Direct sequencing was used to identify EGFR mutations in 180 pairs of lung adenocarcinoma samples from the primary tumor and one metastatic site in 3,071 patients. Correlations between EGFR genetic heterogeneity and patient characteristics were examined.
Keywords: Lung neoplasm, Epidermal growth factor receptor, Metastasis, Heterogeneity
Abstract
Background.
Non-small cell lung cancer patients with epidermal growth factor receptor (EGFR) mutations have mixed responses to tyrosine kinase inhibitors (TKIs). Intertumor heterogeneity in EGFR mutations is one potential explanation for this phenomenon.
Methods.
We performed direct sequencing to identify EGFR mutations in 180 pairs of lung adenocarcinoma samples (from 3,071 patients). The high-resolution melting method was used in discordant cases to confirm EGFR mutation status. Matching samples were divided into four groups: primary lesions detected at different times, primary tumors with matched metastatic lymph nodes, multiple pulmonary nodules, and primary tumors with matched distant metastases. Multivariate analyses were performed to evaluate correlations between heterogeneity and patient characteristics.
Results.
In the study population, the discordance rate was 13.9% (25 of 180). The multiple pulmonary nodules group had the highest discordance rate of 24.4% (10 of 41; odds ratio for heterogeneity in primary lesions detected at different times, 6.37; 95% confidence interval, 1.71–23.72; p = .006). Discordance rates in the metachronous and synchronous settings were 15.7% (22 of 140) and 7.5% (three of 40), respectively. In the 34 patients who developed EGFR TKI resistance, 10 (29.4%) cases exhibited heterogeneity and five (14.7%) patients exhibited a mixed response to the drug. Three (8.8%) of the patients with a mixed response also exhibited discordant EGFR mutations.
Conclusions.
The overall discordance rate of EGFR mutation heterogeneity in Asian patients with pulmonary adenocarcinoma is relatively low, but the rate in patients with multiple pulmonary nodules is significantly higher. This observation may explain the mixed tumor response to EGFR TKIs.
Introduction
Personalized medicine is a promising approach that can improve treatment outcomes in patients with advanced non-small cell lung cancer (NSCLC) [1–3]. The discovery of driver oncogenes, such as activating mutations in the epidermal growth factor receptor gene (EGFR), has made personalized medicine for lung cancer a reality. The dimerization of EGFR induces intrinsic intracellular protein–tyrosine kinase activity, resulting in autophosphorylation and downstream signaling leading to cell proliferation, angiogenesis, and apoptosis [4]. Tyrosine kinase inhibitors (TKIs), including gefitinib and erlotinib, selectively inhibit EGFR signaling by targeting its ATP-binding site and preventing the activation of its intrinsic tyrosine kinase domain [5, 6].
Most patients with EGFR-activating mutations respond well to EGFR TKIs, but a subset of patients exhibit mixed responses. The tumor response rate to first-line EGFR TKIs is in the range of 52%–82% [1, 2, 7]. Among patients who fail to attain a partial or complete response, some tumors respond whereas others progress [8, 9]. This phenomenon may be explained by heterogeneity in the EGFR mutation status between the primary lung tumors and their metastases [10, 11]. To date, only limited information regarding the heterogeneity of EGFR mutations is available, and this hypothesis has remained untested [12–14]. For this reason, we studied discordance in the EGFR mutation status in paired samples of primary pulmonary adenocarcinoma and regional lymph nodes or distant metastases. Our results may help to explain the phenomenon of mixed tumor responses to EGFR TKIs and provide a foundation for future diagnostic and therapeutic approaches to TKI resistance.
Materials and Methods
Patients and Tissue Samples
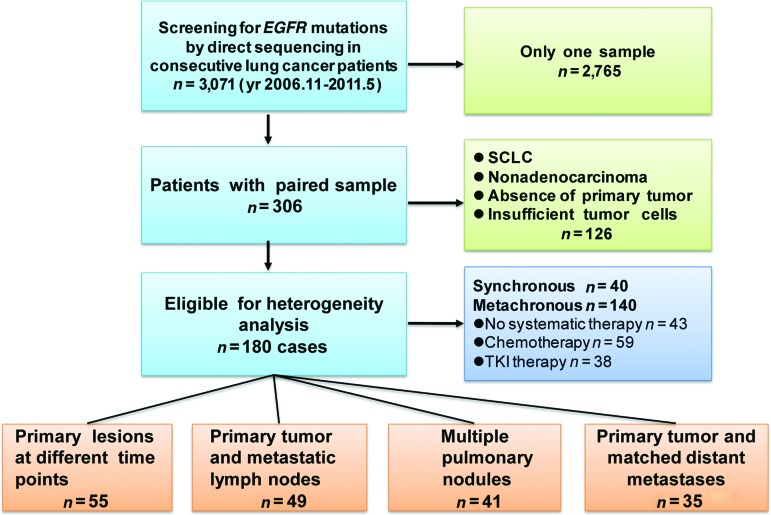
We performed EGFR mutation analyses in 3,071 consecutive lung cancer patients treated at the Guangdong Lung Cancer Institute from November 2006 to May 2011 (Fig. 1). All patients provided informed consent for the use of their tumor samples for molecular and pathologic analyses. The study was approved by the Ethics and Scientific Committees of Guangdong General Hospital. The clinical features of each patient were collected from their medical records. Patients with tumor samples available from two or more disease sites (at least one from the primary tumor) were included. We excluded 126 cases who were diagnosed with small cell lung cancer, did not feature adenocarcinoma in any lesion, showed absence of a primary tumor, or had insufficient tumor tissue for molecular analysis. In total, 180 patients with paired adenocarcinoma samples were eligible, and they were classified into four groups. Group A included patients with paired metachronous primary tumors diagnosed at different times. Group B included patients with a primary tumor paired with regional lymph node metastasis. Group C included patients with multiple pulmonary nodules. Group D included patients with a primary lung tumor paired with a distant metastasis. Furthermore, we categorized patients into synchronous and metachronous groups. The metachronous group included three subgroups: patients who did not undergo systemic therapy, patients who underwent chemotherapy, and patients who underwent TKI therapy. All paired samples were analyzed for activating EGFR mutations through direct DNA sequencing. If the primary tumors and their metastases shared the same mutation, they were considered homogeneous. If they were different, we confirmed the finding using the high-resolution melting method (HRM) to ensure the accuracy of direct sequencing.
Figure 1.
Enrollment and outcomes.
Abbreviations: EGFR, epidermal growth factor receptor; TKI, tyrosine kinase inhibitor; SCLC, small cell lung cancer.
EGFR Mutation Analysis Using DNA Sequencing
EGFR mutation analyses were performed on the 360 tumor samples using direct sequencing. Tumor samples from eligible patients were retrieved from our archives. Genomic DNA was extracted from 226 resection specimens and 134 transthoracic needle aspirations of lung nodules or fiber bronchoscope samples that contained >50% neoplastic cells. Polymerase chain reaction (PCR) was used to amplify exons 18–21 of EGFR. Mutation testing was performed only when the quantity of PCR-amplified DNA was judged to be sufficient through electrophoretic analysis. All detections were performed using a Big Dye Terminator, Version 3.1, Cycle Sequencing Kit (Applied Biosystems, Foster City, CA). Both forward and reverse sequencing reactions were performed with an ABI3100 genetic analyzer. A second round of validation was performed if a novel mutation (other than an exon 19 deletion or L858R mutation) was found or if poor sequence data (twin or sliding peaks) were obtained. If contradictory results were obtained from the paired samples, testing was repeated to exclude the possibility of false-positive or false-negative results [15].
EGFR Mutations Detected Using HRM
HRM is a sensitive genotyping method [16]. The melting profile of a PCR product depends on its guanine and cytosine content, length, and sequence and can therefore be used to detect heterozygosity. Assays were performed using the LightCycler 480 system according to the manufacturer's protocol. Data were analyzed using LightCycler 480 software (version 1.5). PCR was performed in duplicate for each sample, and two investigators blinded to the clinical information analyzed the results.
Statistical Analysis
Multivariate analyses were performed to determine correlation between heterogeneity and the clinical characteristics. In all tests, p ≤ .05 was considered to be statistically significant. All statistical tests were two sided and were performed using SPSS software, version 13.0 (SPSS, Inc., Chicago, IL).
Results
Patient Characteristics
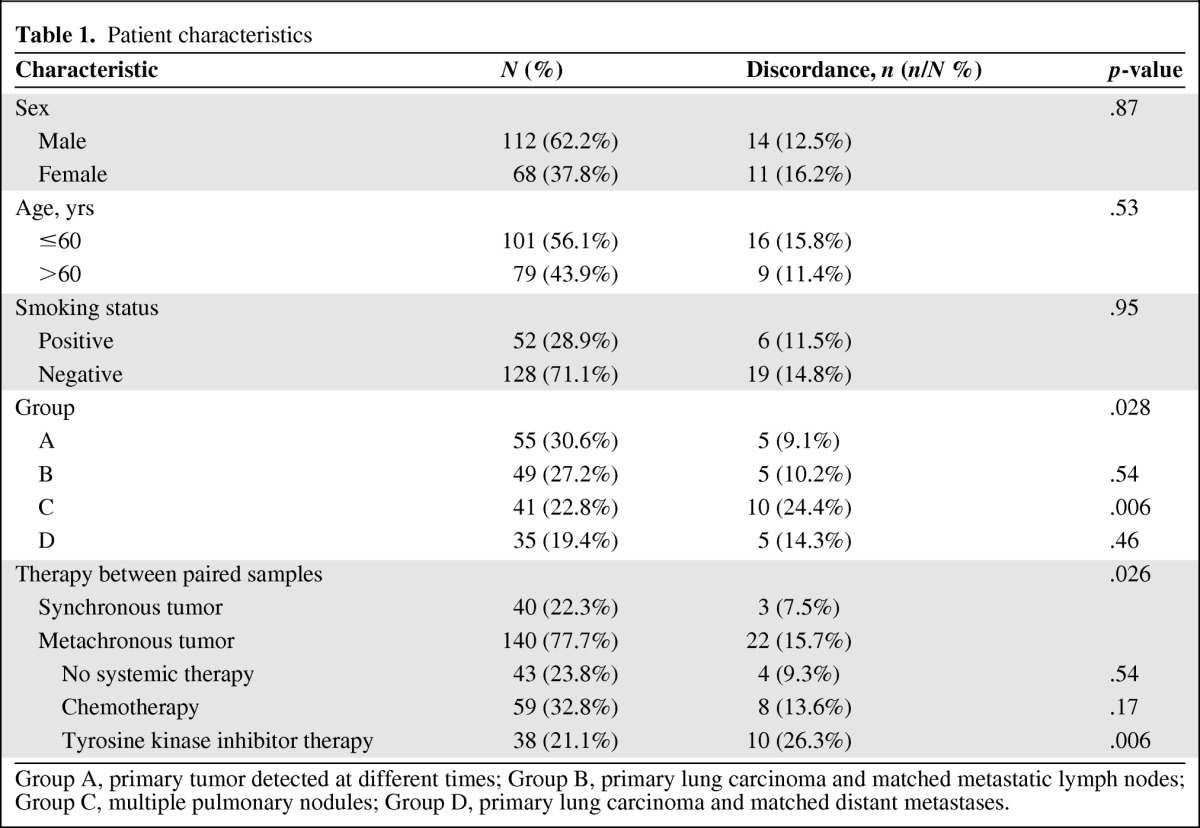
Patient characteristics are summarized in Table 1. The median age of the 180 enrolled patients was 58 years (range, 27–84 years). In total, 38 (21.1%) patients had received TKI therapy and 59 (32.8%) had received chemotherapy between the biopsies of the primary and secondary samples. In total, 360 samples (180 pairs) were analyzed, including 235 primary tumor samples, 49 lymph node metastases, 41 pulmonary nodules, 15 chest-wall metastases, eight pleural metastases, five brain metastases, three liver metastases, three adrenal gland metastases, and one retroperitoneal lymph node metastasis.
Table 1.
Patient characteristics
Group A, primary tumor detected at different times; Group B, primary lung carcinoma and matched metastatic lymph nodes; Group C, multiple pulmonary nodules; Group D, primary lung carcinoma and matched distant metastases.
EGFR Heterogeneity
Activating EGFR mutations were identified in 119 (50.6%) primary tumors, 15 (30.6%) lymph node metastases, 19 (46.3%) pulmonary nodules, and 16 (45.7%) distant metastatic tumors. In total, 27 cases were discordant using direct DNA sequencing and 25 cases were validated using the HRM method. The overall discordance rate in our cohort was 13.9% (25 of 180). The details of each discordant case are summarized in Table 2.
Table 2.
Cases with EGFR mutation heterogeneity
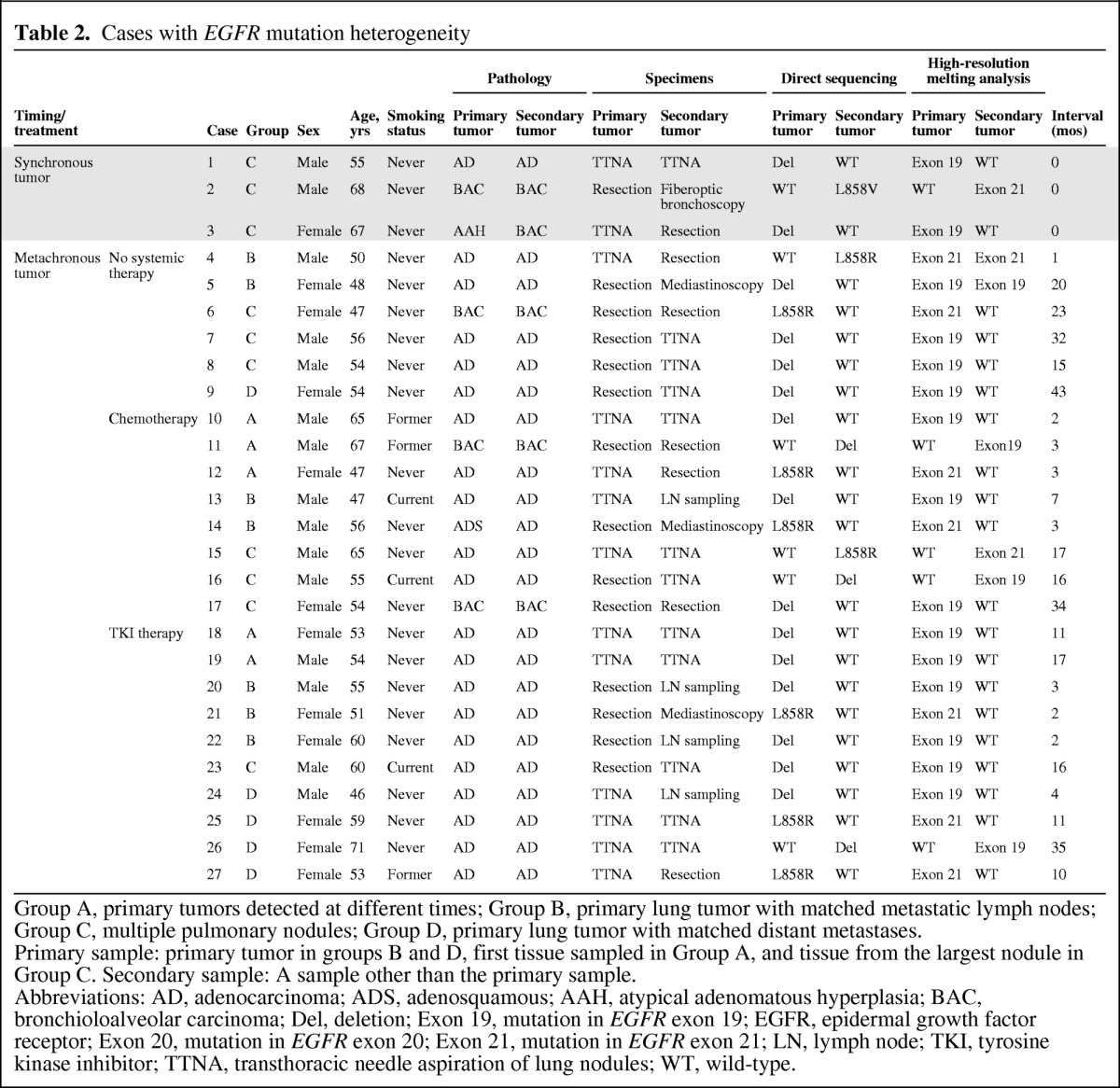
Group A, primary tumors detected at different times; Group B, primary lung tumor with matched metastatic lymph nodes; Group C, multiple pulmonary nodules; Group D, primary lung tumor with matched distant metastases.
Primary sample: primary tumor in groups B and D, first tissue sampled in Group A, and tissue from the largest nodule in Group C. Secondary sample: A sample other than the primary sample.
Abbreviations: AD, adenocarcinoma; ADS, adenosquamous; AAH, atypical adenomatous hyperplasia; BAC, bronchioloalveolar carcinoma; Del, deletion; Exon 19, mutation in EGFR exon 19; EGFR, epidermal growth factor receptor; Exon 20, mutation in EGFR exon 20; Exon 21, mutation in EGFR exon 21; LN, lymph node; TKI, tyrosine kinase inhibitor; TTNA, transthoracic needle aspiration of lung nodules; WT, wild-type.
EGFR Mutation Heterogeneity Between Primary Tumors and Their Metastases
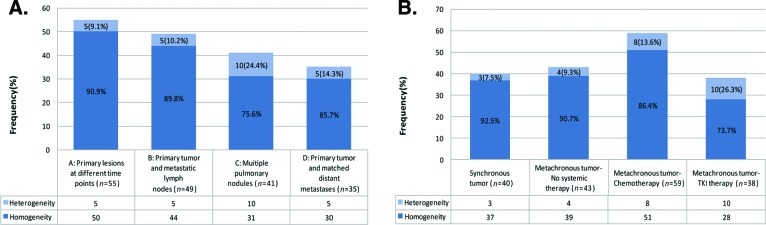
The highest discordance rate of 24.4% (10 of 41) was observed in group C (paired multiple pulmonary nodules), compared with 14.3% (five of 35) in group D (paired primary lung tumor and distant metastasis) and 10.2% (five of 49) in group B (paired primary lung tumor and metastatic lymph nodes). The lowest discordance rate, 9.1% (five of 55), was observed in group A (paired metachronous primary tumors diagnosed at different times) (Fig. 2A).
Figure 2.
EGFR mutation heterogeneity in different spatial and treatment groups. (A): Spatial group and heterogeneity. (B): Treatment group and heterogeneity.
Abbreviations: EGFR, epidermal growth factor receptor; TKI, tyrosine kinase inhibitor.
EGFR Mutation Heterogeneity in Synchronous and Metachronous Groups
Synchronous tumors had a discordance rate of 7.5% (three of 40), whereas metachronous tumors had a discordance rate of 15.7% (22 of 140). In the metachronous group, patients without exposure to any systemic therapy had a discordance rate of 9.3% (four of 43), whereas patients with exposure to chemotherapy had a discordance rate of 13.6% (eight of 59). The highest discordance rate, 26.3% (10 of 38), was observed in patients exposed to TKIs between the biopsies of the two samples (Fig. 2B). Of these 38 patients, 34 developed EGFR TKI resistance. In resistant cases, 10 (29.4%) exhibited heterogeneity and five (14.7%) exhibited mixed responses; three (8.8%) of these cases also exhibited discordant EGFR mutations. Additionally, 10 patients developed acquired resistance with a T790M mutation in exon 20 (Table 3, Fig. 3).
Table 3.
Five cases with mixed response to TKI therapy
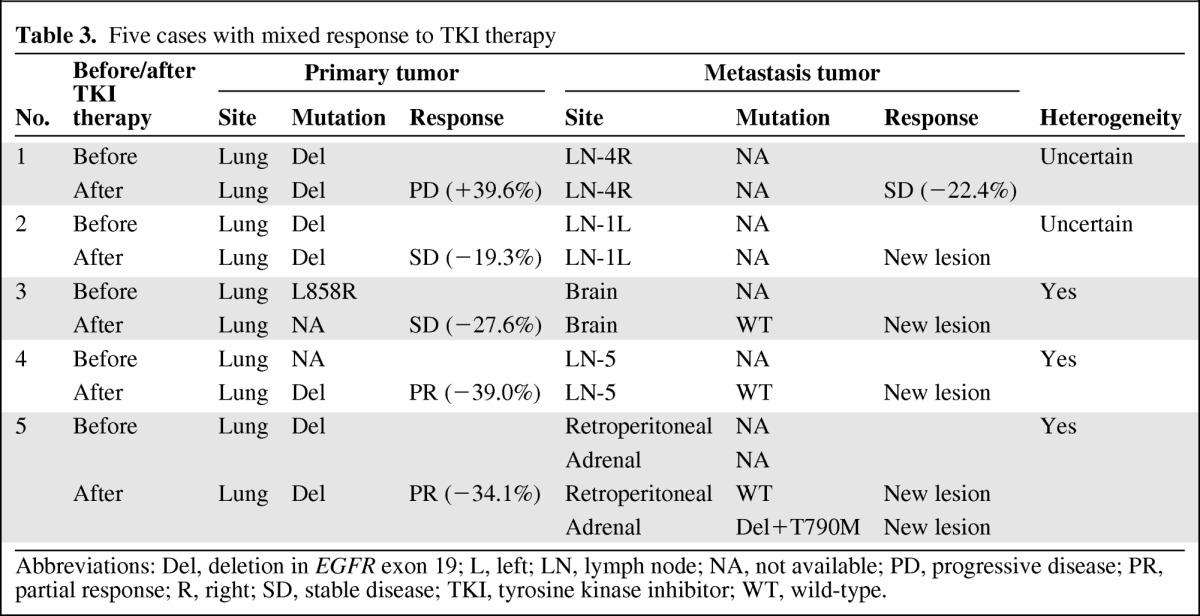
Abbreviations: Del, deletion in EGFR exon 19; L, left; LN, lymph node; NA, not available; PD, progressive disease; PR, partial response; R, right; SD, stable disease; TKI, tyrosine kinase inhibitor; WT, wild-type.
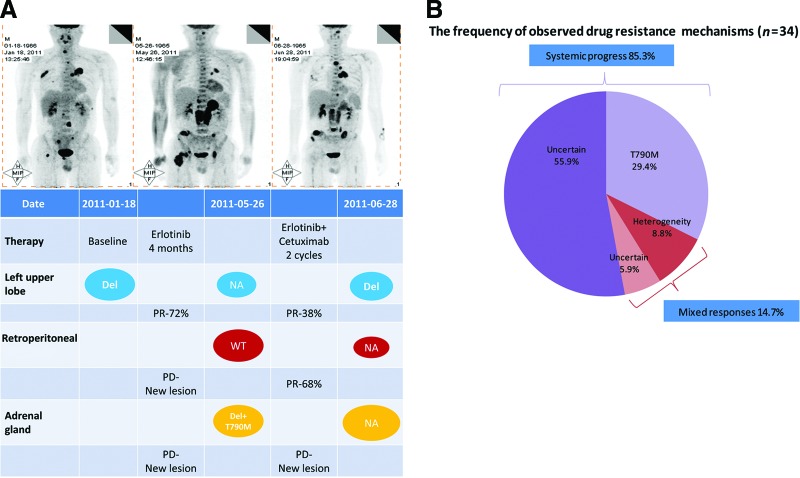
Figure 3.
Heterogeneity and mixed responses. Mixed response indicates shrinkage of the primary tumor but progression of the metastasis (or vice versa). (A): A 46-year-old male (patient 5 in Table 3). PET-CT showed a round mass with a high SUV in the left upper lobe with multiple bone metastases. No suspected lesions were found in the adrenal gland or the retroperitoneal space. A deletion in exon 19 of EGFR was detected in tumor tissue biopsied from the left upper lobe. After 4 months of first-line erlotinib therapy, a second PET-CT scan showed that the SUV and diameter of the primary tumor located in the left upper lobe had decreased significantly, whereas an aggressive novel neoplasm emerged in the retroperitoneal space and adrenal gland. Wild-type EGFR was identified in a tumor retrieved from the retroperitoneal space by laparoscopy, whereas an exon 19 deletion plus a T790M mutation in exon 20 of EGFR was observed in tissue biopsied from the adrenal gland. A third PET-CT scan was conducted after erlotinib plus cetuximab was continued for two cycles. Moderate left upper lobe enlargement was observed, but the EGFR exon 19 deletion was still present in rebiopsied tissue. Additionally, the retroperitoneal lymph node lesion was significantly smaller, but the adrenal gland lesion had grown aggressively. (B): The pie chart illustrates the frequencies of the various mechanisms of EGFR TKI resistance in 34 patients with NSCLC. Pre- and post-treatment images were evaluated to define the role of mixed responses to EGFR TKIs. The mixed-response cases accounted for 14.7% of TKI-resistant cases and include two subsets: mixed response combined with heterogeneity (8.8%) and mixed response combined with uncertain EGFR status (5.9%).
Abbreviations: EGFR, epidermal growth factor receptor; NA, not available; NSCLC, non-small cell lung cancer; PD, progressive disease; PET–CT, positron emission tomography–computed tomography; PR, partial response; SUV, standardized uptake value; TKI, tyrosine kinase inhibitor; WT, wild-type.
Multivariate Analysis: Correlation Between Heterogeneity and Clinical Characteristics
Multivariate analyses were conducted based on data from 180 paired samples. The incidence of heterogeneity was used as the independent variable. Candidate covariates included age (odds ratio for heterogeneity in the age ≤60 years group, 0.74; 95% confidence interval [CI], 0.29–1.91; p = .53), smoking history (odds ratio for heterogeneity in the nonsmoking group, 1.03; 95% CI, 0.34–3.18; p = .95), sex (odds ratio for heterogeneity in the male group, 1.09; 95% CI, 0.41–2.90; p = .87), treatment (p = .026), and four spatial group (p = .028). Heterogeneity was significantly higher in patients with multiple pulmonary nodules (odds ratio for heterogeneity in group A, 6.37; 95% CI, 1.71–23.7; p = .006) and with TKI exposure (odds ratio for heterogeneity in the synchronous tumor group, 8.70; 95% CI, 1.85–40.88; p = .006).
Discussion
This is the largest study of EGFR mutation heterogeneity between primary lung tumors and their metastases to date. We confirmed an overall discordance rate of 13.9% in 180 patients. Other studies with smaller sample sizes have reported variable discordance rates. Kalikaki et al. [17] reported an EGFR mutation heterogeneity rate of 28% in 25 white patients using direct sequencing to identify mutations. Schmid et al. [14] reported a discordance rate of 6.3% in 96 white patients using direct bidirectional sequencing. These differences may be explained by the study populations and the pathological types of the samples. Our larger sample size confirmed a heterogeneity rate of 13.9% in a homogeneous population of Asian patients with pulmonary adenocarcinoma.
We took double diligence to optimize the sensitivity of the mutation analysis. Zhou et al. [18] reported that a more sensitive technique was capable of detecting rare mutated genomes. The apparent absence of any EGFR mutation may simply imply a low abundance of mutated genomes if only the direct DNA sequencing method is used. Other studies have suggested that the detection of changes in EGFR mutation status is primarily related to the sensitivity of the sequencing method. Yatabe et al. [19] studied the EGFR mutation status in 77 paired primary and metastatic tumor samples and did not find discordance in any of the samples. They concluded that the heterogeneous distribution of EGFR mutations is rare and that pseudoheterogeneity is observed as a result of the use of less sensitive methods of detection. In our study, we minimized pseudoheterogeneity by repeating the analysis in discordant cases with an alternative and more sensitive testing method. HRM is an ultrasensitive method that can confirm the accuracy of direct sequencing. The concordance rate between the results of direct sequencing and the HRM method in our study was 92.6% (25 of 27). These results are consistent with the findings reported by Gow et al. [20], who observed heterogeneous EGFR mutations in 38.8% (26 of 67) of Asian patients using direct sequencing and 26.8% of patients using the Scorpion amplified refractory mutation system assay. In contrast, Park et al. [21] found EGFR mutation heterogeneity in 11.9% (12 of 101) of samples with direct sequencing but in 16.8% (17 of 101) of samples tested with heteroduplex analysis.
Although previous studies have reported that heterogeneity occurs prior to patient treatment, there is little evidence regarding the role of treatment (particularly chemotherapy and TKI therapy) in mutagenesis. We therefore compared EGFR heterogeneity between synchronous tumors and tumors from patients who had received TKI therapy, chemotherapy, or no systemic treatment between samplings of metachronous tumors. Our data reflect the potential mutagenic effects of chemotherapy and TKI therapy.
Our previous study found unusual cases in which the primary tumor had a wild-type EGFR sequence but the metastases exhibited a mutated genotype, regardless of the detection method used [22]. Additionally, some patients harbored different histological subgroups coexistent with EGFR heterogeneity. These results might be explained by genomic heterogeneity within the primary carcinomas. Heterogeneity in EGFR mutation was observed within a primary tumor that was composed of mixed atypical adenomatous hyperplasia, bronchoalveolar carcinoma, and adenocarcinoma [23]. Taniguchi et al. [10] evaluated 21 NSCLC patients with EGFR mutations and found six tumor samples containing both mutated and wild-type tumor cells. Another possible explanation for this unusual observation is that the “metastatic” tumor was actually a secondary primary carcinoma, as observed with multiple pulmonary nodules.
Heterogeneity in the EGFR mutation status was higher in patients with multiple pulmonary nodules. This result may be explained by the presence of more than one primary lung cancer [24–27]. Our observation has significant clinical implications for patients who present with multiple pulmonary nodules. Current practice guidelines suggest biopsying only one lesion for the histological diagnosis and molecular analysis. Our data show a discordance rate of 24.4%, suggesting that approximately one in five patients with multiple lung nodules may have heterogeneous disease. This result indicates that serial and/or multiple biopsies for multiple lung nodules may be necessary to understand the biology of the lung cancer. Genomic heterogeneity may also explain why some mutation-negative patients are responsive to TKIs whereas some mutation-positive patients are resistant. Sequist et al. [28] indicated that further mutation of EGFR, MET amplification, phosphatidylinositol 3-kinase mutation, and transition to small cell cancer may be associated with drug resistance. Additionally, we found mixed responses in 14.7% of cases with resistance to TKI therapy, 8.8% of which were coexistent with heterogeneity; thus, up to 8.8% of mixed responses may be explained by EGFR heterogeneity. Based on the genomic changes observed in conjunction with mixed responses, it is rational to suggest that EGFR TKIs should be continued beyond progression and used in combination with other treatment modalities, including radiotherapy, surgery, and chemotherapy, for patients with mixed responses [29]. We demonstrated that selective tumors may still harbor an activating EGFR mutation and be sensitive to EGFR TKIs, whereas the progressing tumor may be sensitive to other treatment modalities. Our findings also support the practice of rebiopsying tumors that progress during TKI treatment, although it is difficult to incorporate synchronous second biopsies into everyday clinical practice [15, 28]. This approach will help to optimize therapy for patients with known activating EGFR mutations. Tumor heterogeneity should also provide the foundation for a multikinase inhibitor and molecular evidence to support clinical treatment strategies for patients with lung cancer [29, 30].
Our study is limited by the availability and quality of archival tumor samples. Some tumor samples were very small and may not have contained sufficient DNA for mutation analysis. Thus, the apparent loss of mutation in some cases could be explained by the quality of the archived material. One other limitation is the lack of multiple sampling from primary tumors. For this reason, we were not able to address the intratumoral heterogeneity issue.
Conclusions
In summary, EGFR mutation heterogeneity exists in Asian patients with pulmonary adenocarcinoma, but the overall discordance rate is relatively low. Only patients with multiple pulmonary nodules have a significantly higher rate of heterogeneity, and this observation may explain the mixed tumor response to EGFR TKIs.
Acknowledgments
We thank the staff of the Pulmonary Department and Research Center, Yan-Hui Liu and Dong-Lan Luo from the Department of Pathology, and Su-Xia Liu and Dan Shao from the Department of PET-CT at Guangdong General Hospital for participating in this study.
This work was supported by grants from the National Natural Science Foundation of China (81001031 and 30772531), the Foundation of Guangdong Science and Technology Department (2006B60101010 and 2007A032000002), the Natural Science Foundation of Guangdong (8151008004000015), the Medical Research and Science Technology Foundation of Guangdong Province (A20100011), and the Chinese Lung Cancer Research Foundation.
Zhi-Yong Chen and Wen-Zhao Zhong contributed equally to this manuscript.
Footnotes
- (C/A)
- Consulting/advisory relationship
- (RF)
- Research funding
- (E)
- Employment
- (H)
- Honoraria received
- (OI)
- Ownership interests
- (IP)
- Intellectual property rights/inventor/patent holder
- (SAB)
- Scientific advisory board
Author Contributions
Conception/Design: Xue-Ning Yang, Xu-Chao Zhang, Wen-Zhao Zhong, Yi-Long Wu, Zhi-Yong Chen, Tony S. Mok
Provision of study material or patients: Xue-Ning Yang, Jian Su, Xu-Chao Zhang, Wen-Zhao Zhong, Yi-Long Wu, Zhi-Yong Chen, Zhi-Hong Chen, Jin-Ji Yang, Qing Zhou, Hong-Hong Yan, She-Juan An, Hua-Jun Chen, Ben-Yuan Jiang, Tony S. Mok
Collection and/or assembly of data: Xue-Ning Yang, Jian Su, Xu-Chao Zhang, Wen-Zhao Zhong, Yi-Long Wu, Zhi-Yong Chen, Zhi-Hong Chen, Jin-Ji Yang, Qing Zhou
Data analysis and interpretation: Jian Su, Xu-Chao Zhang, Wen-Zhao Zhong, Yi-Long Wu, Zhi-Yong Chen, Hong-Hong Yan, Tony S. Mok
Manuscript writing: Wen-Zhao Zhong, Yi-Long Wu, Zhi-Yong Chen, Tony S. Mok
Final approval of manuscript: Xue-Ning Yang, Jian Su, Xu-Chao Zhang, Wen-Zhao Zhong, Yi-Long Wu, Zhi-Yong Chen, Zhi-Hong Chen, Jin-Ji Yang, Qing Zhou, Hong-Hong Yan, She-Juan An, Hua-Jun Chen, Ben-Yuan Jiang, Tony S. Mok
References
- 1.Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin–paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361:947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 2.Rosell R, Moran T, Queralt C, et al. Screening for epidermal growth factor receptor mutations in lung cancer. N Engl J Med. 2009;361:958–967. doi: 10.1056/NEJMoa0904554. [DOI] [PubMed] [Google Scholar]
- 3.Mitsudomi T, Morita S, Yatabe Y, et al. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): An open label, randomised phase 3 trial. Lancet Oncol. 2010;11:121–128. doi: 10.1016/S1470-2045(09)70364-X. [DOI] [PubMed] [Google Scholar]
- 4.Ono M, Kuwano M. Molecular mechanisms of epidermal growth factor receptor (EGFR) activation and response to gefitinib and other EGFR-targeting drugs. Clin Cancer Res. 2006;12:7242–7251. doi: 10.1158/1078-0432.CCR-06-0646. [DOI] [PubMed] [Google Scholar]
- 5.Cohen MH, Williams GA, Sridhara R, et al. United States Food and Drug Administration drug approval summary: Gefitinib (ZD1839; Iressa) tablets. Clin Cancer Res. 2004;10:1212–1218. doi: 10.1158/1078-0432.ccr-03-0564. [DOI] [PubMed] [Google Scholar]
- 6.Cohen MH, Johnson JR, Chen YF, et al. FDA drug approval summary: Erlotinib (Tarceva) tablets. The Oncologist. 2005;10:461–466. doi: 10.1634/theoncologist.10-7-461. [DOI] [PubMed] [Google Scholar]
- 7.Sequist LV, Martins RG, Spigel D, et al. First-line gefitinib in patients with advanced non-small-cell lung cancer harboring somatic EGFR mutations. J Clin Oncol. 2008;26:2442–2449. doi: 10.1200/JCO.2007.14.8494. [DOI] [PubMed] [Google Scholar]
- 8.Ryoo BY, Na II, Yang SH, et al. Synchronous multiple primary lung cancers with different response to gefitinib. Lung Cancer. 2006;53:245–258. doi: 10.1016/j.lungcan.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 9.Park KY, Jung JW, Nam SB, et al. Two lung masses with different responses to pemetrexed. Korean J Intern Med. 2010;25:213–216. doi: 10.3904/kjim.2010.25.2.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taniguchi K, Okami J, Kodama K, et al. Intratumor heterogeneity of epidermal growth factor receptor mutations in lung cancer and its correlation to the response to gefitinib. Cancer Sci. 2008;99:929–935. doi: 10.1111/j.1349-7006.2008.00782.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang SX, Yamashita K, Yamamoto M, et al. EGFR genetic heterogeneity of nonsmall cell lung cancers contributing to acquired gefitinib resistance. Int J Cancer. 2008;123:2480–2486. doi: 10.1002/ijc.23868. [DOI] [PubMed] [Google Scholar]
- 12.Wang X, Wang M, MacLennan GT, et al. Evidence for common clonal origin of multifocal lung cancers. J Natl Cancer Inst. 2009;101:560–570. doi: 10.1093/jnci/djp054. [DOI] [PubMed] [Google Scholar]
- 13.Chang YL, Wu CT, Shih JY, et al. Comparison of p53 and epidermal growth factor receptor gene status between primary tumors and lymph node metastases in non-small cell lung cancers. Ann Surg Oncol. 2011;18:543–550. doi: 10.1245/s10434-010-1295-6. [DOI] [PubMed] [Google Scholar]
- 14.Schmid K, Oehl N, Wrba F, et al. EGFR/KRAS/BRAF mutations in primary lung adenocarcinomas and corresponding locoregional lymph node metastases. Clin Cancer Res. 2009;15:4554–4560. doi: 10.1158/1078-0432.CCR-09-0089. [DOI] [PubMed] [Google Scholar]
- 15.Pirker R, Herth FJ, Kerr KM, et al. Consensus for EGFR mutation testing in non-small cell lung cancer: Results from a European workshop. J Thorac Oncol. 2010;5:1706–1713. doi: 10.1097/JTO.0b013e3181f1c8de. [DOI] [PubMed] [Google Scholar]
- 16.Krypuy M, Newnham GM, Thomas DM, et al. High resolution melting analysis for the rapid and sensitive detection of mutations in clinical samples: KRAS codon 12 and 13 mutations in non-small cell lung cancer. BMC Cancer. 2006;6:295. doi: 10.1186/1471-2407-6-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kalikaki A, Koutsopoulos A, Trypaki M, et al. Comparison of EGFR and K-RAS gene status between primary tumours and corresponding metastases in NSCLC. Br J Cancer. 2008;99:923–929. doi: 10.1038/sj.bjc.6604629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou Q, Zhang XC, Chen ZH, et al. Relative abundance of EGFR mutations predicts benefit from gefitinib treatment for advanced non-small-cell lung cancer. J Clin Oncol. 2011;29:3316–3321. doi: 10.1200/JCO.2010.33.3757. [DOI] [PubMed] [Google Scholar]
- 19.Yatabe Y, Matsuo K, Mitsudomi T. Heterogeneous distribution of EGFR mutations is extremely rare in lung adenocarcinoma. J Clin Oncol. 2011;29:2972–2977. doi: 10.1200/JCO.2010.33.3906. [DOI] [PubMed] [Google Scholar]
- 20.Gow CH, Chang YL, Hsu YC, et al. Comparison of epidermal growth factor receptor mutations between primary and corresponding metastatic tumors in tyrosine kinase inhibitor-naive non-small-cell lung cancer. Ann Oncol. 2009;20:696–702. doi: 10.1093/annonc/mdn679. [DOI] [PubMed] [Google Scholar]
- 21.Park S, Holmes-Tisch AJ, Cho EY, et al. Discordance of molecular biomarkers associated with epidermal growth factor receptor pathway between primary tumors and lymph node metastasis in non-small cell lung cancer. J Thorac Oncol. 2009;4:809–815. doi: 10.1097/JTO.0b013e3181a94af4. [DOI] [PubMed] [Google Scholar]
- 22.Zhong WZ, Wu YL, Yang XN, et al. Genetic evolution of epidermal growth factor receptor in adenocarcinoma with a bronchioloalveolar carcinoma component. Clin Lung Cancer. 2010;11:160–168. doi: 10.3816/CLC.2010.n.020. [DOI] [PubMed] [Google Scholar]
- 23.Nakano H, Soda H, Takasu M, et al. Heterogeneity of epidermal growth factor receptor mutations within a mixed adenocarcinoma lung nodule. Lung Cancer. 2008;60:136–140. doi: 10.1016/j.lungcan.2007.08.021. [DOI] [PubMed] [Google Scholar]
- 24.Chang YL, Wu CT, Lin SC, et al. Clonality and prognostic implications of p53 and epidermal growth factor receptor somatic aberrations in multiple primary lung cancers. Clin Cancer Res. 2007;13:52–58. doi: 10.1158/1078-0432.CCR-06-1743. [DOI] [PubMed] [Google Scholar]
- 25.Ma ES, Cheng PN, Wong CL, et al. Synchronous primary lung cancer and epidermal growth factor receptor mutation. Ann Thorac Surg. 2010;90:e38–e39. doi: 10.1016/j.athoracsur.2010.06.042. [DOI] [PubMed] [Google Scholar]
- 26.Chung JH, Choe G, Jheon S, et al. Epidermal growth factor receptor mutation and pathologic-radiologic correlation between multiple lung nodules with ground-glass opacity differentiates multicentric origin from intrapulmonary spread. J Thorac Oncol. 2009;4:1490–1495. doi: 10.1097/JTO.0b013e3181bc9731. [DOI] [PubMed] [Google Scholar]
- 27.Girard N, Deshpande C, Azzoli CG, et al. Use of epidermal growth factor receptor/Kirsten rat sarcoma 2 viral oncogene homolog mutation testing to define clonal relationships among multiple lung adenocarcinomas: Comparison with clinical guidelines. Chest. 2010;137:46–52. doi: 10.1378/chest.09-0325. [DOI] [PubMed] [Google Scholar]
- 28.Sequist LV, Waltman BA, Dias-Santagata D, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med. 2011;3:75ra26. doi: 10.1126/scitranslmed.3002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oxnard GR, Arcila ME, Chmielecki J, et al. New strategies in overcoming acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in lung cancer. Clin Cancer Res. 2011;17:5530–5537. doi: 10.1158/1078-0432.CCR-10-2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheng H, An SJ, Dong S, et al. Molecular mechanism of the schedule-dependent synergistic interaction in EGFR-mutant non-small cell lung cancer cell lines treated with paclitaxel and gefitinib. J Hematol Oncol. 2011;4:5. doi: 10.1186/1756-8722-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]