Abstract
DEAD-box proteins are vitally important to cellular processes and make up the largest class of helicases. Many DEAD-box proteins function as RNA chaperones by accelerating structural transitions of RNA, which can result in the resolution of misfolded conformers or conversion between functional structures. While the biological importance of chaperone proteins is clear, their mechanisms are incompletely understood. Here we illustrate how the catalytic activity of certain RNAs can be used to measure RNA chaperone activity. By measuring the amount of substrate converted to product, the fraction of catalytically active molecules is measured over time, providing a quantitative measure of the formation or loss of native RNA. The assays are described with references to group I and group II introns and their ribozyme derivatives, and examples are included that illustrate potential complications and indicate how catalytic activity measurements can be combined with physical approaches to gain insights into the mechanisms of DEAD-box proteins as RNA chaperones.
Keywords: catalytic RNA, intron, ribozyme, chaperone, RNA folding, DEAD-box, helicase
1. INTRODUCTION
1.1. Large RNAs become trapped in non-native conformations and fold slowly
To carry out their cellular duties, many RNAs must fold into functional three-dimensional structures. However, the journey to the native state includes opportunities to go astray and populate inactive, misfolded structures (Russell, 2008; Treiber and Williamson, 1999). With only four standard bases, RNA is prone to forming non-native secondary structures (Herschlag, 1995), which can persist on the time scale of hours to give ‘kinetically trapped’ folding intermediates (Treiber and Williamson, 1999), and the same basic issue of local stability can apply to modular tertiary contacts (Russell, 2008). Slow folding to functional structure is a common problem among RNAs, as nearly every large RNA that has been studied misfolds into at least one inactive, non-native structure (Shcherbakova et al., 2008; Treiber and Williamson, 1999). In light of these intrinsic features of RNA, it is not surprising that cells can take action to rescue incorrectly folded RNAs.
1.2. DEAD-box helicase proteins function as RNA chaperones
A major way that cells promote RNA folding is through chaperone proteins, which accelerate escape from kinetically trapped intermediates (Lorsch, 2002; Mohr et al., 2002; Russell, 2008; Schroeder et al., 2004; Zemora and Waldsich, 2010). DEAD-box proteins are a major group of RNA chaperones and comprise the largest family of superfamily 2 helicases (Fairman-Williams et al., 2010; Linder, 2006). They consist of a conserved helicase core made up of two RecA-like domains that may be flanked by additional domains. DEAD-box proteins bind RNA and ATP via the helicase core and are thought to promote RNA structural transitions by using the ATPase cycle to achieve tight, yet regulated RNA binding in a mode that can disrupt secondary structure (Hilbert et al., 2009; Jarmoskaite and Russell, 2011; Potratz et al., 2010). These disruptions are used to rescue RNAs from misfolded non-native structures and to convert RNAs between multiple functional structures. The prominent role played by DEAD-box proteins is highlighted by the fact that essentially every process known to be carried out by a structured RNA requires at least one DEAD-box or related protein (Fairman-Williams et al., 2010; Jankowsky and Bowers, 2006; Linder, 2006, 2008). Some DEAD-box proteins have been shown to act as general RNA chaperones, interacting with multiple RNA substrates nonspecifically to promote native folding (Del Campo et al., 2009; Halls et al., 2007; Huang et al., 2005; Mohr et al., 2002).
In this article we describe how to harness the power of RNA catalytic activity to study protein-assisted RNA folding. Rather than listing detailed protocols, which are likely to be different for individual proteins and RNAs, we describe the basic concepts that underlie the experimental design, and we address major issues that can complicate data analysis. For further details on basic issues that arise when using catalytic activity to measure native RNA folding, we refer readers to an earlier work (Wan et al., 2010a). We also refer to examples that illustrate how catalytic activity has been used, alone and in conjunction with complementary approaches, to gain insights into the mechanisms of DEAD-box proteins as RNA chaperones.
2. CATALYTIC ACTIVITY AS A PROBE OF RNA FOLDING
To track productive folding of an RNA, a signal must exist that allows the native state of the RNA to be distinguished from all other conformations. Functional assays provide a powerful probe, because even folding intermediates with extensive native structure can be readily distinguished if they are unable to function. Catalytic RNAs are well suited for this purpose, as their native states can be readily detected by monitoring chemical conversion of a substrate to its corresponding product.
2.1. Catalytic activity distinguishes the native state from all other conformations
The central goal of using catalytic activity to monitor RNA folding is to measure the fraction of the RNA that is present in the native state unambiguously and quantitatively. This information can then be combined with other approaches to gain insights into the structural properties of folding intermediates. In the case of the ribozyme derived from a group I intron in Tetrahymena thermophila, a misfolded conformation exists that contains the full set of native secondary and tertiary contacts and is difficult to distinguish from the native state by physical approaches (Russell et al., 2006; Wan et al., 2010b). However, it is straightforward to distinguish between the two structures in a catalytic activity assay because the natively folded ribozyme, but not the misfolded ribozyme, can cleave an oligonucleotide substrate in trans. Using this assay, the transition from the misfolded state to the native state has been measured (Chadee et al., 2010; Russell et al., 2006; Russell and Herschlag, 2001; Wan et al., 2010b).
2.2. Catalytic activity can be used to study chaperone-assisted folding
The ability to track native RNA folding allows the influence of chaperone proteins to be monitored. While this chapter highlights group I and group II introns and the DEAD-box helicase chaperone proteins, the general techniques can be applied to a broad range of catalytically active RNAs and chaperone proteins. Indeed, RNA catalytic activity of the hammerhead ribozyme was used nearly two decades ago to study chaperone activities of the HIV NC protein (Herschlag et al., 1994; Tsuchihashi et al., 1993), and group I introns have been used to monitor in vitro and in vivo chaperone activities of a variety of RNA binding proteins (Clodi et al., 1999; Rajkowitsch et al., 2005; Semrad et al., 2004; Semrad and Schroeder, 1998).
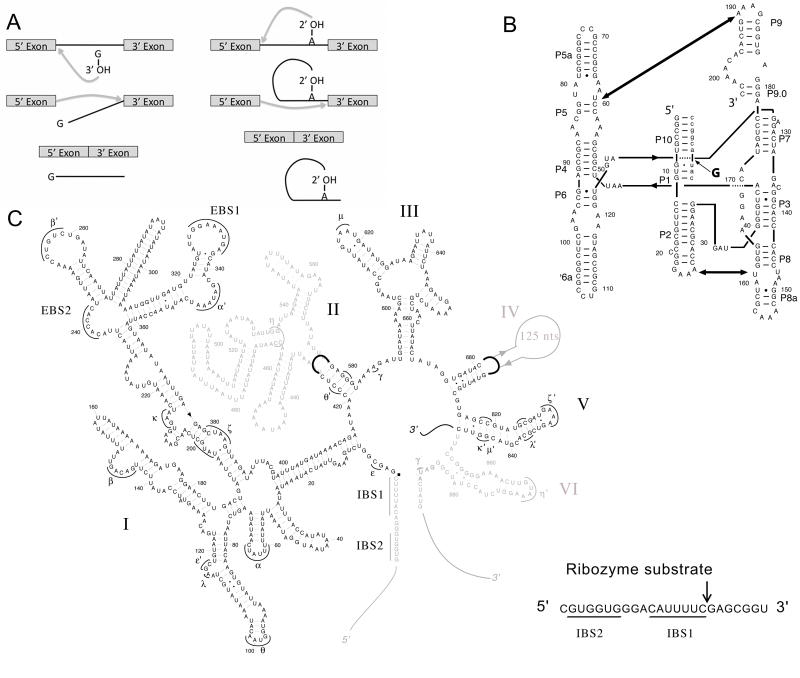
Group I and group II introns are mobile genetic elements that catalyze their own excision from precursor RNA via two transesterification splicing reactions (Fig. 1A). In order for these splicing events to occur, the RNA must fold to an active three-dimensional conformation. Introns can be converted to ribozymes with constructs that lack the exons (Fig. 1B,C). Ribozymes of group I and group II introns cleave oligonucleotide substrates that include a 5′ splice site in reactions that mimic the first step of splicing (Kuo et al., 1999; Qin and Pyle, 1997; Swisher et al., 2001; Tanner and Cech, 1996; Zaug and Cech, 1986; Zaug et al., 1988).
Figure 1.
Group I and group II introns. A, Self-splicing reactions. Left panel, Group I intron splicing reaction. An exogenous guanosine attacks the 5′ splice site in the first step, and the 5′ exon attacks the 3′ splice site in the second step. Right panel, Group II intron splicing reaction. A bulged adenosine near the 3′ end of the intron attacks the 5′ splice site, generating a lariat intermediate. The 5′ exon then attacks the 3′ splice site to ligate the exons together. B, Secondary structure of the Azoarcus group I intron ribozyme. The nine-nucleotide substrate is lowercase. The cleavage site is indicated by a thin arrow, and two tertiary contacts are indicated by thick arrows. C, Secondary structure of the aI5γ group II intron from S. cerevisiae (Swisher et al., 2001). The domains shown in black are present in the D135 ribozyme. Tertiary interactions are indicated by greek letters. Interaction sites between exon and intron sequences are indicated by the abbreviations IBS and EBS (Intron Binding Site and Exon Binding Site, respectively). The 24-nt substrate is shown at the right and the cleavage site is indicated by an arrow.
Group I and group II introns and their corresponding ribozyme constructs are well suited for studying chaperone-assisted RNA folding because they have extensive networks of secondary and tertiary structures (Fig. 1B,C), and RNAs from both groups have been shown to fold slowly and to accumulate intermediates (Emerick et al., 1996; Fedorova et al., 2010; Fedorova et al., 2007; Karunatilaka et al., 2010; Russell and Herschlag, 1999; Sinan et al., 2011; Steiner et al., 2008; Walstrum and Uhlenbeck, 1990; Zhang et al., 2005). These RNAs are sufficiently complex to include diverse sets of intermediates and corresponding kinetic barriers during folding, allowing detailed probing of the abilities of chaperones to assist in overcoming these barriers, while remaining simple enough to deeply probe the folding processes and pathways.
3. SELF-SPLICING AS A READOUT FOR NATIVE STATE FORMATION
A straightforward way of measuring native folding is to follow self-processing of intron-containing constructs after folding is initiated by addition of Mg2+. The reaction can be followed in the format of a continuous assay, in which folding and the catalytic steps take place concurrently and in the same reaction. If the rate of native RNA structure formation is slower than the subsequent catalytic steps, the observed splicing rate provides a good measure of the rate of folding to the native state.
It is common practice to body label the RNA to visualize all the products of intron splicing. This can be accomplished by including a radiolabeled nucleotide, commonly [α-32P] UTP, to label nucleotides throughout the RNA (Huang et al., 2005; Mohr et al., 2006; Pyle and Green, 1994). The reaction is then monitored by denaturing polyacrylamide gel electrophoresis, which allows the unspliced precursor to be separated from the splicing products (Fig. 2A). The loss of precursor RNA over time is quantitated to indicate the observed rate of splicing, and in turn folding to the native state (Del Campo et al., 2009; Mohr et al., 2006; Potratz et al., 2011) (Fig. 2B).
Figure 2.
Self-splicing constructs. A, Denaturing gel showing splicing products for a group II intron: (from top to bottom) lariat intron, unspliced precursor, linear intron, and spliced exons. B, Simulated plot of a splicing reaction showing the fraction of precursor as a function of time. The simulated data are fit by a single exponential curve to obtain rate constants for splicing in the presence (circles) and absence (diamonds) of protein. C, Simulated plot showing how the observed splicing rate varies with protein concentration. The rising linear portion of the data (circles) is fit with a line to obtain a second order rate constant for chaperone-accelerated folding. The plateau and decrease in rate (diamonds in gray area) reflect inhibition by the chaperone at higher concentrations (see section 3.1).
3.1. Interpreting chaperone-promoted changes in observed splicing rate
A simple reaction scheme for intron folding and splicing is shown below and is instructive for understanding how a chaperone protein affects the splicing process.
To identify whether an RNA helicase influences folding, the observed rate constant (kobserved) is compared in the absence and presence of multiple protein concentrations and in the presence and absence of ATP. If the helicase increases the rate of folding to the native state (kfold), most likely by accelerating resolution of one or more intermediates (I), this increase will lead to an increase in kobserved if the splicing rate constant (ksplice) is larger than the folding rate constant. For introns with robust self-splicing activity this condition is generally met because large RNAs usually fold slowly in vitro (Pan and Russell, 2010; Shcherbakova et al., 2008).
The observed rate constant is typically plotted as a function of protein concentration to determine the efficiency with which the chaperone promotes folding (Fig. 2C). The observed rate constant may increase with protein concentration at low concentrations and then level or begin to decrease at higher protein concentrations (Fig. 2C). Possible physical sources for this inhibition are trapping of intermediate conformations in a protein-bound state (shown in Scheme 1 as a single intermediate (I) for simplicity) and chaperone-induced unfolding of the native state. Although unfolding of native RNAs may be physiologically important for RNAs that must cycle between functional structures, this inhibition complicates analysis of the effects of the chaperone on the folding process toward the native state. Thus it is most straightforward to interpret the data in terms of protein-accelerated folding at relatively low protein concentrations where these additional effects are minimized (Fig. 2C).
Scheme 1.
3.2. Potential complications
Since the chaperone is present during the splicing reaction, it could affect the catalytic steps of splicing (ksplice) in addition to the folding steps. If the catalytic steps (ksplice) are rate-limiting, influencing them will alter the overall rate constant (kobserved) even if the protein does not affect the rate of native structure formation (kfold). However, this will not be an issue provided that folding remains slower than splicing. Group I introns allow the rate-limiting step to be determined. A folding incubation is performed in the absence of the exogenous guanosine cofactor and then guanosine is added to permit splicing. If this reaction gives a larger splicing rate than the reaction in which guanosine was present continuously, it indicates that folding is rate limiting.
A second issue concerns the multi-step nature of the splicing process. Although it is shown as a single step for simplicity in scheme 1, some group I and group II introns display rapid, reversible first steps of splicing, giving accumulation of intermediates in a fast phase that is followed by slower completion of the second step and formation of products (Chin and Pyle, 1995; Golden and Cech, 1996; Karbstein et al., 2002; Woodson and Cech, 1989). If native folding is slower than the reversible first step but faster than the second step, the precursor will be lost with an initial rate constant that reflects folding, as in the simple case with a more rapid second step. A slow step will result in additional loss of precursor. This loss could mistakenly be assigned to an additional, slower folding pathway of the precursor, whereas it actually reflects completion of the second splicing step. This behavior can be identified by the appearance and subsequent disappearance of intermediates representing the products of the first step, i.e. the free 5′ exon and the intron-3′-exon.
A final issue concerns the choice of RNA constructs. Although emphasis is typically on the intron for folding, exon length and composition can cause substantial effects (Nolte et al., 1998; Potratz et al., 2011; Zingler et al., 2010). The exons may misfold themselves and/or stabilize structure within the intron, and either of these effects may be important biologically. Care must be taken when comparing results from constructs that differ in the properties of the exons.
4. SUBSTRATE CLEAVAGE AS A READOUT FOR NATIVE STATE FORMATION
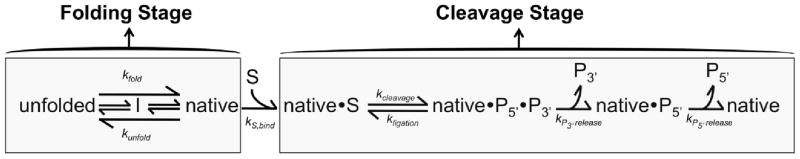
Many of the potential complications associated with using self-splicing constructs in continuous assays can be avoided by using ribozyme versions. Lacking exons, ribozyme constructs allow interpretation of all folding processes to reflect the intron domains. Further, although ribozyme versions can be used in continuous assays, the fact that they cleave oligonucleotide substrates in trans makes them well suited for discontinuous assays, in which folding and catalytic activity are separated into two discrete stages (Fig. 3A). The main advantage of this separation is the ability to directly assess the fraction of native ribozyme during folding. This allows productive folding to be dissected and unfolding of the native structure to be probed (Sect. 5.1). In addition, the discontinuous assay permits the use of folding conditions that do not support robust catalysis, an option not available using a continuous assay.
Figure 3.
The discontinuous assay. A, Reaction schematic. The ribozyme folds in the first stage, and then it cleaves the oligonucleotide substrate (S) in the second stage under reaction conditions that block further native folding. B, Denaturing gel showing the results of cleavage of a 5′ labeled oligonucleotide substrate by native ribozyme. C, Simulated plot of multiple cleavage reactions representing different folding times prior to the cleavage reaction. The curves with larger bursts represent longer folding times, giving greater accumulation of native ribozyme. The amplitude values are shown with filled symbols. D, Simulated plot showing the fraction of native ribozyme (fN) plotted as a function of folding time (t1). The burst amplitudes from simulated cleavage reactions in panel C are plotted as a function of folding time. In this simulated scenario, the folding progress can be fit by a single exponential function, giving a single rate constant for native state formation.
4.1. Setting up a discontinuous assay: folding and catalysis stages
The discontinuous assay is composed of two stages, the folding stage (stage 1) and the catalysis (or cleavage) stage (stage 2) (Fig. 3A). The folding stage contains the ribozyme alone or with the chaperone under conditions desired for chaperone-assisted RNA folding. To prevent catalytic activity at this stage, the oligonucleotide substrate is omitted. In the catalysis stage, conditions are changed such that further folding to the native state is blocked, and radiolabeled oligonucleotide substrate is added, typically in small excess of the ribozyme (2–3-fold over the ribozyme). The cleavage reaction is allowed to proceed for various times (t2), and the substrate and product from each time point are separated on a denaturing polyacrylamide gel (Fig. 3B). The fraction of product, normalized by the substrate and ribozyme concentrations, is plotted against cleavage time (t2) (Fig. 3B,C). Most commonly, the cleavage stage produces a burst of product formation, with the amplitude reflecting the fraction of ribozyme in the native state (Sect. 4.2, below). This fraction increases as a function of time spent in the folding stage (t1), giving a rate constant for native folding of the ribozyme (Fig. 3D). After initial experiments have been performed and the cleavage rate constant is known, a single time point in the cleavage reaction (stage 2) may be sufficient to determine the burst amplitude (see Fig. 4A). This timepoint should be chosen after completion of the burst phase, but before significant contribution from subsequent turnovers (solid symbols in Fig. 3C).
Figure 4.
Examples of catalytic reactions with the D135 and Azoarcus ribozymes. A, Identifying conditions that block folding for the catalysis stage of the discontinuous assay. Comparing burst amplitudes resulting from D135 ribozyme placed into stage 2 conditions with (circles) or without (triangles) a prior incubation in Mg2+-containing buffer to allow prefolding to the native state. The burst is much smaller without the preincubation, indicating that these conditions for stage 2 (pH 8.0, 100 mM Mg2+, 500 mM KCl, 15 °C) effectively block folding. The solid circle indicates how a single time point can be sufficient to determine the fraction of native ribozyme if the kinetics of cleavage are known. B, Ribozyme prefolding to the native state. The burst amplitudes from cleavage reactions of the Azoarcus ribozyme are smaller than they would be for stoichiometric product formation, and further work showed that this results from an equilibrium between substrate cleavage and ligation (Sinan et al., 2011). Note also that the bursts from ribozyme prefolded at 37 °C and 10 mM Mg2+ for 15–45 min are identical, suggesting that 15 minutes is sufficient for complete folding (see section 4.1). Panel A reprinted from (Potratz et al., 2011) with permission from Elsevier. Panel B adapted from (Sinan et al., 2011). This research was originally published in Journal of Biological Chemistry. Selma Sinan, Xiaoyan Yuan, and Rick Russell. The Azoarcus Group I Intron Ribozyme Misfolds and Is Accelerated for Refolding by ATP-dependent RNA Chaperone Proteins. JBC. 2011; 286:37304–37312. © the American Society for Biochemistry and Molecular Biology
A key advantage of the discontinuous assay is that any set of conditions can be used for the folding stage. However, setting up the catalysis stage requires care, as conditions must support enzymatic activity while blocking further native folding. This is because ribozyme that reached the native state during stage 2 would produce cleavage products and cause the calculated fraction of native ribozyme to be artificially high.
The ability of the catalysis stage to prohibit folding is probed by comparing cleavage reactions from a ribozyme that has been prefolded to the native state and a ribozyme that is transferred directly to the catalysis stage, omitting the folding step. (In practice, testing for suitable conditions for the catalysis stage can be undertaken simultaneously with learning how to prefold ribozyme to the native state, as described below.) The burst amplitude of product from ribozyme that skipped the folding stage should be much less than that from the prefolded ribozyme, indicating little formation of native ribozyme during the cleavage reaction. It may not be possible to completely block native folding in stage 2, but a relatively small fraction of native ribozyme produced in this stage can be accounted for by normalization (Potratz et al., 2011).
Although optimal conditions for stage 2 are likely to be different for different catalytic RNAs, some general guidelines are applicable. A study using a ribozyme engineered from the group II intron aI5γ from S. cerevisiae, D135 (Fig. 1C), used a high pH in the catalysis stage to accelerate cleavage, and high Mg2+ concentration (100 mM) and low temperature (15 °C ) to enhance the arrest of folding (Fig. 4A) (Potratz et al., 2011). Cleavage is not particularly fast under these conditions (10−2 min−1). However, slow cleavage is acceptable for the catalysis stage in a discontinuous assay as long as cleavage is significantly faster than folding. In addition, proteinase K is included at the catalysis stage to ensure that protein transferred from the folding stage has no effect on substrate cleavage.
To compare a ribozyme prefolded to the native state with an unfolded ribozyme, it must be known how to fold the ribozyme to the native state. Establishing how to fold the ribozyme to the native state can be undertaken concurrently with exploring conditions to use for the catalysis stage. Folding of a ribozyme (typically by adding Mg2+) is initiated and aliquots are removed at different times from the folding stage. These aliquots are transferred to stage 2 conditions in the presence of a small excess of substrate. The product burst amplitudes are plotted against folding time. When the burst amplitude no longer increases as a function of folding time, the ribozyme has been folded to the native state as fully as it can be under that set of conditions (Fig. 4B). The folding time should be varied widely to determine whether there are slower folding pathways that give additional native ribozyme formation on longer time scales.
4.2. Interpreting results from the catalysis stage
To make optimal use of the discontinuous assay, it is critical to interpret the burst amplitude quantitatively in order to determine the fraction of native ribozyme. Selected examples from work involving chaperones will be covered below, and a more thorough guide to interpreting results from catalytic rate measurements was published in this series last year (Wan et al., 2010a).
The interpretation of the burst amplitudes depends on the relative rate constants of different steps in the catalytic cycle, which is shown in Scheme 2.
Scheme 2.
In general, the cleavage stage is performed under multiple turnover conditions, and the fraction of product is normalized by the substrate and ribozyme concentrations, giving the ratio or product to ribozyme. This ratio is plotted against cleavage time (t2) (Fig. 3B,C). The reaction products are bound by base-pairing, with or without additional tertiary contacts, and for many ribozymes their dissociation rate constants (kP5′release and kP3′release) are small and limit the rates of subsequent turnovers. In all, there are three possible regimes: (1) both products are released quickly relative to the catalytic step (kcleavage); (2) both are released slowly; (3) one is released quickly and the other slowly (Wan et al., 2010a). Regimes 2 and 3 result in bursts of product formation, and the amplitude of this burst can be used to calculate the fraction of native ribozyme, as described in two examples.
The D135 ribozyme was used in a catalytic reaction with three-fold excess substrate. When performed with the ribozyme prefolded to the native state, the cleavage stage resulted in a burst of product with an amplitude approximately equal to one turnover of the ribozyme (Fig. 4A) (Potratz et al., 2011). This is consistent with regime 3, in which there is a burst of product formation with a rate constant equal to the cleavage rate (kcleavage) and a subsequent linear phase dictated by the slow release of one of the products. Because the 5′ product of the oligonucleotide substrate forms twelve Watson-Crick base pairs with the ribozyme, it is likely to be the product that is released slowly. Under this reaction regime, the amplitude of the burst phase is equal to the fraction of D135 ribozyme folded to the native state.
The ribozyme engineered from a group I intron from Azoarcus evansii (Fig. 1B) was used in a catalytic reaction with two-fold excess substrate (Sinan et al., 2011). When the ribozyme was prefolded to the native state, the cleavage stage resulted in a burst of product with an amplitude of half to three-fourths of the ribozyme concentration, depending on solution conditions (Fig. 4B). This is consistent with regime 2, in which the slow release of both products allows equilibrium between the cleavage (kcleavage) and ligation (kligation) to be reached. The burst amplitude is smaller than the fraction of native ribozyme and must be corrected by the value of the internal equilibrium to reveal the fraction of native ribozyme.
4.3. Using the discontinuous assay to probe chaperone-assisted folding
When optimal conditions for stage 2 are established and the relationship of the burst amplitude to the fraction of native ribozyme is understood, the discontinuous assay can provide important insights into chaperone-assisted RNA folding. The most straightforward experiment is to compare the folding reaction in the presence of various concentrations of chaperone, plotting the fraction of native ribozyme against time (t1) (Fig. 3D). A chaperone may increase the rate of native state formation (Sinan et al., 2011; Tijerina et al., 2006), an effect that most likely arises from accelerated resolution of one or more kinetically-trapped intermediates. This interpretation is particularly clear for RNAs that are known to misfold (Pan and Woodson, 1998; Russell and Herschlag, 1999; Sinan et al., 2011; Tijerina et al., 2006). Further confirmation can be obtained by allowing the RNA to misfold first and then adding the chaperone (Sinan et al., 2011; Tijerina et al., 2006).
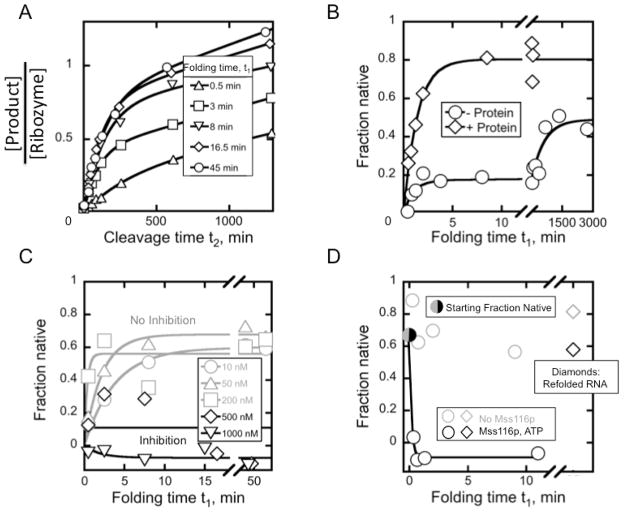
It is possible for a chaperone to increase the fraction of ribozyme that reaches the native state rapidly without having a significant impact on the observed rate constant (Fig. 5A,B) (Potratz et al., 2011). This effect may arise from an influence of the chaperone early in folding, which decreases the probability of misfolding at later folding steps. Alternatively, resolution of an intermediate can become sufficiently fast that this pathway is then indistinguishable from pathways that avoid the intermediate.
Figure 5.
The discontinuous assay with the D135 ribozyme and the DEAD-box helicase Mss116p. A, Cleavage time courses initiated with ribozyme folded in the presence of Mss116p for the times indicated. The increase of the burst amplitude with folding time (t1) indicates productive folding to the native state. B, Comparison of ribozyme folding in the presence (diamonds) and absence (circles) of Mss116p. The protein changes the folding profile from multi-phasic to a single exponential phase. C, High Mss116p concentrations inhibit the native conformation. At 500 nM (diamonds) and 1000 nM (inverted triangles) Mss116p, the fraction of native ribozyme at steady state is lower than in the presence of lower protein concentrations (gray symbols). D, The native conformation of D135 can be unfolded by Mss116p. Ribozyme prefolded to the native state (black/gray solid circle) is divided into reaction tubes containing a high concentration of Mss116p (black circles) or no Mss116p (gray circles). After approximately 10 min, the protein is proteolyzed and the ribozyme is treated to allow it to refold to the native state (black and gray diamonds), demonstrating that the loss of native ribozyme arises from unfolding and not an irreversible process. Panels A–D adapted from (Potratz et al., 2011) with permission from Elsevier.
Analogous to effects on self-splicing constructs, higher protein concentrations can inhibit folding of ribozymes to the native state (Fig. 5C). The physical processes responsible for inhibition are presumably the same for ribozymes and self-splicing constructs, but the discontinuous assay allows the origins of the inhibition to be distinguished. For self-splicing constructs, inhibition by trapping protein-bound nonfunctional intermediates and by unfolding native RNA both lead to a decrease in the observed splicing rate (Fig. 2C). In a discontinuous assay unfolding of native ribozyme decreases the endpoint of the folding curve (Fig. 5C). Trapping of folding intermediates, without unfolding of natively-folded RNA, would result in a decrease in the rate of native ribozyme formation but not a decrease in the endpoint.
5. Other applications of the discontinuous assay
The discontinuous assay is amenable to a diverse set of experiments. It can be used to monitor a decrease in the native ribozyme, and an additional stage can be included to probe the role of ATP in chaperone-mediated folding (Halls et al., 2007; Potratz et al., 2011). Further, the progress of native ribozyme formation obtained from the assay can also complement insight from other powerful physical approaches, and thus provide a more complete understanding of the action of RNA chaperones.
5.1. Unfolding native structure
Many chaperones function non-specifically and are capable of disrupting the native states of RNAs as well as misfolded states (Bhaskaran and Russell, 2007). For probing the mechanisms of chaperone activity in structure disruptions, it can be very useful to monitor the native state because it is relatively homogeneous and the structure may be known. In contrast, folding intermediates may be heterogeneous and their structures poorly defined.
To monitor loss of the native ribozyme, the ribozyme is first pre-folded to the native state, and then the chaperone protein is added. Even for general chaperones, the level of activity may be reduced for the native structure because it is typically highly stable, so it may be necessary to lower the Mg2+ concentration and/or to use relatively high protein concentrations to detect net unfolding. A decrease in the fraction of native ribozyme over time indicates that the chaperone protein has mediated at least partial unfolding of the native ribozyme, giving intermediates that do not readily refold to the native state upon transfer to the stage 2 conditions. To ensure that the loss of native ribozyme reflects reversible unfolding, the protein should be proteolyzed and the ribozyme again folded to the native state (Fig. 5D) (Potratz et al., 2011).
5.2. Integrating results with other methods
The materials and methods required to employ the use of RNA catalytic activity are standard for many laboratories, and the ease of implementing this method makes it a convenient tool for obtaining a kinetic view of the fraction of native ribozyme. Results can be highly complementary to those from methods that provide physical information on folding intermediates.
Three physical probes that have been used extensively for RNA folding studies are chemical footprinting, small angle X-ray scattering (SAXS), and single-molecule FRET. Time-resolved chemical footprinting can be performed with several different probes and provides a highly specific view of nucleotides that are engaged in secondary or tertiary contacts during a folding process (King et al., 1993; Mortimer and Weeks, 2007, 2008, 2009; Sclavi et al., 1997; Tijerina et al., 2007). Hydroxyl radical footprinting has been particularly valuable for probing structured intermediates and elucidating folding pathways (Laederach et al., 2007; Laederach et al., 2006; Sclavi et al., 1998). The orthogonal information provided by catalytic activity measurements – how much of the ribozyme is in the native state – is tremendously valuable because it can be used to place constraints on the folding pathways modeled from footprinting data (Laederach et al., 2007; Mitra et al., 2011). SAXS provides rich information on the overall size and shape of RNA as it folds, which is highly complementary to footprinting (Kwok et al., 2006; Schlatterer and Brenowitz, 2009), and again coordinated activity measurements under the same conditions can assist greatly in constraining physical descriptions of intermediates (Kwok et al., 2006; Laederach et al., 2007; Mitra et al., 2011; Roh et al., 2010; Sinan et al., 2011). Last, single-molecule FRET experiments are uniquely powerful for detecting and characterizing intermediates that do not accumulate in bulk experiments (Bokinsky et al., 2003; Bokinsky and Zhuang, 2005; Karunatilaka et al., 2010; Rueda et al., 2004; Zhuang, 2005; Zhuang et al., 2000), and the concurrent detection of catalytic activity can be used to tremendous advantage for distinguishing the native state from folding intermediates (Bokinsky et al., 2003; Liu et al., 2007; Rueda et al., 2004; Zhuang et al., 2000).
Acknowledgments
Research in the Russell lab is funded by grants from the NIH (GM 070456) and the Welch Foundation (F-1563).
References
- Bhaskaran H, Russell R. Kinetic redistribution of native and misfolded RNAs by a DEAD-box chaperone. Nature. 2007;449:1014–1018. doi: 10.1038/nature06235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokinsky G, Rueda D, Misra VK, Rhodes MM, Gordus A, Babcock HP, Walter NG, Zhuang X. Single-molecule transition-state analysis of RNA folding. Proc Natl Acad Sci U S A. 2003;100:9302–9307. doi: 10.1073/pnas.1133280100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokinsky G, Zhuang X. Single-molecule RNA folding. Acc Chem Res. 2005;38:566–573. doi: 10.1021/ar040142o. [DOI] [PubMed] [Google Scholar]
- Chadee AB, Bhaskaran H, Russell R. Protein roles in group I intron RNA folding: the tyrosyl-tRNA synthetase CYT-18 stabilizes the native state relative to a long-lived misfolded structure without compromising folding kinetics. J Mol Biol. 2010;395:656–670. doi: 10.1016/j.jmb.2009.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin K, Pyle AM. Branch-point attack in group II introns is a highly reversible transesterification, providing a potential proofreading mechanism for 5′-splice site selection. RNA. 1995;1:391–406. [PMC free article] [PubMed] [Google Scholar]
- Clodi E, Semrad K, Schroeder R. Assaying RNA chaperone activity in vivo using a novel RNA folding trap. EMBO J. 1999;18:3776–3782. doi: 10.1093/emboj/18.13.3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Campo M, Mohr S, Jiang Y, Jia H, Jankowsky E, Lambowitz AM. Unwinding by local strand separation is critical for the function of DEAD-box proteins as RNA chaperones. J Mol Biol. 2009;389:674–693. doi: 10.1016/j.jmb.2009.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerick VL, Pan J, Woodson SA. Analysis of rate-determining conformational changes during self-splicing of the Tetrahymena intron. Biochemistry. 1996;35:13469–13477. doi: 10.1021/bi960865i. [DOI] [PubMed] [Google Scholar]
- Fairman-Williams ME, Guenther UP, Jankowsky E. SF1 and SF2 helicases: family matters. Curr Opin Struct Biol. 2010;20:313–324. doi: 10.1016/j.sbi.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorova O, Solem A, Pyle AM. Protein-facilitated folding of group II intron ribozymes. J Mol Biol. 2010;397:799–813. doi: 10.1016/j.jmb.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorova O, Waldsich C, Pyle AM. Group II intron folding under near-physiological conditions: collapsing to the near-native state. J Mol Biol. 2007;366:1099–1114. doi: 10.1016/j.jmb.2006.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden BL, Cech TR. Conformational switches involved in orchestrating the successive steps of group I RNA splicing. Biochemistry. 1996;35:3754–3763. doi: 10.1021/bi952599z. [DOI] [PubMed] [Google Scholar]
- Halls C, Mohr S, Del Campo M, Yang Q, Jankowsky E, Lambowitz AM. Involvement of DEAD-box proteins in group I and group II intron splicing. Biochemical characterization of Mss116p, ATP hydrolysis-dependent and -independent mechanisms, and general RNA chaperone activity. J Mol Biol. 2007;365:835–855. doi: 10.1016/j.jmb.2006.09.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herschlag D. RNA chaperones and the RNA folding problem. J Biol Chem. 1995;270:20871–20874. doi: 10.1074/jbc.270.36.20871. [DOI] [PubMed] [Google Scholar]
- Herschlag D, Khosla M, Tsuchihashi Z, Karpel RL. An RNA chaperone activity of non-specific RNA binding proteins in hammerhead ribozyme catalysis. EMBO J. 1994;13:2913–2924. doi: 10.1002/j.1460-2075.1994.tb06586.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilbert M, Karow AR, Klostermeier D. The mechanism of ATP-dependent RNA unwinding by DEAD box proteins. Biol Chem. 2009;390:1237–1250. doi: 10.1515/BC.2009.135. [DOI] [PubMed] [Google Scholar]
- Huang HR, Rowe CE, Mohr S, Jiang Y, Lambowitz AM, Perlman PS. The splicing of yeast mitochondrial group I and group II introns requires a DEAD-box protein with RNA chaperone function. Proc Natl Acad Sci U S A. 2005;102:163–168. doi: 10.1073/pnas.0407896101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankowsky E, Bowers H. Remodeling of ribonucleoprotein complexes with DExH/D RNA helicases. Nucleic Acids Res. 2006;34:4181–4188. doi: 10.1093/nar/gkl410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarmoskaite I, Russell R. DEAD-box proteins as RNA helicases and chaperones. WIREs RNA. 2011;2:135–152. doi: 10.1002/wrna.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karbstein K, Carroll KS, Herschlag D. Probing the Tetrahymena group I ribozyme reaction in both directions. Biochemistry. 2002;41:11171–11183. doi: 10.1021/bi0202631. [DOI] [PubMed] [Google Scholar]
- Karunatilaka KS, Solem A, Pyle AM, Rueda D. Single-molecule analysis of Mss116-mediated group II intron folding. Nature. 2010;467:935–939. doi: 10.1038/nature09422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King PA, Jamison E, Strahs D, Anderson VE, Brenowitz M. ‘Footprinting’ proteins on DNA with peroxonitrous acid. Nucleic Acids Res. 1993;21:2473–2478. doi: 10.1093/nar/21.10.2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo LY, Davidson LA, Pico S. Characterization of the Azoarcus ribozyme: tight binding to guanosine and substrate by an unusually small group I ribozyme. Biochim Biophys Acta. 1999;1489:281–292. doi: 10.1016/s0167-4781(99)00200-6. [DOI] [PubMed] [Google Scholar]
- Kwok LW, Shcherbakova I, Lamb JS, Park HY, Andresen K, Smith H, Brenowitz M, Pollack L. Concordant exploration of the kinetics of RNA folding from global and local perspectives. J Mol Biol. 2006;355:282–293. doi: 10.1016/j.jmb.2005.10.070. [DOI] [PubMed] [Google Scholar]
- Laederach A, Shcherbakova I, Jonikas MA, Altman RB, Brenowitz M. Distinct contribution of electrostatics, initial conformational ensemble, and macromolecular stability in RNA folding. Proc Natl Acad Sci U S A. 2007;104:7045–7050. doi: 10.1073/pnas.0608765104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laederach A, Shcherbakova I, Liang MP, Brenowitz M, Altman RB. Local kinetic measures of macromolecular structure reveal partitioning among multiple parallel pathways from the earliest steps in the folding of a large RNA molecule. J Mol Biol. 2006;358:1179–1190. doi: 10.1016/j.jmb.2006.02.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder P. Dead-box proteins: a family affair--active and passive players in RNP-remodeling. Nucleic Acids Res. 2006;34:4168–4180. doi: 10.1093/nar/gkl468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder P. mRNA export: RNP remodeling by DEAD-box proteins. Curr Biol. 2008;18:R297–299. doi: 10.1016/j.cub.2008.02.027. [DOI] [PubMed] [Google Scholar]
- Liu S, Bokinsky G, Walter NG, Zhuang X. Dissecting the multistep reaction pathway of an RNA enzyme by single-molecule kinetic “fingerprinting”. Proc Natl Acad Sci U S A. 2007;104:12634–12639. doi: 10.1073/pnas.0610597104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorsch JR. RNA chaperones exist and DEAD box proteins get a life. Cell. 2002;109:797–800. doi: 10.1016/s0092-8674(02)00804-8. [DOI] [PubMed] [Google Scholar]
- Mitra S, Laederach A, Golden BL, Altman RB, Brenowitz M. RNA molecules with conserved catalytic cores but variable peripheries fold along unique energetically optimized pathways. RNA. 2011;17:1589–1603. doi: 10.1261/rna.2694811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr S, Matsuura M, Perlman PS, Lambowitz AM. A DEAD-box protein alone promotes group II intron splicing and reverse splicing by acting as an RNA chaperone. Proc Natl Acad Sci U S A. 2006;103:3569–3574. doi: 10.1073/pnas.0600332103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr S, Stryker JM, Lambowitz AM. A DEAD-box protein functions as an ATP-dependent RNA chaperone in group I intron splicing. Cell. 2002;109:769–779. doi: 10.1016/s0092-8674(02)00771-7. [DOI] [PubMed] [Google Scholar]
- Mortimer SA, Weeks KM. A fast-acting reagent for accurate analysis of RNA secondary and tertiary structure by SHAPE chemistry. J Am Chem Soc. 2007;129:4144–4145. doi: 10.1021/ja0704028. [DOI] [PubMed] [Google Scholar]
- Mortimer SA, Weeks KM. Time-resolved RNA SHAPE chemistry. J Am Chem Soc. 2008;130:16178–16180. doi: 10.1021/ja8061216. [DOI] [PubMed] [Google Scholar]
- Mortimer SA, Weeks KM. Time-resolved RNA SHAPE chemistry: quantitative RNA structure analysis in one-second snapshots and at single-nucleotide resolution. Nat Protoc. 2009;4:1413–1421. doi: 10.1038/nprot.2009.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolte A, Chanfreau G, Jacquier A. Influence of substrate structure on in vitro ribozyme activity of a group II intron. RNA. 1998;4:694–708. doi: 10.1017/s1355838298980165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan C, Russell R. Roles of DEAD-box proteins in RNA and RNP Folding. RNA Biol. 2010;7:667–676. doi: 10.4161/rna.7.6.13571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J, Woodson SA. Folding intermediates of a self-splicing RNA: mispairing of the catalytic core. J Mol Biol. 1998;280:597–609. doi: 10.1006/jmbi.1998.1901. [DOI] [PubMed] [Google Scholar]
- Potratz JP, Del Campo M, Wolf RZ, Lambowitz AM, Russell R. ATP-Dependent Roles of the DEAD-Box Protein Mss116p in Group II Intron Splicing In Vitro and In Vivo. J Mol Biol. 2011;411:661–679. doi: 10.1016/j.jmb.2011.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potratz JP, Tijerina P, Russell R. Mechanisms of DEAD-box proteins in ATP-dependent processes. In: Jankowsky E, editor. RNA helicases. Royal Society of Chemistry; 2010. pp. 61–98. [Google Scholar]
- Pyle AM, Green JB. Building a kinetic framework for group II intron ribozyme activity: quantitation of interdomain binding and reaction rate. Biochemistry. 1994;33:2716–2725. doi: 10.1021/bi00175a047. [DOI] [PubMed] [Google Scholar]
- Qin PZ, Pyle AM. Stopped-flow fluorescence spectroscopy of a group II intron ribozyme reveals that domain 1 is an independent folding unit with a requirement for specific Mg2+ ions in the tertiary structure. Biochemistry. 1997;36:4718–4730. doi: 10.1021/bi962665c. [DOI] [PubMed] [Google Scholar]
- Rajkowitsch L, Semrad K, Mayer O, Schroeder R. Assays for the RNA chaperone activity of proteins. Biochem Soc Trans. 2005;33:450–456. doi: 10.1042/BST0330450. [DOI] [PubMed] [Google Scholar]
- Roh JH, Guo L, Kilburn JD, Briber RM, Irving T, Woodson SA. Multistage collapse of a bacterial ribozyme observed by time-resolved small-angle x-ray scattering. J Am Chem Soc. 2010 doi: 10.1021/ja103867p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueda D, Bokinsky G, Rhodes MM, Rust MJ, Zhuang X, Walter NG. Single-molecule enzymology of RNA: essential functional groups impact catalysis from a distance. Proc Natl Acad Sci U S A. 2004;101:10066–10071. doi: 10.1073/pnas.0403575101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell R. RNA misfolding and the action of chaperones. Front Biosci. 2008;13:1–20. doi: 10.2741/2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell R, Das R, Suh H, Travers KJ, Laederach A, Engelhardt MA, Herschlag D. The paradoxical behavior of a highly structured misfolded intermediate in RNA folding. J Mol Biol. 2006;363:531–544. doi: 10.1016/j.jmb.2006.08.024. [DOI] [PubMed] [Google Scholar]
- Russell R, Herschlag D. New pathways in folding of the Tetrahymena group I RNA enzyme. J Mol Biol. 1999;291:1155–1167. doi: 10.1006/jmbi.1999.3026. [DOI] [PubMed] [Google Scholar]
- Russell R, Herschlag D. Probing the folding landscape of the Tetrahymena ribozyme: commitment to form the native conformation is late in the folding pathway. J Mol Biol. 2001;308:839–851. doi: 10.1006/jmbi.2001.4751. [DOI] [PubMed] [Google Scholar]
- Schlatterer JC, Brenowitz M. Complementing global measures of RNA folding with local reports of backbone solvent accessibility by time resolved hydroxyl radical footprinting. Methods. 2009;49:142–147. doi: 10.1016/j.ymeth.2009.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder R, Barta A, Semrad K. Strategies for RNA folding and assembly. Nat Rev Mol Cell Biol. 2004;5:908–919. doi: 10.1038/nrm1497. [DOI] [PubMed] [Google Scholar]
- Sclavi B, Sullivan M, Chance MR, Brenowitz M, Woodson SA. RNA folding at millisecond intervals by synchrotron hydroxyl radical footprinting. Science. 1998;279:1940–1943. doi: 10.1126/science.279.5358.1940. [DOI] [PubMed] [Google Scholar]
- Sclavi B, Woodson S, Sullivan M, Chance MR, Brenowitz M. Time-resolved synchrotron X-ray “footprinting”, a new approach to the study of nucleic acid structure and function: application to protein-DNA interactions and RNA folding. J Mol Biol. 1997;266:144–159. doi: 10.1006/jmbi.1996.0775. [DOI] [PubMed] [Google Scholar]
- Semrad K, Green R, Schroeder R. RNA chaperone activity of large ribosomal subunit proteins from Escherichia coli. RNA. 2004;10:1855–1860. doi: 10.1261/rna.7121704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semrad K, Schroeder R. A ribosomal function is necessary for efficient splicing of the T4 phage thymidylate synthase intron in vivo. Genes Dev. 1998;12:1327–1337. doi: 10.1101/gad.12.9.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcherbakova I, Mitra S, Laederach A, Brenowitz M. Energy barriers, pathways, and dynamics during folding of large, multidomain RNAs. Curr Opin Chem Biol. 2008;12:655–666. doi: 10.1016/j.cbpa.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinan S, Yuan X, Russell R. The Azoarcus Group I Intron Ribozyme Misfolds and Is Accelerated for Refolding by ATP-dependent RNA Chaperone Proteins. J Biol Chem. 2011;286:37304–37312. doi: 10.1074/jbc.M111.287706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner M, Karunatilaka KS, Sigel RK, Rueda D. Single-molecule studies of group II intron ribozymes. Proc Natl Acad Sci U S A. 2008;105:13853–13858. doi: 10.1073/pnas.0804034105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swisher J, Duarte CM, Su LJ, Pyle AM. Visualizing the solvent-inaccessible core of a group II intron ribozyme. EMBO J. 2001;20:2051–2061. doi: 10.1093/emboj/20.8.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner M, Cech T. Activity and thermostability of the small self-splicing group I intron in the pre-tRNA(lle) of the purple bacterium Azoarcus. RNA. 1996;2:74–83. [PMC free article] [PubMed] [Google Scholar]
- Tijerina P, Bhaskaran H, Russell R. Nonspecific binding to structured RNA and preferential unwinding of an exposed helix by the CYT-19 protein, a DEAD-box RNA chaperone. Proc Natl Acad Sci U S A. 2006;103:16698–16703. doi: 10.1073/pnas.0603127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tijerina P, Mohr S, Russell R. DMS footprinting of structured RNAs and RNA-protein complexes. Nat Protoc. 2007;2:2608–2623. doi: 10.1038/nprot.2007.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treiber DK, Williamson JR. Exposing the kinetic traps in RNA folding. Curr Opin Struct Biol. 1999;9:339–345. doi: 10.1016/S0959-440X(99)80045-1. [DOI] [PubMed] [Google Scholar]
- Tsuchihashi Z, Khosla M, Herschlag D. Protein enhancement of hammerhead ribozyme catalysis. Science. 1993;262:99–102. doi: 10.1126/science.7692597. [DOI] [PubMed] [Google Scholar]
- Walstrum SA, Uhlenbeck OC. The self-splicing RNA of Tetrahymena is trapped in a less active conformation by gel purification. Biochemistry. 1990;29:10573–10576. doi: 10.1021/bi00498a022. [DOI] [PubMed] [Google Scholar]
- Wan Y, Mitchell D, Russell R. Catalytic activity as a probe of native RNA folding. Methods in Enzymology. 2010a;468:195–218. doi: 10.1016/S0076-6879(09)68010-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y, Suh H, Russell R, Herschlag D. Multiple unfolding events during native folding of the Tetrahymena group I ribozyme. J Mol Biol. 2010b;400:1067–1077. doi: 10.1016/j.jmb.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson SA, Cech TR. Reverse self-splicing of the tetrahymena group I intron: implication for the directionality of splicing and for intron transposition. Cell. 1989;57:335–345. doi: 10.1016/0092-8674(89)90971-9. [DOI] [PubMed] [Google Scholar]
- Zaug AJ, Cech TR. The intervening sequence RNA of Tetrahymena is an enzyme. Science. 1986;231:470–475. doi: 10.1126/science.3941911. [DOI] [PubMed] [Google Scholar]
- Zaug AJ, Grosshans CA, Cech TR. Sequence-specific endoribonuclease activity of the Tetrahymena ribozyme: enhanced cleavage of certain oligonucleotide substrates that form mismatched ribozyme-substrate complexes. Biochemistry. 1988;27:8924–8931. doi: 10.1021/bi00425a008. [DOI] [PubMed] [Google Scholar]
- Zemora G, Waldsich C. RNA folding in living cells. RNA Biol. 2010;7:634–641. doi: 10.4161/rna.7.6.13554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Xiao M, Lu C, Zhang Y. Fast formation of the P3–P7 pseudoknot: a strategy for efficient folding of the catalytically active ribozyme. RNA. 2005;11:59–69. doi: 10.1261/rna.7145105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang X. Single-molecule RNA science. Annu Rev Biophys Biomol Struct. 2005;34:399–414. doi: 10.1146/annurev.biophys.34.040204.144641. [DOI] [PubMed] [Google Scholar]
- Zhuang X, Bartley LE, Babcock HP, Russell R, Ha T, Herschlag D, Chu S. A single-molecule study of RNA catalysis and folding. Science. 2000;288:2048–2051. doi: 10.1126/science.288.5473.2048. [DOI] [PubMed] [Google Scholar]
- Zingler N, Solem A, Pyle AM. Dual roles for the Mss116 cofactor during splicing of the ai5gamma group II intron. Nucleic Acids Res. 2010;38:6602–6609. doi: 10.1093/nar/gkq530. [DOI] [PMC free article] [PubMed] [Google Scholar]