Abstract
OBJECTIVE
To identify a disease locus for autosomal recessive retinitis pigmentosa in a consanguineous Pakistani family.
DESIGN
Prospective linkage study.
METHODS
Blood samples were collected and genomic DNA was extracted. A genome-wide scan was performed using 382 polymorphic microsatellite markers on genomic DNA from 4 affected and 5 unaffected family members, and logarithm of odds scores were calculated.
RESULTS
A maximum 2-point logarithm of odds score of 3.14 at θ = 0 was obtained for marker D2S165 during the genome-wide scan. Fine mapping markers identified a 20.92-cM (19.98-Mb) interval flanked by D2S149 and D2S367 that cosegregates with the disease phenotype. Haplotype analyses further refined the critical interval, distal to D2S220 in a 12.31-cM (13.35-Mb) region that does not harbor any genes that previously have been associated with retinitis pigmentosa.
CONCLUSIONS
Linkage analysis identified a new locus for autosomal recessive retinitis pigmentosa that maps to chromosome 2p22.3-p24.1 in a consanguineous Pakistani family.
The term retinitis pigmentosa (rp) was used first by the german physician Donders in 1857.1 It refers to bone spicule pigmentation in the mid-peripheral fundus simulating inflammation. Patients experience night blindness, followed by loss of peripheral visual field, and later loss of central vision often leading to complete blindness. RP primarily affects the rod photoreceptors, whereas the function of the cone receptors is compromised as the disease progresses.2 Typical fundus appearance includes attenuated arterioles, bone spicule pigmentation, and waxy pallor of the optic disc. Affected individuals often have severely abnormal or nondetectable rod responses in the electroretinography (ERG) recordings, even in the early stage of the disease.2
RP is the most common inherited retinal dystrophy, affecting approximately 1 in 5000 individuals worldwide.3,4 It may be inherited as an autosomal recessive, an autosomal dominant, or an X-linked recessive trait. According to the RetNet database (www.sph.uth.tmc.edu/Retnet), more than 50 loci have been implicated in nonsyndromic RP, of which pathogenic mutations in 38 genes have been associated with RP. These include genes encoding components of the phototransduction cascade, proteins involved in retinoid metabolism, cell–cell interaction proteins, photoreceptor structural proteins, transcription factors, intracellular transport proteins, and splicing factors. Autosomal recessive retinitis pigmentosa comprises 20% to 25% of all RP cases.5–8
Herein we report a consanguineous Pakistani family with multiple members affected by autosomal recessive RP. Initially, a genome-wide scan including exclusion of known recessive RP loci was completed. Linkage analysis provided evidence for a new locus for autosomal recessive RP on chromosome 2p22.3-p24.1 with a 2-point logarithm of odds (LOD) score of 3.14, at θ = 0 with D2S165. Fine mapping identified a 20.92-cM (19.98-Mb) interval flanked by markers D2S305 and D2S367. Lack of homozygosity in affected individuals for D2S2150 and D2S220 refined the critical interval distal to marker D2S220 in a 12.31-cM (13.35-Mb) region flanked by markers D2S220 and D2S367. This region does not harbor any genes that have been associated with RP.
METHODS
CLINICAL ASCERTAINMENT
Seventy-five consanguineous Pakistani families with nonsyndromic RP were recruited to participate in a collaborative study between the Center of Excellence in Molecular Biology, Lahore, Pakistan, and the National Eye Institute, Bethesda, Maryland. Institutional review board approval was obtained for this study from the Centre of Excellence in Molecular Biology, Lahore, Pakistan, and National Eye Institute, Bethesda, Maryland. The participating subjects gave informed consent consistent with the tenets of the Declaration of Helsinki. The family described in this study is from the Punjab province of Pakistan. A detailed medical history was obtained by interviewing family members.
All of the ophthalmologic examinations were completed either at the Rehmatullah Benevolent Trust Hospital or at the Centre of Excellence in Molecular Biology, Lahore, Pakistan. Fundus photographs were acquired by the camera manufactured by Fuji. ERG responses were recorded using ERG equipment manufactured by LKC (Gaithersburg, Maryland, USA). Scotopic responses were recorded under dark adapted conditions using a single bright flash stimulus, whereas the photopic responses were recorded under light adapted conditions using a 30-Hz flicker stimulus. Blood samples were collected from affected and unaffected family members. DNA was extracted by a nonorganic method as described by Grimberg and associates.9
GENOTYPE ANALYSIS
A genome-wide scan was performed with 382 highly polymorphic fluorescent markers from the ABI PRISM Linkage Mapping Set MD-10 (Applied Biosystems, Foster City, California, USA) having an average spacing of 10 cM. Multiplex polymerase chain reactions (PCRs) were carried in a GeneAmp PCR System 9700 thermocycler (Applied Biosystems). Briefly, each reaction was carried out in a 5 μL mixture containing 40 ng genomic DNA, various combinations of 10-μM dye-labeled primers pairs, 0.5 μL 10X GeneAmp PCR Buffer II (Applied Biosystems), 0.5 μL 10-mM dNTP mix, 2.5 mM MgCl2, and 0.2 U Taq DNA polymerase (Applied Biosystems). Initial denaturation was carried out for 5 minutes at 95 C, followed by 10 cycles of 15 seconds at 94 C, 15 seconds at 55 C, and 30 seconds at 72 C, and then 20 cycles of 15 seconds at 89 C, 15 seconds at 55 C, and 30 seconds at 72 C. The final extension was performed for 10 minutes at 72 C and was followed by a final hold at 4 C. PCR products from each DNA sample were pooled, mixed with cocktail containing HD-400 size standards (Applied Biosystems), and separated in an ABI 3100 DNA analyzer (Applied Biosystems), and alleles were assigned using GeneMapper (version 4.0; Applied Biosystems).
LINKAGE ANALYSIS
Two-point linkage analyses were performed using the FASTLINK version of MLINK from the LINKAGE program package (available at http://linkage.rockefeller.edu/ott/linkhelp.htm).10,11 Maximum LOD scores were calculated using ILINK. Autosomal recessive RP was analyzed as a fully penetrant trait with an affected allele frequency of 0.001. The marker order and distances between the markers were obtained from the Marshfield database (http://research.marshfieldclinic.org/) and the National Center for Biotechnology Information chromosome 2 sequence maps (http://www.ncbi.nlm.nih.gov). For the initial genome scan, equal allele frequencies were assumed, whereas for fine mapping, allele frequencies were estimated from 96 unrelated and unaffected individuals from the Punjab province of Pakistan.
MUTATION SCREENING
Primer pairs for individual exons were designed using the primer3 program (http://primer3.sourceforge.net/). The sequences and annealing temperatures are available on request. Amplifications were performed in a 25-μl reaction containing 50 ng genomic DNA, 400 nM each primer, 2.5 mM dNTP, 2.5 mM MgCl2, and 0.2 U Taq DNA polymerase in the standard 1x PCR buffer provided by the manufacturer (Applied Biosystems). PCR amplification consisted of a denaturation step at 96 C for 5 minutes, followed by 40 cycles, each consisting of 96 C for 45 seconds, followed by 57 C for 45 seconds and at 72 C for 1 minute. PCR products were analyzed on 2% agarose gel and were precipitated and purified by ethanol precipitation. The PCR primers for each exon were used for bidirectional sequencing using Big Dye Terminator Ready reaction mix according to manufacturer instructions (Applied Biosystems). Sequencing products were resuspended in 10 μL formamide (Applied Biosystems), denatured at 95 C for 5 minutes, and separated in an ABI PRISM 3100 automated sequencer (Applied Biosystems). Sequencing results were assembled using ABI PRISM sequencing analysis software (version 3.7; Applied Biosystems) and were analyzed with SeqScape software (version 2.5; Applied Biosystems).
RESULTS
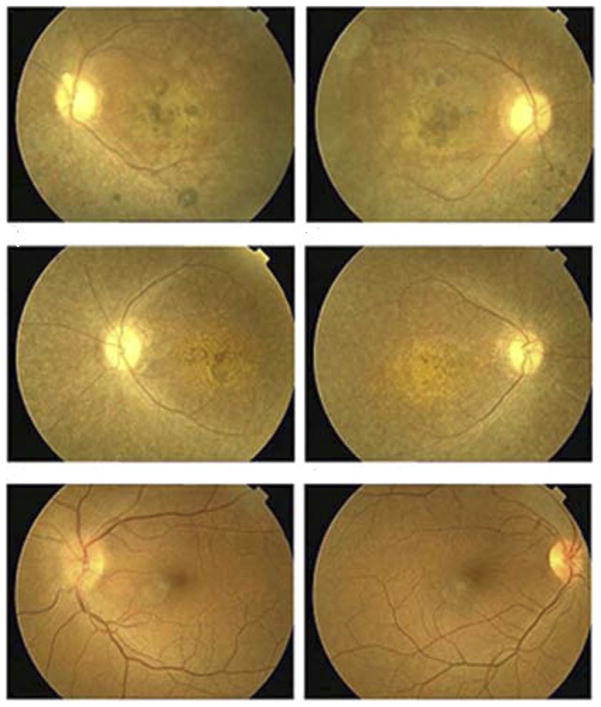
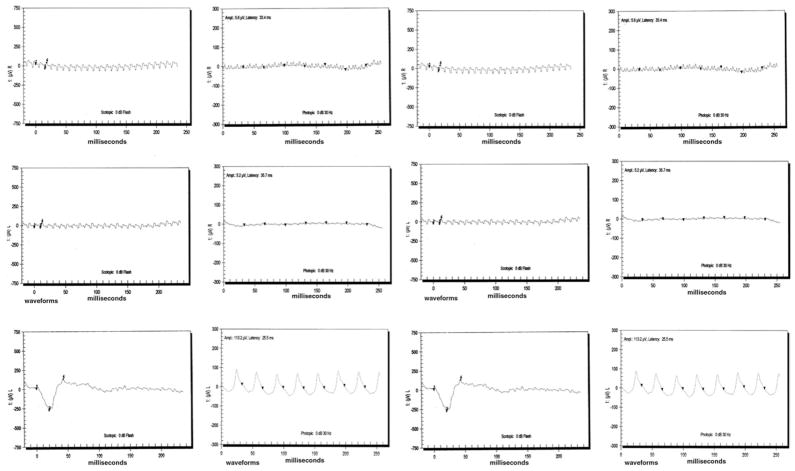
A detailed medical history was obtained by interviewing family members of PKRP115. All affected individuals have had night blindness since early childhood. Vision in affected individuals is limited to light perception or hand movements with no peripheral vision (Table 1). Fundus photographs of affected individuals show typical changes of RP, including a waxy pale optic disc, attenuation of retinal arteries, and bone spicule pigment deposits in the mid periphery of the retina (Figure 1). No attenuation of retinal arteries and bone spicule pigment deposits were detected in the fundus photographs of unaffected individuals, as shown in Figure 1. Affected individuals have typical RP changes on ERG, including loss of both the rod and cone responses as shown in Figure 2, whereas ERG readings of unaffected individuals show no changes in the rod and cone responses as shown in Figure 2.
TABLE 1.
Clinical Characteristics of the Affected Individuals of Family PKRP115 Diagnosed with Autosomal Recessive Retinitis Pigmentosa
| Individual No. | Current Age (yrs) | Age at Onset (yrs) | First Symptoms | Night Blindness | Fundus Examination Results | Visual Acuity |
|---|---|---|---|---|---|---|
| 8 | 28 | 8 | NB | Progressive | MD, artery attenuation, pigment deposit, PD | LP |
| 9 | 25 | 10 | NB | Progressive | MD, artery attenuation, PD | 6/60 |
| 11 | 22 | 10 | NB | Progressive | MD, artery attenuation, PD | 6/50 |
| 13 | 32 | 8 | NB | Progressive | MD, artery attenuation, pigment deposit, PD | 6/60 |
LP = light perception; MD = macular degeneration; NB = night blindness; PD = pale optic disc.
FIGURE 1.
Fundus photographs of members of family PKRP115 diagnosed with autosomal recessive retinitis pigmentosa (RP). (Top row) Images obtained from Individual 8, affected: (Top left) left eye and (Top right) right eye. (Middle row) Images obtained from Individual 13, affected: (Middle left) left eye and (Middle right) right eye. (Bottom row) Images obtained from Individual 7, unaffected: (Bottom left) left eye and (Bottom right) right eye. Fundus photographs of both affected individuals show several features associated with RP, including a waxy pallor of the optic disc, attenuated arterioles, atrophy of the retinal pigment epithelium, and peripheral bone spicules. Notably macular atrophy with pigment clumping is shown in the central retina.
FIGURE 2.
Electroretinogram responses of members of family PKRP115 diagnosed with autosomal recessive retinitis pigmentosa (RP). (First row) Responses from Individual 11 (from left to right): left eye combined rod and cone response, left eye cone response, right eye combined rod and cone response, and right eye cone response. (Second row) Responses from Individual 9 (from left to right): left eye combined rod and cone response, left eye cone response, right eye combined rod and cone response, and right eye cone response. (Third row) Responses from Individual 7 (from left to right): left eye combined rod and cone response, left eye cone response, right eye combined rod and cone response, and right eye cone response. Affected individuals have typical RP changes including loss of both rod and cone responses, whereas electroretinography readings of unaffected persons show no changes in the rod and cone response.
Initially, the reported autosomal recessive RP loci were excluded for linkage using primer pairs specific for known loci (data not shown). During the genome scan, LOD scores of more than 1.0 were obtained only for markers D2S165 and D2S367; both are adjacent markers in the MD-10 mapping set, yielding LOD scores of 3.14 and 1.49 at θ = 0 and θ = 0.09, respectively. Additional short tandem repeats (STR) markers were selected for fine mapping from the Marshfield map (http://research.marshfieldclinic.org/). D2S2144, D2S2223, D2S2350, D2S174, D2S2247, D2S223, and D8S352 yielded LOD scores of 1.07, 1.97, 1.07, 1.07, 3.14, 2.87, and 1.97 at θ = 0, respectively, as shown in Table 2. The highest multi-point LOD score of 3.35 was obtained with markers D2S165 and D2S352 (data not shown).
TABLE 2.
Two-Point Logarithm of Odds Scores for Chromosome 2p Markers of Family PKRP115 Calculated at Different Recombination Fractions
| Marker | cM | Mb | 0.00 | 0.01 | 0.05 | 0.09 | 0.10 | 0.20 | 0.30 | Zmax | θmax |
|---|---|---|---|---|---|---|---|---|---|---|---|
| D2S149a | 34.04 | 14.31 | −∞ | −0.71 | 0.45 | 0.73 | 0.76 | 0.77 | 0.55 | 0.77 | 0.20 |
| D2S2150 | 40.47 | 20.39 | −2.34 | −0.75 | −0.23 | −0.14 | −0.13 | −0.13 | −0.08 | −0.08 | 0.30 |
| D2S220 | 42.65 | 20.94 | −0.98 | 0.59 | 1.06 | 1.08 | 1.07 | 0.80 | 0.44 | 1.09 | 0.07 |
| D2S2144 | 46.37 | 25.79 | 1.07 | 1.05 | 0.97 | 0.88 | 0.86 | 0.63 | 0.39 | 1.07 | 0.00 |
| D2S2223 | 46.37 | 26.41 | 1.97 | 1.93 | 1.77 | 1.61 | 1.57 | 1.15 | 0.74 | 1.97 | 0.00 |
| D2S2350 | 46.37 | 26.58 | 1.07 | 1.05 | 0.97 | 0.88 | 0.86 | 0.63 | 0.39 | 1.07 | 0.00 |
| D2S174 | 46.90 | 26.69 | 1.07 | 1.05 | 0.97 | 0.88 | 0.86 | 0.63 | 0.39 | 1.07 | 0.00 |
| D2S2247 | 46.90 | 27.15 | 3.14 | 2.97 | 2.72 | 2.46 | 2.39 | 1.74 | 1.09 | 3.14 | 0.00 |
| D2S223 | 45.83 | 28.19 | 2.87 | 2.81 | 2.56 | 2.30 | 2.23 | 1.58 | 0.93 | 2.87 | 0.00 |
| D2S165a | 47.13 | 28.45 | 3.14 | 2.98 | 2.74 | 2.49 | 2.43 | 1.80 | 1.15 | 3.14 | 0.00 |
| D2S352 | 50.65 | 31.35 | 1.97 | 1.93 | 1.77 | 1.61 | 1.57 | 1.15 | 0.74 | 1.97 | 0.00 |
| D2S367a | 54.96 | 34.29 | −∞ | 0.98 | 1.46 | 1.49 | 1.48 | 1.20 | 0.79 | 1.49 | 0.07 |
Autosomal recessive retinitis pigmentosa was analyzed as a fully penetrant trait with an affected allele frequency of 0.001.
Marker included in genome wide scan.
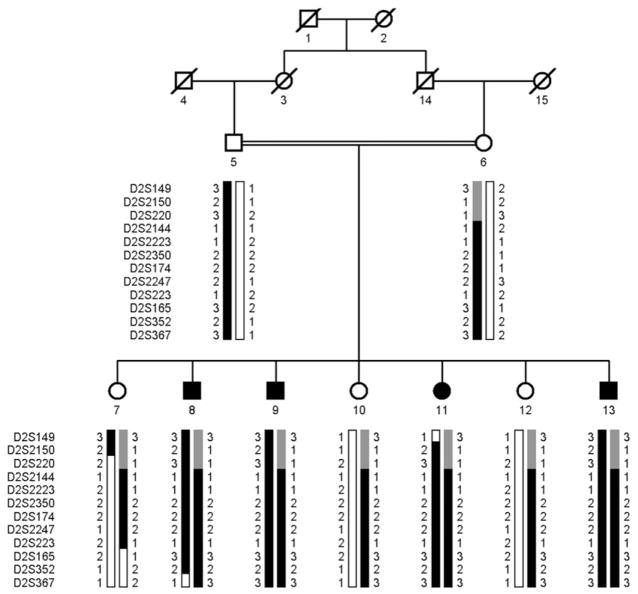
Visual inspection of the haplotypes supports the results of linkage analysis. There is a proximal recombination in Individual 11 at D2S149 (Figure 3). Similarly, there is distal recombination in Individual 8 at D2S367 (Figure 3). Taken together, this places the disease locus in a 20.92-cM (19.98-Mb) interval flanked by markers D2S149 and D2S367. Lack of homozygosity in affected individuals for markers D2S2150 and D2S220 further suggests that the disease-causing gene lies distal to D2S220 in a 12.31-cM (13.35-Mb) interval flanked by markers D2S220 and D2S367 on chromosome 2p22.3-p24.1. Alleles for D2S2144, D2S2223, D2S2350, D2S174, D2S2247, D2S223, D2S165 and D2S352 are homozygous for all affected individuals.
FIGURE 3.
Pedigree drawing of family PKRP115 with multiple individuals diagnosed with autosomal recessive retinitis pigmentosa (RP). Squares are males, circles are females, filled symbols are affected individuals, double line between individuals indicates consanguinity, and diagonal line through a symbol is deceased family member. The haplotypes of 12 adjacent chromosome 2p22.3-p24.1 microsatellite markers are shown with alleles forming the risk haplotype are shaded black, alleles cosegregating with RP but not showing homozygosity are shaded grey, and alleles not cosegregating with RP are shown in white. The minimal region shared by affected individuals is flanked by markers D2S220 and D2S367.
Most of the genes in the critical interval are predicted genes, and only a few annotated genes have functional similarity with reported RP genes. Among these, KIAA1240, RAB10, and CENTG2 were chosen for sequencing. We sequenced all coding exons, exon–intron boundaries, and 5′ and 3′ untranslated regions of KIAA1240, RAB10, and CENTG2. We did not find any pathogenic mutations in these genes, although we identified rs62125899 in KIAA1240, rs4665301 and rs6705 in RAB10, and rs2034648 in CENTG2. These polymorphisms are believed to be nonpathogenic.
DISCUSSION
Herein we report linkage of autosomal recessive rp in a consanguineous Pakistani family to markers on chromosome 2p22. A maximum 2-point LOD score of 3.14 was obtained with D2S165 at θ = 0 during the genome-wide scan, and the autosomal recessive RP locus cosegregated in a 20.92-cM (19.98-Mb) interval flanked by markers D2S149 and D2S367. Haplotype analyses further refined a critical interval distal to D2S220 in a 12.31-cM (13.35-Mb) interval flanked by markers D2S220 and D2S367. Although the maximum LOD score of 3.14 is only slightly higher than the traditional limiting value of 3.0, it represents the maximum value obtainable with this family. Lack of any suggestive LOD scores, except with marker D2S165 during the genome-wide scan, provides additional support for localization of the autosomal recessive RP locus to chromosome 2p22.3-p24.1.
More than 80 genes are located in this region, with variable expression in ocular tissues. KIAA1240, RAB10, and CENTG2 were selected for analysis based on their expression pattern and presumed functional similarity to reported RP genes. KIAA1240 has a similar activity to ABCA4, a member of an ATP-binding cassette proteins family, whereas CENTG2 and RAB10 belong to the family of Ras-like GTPases that regulate membrane traffic between the cellular compartments.12–14 We sequenced all coding exons as well as the 5′ and 3′ untranslated regions, but we did not find any disease-causing mutation in these genes.
We currently are sequencing other candidate genes to identify the pathogenic mutation. Identification of the specific mutation and gene associated with RP will increase our understanding of retinal biology and our knowledge of the cause of retinal dystrophies. Potential candidate genes present in this region currently are being screened for a possible role in the congenital RP in this family.
Acknowledgments
Supported in part by the higher education commission and ministry of science and technology, islamabad, Pakistan.
Footnotes
The authors indicate no financial support or financial conflict of interest.
Involved in design and conduct of study (S.N., S.A.R.); Data collection (S.N., L.L.); Statistical analysis (S.N., S.A.R., L.L.); Analysis and interpretation of data (S.N., S.A.R., L.L., M.S., S.K.); Preparation, review, and approval of manuscript (S.N., S.A.R., L.L., P.A.S., J.F.H., S.R.); Obtaining funding (J.F.H., S.R.); and Administrative, technical, and logistic support (J.F.H., S.R.). Authors (S.N., S.A.R., J.F.H., S.R.) contributed equally towards the study. Institutional review board approval was obtained for this study from the Centre of Excellence in Molecular Biology, Lahore, Pakistan, and the National Eye Institute, Bethesda, Maryland. The participating subjects gave informed consent consistent with the tenets of the Declaration of Helsinki.
References
- 1.Donders F. Beiträge zur pathologischen Anatomie des Auges. 2. Pigmentbildung in der Netzhaut. Arch Ophthalmol. 1857;3:139–165. [Google Scholar]
- 2.Bird AC. Retinal photoreceptor dystrophies LI. Edward Jackson Memorial Lecture. Am J Ophthalmol. 1995;119:543–562. doi: 10.1016/s0002-9394(14)70212-0. [DOI] [PubMed] [Google Scholar]
- 3.Bunker CH, Berson EL, Bromley WC, et al. Prevalence of retinitis pigmentosa in Maine. Am J Ophthalmol. 1984;97:357–365. doi: 10.1016/0002-9394(84)90636-6. [DOI] [PubMed] [Google Scholar]
- 4.Bundey S, Crews SJ. A study of retinitis pigmentosa in the city of Birmingham. J Med Genet. 1986;23:188. doi: 10.1136/jmg.23.2.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boughman JA, Conneally PM, Nance WE. Population genetic studies of retinitis pigmentosa. Am J Hum Genet. 1980;32:223–235. [PMC free article] [PubMed] [Google Scholar]
- 6.Boughman JA, Caldwell RJ. Genetic and clinical characterization of a survey population with retinitis pigmentosa BOUGHMAN1982A. Prog Clin Biol Res. 1982;82:147–166. [PubMed] [Google Scholar]
- 7.Jay M. Figures and fantasies: the frequencies of the different genetic forms of retinitis pigmentosa. Birth Defects Orig Artic Ser. 1982;18:167–173. [PubMed] [Google Scholar]
- 8.Inglehearn CF. Molecular genetics of human retinal dystrophies. Eye. 1998;12(Pt 3b):571–579. doi: 10.1038/eye.1998.147. [DOI] [PubMed] [Google Scholar]
- 9.Grimberg J, Nawoschik S, Belluscio L, et al. A simple and efficient non-organic procedure for the isolation of genomic DNA from blood. Nucleic Acids Res. 1989;17:8390. doi: 10.1093/nar/17.20.8390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lathrop GM, Lalouel JM. Easy calculations of LOD scores and genetic risks on small computers. Am J Hum Genet. 1984;36:460–465. [PMC free article] [PubMed] [Google Scholar]
- 11.Schaffer AA, Gupta SK, Shriram K, Cottingham RW., Jr Avoiding recomputation in linkage analysis. Hum Hered. 1994;44:225–237. doi: 10.1159/000154222. [DOI] [PubMed] [Google Scholar]
- 12.Nie Z, Stanley KT, Stauffer S, et al. AGAP1, an endosome-associated, phosphoinositide-dependent ADP-ribosylation factor GTPase-activating protein that affects actin cytoskeleton. J Biol Chem. 2002;277:48965–48975. doi: 10.1074/jbc.M202969200. [DOI] [PubMed] [Google Scholar]
- 13.Moss J, Vaughan M. Molecules in the ARF orbit. J Biol Chem. 1998;273:21431–21434. doi: 10.1074/jbc.273.34.21431. [DOI] [PubMed] [Google Scholar]
- 14.Takai Y, Sasaki T, Shirataki H, Nakanishi H. Rab3A small GTP-binding protein in Ca(2+)-dependent exocytosis. Genes Cells. 1996;1:615–632. doi: 10.1046/j.1365-2443.1996.00257.x. [DOI] [PubMed] [Google Scholar]