Abstract
The plant cell wall is composed of a matrix of cellulose fibers, flexible pectin polymers, and an array of assorted carbohydrates and proteins. The receptor-like Wall-Associated Kinases (WAKs) of Arabidopsis bind pectin in the wall, and are necessary both for cell expansion during development and for a response to pathogens and wounding. Mitogen Activated Protein Kinases (MPKs) form a major signaling link between cell surface receptors and both transcriptional and enzyme regulation in eukaryotes, and Arabidopsis MPK6 and MPK3 indeed have important roles in development and the response to stress and pathogens. A dominant allele of WAK2 requires kinase activity and activates a stress response that includes an increased ROS accumulation and the up-regulation of numerous genes involved in pathogen resistance, wounding, and cell wall biogenesis. This dominant allele requires a functional pectin binding and kinase domain, indicating that it is engaged in a WAK signaling pathway. A null mutant of the major plasma membrane ROS-producing enzyme complex, rbohd/f does not suppress the WAK2cTAP-induced phenotype. A mpk6, but not a mpk3, null allele is able to suppress the effects of this dominant WAK2 mutation, thus distinguishing MPK3 and MPK6, whose activity previously was thought to be redundant. Pectin activation of gene expression is abated in a wak2-null, but is tempered by the WAK-dominant allele that induces elevated basal stress-related transcript levels. The results suggest a mechanism in which changes to the cell wall can lead to a large change in cellular responses and help to explain how pathogens and wounding can have general effects on growth.
Keywords: Wall Associated Kinases, MPK3, MPK6, pectin, oligogalacturonides
INTRODUCTION
The Wall Associated Kinases of Arabidopsis bind to pectin in the extracellular space, traverse the plasma membrane, and have a functional serine threonine kinase domain in the cytoplasm (He et al., 1996; Kohorn 2000, 2001; Wagner and Kohorn, 2001; Seifert and Blaukopf, 2010). WAKs are required for cell elongation and for the response to pathogens (He et al., 1998; Wagner and Kohorn, 2001; Kohorn et al., 2006b, 2009). There are five WAK genes that are clustered in one 30-kb locus in Arabidopsis, and their collective expression patterns distribute across most cell types (He et al., 1999; Wagner and Kohorn, 2001). While these receptors are highly similar in their kinase domains, the extracellular domains diverge slightly but all contain several epidermal growth factor repeats (Sampoli Benitez and Komives, 2000) of undefined function in plants. WAKs are required for cell elongation (Lally et al., 2001; Wagner and Kohorn, 2001), which is, in part, driven by the WAK2-dependent pectin activation of a set of genes that includes vacuolar invertase (Kohorn et al., 2006b, 2009). The resulting increase in turgor due to invertase activity can drive cell elongation. However, there are numerous other genes activated by pectin in a WAK2-dependent fashion that help to fashion the expanding cell and these include cell wall biosynthesis enzymes (Kohorn et al., 2009). WAKs are known to bind to pectin in the cell wall (Wagner and Kohorn, 2001; Kohorn et al., 2006a, 2009) and in vitro WAK1 and WAK2 have a preference for short oligogalaturonides (OGs) with a degree of polymerization (dp) of 9–15, but also bind with less affinity to longer pectin polymers in a calcium-dependent fashion (Decreux and Messiaen, 2005; Decreux et al., 2006). Together, these genetic, in planta and in vitro results suggest that WAK1 and 2 are pectin receptors, and this has been further supported by the regulation by pectin of a WAK-kinase chimera containing only the WAK1 extracellular domain (Brutus et al., 2010).
WAKs have also been implicated in the response to pathogens but their exact role has not been clear (He et al., 1999; Kohorn et al., 2009). In Arabidopsis, over 25 WAK-like genes (WAKLs) appear to have gone through a further large expansion in family size in crop plants such as rice, leading to the suggestion of some role in pathogen resistance (Verica et al., 2003; Zhang et al., 2005). Indeed, mutant alleles of WAKLs in Arabidopsis and rice can provide different sensitivities to pathogens (Diener and Ausubel, 2005; Li et al., 2009). Recently, a dominant allele of WAK2, created by the fusion to the WAK carboxyl terminus of the epitope tag TAP (WAK2cTAP), was shown to cause a dramatic loss of cell expansion and hence dwarf leaves, a greatly reduced inflorescence stem, and a hedgehog-like phenotype upon flowering and, in addition, a curling of leaves and necrotic lesions in the absence of pathogens (Kohorn et al., 2009). The WAKcTAP fusion is assembled with pectin in the Golgi and then transported to the cell surface (Kohorn et al., 2006a), and WAK2cTAP alleles lacking the extracellular or kinase domains do not induce phenotypes, indicating that active receptor function is required (Kohorn et al., 2009). Fusion of GFP or HA tags to the WAK carboxy terminus does not induce a phenotype, but these alleles are silenced at the early seedling stage (Kohorn et al., 2006a, 2006b, 2009).
Evidence points to a role of MPKs in WAK signaling. Protoplasts from plants homozygous for the null allele wak2-1 showed a reduction in the activation of MPK3, and MPK3 activity is elevated in cells treated with WAK-binding pectin (Kohorn et al., 2009). However, these experiments failed to distinguish whether WAK2 regulates MPK6, or indeed other MPKs. Importantly, MPK3 and 6 have been implicated in the response to pathogen and stress, and are known to play a role in pectin and OG-induced plant responses (Andreasson and Ellis, 2010). Thus, WAKs are required during normal cellular development, but also during the pathogen and stress response, and this dual role remains unexplained. Given that there is ample evidence to suggest that WAKs are pectin receptors, we further explored the relationship between WAKs, pectin, and MPK-dependent growth and stress responses. Here, we show that a dominant allele of WAK2 constitutively activates a stress response that includes numerous genes involved in pathogen resistance and stress, cell wall biosynthesis, and the accumulation of ROS. We find that ROS accumulation by plasma membrane enzymes is not required for the WAKcTAP phenotype and that a genetic interaction between MPK and WAK2 alleles distinguishes MPK3 from MPK6. While pectin can activate MPK3 and genes required for cell elongation in a WAK2-dependent manner (Kohorn et al., 2009), short pectin fragments (oligogalacturonides (OGs)) generated by wounding or pathogen can instead lead to a stress response (Denoux et al., 2008; Willats et al., 2001). Evidence presented here suggests that WAKs provide a mechanism for this pectin and OG-mediated response, where changes to the cell wall can lead to a large change in cellular responses, and help to explain how pathogens and wounding can have general effects on growth.
RESULTS
Dominant WAK Allele
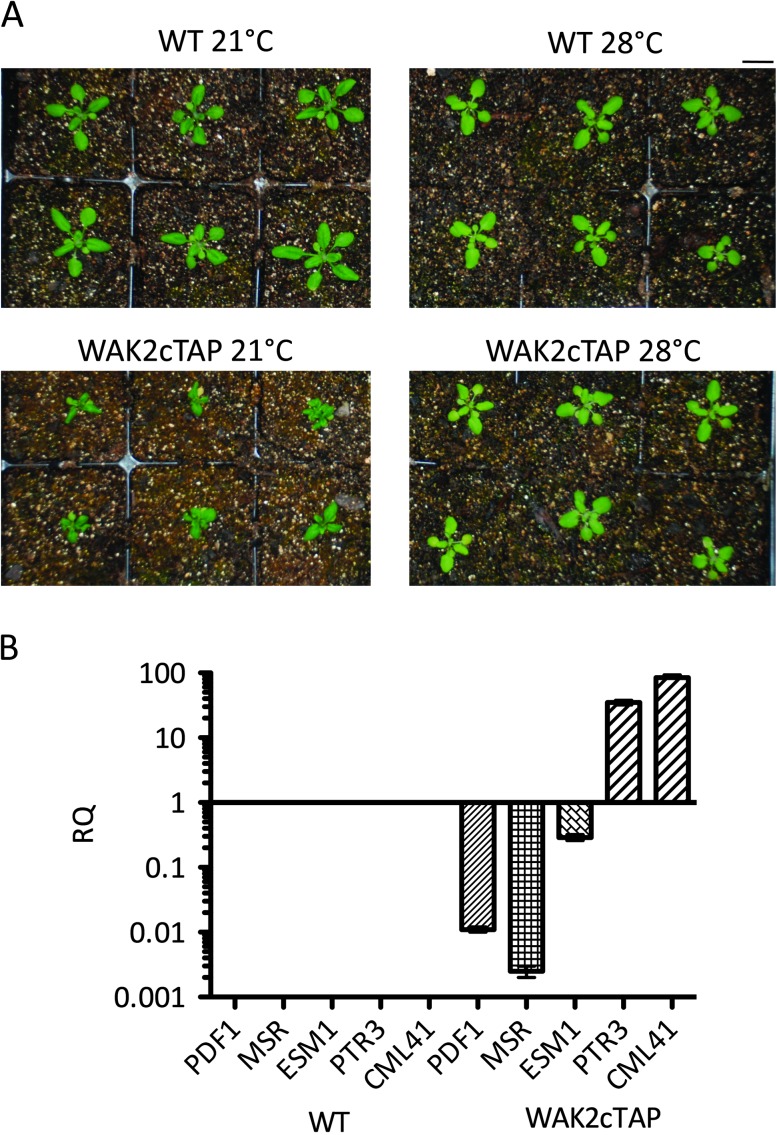
Expression of the WAK2cTAP allele leads to dwarf plants, with leaves that are curled and have lesions, and with short inflorescence stems (Kohorn et al., 2009). Growth at 28°C rather than 21°C suppresses the phenotype (Figure 1), which is also dependent upon 8-h dark/16-h light and is not apparent under 24-h light (data not shown). Within a given growth condition and planting time, there is little variation in phenotype, yet the extent of dwarfism and necrosis does vary between flats of plants grown at different times in different soil batches. We were unable to further define the influencing parameters but subsequent analysis took care to compare plants grown at the same time and soils. Such variation and suppression of appearance are often typical of pathogen and stress responses (Jones and Dangl, 2006). To explore the genetic effect of WAK2cTAP, we performed a gene expression analysis using Arabidopsis Affymetrix arrays with RNA from wild-type (WT) or WAK2cTAP 20-day-old leaf tissue, where strong phenotype was evident in tissue expressing the dominant allele, and all biological replicates were grown at the same time and environmental conditions. Total RNA was extracted and used as template for a probe of Affymetrix gene chips. The differences in gene expression between WT and WAK2cTAP samples were analyzed with GeneSpring GX 11, and the results are shown in Supplemental Table 1. More than 2600 genes showed a significant change in expression (higher than twofold and up to 100-fold) in a WAK2cTAP background compared to wild-type. Overall, genes up-regulated in WAK2cTAP are predominantly involved in cell defense and apoptosis, and cell wall synthesis and maintenance. The stress, wounding, signaling, cell wall-related, and membrane protein classes of genes are also grouped into tables and presented according to their associated biological function (Supplemental Table 2A–2E). These profiles are consistent with the observed phenotype and a role for the WAKs in regulating cell expansion and the stress response. However, there are a number of genes that fall into these classes that are down-regulated in the mutant, indicating that the response is quite specific and complex and does not follow a typical defense response. The general trend of stress induction and cell wall biosynthesis is clear, but the complexity shows no clear patterns. In addition, WAK1, WAK2, and WAK3 expression is increased in WAK2cTAP plants (fivefold, 3.8-fold and 16.3-fold, respectively), while the expression of the other members of the WAK family (WAK4 and WAK5) did not change significantly compared to wild-type. Vacuolar invertase1 (AT1G62660) transcription was reduced by twofold in WAK2cTAP compared to wild-type, as it was for a wak2-null mutant (Kohorn et al., 2006b, 2009). This suggests that, while some stress genes are activated, those involved in cell expansion are down-regulated, consistent with the WAK2cTAP phenotype.
Figure 1.
WAK2cTAP phenotype is affected by temperature.(A) Wild-type (WT) and WAK2cTAP-expressing plants grown at 21 or 28°C. Six plants are shown for each condition. Bar top right indicates 14 mm.
(B) QPCR of five of the 2600 genes that showed changes in expression levels higher than twofold in a WAK2cTAP background compared to wild-type (WT). The y-axis in log scale shows expression relative (RQ) to wild-type that was set to a value of 1, n = 3. The x-axis indicates the genotype (WT or WAK2cTAP) and the gene (PTR3, PDF1, MSR, CML41, or ESM1).
Quantitative real-time PCR (QPCR) was used to measure the fold change in expression (RQ) for five genes—PTR3 (peptide transporter), MSR (methionine sulfoxide reductase), PDF1 (plant defensin 1), ESM1 (carboxylesterase), and CML4 (calmodulin-like protein), whose expression was indicated by the chips to be altered. Figure 1B shows that, compared to wild-type (set as an RQ of 1), the relative expression patterns follow that indicated by the Affymetrix chips, although the fold change varied between the chips and the QPCR. This was not unexpected, given that the RNA from the QPCR was from plants grown at different times from those used for the chips, although both showed strong phenotype. One additional difference was that the chips indicated a fivefold induction while the QPCR showed an 80-fold increase for CML41. The reason for this discrepancy is not known, but may reflect a limitation of the chip analysis. Since all parameters have been controlled and verified for the QPCR, we expect this type of analysis to be more reliable and the chips more of an initial indication of genes that should be investigated further. What is clear is that the WAKcTAP response is not that of a typical one found with pathogens or wounding, as not all defense genes are induced, and we have been unable to define a pattern other than a select induction and repression of stress-related and cell wall biosynthesis gene expression.
WAK2cTAP Is Dominant Active
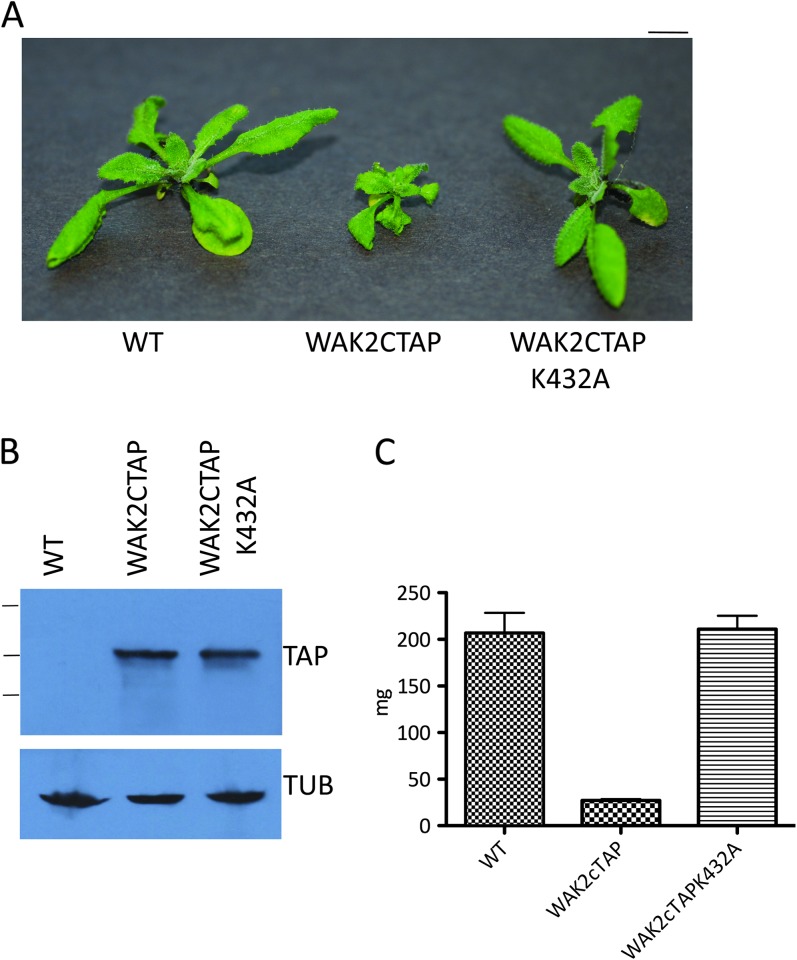
Previous reports described WAK2cTAP as a dominant negative allele (Kohorn et al., 2009), but it was not clear that function was actually blocked. Both the WAK2 extracellular and cytoplasmic domain are required for the phenotype (Kohorn et al., 2009), indicating that the whole receptor is needed. To determine whether an active kinase is required, a mutation in the catalytic site of the kinase (a K to an A substitution at position 432) that eliminates kinase activity (Anderson et al., 2001) was created and expressed in wild-type Arabidopsis in five independent transformants. All transformants were wild-type in appearance. Figure 2A shows representative wild-type, WAK2cTAP, and the kinase inactive mutant (WAK2cTAPK432A) plants grown at the same time and under the same conditions, and indicates the kinase inactive mutant indeed appears wild-type. Equal amounts of total cell protein from these plants were analyzed by Western blot for levels of WAK2cTAP or WAK2cTAP K432A expression and the results are shown in Figure 2B, which shows that the expression levels of WAKcTAP are similar. Westerns were also probed with anti-Tubulin antiserum to verify equal loadings of samples. The fresh weight of five individuals of each type shown in Figure 2A were measured, and there is no difference between the wild-type and the WAK2cTAP K432A (Figure 2C). Thus, active kinase is required for the dominant effect of the WAK2cTAP. This might indicate that the WAK2cTAP is active in signal transduction and is a dominant positive allele. However, an active kinase does not necessarily indicate activated (versus inhibited) signaling downstream and thus we prefer to call this a dominant active allele.
Figure 2.
An Active Kinase Domain Is Required for the Dominant Effect of WAK2cTAP.
(A) Wild-type (WT) plants, or WT expressing WAK2cTAP or WAK2cTAPK432A (mutation in kinase catalytic site) grown under the same conditions for 20 d. Bar top right indicates 7 mm.
(B) Extracts of equal protein amounts from plants shown in (A), probed by Western blot for TAP tag or Tubulin (Tub). Lines on left indicate (top down) 170, 130, and 90 kDa markers.
(C) Mass in mg of five individuals of the indicated genotype.
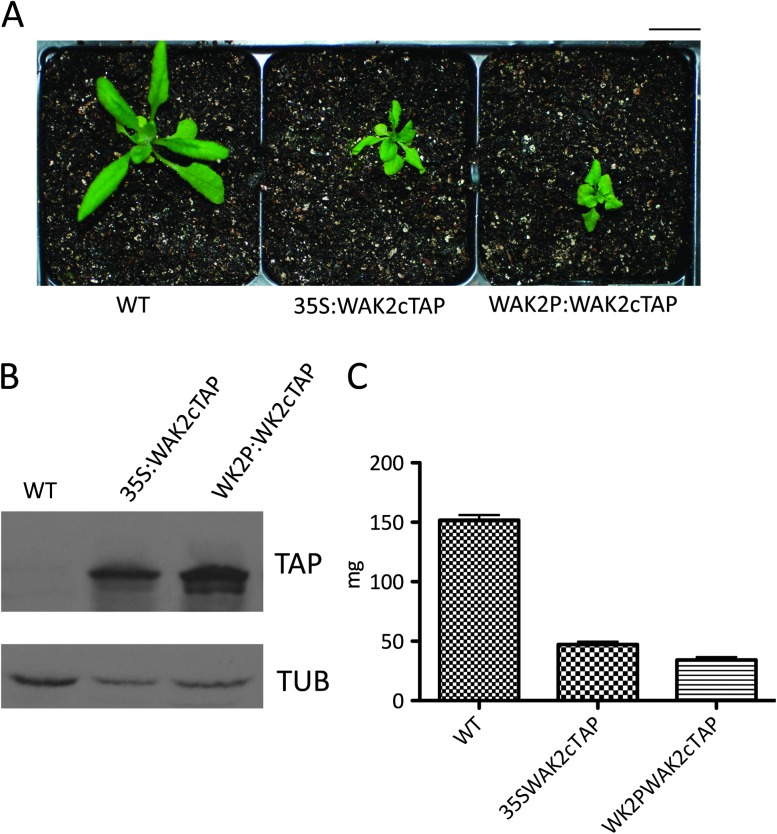
To further characterize the dominant phenotype, WAK2cTAP expression was driven by the WAK2 promoter in Arabidopsis and these plants also developed the dwarf, curled, and necrotic leaves. Figure 3A shows WT and a plant expressing WAK2cTAP under the 35S or WAK2 promoter grown at the same time. Three independent transformants showed similar phenotypes when expressing equivalent levels of WAKcTAP and a Western blot of representative isolates is shown in Figure 3B. The average mass of the three transformants is shown in Figure 3C, indicating that the WAK2 promoter also induces dwarfism. Thus, the WAK2cTAP phenotype can result from expression in the appropriate tissue, similar to the endogenous WAK2.
Figure 3.
The WAK2 and 35sCaMV Promoters Both Drive WAKcTAP to Produce Phenotype.
(A) Representative plants of the indicated genotype grown under the same conditions. Bar top right indicates 14 mm.
(B) Extracts of equal protein amounts from plants shown in (A), probed by Western blot for TAP tag or Tubulin (Tub).
(C) Mass in mg of three individuals of the indicated genotype.
WAK2cTAP Is a Functional Receptor
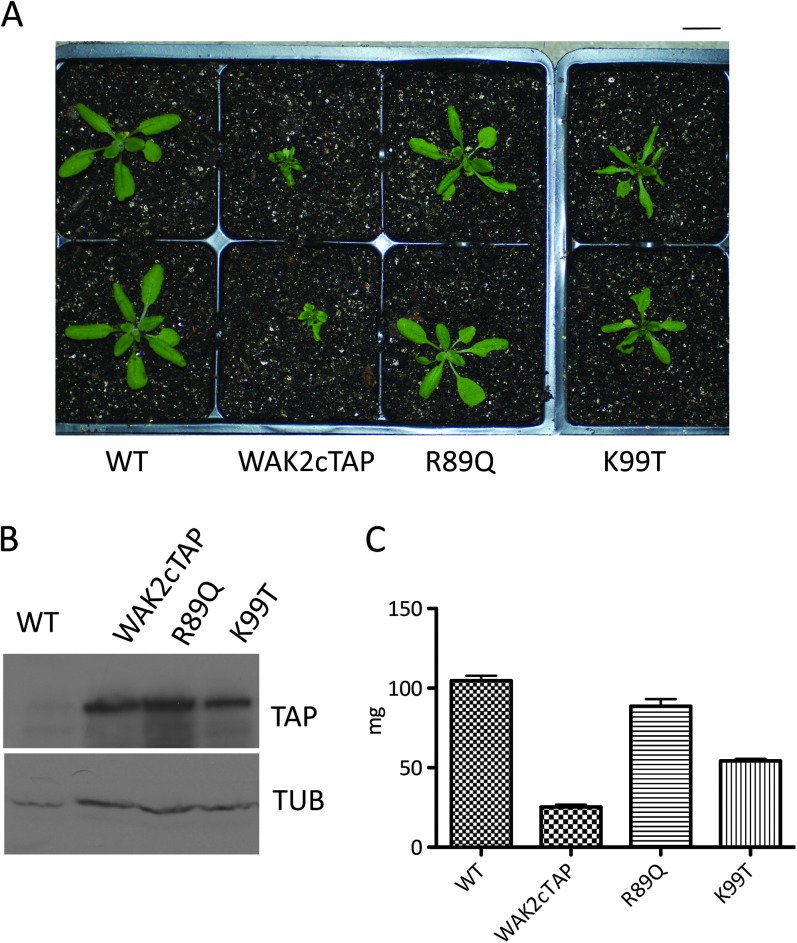
If WAK2cTAP is engaged in signaling in a similar path to that of the endogenous WAKs, then one would expect mutations in the amino acids of the extracellular domain required for pectin binding to affect WAK2cTAP function. An arginine at position 89 and a lysine at 99 were altered to a glutamine and a threonine, respectively, as these changes do inhibit pectin binding (R89Q, K99T, Decreux et al., 2006). These mutant genes were fused to cTAP and expressed in Arabidopsis using the 35S promoter. Two independent transformants expressing equivalent WAK2cTAP levels (relative to Tubulin) were isolated and one of each is shown in Figure 4. WAK2R89QcTAP-expressing plants appear WT, while WAK2K99TcTAP plants show some leaf curling and dwarfism (t-test p < 0.01) but not as pronounced as WAK2cTAP, as indicated by the appearance (Figure 4A), and the mass (Figure 4C). Thus, amino acids required for pectin binding are required or compromise WAK2cTAP function. We also created point mutations in the WAK2 extracellular domain conserved cysteines that are required in mammalian Epidermal Growth Factor (EGF) repeats to create the EGF repeat structure (Sampoli Benitez and Komives, 2000), and these, too, were expressed in Arabidopsis and eliminate the ability of WAKcTAP to provide a dominant phenotype (Kohorn, unpublished). These data are consistent with the view that WAK2cTAP is a functional, albeit partly deregulated, receptor. However, we can not exclude the possibility that WAK2cTAP is also activating additional paths not experienced by native WAK2. Given that WAKs are likely pectin and OG receptors, and pectins activate a stress response, the constitutive activation of a stress response by WAK2cTAP argues for its involvement in a WAK pathway.
Figure 4.
Residues Required for Pectin Binding Affect WAK2cTAP Phenotype.
(A) Representative plants (two isolates each) of the indicated genotype grown under the same conditions. Bar top right indicates 14 mm.
(B) Extracts of equal protein amounts from plants shown in (A), probed by Western blot for TAP tag or Tubulin (Tub).
(C) Mass in mg of three individuals of the indicated genotype.
MPKs
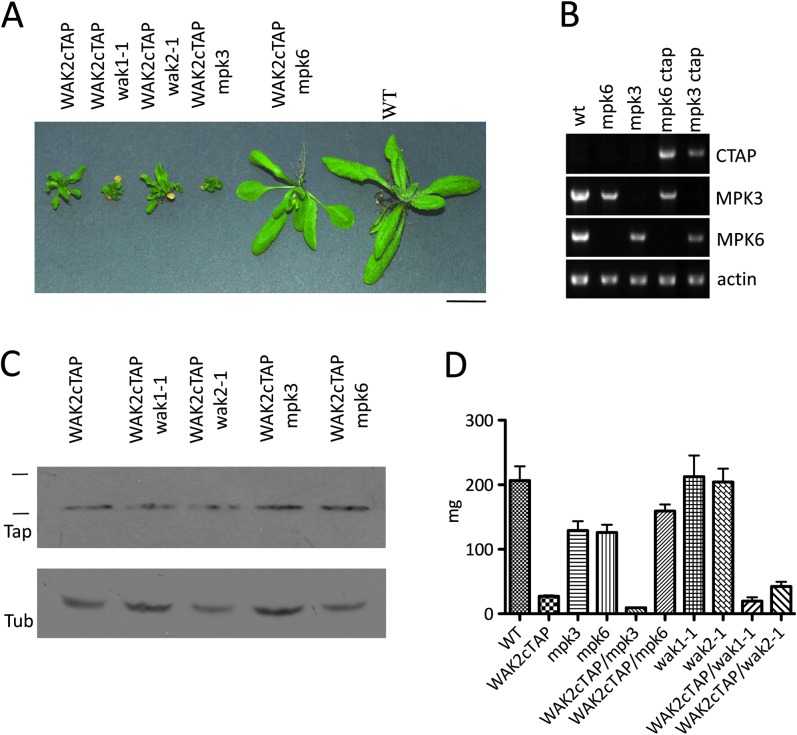
Evidence points to a role of MPKs in WAK signaling. Protoplasts from plants homozygous for the null allele wak2-1 showed a reduction in the activation of MPK3, and MPK3 activity is elevated in cells treated with WAK-binding pectin (Kohorn et al., 2009). However, these experiments were unable to distinguish whether WAK2 regulates MPK6, and indeed other MPKs. In vitro MPK assays lack cellular context and may not be sensitive enough to distinguish multiple concurrent MPK regulatory events. The effect of mpk3 or mpk6-null alleles on Wak2 alleles was therefore examined. Plants homozygous for the null mpk6-2 or mpk3 alleles that alone give only subtle phenotypes (Bush and Krysan, 2007; Wang et al., 2007) were transformed with a Wak2cTAP gene driven by the CaMv 35S constitutive promoter. Five independent transformants for each background were identified, each expressing WAK2cTAP at similar levels to that of the published WAK2cTAP expresser (Kohorn et al., 2009). One F2 progeny representative of each genotype grown in the same flat are shown in Figure 5A. PCR of genomic DNA from theses plants using primers for the indicated WT gene showed that each is homozygous for the appropriate mpk allele (Figure 5B). These transformants express similar levels of WAK2cTAP, as determined by Western (Figure 5C) of equivalent amount of total leaf protein per lane (Tubulin was used as a reference).
Figure 5.
WAK2cTAP Phenotype Is Suppressed by mpk6-2 But Not mpk3-Null Alleles.
(A) Representative individual plants of the indicated genotype grown under identical conditions. Bar bottom right indicates 14 mm.
(B) PCR analysis of genomic DNA, in an ethidium bromide-stained gel, from plant lines indicated above the gels, using primers for wild-type genes indicated on the right.
(C) Western blot detecting TAP tagged protein (Tap) or Tubulin (Tub) in equal amounts of protein extract from the plants indicated above the lanes. Lines on left indicate (top down) 170 and 130-kDa markers.
(D) Average mass of five 20-day-old plants in mg (y-axis) of indicated genotype shown on x-axis.
To quantify the size difference of these plants, the mass of five individuals of each genotype grown under the same conditions at the same time was determined, and the results are shown in Figure 5D. Wild-type plants expressing WAK2cTAP are greatly reduced in size and have curly leaves. In a mpk3 –/– background, this phenotype is more severe, as previously reported (Kohorn et al., 2009). In a mapk6-2 –/– background, the phenotype induced by WAK2cTAP expression is completely suppressed as the appearance and average mass are near that of mapk6-2 –/– (t-test p > 0.05, Figure 5A and 5D). Thus, MPK6, but not MPK3, is required for the stress response induced by WAK2cTAP. While some previous reports have suggested that MPK3 and MPK6 provide overlapping functions (Wang et al., 2007; Colcombet and Hirt, 2008), only limited data from in vitro experiments have suggested that their interacting proteins and substrates might be non-overlapping (Andreasson and Ellis, 2010). The results here show that MPK6 has a distinct function from that of MPK3 as revealed by the dominant WAK2cTAP allele.
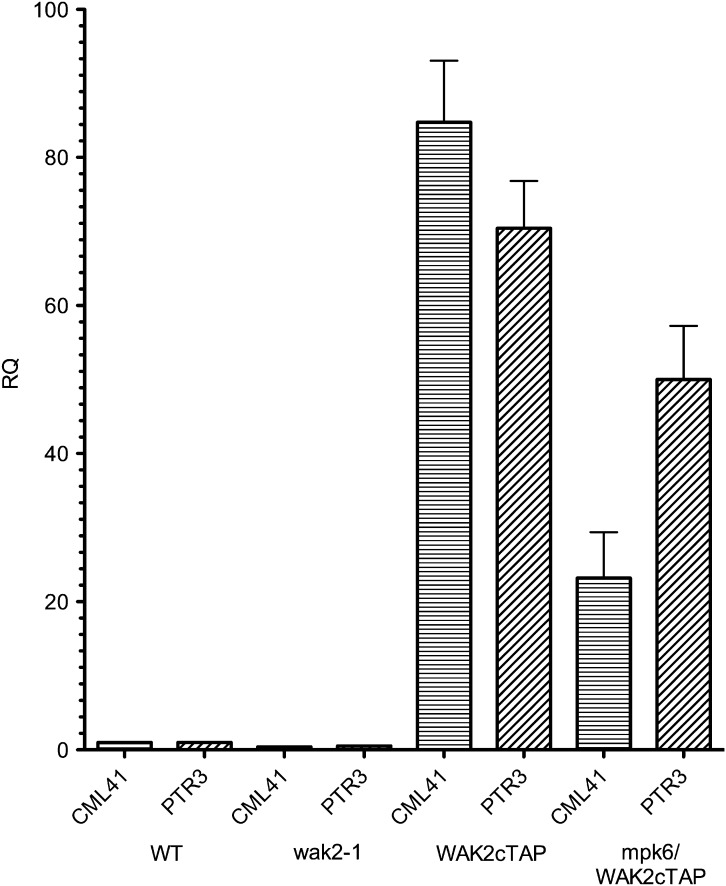
To determine whether the up-regulation of stress-related genes by WAK2cTAP was affected in the mapk6-suppressed WAK2cTAP line, QPCR was used to measure the levels of transcripts from WT, WAK2cTAP, and mapk6-2/WAK2cTAP leaves. Leaves of wak2-1-null plants were also included to determine whether WAK2 expression affects these transcripts and the results are shown in Figure 6, where the fold change (RQ) is reported relative to WT, which is set at a value of 1. CML41 and PTR3 were chosen, as they are good markers of the stress and pathogen response (Denoux et al., 2008), and are up-regulated in WAK2cTAP, relative to WT. In mapk6/WAK2cTAP leaves, CML41 and PTR3 are reduced in their expression relative to WAK2cTAP (t-test p < 0.01), but still elevated relative to WT. We note an increase (two times) in the fold induction of PTR3 relative to that measured in Figure 1, which is likely due to variation between plants grown at different times. Nevertheless, since the samples in Figure 6 were taken from plants grown together, these results do show that the mapk6-2 allele suppresses the visible phenotype induced by WAK2cTAP, and it only partially suppresses the induced gene expression. This might indicate that only some of the WAK2cTAP induction is mediated by MPK6, and that this suppressed level of gene expression is insufficient to provide a visible phenotype. The WAK2cTAP phenotype shows a complex gene expression pattern, and it remains to be determined how extensive the suppression by the mapk6-2-null allele is. Nevertheless, the results do indicate that MPK6 can be distinguished from MPK3, and is likely a target of WAK2cTAP.
Figure 6.
Gene Expression in WAK Mutants.
QPCR of reverse-transcribed RNA isolated from 20-day-old leaves from the indicated genotype (bottom of x-axis) and indicated gene (x-axis). Expression levels (RQ) are shown as relative to wild-type (WT), which is set to a standard RQ vale of 1, n = 3. Actin was used as an internal reference.
Double WAK Mutations
A genetic interaction between mutant receptor alleles could indicate signaling interaction. To determine what WAK partners may participate in WAK2 signaling, WAK2cTAP was expressed in plants homozygous for either a wak1 or wak2-null allele (Kohorn et al., 2006b), and the results are also shown in Figure 5. Plants homozygous for wak2-1 but not wak1-1 and that express equivalent amounts of WAK2cTAP (Figure 5C) were only slightly decreased in the severity of the WAK2cTAP-induced phenotype, as the plants were larger than WAK2cTAP (Figure 5D, p < 0.01), although the leaves were still curled. The slight reduction could be due to the additive effect of less WAK2 protein, suggesting that WAK2 is in part required for the effect of WAK2cTAP, but, since there is so little suppression, it is unlikely that a wak2-null has a significant affect on WAK2cTAP, and there are likely other partners at the membrane to be defined.
Induction of ROS
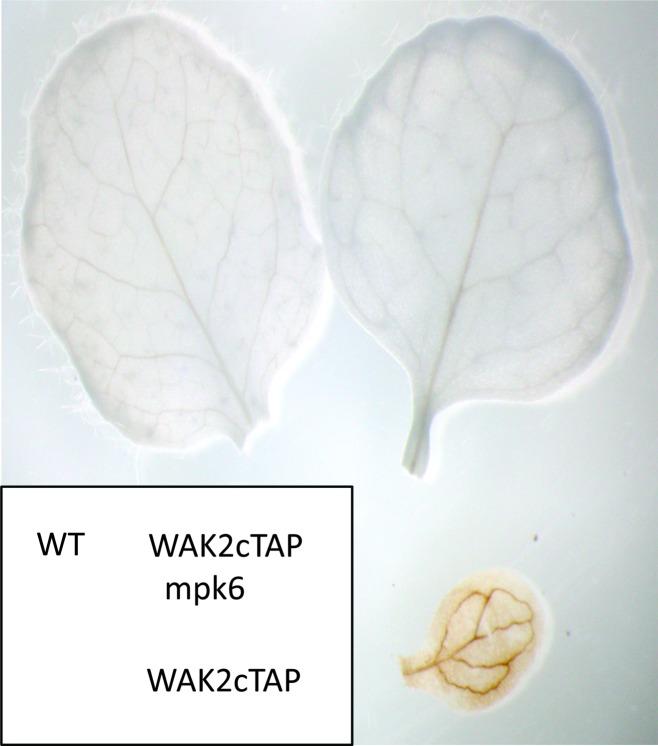
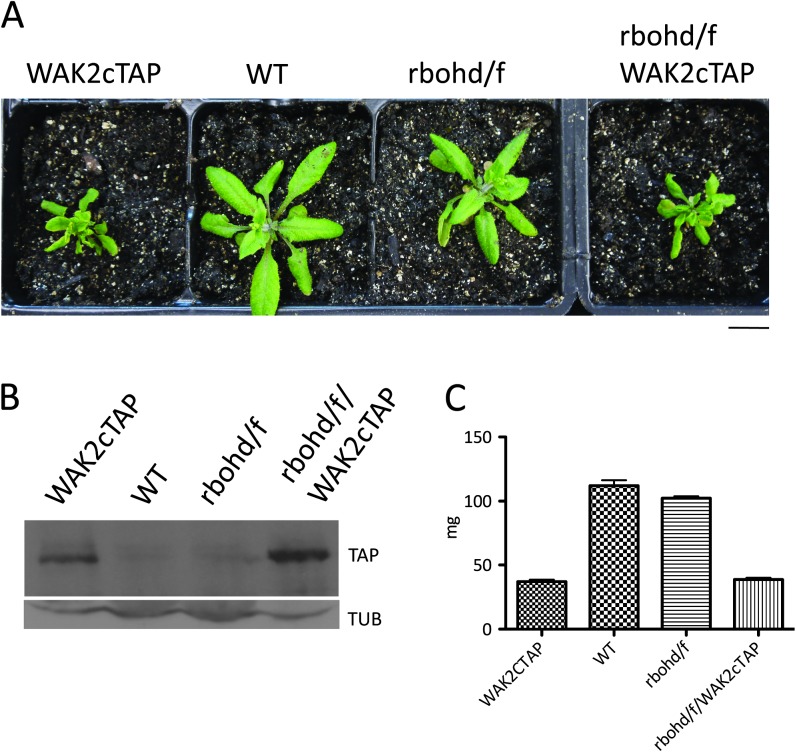
The leaves of plants transformed with WAK2cTAP show a stress response and thus might be expected to have elevated levels of reactive oxygen species (ROS). Figure 7 shows that the WAK2cTAP allele does induce a constitutive production of reactive oxygen species (ROS), as detected by Diaminobenzidine staining (DAB, Clarke, 2002) and this is consistent with the visual phenotype. In addition to suppressing the dwarf and curled phenotype induced by WAK2cTAP, the mapk6-2 allele also suppressed the constitutive production of ROS, as there is no DAB staining in WAK2cTAP-expressing plants homozygous for mapk6 (Figure 7). We crossed a double mutant of the major plasma membrane ROS-producing enzyme complex, AtrbohD/F (Torres, 2010), and WAK2cTAP and screened the F2 progeny by PCR and Western for plants homozygous for rbohd/f –/– and expressing WAK2cTAP. Figure 8 shows a representative F2 rbohd/f –/–; WAK2cTAP triple mutant compared to WAK2cTAP, WT, and rbohd/f –/– grown under the same conditions. Figure 8B shows that WAK2cTAP is still expressed in the rbohd/f –/–; WAK2cTAP triple mutant, and Figure 8C provides the average mass of three plants of each genotype. rbohd/f –/– shows no obvious phenotype under these growth conditions, and does not suppress the WAK2cTAP phenotype, indicating that the accumulation of ROS at the membrane is not a causative event in creating phenotype. This finding is consistent with the observation that ROS accumulation by the membrane complex is not required for OG activation of gene expression (Galletti et al., 2008). Cytoplasmic ROS production is not affected by the rbohd/f mutations and, indeed, the rbohd/f –/–; WAK2cTAP triple mutant still accumulates ROS, as shown by DAB staining (Supplemental Figure 1). Under the growth condition used, ROS is also detected in rbohd/f –/– plants, similar to that found in rbohd/f –/–; WAK2cTAP plants (Supplemental Figure 1).
Figure 7.
WAK2cTAP Activate ROS.
Shown are DAB staining for ROS, where identity of plant is in lower left insert; wild-type (WT), WAK2cTAP, and mpk6-2/WAK2cTAP leaves of the same age.
Figure 8.
Plasma Membrane ROS Accumulation Is Not Required for WAK2cTAP Effect.
(A) Plants of the indicated genotype were grown under identical conditions. Bar bottom right indicates 14 mm.
(B) Extracts of equal protein amounts from plants shown in (A), probed by Western blot for TAP tag or Tubulin (Tub).
(C) Average mass of three 20-day-old plants in mg (y-axis) of indicated genotype shown on x-axis.
Pectin and OG Regulation
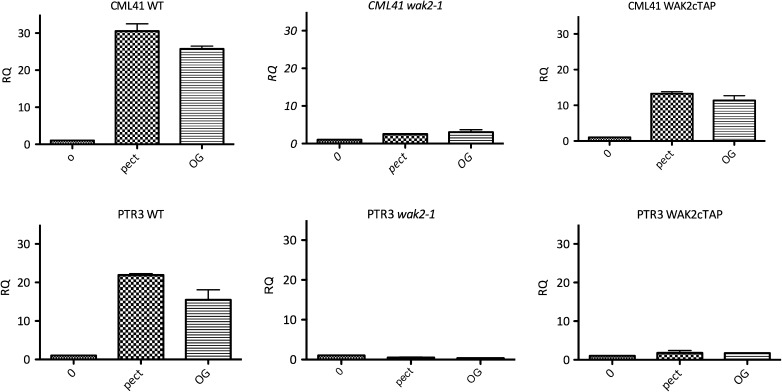
Pectin binds to WAKs and activates MPKs and downstream genes (Wagner and Kohorn, 2001; Kohorn et al., 2006b; Denoux et al., 2008; Kohorn et al., 2009) and, since WAK2cTAP constitutively activates a stress response, we asked whether the regulation by pectin was altered by this dominant allele. The expression of two genes (CML41 and PTR3) that are induced by WAK2cTAP (Figure 6) and by pectin (Denoux et al., 2008) was followed by QPCR in a protoplast assay that provides WAK-dependant activation of genes by pectin (Kohorn et al., 2009). Protoplasts have no cell wall, yet still express active WAK and WAKcTAP protein on their plasma membrane (He et al., 1996; Kohorn et al., 2006a). The induced expression (RQ) is presented as relative to the untreated sample of each genotype, which is standardized to a level of 1, and the results are shown in Figure 9. (Note that this figure compares the pectin induction for each genotype, but not the absolute levels between genotypes.) The addition of either long pectin fragments (citrus pectin) or OGs of dp 9–15 activate CML41 and PTR3 in WT cells, but far less so or not at all in wak2-1 mutants. Pectins still activate CML41 in WAK2cTAP cells, but the fold activation is less than that of WT. However, PTR3 is not induced further by pectin in WAK2cTAP cells. The general level of expression of both genes is far higher in WAK2cTAP than in WT (Figure 6) and thus the reduced fold pectin induction may be due to the fact that these genes are already so highly activated in WAK2cTAP cells that further stimulation is limited. Since the extent of pectin induction of CML41 and PTR3 is different in WAKcTAP cells and WT, it indicates that the observed induction is due not only to the endogenous WAKs, but also to the influence of WAK2cTAP expression. Thus, WAK2cTAP cells are still pectin responsive, consistent with the observation that this allele requires a functional extracellular domain for its effect (Kohorn et al., 2009) and that the kinase is a dominant active allele requiring kinase activity.
Figure 9.
Gene Activation by Pectin in WAK Mutants. Protoplasts from the indicated plant line (above each graph) were aliquoted equally and incubated with the indicated pectin (pect, mixed length citrus pectin; OG, purified dp 9–15) for 6 h, and then the expression of the indicated gene (above each graph) determined by QPCR of reverse-transcribed RNA. Fold induction is provided as a relative RQ value (y-axis), where untreated of each genotype serves as the reference and has a value of 1, n = 3. Actin was used as an internal reference.
DISCUSSION
Our results show that WAK2cTAP requires a functional pectin binding extracellular domain and an active kinase to induce a dwarf phenotype. This dominant active allele is characterized by a change in gene expression that includes cell wall synthesis enzymes and some defense response proteins, but this response is complex and not identical to a typical pathogen response. The accumulation of ROS by the plasma membrane complex is not required for this phenotype, consistent with that found for the response to OGs (Galletti et al., 2008), although cytoplasmic ROS does accumulate. A mapk6 but not mpk3-null allele suppresses the WAK2cTAP-induced phenotype and the accumulation of ROS, and abates some of the induced gene expression changes. We also show that WAK2cTAP cells are still partially responsive to pectin.
The results are consistent with a model in which WAK2 activates MPK3 to mediate cell expansion. This is likely only one of many inputs required, perhaps including other redundant WAKs, and this explains the subtle or lack of phenotype of the single wak-null alleles. At the same time, a WAK-influenced MPK6 path may not be activated, thereby keeping the stress response off. By an undefined mechanism, OGs cause WAKs to lead to the activation of MPK6, thereby activating the stress response. WAKcTAP, while still pectin-responsive, may also activate MPK6. MPKs receive multiple concurrent stimulations, only one of which originates from WAKs and pectin, and thus they appear activated in vitro and subtle changes from one input can be hard to detect (Andreasson and Ellis, 2010). There is clearly interaction and some redundancy between MPK3 and 6, but WAK2cTAP is able to distinguish these, as detected genetically. Clearly, there are alternate complexities of this model, but it presents one explanation for our results.
These results raise the question as to how WAKs can be required on the one hand for cell expansion during development (Kohorn et al., 2006b, 2009) and on the other be responsive to pectin fragments and induce a stress response (Kohorn et al., 2009; Brutus et al., 2010). WAKs are bound to pectin in native cell walls and their activity is required for normal cell expansion, yet OGs also bind to WAKs and mediate a response to pathogens and wounding. It follows that the type and concentration of pectin present in the wall could lead to a WAK-dependant activation of different signaling pathways, and this could be what is represented by the MPK3 activation by pectin and effect on cell growth, and the mapk6 suppression of WAKcTAP stress-induced phenotype. How this differentiation is achieved is not known, but clues may lie in the observation that in vitro binding assays reveal a higher binding affinity of OGs than longer polymers for WAK (Decreux and Messiaen, 2005; Decreux et al., 2006). Results shown here indicate that both citrus pectin and OGs independently are capable of activating target downstream genes in a protoplast assay in which there is no other preexisting pectin structure. Activation of different cytoplasmic pathways, perhaps distinguished by MPK6 and MPK3, could be effected by different types of pectins created by pathogens or wounding. Whether this differentiation is achieved by homo-complexes of WAKs or associations with other receptors remains to be determined, but the latter is more likely, given the lack of genetic interactions between WAK alleles shown here. Understanding the identity and interactions of WAK and WAK2cTAP partners will be important in this evaluation.
It has been observed by many that pathogen presentation and stress responses in plants lead to stunted growth (Jones and Dangl, 2006), and the results presented here suggest that WAKs are involved in this mechanism to switch from cell expansion to cell stress, which is in part effected by different MPKs and influenced by pectins. It will be of interest to see whether WAKs form complexes with a variety of receptors, including themselves, to mediate a general growth response phenotype upon stimulation of those receptors or whether these cascades are independent.
METHODS
Plant Materials
Arabidopsis thaliana columbia plants were grown in soil in a growth chamber, with a 16-h light/8-h dark photoperiod.
PCR Analysis of Alleles
PCR analysis of alleles was carried out using the following primers and Titanium™ Taq (Clontech 639208) using the manufacturers’ buffers and conditions, and genomic DNA extracted as described (Kohorn et al., 2006b). Amplified DNA using primers listed below was analyzed by agarose gel electrophoresis and staining with ethidium bromide:
Actin: Actin-F 5'-AAAGGATGCTTATGTTGGCG-3', Actin-Rev 5'-GAAAGAGTAACCACGCTCGG-3';
MPK3 wild-type: MPK3F 5'-CCGAGCAATCTTCTGTTGAACGCGAATTG-3', MPK3R 5'-TGCTGCACTTCTAACCGTATGTTGGATTG-3';
MPK3null: MPK3F 5'-CCGAGCAATCTTCTGTTGAACGCGAATTG-3', P745 5'-AACGTCCGCAATGTGTTATTAAGTTGTC-3';
MPK6 wild-type: MPK6-F2 5'-GCCTCAGATGCCTGGGATTGAGAATATTC-3', MPK6-RT-R2 5'-AGAGTGGCTTACGGTCCATTAACTCCAATG-3';
MPK6null: MPK6-F2 and p745;
WAK2cTAP: WAK2-2405F 5'-GAGCGATGTTTATAGTTTTGGGGTCGTCC-3', CTAP303R 5'-GGTTTGCGGCGCTCACTG-3'.
QPCR
RNA was isolated from 20-day-old leaves or from protoplasts from 20-day-old leaves of the indicated mutant line using the RNA RNeasy Plant Mini Kit (QIAGEN, www.qiagen.com). 1 μg of RNA was used for a reverse transcription assay using oligo dT for first-strand synthesis in a Invitrogen Superscript III RTPCR Kit (Invitrogen 18080–051). cDNA was then used for QPCR using Power SYBR Green and an Applied Biosystems StepOne system, version 2.1, which calculated the comparative CT (ΔΔCT) with the following cycles; 95°C for 15 s, 56°C for 1 m, repeated 38 times. Actin expression served as an internal standard and wild-type untreated samples were set as the standard to which other samples were compared, in triplicate. Bar graphs in figures show standard deviation. Primers were as follows:
CML41 (At3g50770) 5'-CGGACGAAGATCACCAAAAT-3' and TGTCTGAGCTCAAAGGCTGA-3'; PAD3 (At3g26830) 5'-ACGAGCATCTTAAGCCTGGA-3' and 5'-TCGGTCATTCCCCATAGTGT-3';
Actin (AT3G18780) 5'-AGTGGTCGTAGAACCGGTATTGT-3' and 5'-GATGGCATGAGGAAGAGAGAAAC-3';
PTR3 (AT5G46050) 5'-ATCTTGGGTGCTTACGTTGGAG-3' and 5'-CCGAACTTTTTCGCAATTTTCCAC-3';
ESM1 (AT3G14210) 5'-CTTACTACGACGCCGGAAAC-3' and 5'-TTTAAGCACCGGTGGGAGTA-3';
MSR (At4g 04840) 5'-CGAAGTGTTCATTGTAGATCTGAGCTTGGACTTTGG-3' and 5'-GATTTCCGGCCATGGGTAAATACCCCG-3';
PDF1 (At5g44420) 5'-GATAAAGATAGGGATTTACCATGGTAAATGTAAAACGGGCG-3' and 5'-GGGTGATGCAAACTTAGCCATACTAGTGATGATTATTACTATTTTG-3'.
Mutagenesis
The WAK2 kinase catalytic site at amino acid 432 was altered from a K to an A using QuickChange Lightning kit (Strategene, La Jolla, CA), with the following oligonucleotide: 5'-CCGGACAACTCCATAGTTGCTATAAGCAAAGCTCGGCTTGG-3'. An R at position 89 was changed to a Q, and a K at position 99 to a T using the following oligonucleotides (respectively): 5'-GCCAGCTTCGTGTTCGGCTAGTTGAATCCAGAGTTTGCT-3' and 5'-TTTGCTACGATAGTCAAGGAACACAGACTGACTACATTGC-3'.
The mutant gene was cloned into the XhoI and SpeI sites of plasmid p289cTAP such that expression was driven by the 35SCaMV promoter as described (Kohorn et al., 2009) and the TAP tag was a carboxyl-terminal fusion (Kohorn et al., 2009).
Wild-typeA. thaliana plants were infected with the Agrobacteria culture carrying the plasmid, according to the Agrobacterium-mediated floral dip method (Clough and Bent, 1998).
Differential Gene Expression
RNA was extracted from 20-day-old WT or WAK2cTAP leaves using the RNeasy Plant Mini Kit (QIAGEN, www.qiagen.com). RNA was submitted to the Duke University Microarray Facility (Duke Institute for Genome Sciences and Policy, Durham, NC, USA). RNA quality assessments were performed on a Bioanalyzer and chip hybridization was performed according protocols available at www.genome.duke.edu/cores/microarray/services/affymetrix/. Affymetrix gene chip files were analyzed with GENESPRING GX 11 using Guided Workflow. Three biological replicates were analyzed. The robust multiple chip average (RMA) algorithm and baseline transformation to the median of all samples were performed to obtain normalized data. To control for the quality of the samples and for any possible degradation, the 3'/5' ratios for internal controls were calculated. The data were accepted if the ratios did not exceed 3. The quality of hybridization was controlled for with the hybridization controls bioB, bioC, bioD, and cre. These probes hybridize to artificial, non-Arabidopsis RNA added to each sample. Each is represented on the chip by four repeats of different concentrations. Data were used if the bioB signal was present at least 50% of the time, and if bioC, bioD, and cre were present all the time, and appeared in increasing concentrations. All chips satisfied these quality standards. To minimize error, the lowest 20 percentile of the row intensity values was removed from further analysis. This excluded unreliable fluorescence levels from the analysis, ensuring that only real readings were included in further data interpretation. Fold change analysis was performed on the normalized intensity values. The cut-off for significance that we used in our analysis was 2.0-fold. Gene onthology (GO) analysis was performed to identify genes of similar function or cellular localization within a large subset of genes. A P-value identifies the significance of the GO term within the subset compared with the whole dataset. The cut-off for the GO analysis that we used was 0.01, unless stated otherwise.
Western Blotting
Leaves were ground in 10 mM Tris 7, 3% SDS, 100 mM DTT, 10% glycerol, centrifuged at 10 000 g for 5 min, measured for chlorophyll content by spectrophotometry at 660 nm, adjusted for equal protein concentration, bromophenol blue was added, the sample was heated at 80°C for 10 min, and then separated by SDS–PAGE using 10% acrylamide gel, and transferred to nitrocellulose membrane for 1500 mA hours. Western blots were blocked with 5% (w/v) non-fat dry milk in Tris-buffered saline (TBS) supplemented with 3% Tween 20, incubated with peroxidase-antiperoxidase (PAP)-soluble complex (Sigma Corp.) at 1:2500 dilution for 2 h, and detected with chemiluminescence.
DAB staining was as described (Clarke, 2002). Young leaves were excised and placed in a 2-ml tube containing 50 μl DAB solution (1 mg ml−1 DAB, 0.05% Tween 20, 10 mM NaHPO4, pH 7). Vacuum was applied to facilitate the diffusion of the dye solution into the leaf, after which leaves were placed in a 80–90°C solution of 3:1:1 ethanol:acetic acid: glycerol (volume ratios) for 10 min to extract the chlorophyll pigment and visualize the DAB staining. Leaves were stored in the cooled-down solution overnight and observed under a Leica Wild M3Z dissecting microscope at 6–10× magnification. Digital images were taken with a Q-Imaging camera Retiga EX, 32–0029B-301.
Protoplast preparation was as described (Kohorn et al., 2006b, 2009). Protoplasts were treated with 40 μg ml−1 citrus pectin (mixed length from Sigma-Aldrich Corp P9561, St Louis, USA) and/or 100 μg ml−1 OGs dp 9–15 (purified by M. O’Niel, Univ. Georgia) for 6 h. RNA isolation and QPCR were as above.
Graphing and statistical analysis of plant mass (T test and standard error bars) were calculated and displayed with Prism software. Quantification for QPCR is described above.
SUPPLEMENTARY DATA
Supplementary Data are available at Molecular Plant Online.
FUNDING
This work is supported by an NSF grant MCB 0717983 to B.D.K. Students were supported in part by grants from NSF, the Howard Hughes Medical Institute, and NIH INBRI to Bowdoin College.
Supplementary Material
Acknowledgments
We would like to thank Patrick Krysan (University of Wisconsin) and John Walker (University of Missouri) for supplying the MPK mutant plants. OGs were a gift from Malcolm O’Niel (University of Georgia). We thank Barry Logan, Nat Wheelwright, and Jack Bateman for helpful comments. No conflict of interest declared.
References
- Anderson CM, Wagner TA, Perret M, He ZH, He D, Kohorn BD. WAKs: cell wall-associated kinases linking the cytoplasm to the extracellular matrix. Plant Mol. Biol. 2001;47:197–206. [PubMed] [Google Scholar]
- Andreasson E, Ellis B. Convergence and specificity in the Arabidopsis MAPK nexus. Trends Plant Sci. 2010;15:106–113. doi: 10.1016/j.tplants.2009.12.001. [DOI] [PubMed] [Google Scholar]
- Brutus A, Sicilia F, Macone A, Cervone F, De Lorenzo G. A domain swap approach reveals a role of the plant wall-associated kinase 1 (WAK1) as a receptor of oligogalacturonides. Proc. Natl Acad. Sci. U S A. 2010;107:9452–9457. doi: 10.1073/pnas.1000675107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush SM, Krysan PJ. Mutational evidence that the Arabidopsis MAP kinase MPK6 is involved in anther, inflorescence, and embryo development. J. Exp. Bot. 2007;58:2181–2191. doi: 10.1093/jxb/erm092. [DOI] [PubMed] [Google Scholar]
- Clarke J. Phenotypic analysis of Arabidopsis mutants: diaminobenzidine stain for hydrogen peroxide. In: Weigel D, Glazebrook J, editors. Arabidopsis: A Laboratory Manual. Cold Spring Harbor, NY, USA: CSHL Press; 2002. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana . Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Colcombet J, Hirt H. Arabidopsis MAPKs: a complex signalling network involved in multiple biological processes. Biochem. J. 2008;413:217–226. doi: 10.1042/BJ20080625. [DOI] [PubMed] [Google Scholar]
- Decreux A, Messiaen J. Wall-associated kinase WAK1 interacts with cell wall pectins in a calcium-induced conformation. Plant Cell Physiol. 2005;46:268–278. doi: 10.1093/pcp/pci026. [DOI] [PubMed] [Google Scholar]
- Decreux A, Thomas A, Spies B, Brasseur R, Van Cutsem P, Messiaen J. In vitro characterization of the homogalacturonan-binding domain of the wall-associated kinase WAK1 using site-directed mutagenesis. Phytochemistry. 2006;67:1068–1079. doi: 10.1016/j.phytochem.2006.03.009. [DOI] [PubMed] [Google Scholar]
- Denoux C, Galletti R, Mammarella N, Gopalan S, Werck D, De Lorenzo G, Ferrari S, Ausubel FM, Dewdney J. Activation of defense response pathways by OGs and Flg22 elicitors in Arabidopsis seedlings. Mol. Plant. 2008;1:423–445. doi: 10.1093/mp/ssn019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diener AC, Ausubel FM. RESISTANCE TO FUSARIUM OXYSPORUM 1, a dominant Arabidopsis disease-resistance gene, is not race specific. Genetics. 2005;171:305–321. doi: 10.1534/genetics.105.042218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galletti R, Denoux C, Gambetta S, Dewdney J, Ausubel FM, De Lorenzo G, Ferrari S. The AtrbohD-mediated oxidative burst elicited by oligogalacturonides in Arabidopsis is dispensable for the activation of defense responses effective against Botrytis cinerea . Plant Physiol. 2008;148:1695–1706. doi: 10.1104/pp.108.127845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He ZH, Cheeseman I, He D, Kohorn BD. A cluster of five cell wall-associated receptor kinase genes, Wak1-5, are expressed in specific organs of Arabidopsis . Plant Mol. Biol. 1999;39:1189–1196. doi: 10.1023/a:1006197318246. [DOI] [PubMed] [Google Scholar]
- He ZH, Fujiki M, Kohorn BD. A cell wall-associated, receptor-like protein kinase. J. Biol. Chem. 1996;271:19789–19793. doi: 10.1074/jbc.271.33.19789. [DOI] [PubMed] [Google Scholar]
- He ZH, He D, Kohorn BD. Requirement for the induced expression of a cell wall associated receptor kinase for survival during the pathogen response. Plant J. 1998;14:55–63. doi: 10.1046/j.1365-313x.1998.00092.x. [DOI] [PubMed] [Google Scholar]
- Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- Kohorn BD. Plasma membrane-cell wall contacts. Plant Physiol. 2000;124:31–38. doi: 10.1104/pp.124.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohorn BD. WAKs: cell wall associated kinases. Curr. Opin. Cell Biol. 2001;13:529–533. doi: 10.1016/s0955-0674(00)00247-7. [DOI] [PubMed] [Google Scholar]
- Kohorn BD, Johansen S, Shishido A, Todorova T, Martinez R, Defeo E, Obregon P. Pectin activation of MAP kinase and gene expression is WAK2 dependent. Plant J. 2009;60:974–982. doi: 10.1111/j.1365-313X.2009.04016.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohorn BD, Kobayashi M, Johansen S, Friedman HP, Fischer A, Byers N. Wall-associated kinase 1 (WAK1) is crosslinked in endomembranes, and transport to the cell surface requires correct cell-wall synthesis. J. Cell Sci. 2006a;119:2282–2290. doi: 10.1242/jcs.02968. [DOI] [PubMed] [Google Scholar]
- Kohorn BD, Kobayashi M, Johansen S, Riese J, Huang LF, Koch K, Fu S, Dotson A, Byers N. An Arabidopsis cell wall-associated kinase required for invertase activity and cell growth. Plant J. 2006b;46:307–316. doi: 10.1111/j.1365-313X.2006.02695.x. [DOI] [PubMed] [Google Scholar]
- Lally D, Ingmire P, Tong HY, He ZH. Antisense expression of a cell wall-associated protein kinase, WAK4, inhibits cell elongation and alters morphology. Plant Cell. 2001;13:1317–1331. doi: 10.1105/tpc.13.6.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Zhou SY, Zhao WS, Su SC, Peng YL. A novel wall-associated receptor-like protein kinase gene, OsWAK1, plays important roles in rice blast disease resistance. Plant Mol. Biol. 2009;69:337–346. doi: 10.1007/s11103-008-9430-5. [DOI] [PubMed] [Google Scholar]
- Sampoli Benitez BA, Komives EA. Disulfide bond plasticity in epidermal growth factor. Proteins. 2000;40:168–174. doi: 10.1002/(sici)1097-0134(20000701)40:1<168::aid-prot180>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- Seifert GJ, Blaukopf C. Irritable walls: the plant extracellular matrix and signaling. Plant Physiol. 2010;153:467–478. doi: 10.1104/pp.110.153940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres MA. ROS in biotic interactions. Physiol Plant. 2010;138:414–429. doi: 10.1111/j.1399-3054.2009.01326.x. [DOI] [PubMed] [Google Scholar]
- Verica JA, Chae L, Tong H, Ingmire P, He ZH. Tissue-specific and developmentally regulated expression of a cluster of tandemly arrayed cell wall-associated kinase-like kinase genes in Arabidopsis . Plant Physiol. 2003;133:1732–1746. doi: 10.1104/pp.103.028530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner TA, Kohorn BD. Wall-associated kinases are expressed throughout plant development and are required for cell expansion. Plant Cell. 2001;13:303–318. doi: 10.1105/tpc.13.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Ngwenyama N, Liu Y, Walker JC, Zhang S. Stomatal development and patterning are regulated by environmentally responsive mitogen-activated protein kinases in Arabidopsis . Plant Cell. 2007;19:63–73. doi: 10.1105/tpc.106.048298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willats WG, McCartney L, Mackie W, Knox JP. Pectin: cell biology and prospects for functional analysis. Plant Mol. Biol. 2001;47:9–27. [PubMed] [Google Scholar]
- Zhang S, Chen C, Li L, Meng L, Singh J, Jiang N, Deng XW, He ZH, Lemaux PG. Evolutionary expansion, gene structure, and expression of the rice wall-associated kinase gene family. Plant Physiol. 2005;139:1107–1124. doi: 10.1104/pp.105.069005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.