessitate the accumulation of multiple amino acid substitutions; second, as most resistance mutations impair viral replication, the emergence of compensatory mutations that attenuate fitness loss usually follows the appearance of primary mutations. One of most remarkable features of our dataset was the unequal distribution of NRTI resistance mutations among adult and children, a situation that was also observed, though to a lesser extent, for PI resistance mutations (Table 4). A factor that could be responsible for this situation is vertical transmission, which results in the presence of primary resistance mutations to which more mutations may be readily added during suboptimal antiretroviral treatment. Another one is adherence, which is related to virological response and thus is crucial for the development of antiretroviral resistance. This is specially challenging in infants and children because adherence is frequently jeopardized by factors such as psychological issues, lack of pediatric formulations, poor palatability, high pill burden or liquid volume, frequent dosing requirements, and side effects. Thus, we think that PI and NRTI resistance mutations could have more opportunities to accumulate in children than in adults due to adherence issues. This is consistent with the fact that the number of NNRTI resistance mutations was similar among adult and children, as there is a genetic barrier of one or two mutations to NNRTI resistance, and thus the accumulation of further mutations would have minimal or no effect on antiretroviral resistance.8
In general, sequences from men and women displayed different amounts of DRMs (Tables 5 and 6). In the case of NVP resistance ones, which were more prevalent among subtype B strains from adult women, a very plausible explanation is the use of this antiretroviral for preventing mother-to-child transmission. A single mutation can confer resistance to NVP. Furthermore, it has a long half-life, and drug levels persist for weeks after women receive single-dose NVP, a situation that constitutes the ideal scenario for the emergence of resistant viral variants. That is why this antiretroviral readily selects for NNRTI resistance in postpartum women and infants where transmission does occur. Thus, our results reinforce the idea that in order to decrease the incidence of NVP resistance, combinations and longer antiretroviral prophylaxis regimens should be preferred to single-dose NVP ones.18 Also, these observations support current guidelines recommending PI-based, non-NVP-containing regimens for infants who do become infected despite single-dose NVP or extended NVP prophylaxis.19 The fact that only subtype B sequences displayed statistically significant differences regarding the degree of NVP resistance in women compared to men points out that differences in the viral genetic background could be important when deciding which are the better treatment options.
The degrees of resistance to many PIs and NRTIs were higher among strains from men than among strains from women (Tables 5 and 6). Although we did not expect to observe such marked differences among sequences from men and women, this is not the first time that this situation has been observed, as a previous investigation performed in Puerto Rico also showed that the average number of resistance mutations was higher among viruses from men than among viruses from women.20 So far, some investigations have shown that gender could influence biological issues such as the characteristics of transmitted virus population,21,22 the levels of HIV RNA,23–25 and HIV-specific CD8+ T cell response.26 Nevertheless, these and other similar reports have led to intense debates without reaching a consensus on whether gender can determine the characteristics of HIV infection.
Although we do not completely discard this last possibility, we think that lurking epidemiological variables could better explain the differences in the number of PI and NRTI resistance mutations observed among viruses from men and women. For example, previous studies have shown that in Argentina, the incidence of HIV-1 genotypes varies among vulnerable groups such as drug users, sex workers, men who have sex with men, and heterosexuals,27,28 suggesting that viral populations circulating in Argentina are structured. Also, it is known that men are less adherent to treatment than women, which, as mentioned above, favors the nonsuppressive conditions that allow the emergence of resistant variants. Regardless of the preferred explanation for the gender-related differences observed here, our results indicate that there is a strong need for further epidemiological studies to understand the antiretroviral resistance epidemics in Argentina. Ideally, these studies must integrate large amounts of sequence data together with risk groups and demographic and clinical information.
Acknowledgments
Continuous support from National Council of Scientific and Technical Research (CONICET, Argentina) is most appreciated. This work was partially supported by Grants PICT-PRH 2008-120 from the National Scientific and Technical Research Agency (Argentina) and PIP 11420090100254 from CONICET.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Rhee SY. Taylor J. Wadhera G. Ben-Hur A. Brutlag DL. Shafer RW. Genotypic predictors of human immunodeficiency virus type 1 drug resistance. Proc Natl Acad Sci USA. 2006;103(46):17355–17360. doi: 10.1073/pnas.0607274103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.WHO. UNAIDS report on the global AIDS epidemic. 2010.
- 3.Gomez-Carrillo M. Quarleri JF. Rubio AE, et al. Drug resistance testing provides evidence of the globalization of HIV type 1: A new circulating recombinant form. AIDS Res Hum Retroviruses. 2004;20(8):885–888. doi: 10.1089/0889222041725172. [DOI] [PubMed] [Google Scholar]
- 4.Vignoles M. Barboni G. Agosti MR, et al. High frequency of primary mutations associated with antiretroviral drug resistance in recently diagnosed HIV-infected children. Antivir Ther. 2007;12(7):1133–1137. [PubMed] [Google Scholar]
- 5.Salminen MO. Carr JK. Burke DS. McCutchan FE. Identification of breakpoints in intergenotypic recombinants of HIV type 1 by bootscanning. AIDS Res Hum Retroviruses. 1995;11(11):1423–1425. doi: 10.1089/aid.1995.11.1423. [DOI] [PubMed] [Google Scholar]
- 6.Rhee SY. Gonzales MJ. Kantor R. Betts BJ. Ravela J. Shafer RW. Human immunodeficiency virus reverse transcriptase and protease sequence database. Nucleic Acids Res. 2003;31(1):298–303. doi: 10.1093/nar/gkg100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.R-Development-Core-Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2010. [Google Scholar]
- 8.Shafer RW. Schapiro JM. HIV-1 drug resistance mutations: An updated framework for the second decade of HAART. AIDS Rev. 2008;10(2):67–84. [PMC free article] [PubMed] [Google Scholar]
- 9.Ghosn J. Chaix ML. Delaugerre C. HIV-1 resistance to first- and second-generation non-nucleoside reverse transcriptase inhibitors. AIDS Rev. 2009;11(3):165–173. [PubMed] [Google Scholar]
- 10.Hirsch MS. Gunthard HF. Schapiro JM, et al. Antiretroviral drug resistance testing in adult HIV-1 infection: 2008 recommendations of an International AIDS Society-USA panel. Clin Infect Dis. 2008;47(2):266–285. doi: 10.1086/589297. [DOI] [PubMed] [Google Scholar]
- 11.Geretti AM. Epidemiology of antiretroviral drug resistance in drug-naive persons. Curr Opin Infect Dis. 2007;20(1):22–32. doi: 10.1097/QCO.0b013e328013caff. [DOI] [PubMed] [Google Scholar]
- 12.Clavel F. Hance AJ. HIV drug resistance. N Engl J Med. 2004;350(10):1023–1035. doi: 10.1056/NEJMra025195. [DOI] [PubMed] [Google Scholar]
- 13.Dilernia DA. Gomez AM. Lourtau L, et al. HIV type 1 genetic diversity surveillance among newly diagnosed individuals from 2003 to 2005 in Buenos Aires, Argentina. AIDS Res Hum Retroviruses. 2007;23(10):1201–1207. doi: 10.1089/aid.2007.0068. [DOI] [PubMed] [Google Scholar]
- 14.Kijak GH. Pampuro SE. Avila MM, et al. Resistance profiles to antiretroviral drugs in HIV-1 drug-naive patients in Argentina. Antivir Ther. 2001;6(1):71–77. [PubMed] [Google Scholar]
- 15.Perno CF. Cozzi-Lepri A. Balotta C, et al. Secondary mutations in the protease region of human immunodeficiency virus and virologic failure in drug-naive patients treated with protease inhibitor-based therapy. J Infect Dis. 2001;184(8):983–991. doi: 10.1086/323604. [DOI] [PubMed] [Google Scholar]
- 16.Ariyoshi K. Matsuda M. Miura H. Tateishi S. Yamada K. Sugiura W. Patterns of point mutations associated with antiretroviral drug treatment failure in CRF01_AE (subtype E) infection differ from subtype B infection. J Acquir Immune Defic Syndr. 2003;33(3):336–342. doi: 10.1097/00126334-200307010-00007. [DOI] [PubMed] [Google Scholar]
- 17.Kantor R. Katzenstein DA. Efron B, et al. Impact of HIV-1 subtype and antiretroviral therapy on protease and reverse transcriptase genotype: Results of a global collaboration. PLoS Med. 2005;2(4):e112. doi: 10.1371/journal.pmed.0020112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.WHO. Antiretroviral drugs for treating pregnant women and preventing HIV infection in infants: Recomendations for a public health approach. 2010. [PubMed]
- 19.WHO. Antiretroviral therapy for HIV infection in infants and children (2010 revision) 2010.
- 20.Cubano LA. Sepulveda-Torres Ldel C. Sosa G, et al. Prevalence of drug resistance and associated mutations in HIV-positive Puerto Ricans: Sex variations. Ethn Dis. 2008;18(2 Suppl 2):S2-S132–S136. [PubMed] [Google Scholar]
- 21.Long EM. Martin HL., Jr. Kreiss JK, et al. Gender differences in HIV-1 diversity at time of infection. Nat Med. 2000;6(1):71–75. doi: 10.1038/71563. [DOI] [PubMed] [Google Scholar]
- 22.Ray SC. Quinn TC. Sex and the genetic diversity of HIV-1. Nat Med. 2000;6(1):23–25. doi: 10.1038/71487. [DOI] [PubMed] [Google Scholar]
- 23.Farzadegan H. Hoover DR. Astemborski J, et al. Sex differences in HIV-1 viral load and progression to AIDS. Lancet. 1998;352(9139):1510–1514. doi: 10.1016/S0140-6736(98)02372-1. [DOI] [PubMed] [Google Scholar]
- 24.Sterling TR. Lyles CM. Vlahov D. Astemborski J. Margolick JB. Quinn TC. Sex differences in longitudinal human immunodeficiency virus type 1 RNA levels among seroconverters. J Infect Dis. 1999;180(3):666–672. doi: 10.1086/314967. [DOI] [PubMed] [Google Scholar]
- 25.Sterling TR. Vlahov D. Astemborski J. Hoover DR. Margolick JB. Quinn TC. Initial plasma HIV-1 RNA levels and progression to AIDS in women and men. N Engl J Med. 2001;344(10):720–725. doi: 10.1056/NEJM200103083441003. [DOI] [PubMed] [Google Scholar]
- 26.Sterling TR. Pisell-Noland T. Perez JL, et al. Sex-based differences in T lymphocyte responses in HIV-1-seropositive individuals. J Infect Dis. 2005;191(6):881–885. doi: 10.1086/427827. [DOI] [PubMed] [Google Scholar]
- 27.Avila MM. Pando MA. Carrion G, et al. Two HIV-1 epidemics in Argentina: Different genetic subtypes associated with different risk groups. J Acquir Immune Defic Syndr. 2002;29(4):422–426. doi: 10.1097/00126334-200204010-00015. [DOI] [PubMed] [Google Scholar]
- 28.Pando MA. Gomez-Carrillo M. Vignoles M, et al. Incidence of HIV Type 1 infection, antiretroviral drug resistance, and molecular characterization in newly diagnosed individuals in Argentina: A global fund project. AIDS Res Hum Retroviruses. 2011;27(1):17–23. doi: 10.1089/aid.2010.0013. [DOI] [PMC free article] [PubMed] [Google Scholar]