Abstract
Plants synchronize their cellular and physiological functions according to the photoperiod (the length of the light period) in the cycle of 24 h. Photoperiod adjusts several traits in the plant life cycle, including flowering and senescence in annuals and seasonal growth cessation in perennials. Photoperiodic development is controlled by the coordinated action of photoreceptors and the circadian clock. During the past 10 years, remarkable progress has been made in understanding the molecular mechanism of the circadian clock, especially with regard to the transition of Arabidopsis from the vegetative growth to the reproductive phase. Besides flowering photoperiod also modifies plant photosynthetic structures and traits. Light signals controlling biogenesis of chloroplasts and development of leaf photosynthetic structures are perceived both by photoreceptors and in chloroplasts. In this review, we provide evidence suggesting that the photoperiodic development of Arabidopsis leaves mimics the acclimation of plant to various light intensities. Furthermore, the chloroplast-to-nucleus retrograde signals that adjust acclimation to light intensity are proposed to contribute also to the signaling pathways that control photoperiodic acclimation of leaves.
Keywords: acclimation, chloroplast biology, circadian clock, leaf/vegetative development, light signaling, photomorphogenesis, plastid signaling
INTRODUCTION
Plant development is controlled by numerous external factors that coordinate the timing of developmental and adaptive processes to meet the requirements of the environment. The quantity and quality of light and the length of the diurnal light period in a day cycle of 24 h (photoperiod) together with the temperature and availability of nutrients adjust the morphology and extent of plant growth as well as the timing of the annual developmental phases in nature. From these variables, the day length is the most reliable indicator for the annual season because of its high predictability. Strict response to photoperiod is critical for perennial overwintering plants in temperate latitudes to adjust their yearly development with favorable growth conditions and to initiate bud formation and growth cessation before the cold season. Length of photoperiod is important also for annual plant species in adjusting the transitions of developmental phases from juvenile to vegetative and from vegetative to reproductive phase during their lifecycle.
Plant photoperiodic responses are classified into three categories: short-day (SD) responses, in which the response occurs in photoperiod shorter than the critical photoperiod; long-day (LD) responses, in which the response occurs in photoperiod longer than the critical photoperiod; and day-neutral (DN) responses. Plants showing the response under distinct photoperiod are called SD, LD, or DN plants, respectively. Obligate SD or LD plant species show the response only under inducing photoperiod. In facultative SD or LD plants, the response is promoted by a short or long photoperiod, respectively, but it can be induced also by other photoperiods. In the latter case, the intensity of the response is weaker and/or the initiation of the response is delayed. Timing of flowering is one of the few photoperiodic responses that have been minutely characterized at the molecular level (recent reviews, see Turck et al., 2008; Imaizumi, 2010). Recently, however, growing interest has been paid to the initiation of bud dormancy and the cessation of growth in trees (Jimenez et al., 2010; Kozarewa et al., 2010; Olsen, 2010). Molecular dissection of the initiation, transition, and development of the photoperiodic responses is crucial, since the photoperiod contributes to the control of several scientifically and economically important plants traits, including leaf morphology, vegetative production, seed production, stress tolerance, and dormancy.
Light is the major environmental factor that adjusts the photosynthetic traits in plant species. Light signaling pathways associated with de-etiolation of seedlings and with acclimation to light intensity have been actively studied in plants (Nagy and Schafer, 2002; Sullivan and Deng, 2003; Jung and Chory, 2010), while photoperiodic adjustments in chloroplast structure and function are less well characterized. Recently, it was proposed that the photoreceptor-dependent signaling pathways interact with chloroplast retrograde signaling pathways by either promoting or antagonizing each other, depending on the processes dissected (Ruckle et al., 2007; Ruckle and Larkin, 2009). Here, we review the photosynthetic traits and structures controlled by the length of the photoperiod. Specific focus is put on chloroplast biogenesis and plastid-derived signals in the control of light intensity-dependent and photoperiodic growth in Arabidopsis.
ACCLIMATION OF ARABIDOPSIS ACCORDING TO THE LENGTH OF THE PHOTOPERIOD
Arabidopsis Col-0 is a facultative LD plant, in which photoperiods longer than 12 h (LD) accelerate flowering by several weeks in comparison with photoperiods shorter than 12 h (SD). SD distinctly extends the vegetative phase of Arabidopsis and delays senescence. The number of leaves in mature rosette is on average 40% higher in SD than in LD plants (Cookson et al., 2007). This is opposite to perennial deciduous trees, in which the SD promotes leaf senescence (Zhao et al., 2009) that is related to the growth cessation at the end of the growing season.
The ability of Arabidopsis to react to the daily light rhythm increases growth, whereas incorrect matching of endogenous rhythms with environmental rhythms reduces plant fitness. For example, the extension of external light–dark cycle of 24 to 28 h (14-h L/14-h D) reduced the areal biomass production by ∼50% (Dodd et al., 2005). Likewise, the growth of the short- and long-circadian period mutants with altered endogenous clock periods was promoted by the external day–night cycle corresponding to their own endogenous circadian rhythms (Dodd et al., 2005). Furthermore, the arrhythmic plants overexpressing the molecular oscillation component CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) of the circadian clock and grown under a normal 24-h cycle had distinctly lower net CO2 assimilation and biomass production than wild-type plants (Dodd et al., 2005). Adjustment of the growth with the external light–dark cycle is partially attained by circadian-clock-dependent control of global gene expression. Indeed, 5.5–15.4% of Arabidopsis genes have been estimated to be regulated by the circadian clock (Covington et al., 2008).
Length of the photoperiod has a distinct influence on biomass production, leaf and cell structure, and on the ultrastructure of chloroplasts. In general, Arabidopsis plants grown under SD or LD photoperiod with similar light intensity show both structural and photosynthetic characteristics typical of shade or sun plants, respectively (Figure 1; Lepistö et al., 2009). Like sun plants (Walters and Horton, 1995; Lake et al., 2001), LD-grown plants and plants grown under continuous light have thicker leaves, long-shaped palisade cells, high stomatal index in leaf epidermis, and smaller grana stacks in chloroplasts when compared to SD-grown plants (Figure 1; Lepistö et al., 2009). For example, growth of Arabidopsis in LD substantially increased the stomatal index (the ratio of the number of stomata to the total number of epidermal cells) about 40% as compared with SD plants (Lepistö et al., 2009). Furthermore, the net CO2 assimilation per rosette area (measured at ambient CO2 and saturating light intensity) is about 20% higher in LD-leaves, whereas the mitochondrial respiration rate is only 50% of that measured in SD-leaves (Lepistö et al., 2009). LD-leaves also have a higher chlorophyll (Chl) content per leaf area due to thicker leaves compared to SD-leaves and the Chl a/b ratio in LD-leaves is similar to plants grown at medium or high light (Walters and Horton, 1995; Lepistö et al., 2009).
Figure 1.
Light Micrographs of Leaf Cross-Sections and Electron Micrographs of Chloroplasts in Col-0 (A) and ntrc (B).
Plants were grown under short day (SD) for 4 weeks and under long day (LD) for 3 weeks. Arrows indicate the irregular shape of the ntrc cells. * indicates a plastid-like organelle in ntrc cell. Scale bars: 100 μm for light micrographs and 2 μm for electron micrographs.
Substantial increase in Chl a/b ratio of LD-leaves implies photoperiodical changes in the composition of the light-harvesting complexes of the thylakoid membranes. The grana stacks are smaller in chloroplasts of LD-leaves in comparison to SD-leaves (Figure 1). Accordingly, the amount of the major trimeric chlorophyll a/b-binding proteins of the Photosystem II (PSII) antenna (LHCII) in thylakoid membranes is declined in LD-leaves as compared to SD-leaves (Lepistö et al., 2009; Victor et al., 2010). The relative proportion of the representative subunits of PSII, PSI, and cytochrome bf complexes did not, however, differ significantly in SD- and LD-leaves (A. Lepistö, E. Pakula, E. Rintamäki, unpublished results), neither did the maximal electron transport rates estimated for the SD- and LD-grown plants (Lepistö et al., 2009).
METABOLIC AND TRANSCRIPTOMIC MODIFICATIONS IN ARABIDOPSIS LEAVES GROWN UNDER SD AND LD PHOTOPERIODS
Anatomical and photosynthetic traits of leaves indicate that the acclimation of Arabidopsis to SD and LD photoperiods mimics the responses detected in leaves acclimated to low light and medium/high light, respectively. The question is whether the photoperiodic development is controlled by the same signaling network mediating the light-intensity-dependent acclimation of plants. Redox signals that arise from chloroplasts play a major role in the development of high-light structures in leaves (Nott et al., 2006; Piippo et al., 2006; Pfannschmidt et al., 2009). Short-term transfer of Arabidopsis to high light enhances the production of reactive oxygen species that has been suggested to initiate high-light acclimation (Vanderauwera et al., 2005; Muhlenbock et al., 2008; Foyer and Noctor, 2009). In the course of high-light acclimation, elevated ROS production is compensated for by induction of antioxidant systems in leaves (Mittler et al., 2004), which in turn prevents the oxidation of leaf cells. In Arabidopsis acclimated to LD photoperiods, no substantial amounts of superoxide or H2O2 were found to accumulate in illuminated leaves (Figure 2). Furthermore, the growth in the LD photoperiod was shown to modify only slightly the antioxidant levels in Arabidopsis leaves. Catalase activity has been reported to rise in LD-grown leaves in comparison to SD-leaves, whereas the steady-state contents and the oxidation level of ascorbate and glutathione were not markedly different in SD- and LD-leaves (Queval et al., 2007). These reports suggest that the production and detoxification of ROS are balanced in plants acclimated to LD.
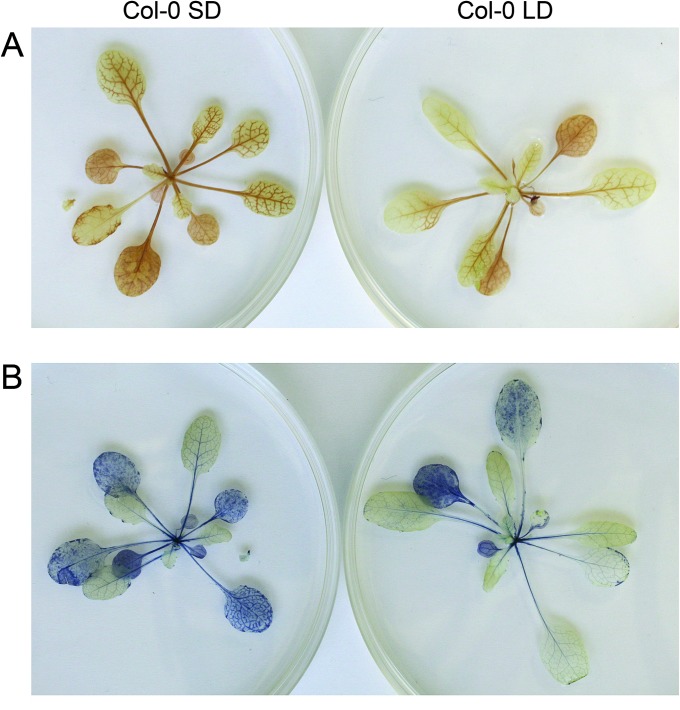
Figure 2.
Accumulation of H2O2 (A) and Superoxide (B) in Col-0 Leaves Grown under SD or LD Conditions.
Accumulation of H2O2 and superoxide was detected using DAB (diaminobenzidine; Sigma-Aldrich) and NBT (nitroblue tetrazolium; Sigma-Aldrich) substrates, respectively. Rosettes were excised at the end of the light period, and incubated on Petri dishes containing 0.1 mg ml−1 solution of DAB (pH 3.8) or a 5 mg ml−1 solution of NBT overnight in darkness. In the subsequent morning, the dishes were transferred to growth light (130 μmol photons m−2 s−1 at 20°C) for 1 h and, thereafter, the rosettes were incubated in ethanol until chlorophyll was bleached.
In contrast to LD conditions, H2O2 accumulated in Arabidopsis leaves upon acclimation to SD photoperiod (Figure 2). Chloroplasts may contribute to increased accumulation of ROS in SD-leaves, since the thylakoid membranes isolated from SD-acclimated Arabidopsis (A. Lepistö, E. Pakula, J. Toivola, A. Krieger-Liszkay, F. Vignols, E. Rintamäki, unpublished results) and tobacco leaves (Michelet and Krieger- Liszkay, 2011) produced more ROS than thylakoids isolated from LD-acclimated plants. Furthermore, the abundance of photorespiratory enzymes, except peroxisomal catalase, increased in SD-acclimated plants (Victor et al., 2010). This suggests an elevation in peroxisomal H2O2 production in leaves as well. Accordingly, acclimation to SD conditions has been shown to result in increased expression of H2O2 marker genes (Queval et al., 2007). The growth in SD conditions also promotes the ascorbate metabolism in leaves. The abundances of the enzymes related to ascorbate biosynthesis, monodehydroascorbate reductase, and dehydroascorbate reductase were three to fourfold higher in SD-acclimated shoot tips of grapevine in comparison to plants acclimated to LD (Victor et al., 2010). Regardless of the changes in antioxidant components, higher accumulation of ROS in illuminated SD-leaves (Figure 2) suggests that ROS production is controlled in SD-leaves instead of complete elimination of oxidants. The elevated oxidative state of SD-cells likely operates as a control loop in adjusting the redox-controlled metabolism to the photoperiod during growth.
Length of the photoperiod modifies not only the ROS metabolism, but also the sugar metabolism of leaves. Control of metabolism and growth by photoperiod has been tested by transferring 21-day-old Arabidopsis plants to various light–dark regimes with 2–12 h of light in 24-h cycles and by analyzing the growth rate of rosettes, the metabolites (sugars, amino acids, organic acids) and metabolic enzymes in leaves 3 weeks after the transfer (Gibon et al., 2009). In this experiment, the highest positive correlation was found between the growth rate of rosettes and the degradation rate of starch in the dark. Growth under the SD photoperiod increased the synthesis rate of starch in the light period, whereas the degradation rate of starch in the dark period was strongly decreased in comparison to LD (Lu et al., 2005; Gibon et al., 2009). The molecular mechanism controlling the transient formation of starch under various light–dark regimes is not known, but several mechanisms including feedback inhibition from carbohydrate metabolism, redox regulation, and transcriptional control of chloroplast enzymes have been proposed (Zeeman et al., 2007). Accumulation of sucrose and maltose in night correlated positively with the starch degradation rate (Lu et al., 2005), suggesting that the feedback inhibition from end products may not be a primary cause for slow starch degradation rate in SD-leaves. The expression of the genes encoding starch-metabolizing enzymes is under light-dependent circadian control (Lu et al., 2005). However, the degradation rate of starch declined/increased already in the first night after a change of photoperiod from LD to SD and vice versa, respectively (Lu et al., 2005), suggesting that, if the transcriptional control is involved in the regulation of starch breakdown, the signal should preferably come directly from chloroplast to nucleus rather than from the external input.
Redox regulation of enzymes in starch metabolism likely is a key mechanism that controls the differential starch turnover in plants acclimated to SD and LD photoperiods. ADP-glucose pyrophosphorylase (AGPase) is a key enzyme in starch synthesis that controls the flux from photosynthates to starch. AGPase is a heterotetrameric enzyme that consists of large and small subunits, and is redox-activated in light by thioredoxin that reduces the disulphide bridge between small subunits (Hendriks et al., 2003). Also, the enzymes involved in starch degradation, glucan, water dikinase (GWD), dual specificity protein phosphatase (DSP4), and β-amylase 1 (BAM1) have been shown to be under redox control (Mikkelsen et al., 2005; Sokolov et al., 2006; Sparla et al., 2006). Prior to degradation by amylases, starch granules are reversibly phosphorylated by GWD and DSP4 (Zeeman et al., 2007). This reversible phosphorylation is proposed to disrupt the crystalline structure of amylopectin and mutant analyses have shown that both enzymes are necessary for efficient remobilization of starch in Arabidopsis (Yu et al., 2001; Ritte et al., 2002; Zeeman et al., 2010). All these enzymes are reported to be regulated by thioredoxins (Hendriks et al., 2003; Mikkelsen et al., 2005; Sokolov et al., 2006), pointing to the importance of the thioredoxin system in the regulation of starch metabolism. Besides controlling enzyme activities, thioredoxins are involved in ROS scavenging (Mittler et al., 2004). Thus, the elevated accumulation of ROS in illuminated SD-leaves (Figure 2) may impact on the activity of the enzymes in starch metabolism by challenging the thioredoxin systems in chloroplast. Photosynthetic carbon fixation is feedback-regulated by starch metabolism (Stettler et al., 2009). It is thus likely that the redox-dependent regulation of starch metabolism adjusts the rate of photosynthetic carbon fixation with the growth potential of SD-acclimated Arabidopsis.
As reviewed in the previous chapters, particularly short photoperiods induce structural and metabolic changes in Arabidopsis leaves. Global transcript profiling approaches have been used to reveal the specific gene clusters related to the acclimation of Arabidopsis to the SD photoperiod and to the maintenance of the metabolic state in the SD photoperiod (Queval et al., 2007; Tables 1 and 2). When Arabidopsis plants grown for 2 weeks under a 12-h/12-h photoperiod were transferred to SD, the majority of genes differentially expressed in leaves in the second day after the transfer were up-regulated (Cluster 1 genes, Table 1). These Cluster 1 genes are postulated to be important for acclimation of Arabidopsis to the SD photoperiod, based both on the biological process assigned to the differentially expressed gene and on the previous microarray analyses (the Genevestigator database of 6100 ATH1 experiments, www.genevestigator.com/gv/index.jsp; Hruz et al., 2008). Stimulated Cluster 1 genes include a gene associated with the cell cycle as well as genes involved in the regulation of transcription and circadian rhythm. Furthermore, ∼50% of Cluster 1 genes were moderately or strongly repressed in Arabidopsis shoot apex after transfer of 5-week-old plants from the SD to LD photoperiod promoting flowering (accession number AT-00326 in Genevestigator database; Balasubramanian et al., 2006). This indicates that the Cluster 1 genes are important in maintenance of the vegetative phase of the shoot apex in SD conditions. Although the SD-grown leaves structurally and functionally resemble the leaves acclimated to low light intensity (Figure 1; Lepistö et al., 2009), only five genes differentially expressed after transfer of plants from 12L/12D rhythm to SD respond to light intensity (Table 1). From these genes, COLD, CIRCADIAN RHYTHM, AND RNA BINDING2 -LIKE gene (CCR-LIKE) is an interesting one, since its expression is controlled by circadian clock, photoperiod, and light intensity (Table 1). The expression of CCR-LIKE is stimulated in leaves transferred to short photoperiod, whereas the gene is repressed in shoot apex after transfer of plants to long photoperiod and also in leaves exposed to high light. CCR-LIKE shows homology to CCR2 gene that is also up-regulated in Arabidopsis leaves under the SD photoperiod (Table 1). CCR2 controls the stability of its own and other target transcripts (Staiger et al., 2003), while CCR-LIKE gene encodes chloroplast-localized protein with unknown function. Despite the accumulation of ROS in SD-leaves, the Cluster 1 genes do not respond to treatment of leaves with H2O2 (accession number AT-00185 in Genevestigator database), suggesting that the expression of these genes is not primarily controlled by H2O2 signaling cascade. Therefore, the enhanced accumulation of H2O2 in SD-acclimated plants is likely not a factor that induces acclimation to the SD photoperiod.
Table 1.
Differentially Expressed Genes (Cluster 1) in Arabidopsis Leaves after Transfer from 12L/12D Rhythm to Short-Day Conditions.
| AGI code | Fold change | Description | Location | Biological process |
| AT3G27060* | 7,12 | ATTSO2 | Cell cycle | |
| AT1G28160 | 3,69 | Member of the ERF subfamily B-1 of ERF/AP2 transcription factor family | Nucleus | Transcription |
| AT2G40350* | 3,67 | Member of the DREB subfamily A-2 of ERF/AP2 transcription factor family | Regulation of transcription | |
| AT4G30650* | 3,23 | Unknown protein | ||
| AT1G69190 | 2,96 | Bifunctional cytosolic hydroxymethyldihydropterin pyrophosphokinase/dihydropteroate synthase (HPPK/DHPS) | Cytosol | Tetrahydrofolate biosynthesis |
| AT1G28520 | 2,92 | VASCULAR PLANT ONE ZINC FINGER PROTEIN | ||
| AT2G15970* | 2,82 | ARABIDOPSIS THALIANA COLD-REGULATED413 PLASMA MEMBRANE 1 | Plasma membrane, vacuole | |
| AT1G53290 | 2,79 | Galactosyltransferase family protein | Protein glycosylation | |
| AT2G24330 | 2,74 | Unknown protein | ||
| AT2G21660* | 2,70 | COLD, CIRCADIAN RHYTHM, AND RNA BINDING 2 (CCR2) (AtGRP7) | Circadian rhythm | |
| AT3G13500 | 2,64 | Unknown protein | ||
| AT5G15960↑ | 2,62 | Cold and ABA inducible protein kin1 | ||
| AT2G30720 | 2,61 | Thioesterase family protein | ||
| AT2G24290 | 2,38 | Na+- and K+-sensitive 1 | ||
| AT3G26470 | 2,37 | Unknown protein | ||
| AT1G13930* | 2,33 | Involved in response to salt stress | ||
| AT2G35733 | 2,33 | Unknown protein | ||
| AT2G47070 | 2,32 | SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 1 | ||
| AT2G42070 | 2,31 | ARABIDOPSIS THALIANA NUDIX HYDROLASE HOMOLOG 23 | Chloroplast | |
| AT3G26740↓* | 2,26 | CCR-LIKE | Chloroplast | Circadian clock |
| AT1G10760* | 2,13 | STARCH EXCESS1, starch degradation | Chloroplast | Carbohydrate metabolism |
| AT2G40840* | 2,12 | DISPROPORTIONATING ENZYME 2 | Cytosol | Carbohydrate metabolism |
| AT1G20440↑* | 2,10 | COLD-REGULATED 47 Dehydrin | ||
| AT4G11600* | 2,08 | GLUTATHIONE PEROXIDASE 6 | Chloroplast, mitochondria, cytosol | Oxidative stress defense |
| AT4G26820 | 2,06 | Unknown protein | ||
| AT1G05170↑ | 2,04 | Galactosyltransferase family protein | Protein glycosylation | |
| AT1G52870 | 2,04 | Peroxisomal membrane protein-related | ||
| AT3G18080* | 2,04 | B-S GLUCOSIDASE 44 | Cell wall | Carbohydrate metabolism |
| AT5G62350* | 2,04 | Invertase/pectin methylesterase inhibitor family protein | ||
| AT5G01370 | 2,03 | ALC-INTERACTING PROTEIN1 | Nucleus | |
| AT1G20620* | 2,01 | CATALASE 3 | Mitochondrion, peroxisome | Hydrogen peroxide catabolic processes |
| AT5G13930↑ | 0,35 | CHALCONE SYNTHASE | ER | Flavonoid biosynthesis |
Arabidopsis was grown for 2 weeks in 12-h photoperiod and then transferred to 8-h SD photoperiod for 2 d. Gene expression is indicated as a ratio of transcript level in leaves transferred to SD in comparison to leaves before the transfer. The fold-change values are means of three independent biological replicates. ↑↓, genes induced or repressed by high-light treatment, respectively (accession AT-00246 in Genevestigator database, Kleine et al., 2007). * Genes repressed in Arabidopsis shoot apex after transfer of 5-week-old plant from SD to LD photoperiod (see text for details).
Rosette leaves were harvested from plants grown under 100 μmol photons m−2 s−1 at 20°C under 12L/12D for 2 weeks and thereafter transferred to SD (8L/16D) for 2 d. Total RNA was isolated with Trizol reagent and labeled by the aminoallyl method with Cy3 or Cy5 fluorescent dyes. RNA isolation, cDNA synthesis, labeling, hybridization, and the data analysis were performed as described in Lepistö et al. (2009). Genes up-regulated more than twofold or down-regulated more than 0.5-fold with P < 0.10 are shown in the table.
Table 2.
Differentially Expressed Genes (Cluster 1) in SD-Grown Arabidopsis Leaves in Comparison to LD-Grown Leaves.
| AGI code | Fold change | Description | Location | Biological process |
| AT4G27440↓ | 16,71 | PROTOCHLOROPHYLLIDE OXIDOREDUCTASE B | Chloroplast | Chlorophyll biosynthesis |
| AT2G24330 | 5,14 | Unknown protein | ||
| AT3G52610 | 5,04 | Unknown protein | ||
| AT1G28520 | 5,01 | VASCULAR PLANT ONE ZINC FINGER PROTEIN (VOZ1) | ||
| AT2G42070 | 4,83 | ARABIDOPSIS THALIANA NUDIX HYDROLASE HOMOLOG 23 | Chloroplast | |
| AT4G26820 | 4,43 | Unknown protein | ||
| AT5G03350↓ | 4,18 | Legume lectin family protein | ||
| AT1G53290 | 4,06 | Galactosyltransferase family protein | Protein glycosylation | |
| AT5G02160↓ | 3,81 | Unknown protein | Chloroplast | |
| AT1G12090 | 3,57 | EXTENSIN-LIKE PROTEIN | Lipid transport | |
| AT2G15970 | 3,41 | COLD REGULATED 413 PLASMA MEMBRANE 1 | ||
| AT2G26830 | 3,38 | EMBRYO DEFECTIVE 1187 | ||
| AT2G16030 | 3,22 | Methyltransferase | ||
| AT1G33850 | 3,13 | 40S ribosomal protein S15 | Translation | |
| AT1G28160 | 3,13 | Member of the ERF subfamily B-1 of ERF/AP2 transcription factor family | Nucleus | Transcription |
| AT3G15000 | 3,10 | DAG (differentiation and greening)-like | Mitochondria | |
| AT2G44930 | 3,04 | Unknown protein | ||
| AT2G20420 | 2,98 | Succinyl-CoA ligase | Mitochondrion | |
| AT1G49500↓ | 2,90 | Unknown protein | ||
| AT5G50890 | 2,86 | Unknown protein | ||
| AT5G62350 | 2,84 | Invertase/pectin methylesterase inhibitor family protein | ||
| AT1G73770 | 2,79 | Unknown protein | ||
| AT4G01210 | 2,69 | Glycosyltransferase family protein | ||
| AT3G08010 | 2,63 | ATAB2 | Chloroplast | Biogenesis of Photosystem I and II |
| AT2G30720 | 2,47 | Thioesterase family protein | ||
| AT1G48920 | 2,43 | NUCLEOLIN LIKE 1 | Nucleolus | rRNA processing |
| AT2G45170↓ | 2,40 | AUTOPHAGY 8E | Autophagy | |
| AT2G26135 | 2,36 | Zinc finger family protein | ||
| AT4G14230 | 2,29 | Unknown protein | ||
| AT1G20620 | 2,25 | CATALASE 3 | Mitochondrion, peroxisome | Hydrogen peroxide catabolic processes |
| AT5G01370 | 2,22 | ALC-INTERACTING PROTEIN1 | Nucleus | |
| AT1G20020 | 2,13 | LEAF FNR 2 | Chloroplast | Photosynthesis |
| AT3G15800 | 2,07 | Glycosyl hydrolase family 17 protein | Carbohydrate metabolism | |
| AT5G58250 | 2,06 | Unknown protein | ||
| AT5G45300 | 2,04 | BETA-AMYLASE 2 | Carbohydrate metabolism | |
| AT5G14200 | 0,48 | ISOPROPYLMALATE DEHYDROGENASE 1 | Leucine biosynthesis | |
| AT3G07440 | 0,48 | Unknown protein | ||
| AT2G15020 | 0,47 | Unknown protein | ||
| AT4G13770 | 0,45 | CYTOCHROME P450 83A1 | Glucosinolate biosynthetic process | |
| AT5G04140 | 0,43 | FERREDOXIN-DEPENDENT GLUTAMATE SYNTHASE 1 | Chloroplast, mitochondrion, apoplast | Photorespiration |
| AT2G38170 | 0,43 | ATCAX1, RARE COLD INDUCIBLE 4 | Vacuole | Cellular manganese and zink ion homeostasis |
| AT2G38230 | 0,42 | PYRIDOXINE BIOSYNTHESIS 1.1 | Cytosol, chloroplast | Vitamin biosynthesis |
| AT3G22890 | 0,42 | ATP SULFURYLASE 1 | Chloroplast | Sulfate assimilation |
| AT1G64500 | 0,33 | Glutaredoxin family protein | Cell redox homeostasis | |
| AT1G23130 | 0,33 | Polyketide cyclase/dehydrase and lipid transport superfamily protein | ||
| AT1G67865 | 0,33 | Unknown protein | ||
| AT2G21970 | 0,32 | STRESS ENHANCED PROTEIN 2, chlorophyll a/b-binding protein | Chloroplast | Photosynthesis |
| AT4G35090 | 0,32 | CATALASE 2 | Peroxisome | Photorespiration |
| AT1G37130 | 0,29 | ARABIDOPSIS NITRATE REDUCTASE 2 | Plasma membrane, vacuole | Nitrate assimilation |
| AT3G09390 | 0,28 | ARABIDOPSIS THALIANA METALLOTHIONEIN-1 | Cellular copper ion homeostasis |
The fold-change values are means of three independent biological replicates. ↓ Genes repressed by high-light treatment (accession AT-00246 in Genevestigator database; Kleine et al., 2007).
Rosette leaves were harvested from plants grown under 100 μmol photons m−2 s−1 at 20°C under SD for 4 weeks and under LD for 3 weeks. Total RNA was isolated with Trizol reagent and labeled by the aminoallyl method with Cy3 or Cy5 fluorescent dyes. RNA isolation, cDNA synthesis, labeling, hybridization, and the data analysis were performed as described in Lepistö et al. (2009). Genes up-regulated more than twofold or down-regulated more than 0.5-fold with P < 0.10 are shown in the table.
The transcript profiling of plants shifted to the SD photoperiod did not highlight any distinct metabolic pathway (Table 1). The genes involved in sugar and starch metabolisms were induced, which is likely linked with the modification of the diurnal cycle of starch metabolism in SD-plants. The key enzyme in flavonoid biosynthesis, CHALCONE SYNTHASE, was strongly repressed after transfer to the SD photoperiod, being in accordance with the low accumulation of anthocyanins in SD-grown Arabidopsis leaves (Lepistö et al., 2009).
Comparison of transcript levels in SD- and LD-acclimated leaves did not reveal either any drastic differences in the expression of genes involved in primary metabolism or stress responses (Table 2; Queval et al., 2007). The majority of differentially expressed genes were activated under SD conditions in comparison to LD-grown leaves (Table 2). Thirty-four percent of the Cluster 1 genes (Table 1) were also differentially expressed in leaves grown in SD conditions (Table 2). Repressed genes in SD include genes connected to nitrogen (NITRATE REDUCTASE 2 and FERREDOXIN-DEPENDENT GLUTAMATE SYNTHASE 1) and sulfate assimilation (ATP SULFURYLASE 3), implying reduced growth capacity of SD-grown Arabidopsis. Also, some distinct genes related to cellular redox control (CATALASE 2, THIOREDOXIN H3, METALLOTHIONEIN-1, GLUTAREDOXIN) were repressed in SD-acclimated plants. CATALASE 2 encodes a peroxisomal isoform of catalase that detoxifies H2O2 produced in photorespiration. Interestingly, the abundances of other photorespiratory enzymes except CATALASE2 were higher in SD-acclimated plants (Victor et al., 2010). This indicates that photorespiration is enhanced in SD-leaves, while the scavenging machinery in peroxisome is likely down-regulated. This provides further evidence for the hypothesis that elevated production of ROS in SD-leaves is an inductive regulatory mechanism that controls metabolism in SD-acclimated leaves and not a consequence of oxidative stress.
Acclimation of plants to low light increases the light-harvesting capacity in chloroplasts, especially in Photosystem II (Walters and Horton, 1995). Acclimation of Arabidopsis to SD induced identical modifications in thylakoid LHCII complexes as acclimation to low light intensity (Figure 1; Lepistö et al., 2009). The high accumulation of PROTOCHLOROPHYLLIDE OXIDOREDUCTASE B (PORB) transcripts in SD-grown leaves is likely associated with the tendency of the SD photoperiod to maintain high light-harvesting capacity compared to LD-grown plants (Table 2). POR catalyzes the light-dependent reaction of chlorophyll biosynthesis and this enzyme is encoded by three genes in Arabidopsis (PORA, PORB, PORC) (Reinbothe et al., 1996). From POR genes, both PORB and PORC are expressed in Arabidopsis rosette leaves under rhythmic growth conditions (Matsumoto et al., 2004), whereas only PORB gene is repressed by high-light treatment (accession AT-00246 in Genevestigator database; Kleine et al., 2007). The other genes of chlorophyll biosynthesis were not up-regulated in SD-grown Arabidopsis (Table 2). This discrepancy may be due to the lower response of the other chlorophyll biosynthesis genes to changes in light intensity (accession AT-00246 in Genevestigator database; Kleine et al., 2007). Higher level of PORB transcript in SD-grown leaves in comparison to LD-leaves is likely related to a tendency to maintain higher LHCII capacity in SD-chloroplasts in comparison to LD-chloroplast.
LIGHT SIGNALING PATHWAYS CONTROLLING THE DEVELOPMENT OF PHOTOSYNTHETIC TRAITS
Light controls the entire plant lifecycle from the germination of seeds to the production of the new generation (Sullivan and Deng, 2003). The structural and functional characterization of photosynthetic traits in Arabidopsis leaves acclimated to various photoperiods indicates that the photoperiod-induced modifications in leaf anatomy, photosynthetic parameters, and ultrastructure of chloroplasts mimic the changes observed in leaves acclimated to different light quantities (Lepistö et al., 2009). Thereby, the question is how the light-intensity-dependent and photoperiod-dependent signaling pathways are interacting with each other upon leaf development. Light is directly perceived by blue (cryptochromes CRY, phototropins, and zeitlupe ZTL) and red (phytochromes PHY) light receptors that, in turn, activate the complex signaling networks inducing a high number of light responses in plants, including photomorphogenetic and photoperiodic development. Besides light receptors, chloroplasts also mediate light-induced signals that control the biogenesis of chloroplast and acclimation of plants to light intensity.
Molecular Bases of Plant Circadian Clock
The photoperiodic signaling pathway has mainly been dissected in the transition from vegetative phase to flowering phase, whereas less attention has been paid to the photoperiodic control of the vegetative development. The regulatory pathway leading to induction of flowering in Arabidopsis under LD comprises extremely complex networks of multiple functionally redundant regulators within a circadian clock (for recent comprehensive reviews, see Turck et al., 2008; Harmer, 2009; Imaizumi, 2010; Song et al., 2010). In brief, the ability to respond to photoperiod requires the mechanism to measure the day length via the action of circadian clock. Under conditions promoting flowering, light receptors entrain the circadian clock to a 24-h cycle. The light signaling pathway that resets the clock is still not clear but light induces the expression of the genes within a clock, CIRCADIAN CLOCK ASSOCIATED 1 (CCA1), LATE ELONGATED HYPOCOTYL (LHY), and PSEUDO RESPONSE REGULATORs (PRRs) (Harmer, 2009). These genes are proposed to act in the clock transcriptional feedback loops together with TIMING OF CAB EXPRESSION 1 (TOC1) and other clock genes (Imaizumi, 2010; Song et al., 2010). The feedback loops control the interaction of clock components, ZTL and GIGANTEA (GI), which, in turn, are involved in the regulation of CONSTANS (CO) expression, a master clock-dependent transcription regulator (reviewed by Imaizumi, 2010). Furthermore, light also affects posttranscriptional regulation of CO protein by red-light-dependent (PHYB) destabilization and far-red-light (PHYA) and blue-light-induced (CRY2) stabilization of CO protein (Valverde et al., 2004; Jang et al., 2008; Liu et al., 2008). Accordingly, CO protein accumulates only at the end of the LD photoperiod. CO protein promotes flowering by inducing the expression of a floral integrator gene FLOWERING LOCUS T (FT). The photoperiod is perceived in leaf vascular tissues, in which the CO and FT proteins accumulate only under favorable photoperiod. FT protein is transported to shoot apex to promote induction of genes inducing flower development (Corbesier et al., 2007; Turck et al., 2008). Importantly, this simplified summary depicts only the main streams of photoperiodic regulatory systems in flowering. Besides the interaction with clock components, light signaling has multiple independent targets in the regulatory photoperiodic networks. Furthermore, circadian clock outputs also control the light signaling input to the clock (Harmer, 2009). Many known clock genes have also a discrete role in light signaling (see the references in Harmer, 2009), indicating the intimate relationship between the clock and light signaling in plants. Recently demonstrated epigenetic control of flowering further inserts the complexity of the photoperiodic regulatory network in Arabidopsis (Jiang et al., 2008; He, 2009; Jackson, 2009).
Tuberization in potatoes as well as bud formation and growth cessation in trees are photoperiodic responses that have been less distinctly dissected at the molecular level compared to induction of flowering. Nevertheless, SD-induced tuberization in potato and dormancy in trees seem to recruit molecular components identical to those involved in the induction of flowering in Arabidopsis, namely CO and FT orthologs in potato and Populus (reviewed by Lagercrantz, 2009; Olsen, 2010). For example, CO–FT regulon controls the active shoot elongation of Populus under the LD photoperiod (Bohlenius et al., 2006; Olsen, 2010), suggesting the existence of a general mechanism involving FT as a final target in various photoperiodic signaling networks in different plant species.
Light Signaling Pathways
A number of comprehensive reviews on chloroplast biogenesis, signaling networks of light receptors, and chloroplast-to-nucleus retrograde signaling pathways have recently been published (Bae and Choi, 2008; Larkin and Ruckle, 2008; Pogson et al., 2008; Woodson and Chory, 2008; Kleine et al., 2009; Inaba, 2010; Jung and Chory, 2010). Here, only an overview on these signaling pathways is presented. Light is a primary environmental cue that controls the biogenesis of chloroplasts in angiosperm species. In dark-germinated seedlings, proplastids differentiate into etioplasts, in which a substantial number of photosynthetic proteins are already present, including POR enzyme, protease complexes, ATPase, Rubisco, Cyt b6f, and individual subunits of photosystems (Kanervo et al., 2008). Upon light treatment of the etiolated seedlings, a large number of photosynthetic proteins accumulate rapidly in 24 h (Kanervo et al., 2008). The development of etioplast to chloroplast is triggered by light by two primary mechanisms. First, the phytochromes and cryptochromes induce a removal of the repressor molecules from nucleus, such as CONSTITUTIVE PHOTOMORPHOGENIC 1 (COP1) and PHYTOCHROME-INTERACTING FACTORs (PIFs), that maintain plant scotomorphogenic development in darkness (Bae and Choi, 2008; Bu et al., 2011). These repressors of photomorphogenesis prevent the accumulation of the positive transcription factors of light-induced genes by triggering their proteolytic degradation in the 26S proteasome. After removal of the repressors from the nucleus, the positive transcription factors, including HY 5 (LONG HYPOCOTYL 5), LAF1 (LONG AFTER FAR-RED LIGHT 1), HFR1 (LONG HYPOCOTYL IN FARRED 1), and GOLDEN2-LIKEs (GLKs) (Bae and Choi, 2008; Waters et al., 2009), accumulate, which, in turn, activate the expression of photosynthesis-associated nuclear genes (PhaNGs). Second, in angiosperms, the chlorophyll synthesis depends on light (Reinbothe et al., 1996). The reduction of protochlorophyllide to chlorophyllide is energized by photons absorbed by protochlorophyllide bound to the POR enzyme.
Besides photoreceptor-mediated pathways, the retrograde signals from chloroplast to nucleus have also been shown to modify the expression of PhaNGs. The transcription of PhaNGs is down-regulated if the biogenesis of chloroplast is restrained or the chloroplast function is severely defective (Pogson et al., 2008; Woodson and Chory, 2008; Inaba, 2010). To dissect the nature of the retrograde signals, a genetic screen for gun mutants (genomes uncoupled) was employed in Arabidopsis (Mochizuki et al., 2001). The isolated gun mutants had a higher number of PhaNGs transcripts in seedlings treated with plastid-bleaching-inducing herbicide, norflurazon compared to wild-type line, indicating a weakened repression signal from chloroplast to nucleus. All but one (gun1) gun line had mutations in genes encoding the enzymes of the tetrapyrrole pathway that produces chlorophyll, heme, and the chromophore of phytochromes in chloroplasts (Nott et al., 2006). Mg-protoporphyrin, a first intermediate of the chlorophyll branch of the tetrapyrrole pathway was identified as a promising signaling component (Strand et al., 2003). However, the re-analyses of the accumulation of chlorophyll intermediates in gun mutants have challenged the hypothesis of Mg-protoporphyrin as a repressing signal for PhaNGs transcription (Mochizuki et al., 2008; Moulin et al., 2008). On the other hand, it has been reported that the incubation of Chlamydomonas cells in darkness with Mg-protoporphyrin or hemin activated a set of light-responsive nuclear genes (Vasileuskaya et al., 2004; von Gromoff et al., 2006, 2008). Furthermore, a recent report by Woodson et al. (2011) shows that transgenic plants overexpressing plastid FERROCHELATASE 1 (FC1) have a gun phenotype in the presence of norfluranzon. FC1 catalyzes heme synthesis in chloroplast. According to the authors’ conclusion, heme that is exported from chloroplast may be used as a signal to control PhaNG expression in nucleus via an unknown mechanism. Finally, the intermediates of chlorophyll biosynthesis were also recently demonstrated to act as a positive plastidial signal in the regulation of the nuclear DNA replication in unicellular red alga and in synchronized plant suspension culture during the cell division (Kobayashi et al., 2009, 2011). These examples suggest that tetrapyrrole intermediates can, indeed, initiate the signal from chloroplast to nucleus.
Besides scoto/photomorphogentic differentiation of plants, the development of the leaf photosynthetic structures depends on the light intensity in the plant habitat. Plants adjust the leaf and cell morphology as well as the molecular composition and the number of chloroplasts to the incident light conditions to optimize the absorption and conversion of solar energy to biomass. This acclimation includes the modulation of the stoichiometry of photosystems and the light-harvesting antenna size in thylakoids, changes in the number of stromal enzymes, and the induction of a complex set of antioxidant systems in high light (Walters and Horton, 1995; Vanderauwera et al., 2005; Bartoli et al., 2006; Li et al., 2009). It has been suggested that the photoreceptors do not play a major role in the acclimation of photosynthesis to light intensity (Walters et al., 1999). Instead, a number of studies point to the contribution of chloroplast-to-nucleus retrograde signals in the light intensity-dependent modification of the chloroplast ultrastructure (Pfannschmidt et al., 1999; Pursiheimo et al., 2001; Piippo et al., 2006; Muhlenbock et al., 2008; Foyer and Noctor, 2009). Vivid debate has been raised on the origin of the chloroplast signals in the light acclimation process. The altered redox state of the photosynthetic electron transport chain (PET) is essential for the initiation of acclimatory processes, whereas both the redox state of the plastoquinone pool (Pfannschmidt et al., 1999) and the acceptor side of PSI (Piippo et al., 2006) have been proposed to be the primary source of a PET signal. In the latter case, both the reactive oxygen species (Muhlenbock et al., 2008) and the thylakoid-bound STN7 kinase (Pursiheimo et al., 2001; Pesaresi et al., 2007), the activity of which is controlled both by PET and thioredoxin (Vener et al., 1997; Rintamäki et al., 2000), are conceivable signaling candidates.
Only a few downstream components involved in the chloroplast-to-nucleus retrograde signaling have been identified so far. In contrast to the other gun mutants, gun1 did not exhibit lesions in tetrapyrrole metabolism. GUN1 encodes a chloroplast pentatricopeptide repeat-containing protein (Koussevitzky et al., 2007) that is proposed to act as a switchboard mediating the signal inside the chloroplast from tetrapyrrole intermediates, from chloroplast translation machinery (Koussevitzky et al., 2007; Woodson and Chory, 2008; Cottage et al., 2010), and probably also from the redox state of PET (Inaba, 2010; Sun et al., 2011) to an unknown component. A recent paper reported on the identification of a highly promising component that mediates the signal from chloroplast to nucleus. Sun et al. (2011) demonstrated that a chloroplast signal triggered a proteolytic cleavage of an envelope-bound plant homeodomain transcription factor PTM. The N-terminal fragment of PTM was transmitted to the nucleus, where it activated the expression of ABI4, an AP2-type transcription factor that has been previously shown to act downstream from GUN1 in the plastid-derived signaling pathway (Koussevitzky et al., 2007). Demonstration that PTM indeed acts downstream of GUN1 would significantly further elucidate the plastid-to-nucleus signaling pathway in plant cells.
ABI4 represses the expression of PhaNGs by binding to the CCAC motif upstream of light-responsive genes (Koussevitzky et al., 2007). Two positive transcription factors (GLK1 and GLK2) are essential for proper biogenesis of chloroplasts and influence the acclimation of plant to light intensity (Waters et al., 2009). GLKs preferably induce genes encoding enzymes of the tetrapyrrole pathway and nuclear encoded photosystem components (Waters et al., 2009). GLKs may act as a shared component of both photoreceptor-dependent and plastid signal-dependent signaling, since the expression of GLKs is regulated by PhyA and PhyB (Tepperman et al., 2006), while GLK2 has been shown to be sensitive also to plastid-derived signals (Waters et al., 2009).
COORDINATION OF LIGHT INTENSITY-DEPENDENT, PHOTOPERIODIC, AND CHLOROPLAST SIGNALING PATHWAYS IN THE DIFFERENTIATION AND ENVIRONMENTAL ACCLIMATION OF LEAVES
Both light receptors and chloroplast signals contribute to the control of leaf acclimation to light quantity, and an interaction between these signaling pathways has recently been proposed (Ruckle et al., 2007; Ruckle and Larkin, 2009). In this review, we have demonstrated that shortening of the photoperiod also alters the photosynthetic structures resembling the acclimation to low light. An interesting question is how closely the different signaling pathways are interconnected in guiding of leaf differentiation under various light regimes: the quantity, quality, and duration of light per day. Today, only fragments of the interconnected light signaling networks are known. Shading experiments have demonstrated that the light-intensity-dependent development of leaf anatomy is controlled by a systemic signal from mature leaves to developing leaves (Lake et al., 2001; Yano and Terashima, 2001), whereas chloroplasts differentiate according to local signal perceived in the developing leaves (Yano and Terashima, 2001). Accordingly, high-light-illuminated developing leaves have shade-type leaf anatomy with sun-type chloroplasts, if the mature leaves were shaded during differentiation of the young leaves. Thus, the signal determining the chloroplast ultrastructure may be perceived locally in chloroplasts of developing leaves, while the light-intensity-dependent systemic signaling arising from mature leaves resembles the mobile signal that controls photoperiodic flowering in Arabidopsis. This unidentified systemic signal may contribute both to light-intensity and photoperiod-dependent pathways.
Mutation in Chloroplast Proteins Alters the Plant Developmental Program by Modifying Chloroplast-to-Nucleus Retrograde Signaling
Arabidopsis mutants with defects in genes encoding chloroplast components show mutant phenotype only under specific environmental conditions (Yu et al., 2007; Kim et al., 2008; Sirpiö et al., 2008; Lepistö et al., 2009; Rosso et al., 2009; Tikkanen et al., 2010). The flu mutant is an elegant example of the case in which the phenotype can be caused by the activation of signaling cascade by chloroplast signal and not directly by the physiochemical effects of a compound accumulating in mutant plants. The flu mutant is defective in feedback control of chlorophyll biosynthesis and accumulates protochlorophyllide in darkness (Meskauskiene et al., 2001). The flu mutant is viable under continuous light, but, if the light-germinated flu seedlings are transferred to the dark, a subsequent illumination of seedlings induces the production of 1O2 by protochlorophyllide and results in photobleaching of the plant (Kim et al., 2008). Under these conditions, the photobleaching is not directly due to the oxidative damage caused by 1O2. Instead, 1O2 initiates chloroplast-to-nucleus signaling that activates the suicidal program in flu seedlings (Kim et al., 2008). This plastid-initiated and 1O2-mediated cell death is controlled by two chloroplast proteins, EXECUTER 1 and 2 (EX1, EX2), mutations of which in flu background totally suppress flu phenotype in dark/light transition of ex1 ex2 flu seedlings (Wagner et al., 2004; Kim et al., 2008). Nevertheless, the conditions causing high accumulation of protochlorophyllide in the dark and drastic production of 1O2 upon subsequent light period induce oxidative damage both in flu and in ex1 ex2 flu seedlings, indicating that the output responses to 1O2 (signaling or damage) depend on the concentration of the effector produced in the cell.
Mutations in genes encoding chloroplast components also modify the morphogenetic development of leaves, especially the differentiation of mesophyll cells (Figure 1; Knappe et al., 2003; Hricova et al., 2006). Perturbation of leaf differentiation may be due to the lack or deficient function of a mutated chloroplast protein. Alternatively, if the chloroplast-generated signals interfere with other signaling networks, the signal from malfunctional chloroplast may impact on the developmental processes. A variegated mutant chlorophyll a/b-binding protein underexpressed 1 (cue1) is an example of a signal from malfunctional chloroplast that interferes with the developmental processes in the Arabidopsis leaf (Knappe et al., 2003). The chloroplasts in bundle sheath cells have unique redox, hormonal, and carbon metabolism (especially shikimate pathway) (recent review by Kangasjärvi et al., 2009), suggesting that bundle sheath cell chloroplasts likely have a minor role in the photosynthetic yield of leaves and, instead, they receive environmental signals to control the development of young leaves. The cue1 is deficient in plastidic phosphoenolpuryvate (PEP) phosphate translocator 1 (PPT1) that provides PEP to shikimate pathway. The cue1 has abnormal mesophyll cells with undeveloped chloroplast and green paraveinal region with properly developed chloroplast (Streatfield et al., 1999; Knappe et al., 2003). Thereby, it is surprising that PPT1 is not present in wild-type mesophyll cell chloroplasts, since the PPT1 gene is mainly expressed in parenchyma cells of vascular tissues (Knappe et al., 2003). It was proposed that the signal generated in plastids of vascular tissue is crucial for proper differentiation of mesophyll cells and for biogenesis of chloroplasts in the interveinal mesophyll region of leaves (Figure 3A).
Figure 3.
Diagrams Depicting the Proposed Mechanisms How Light, Perceived by Chloroplasts and Photoreceptors, Is Mediated to Signals that Interfere with the Morphological Development of Plant and with the Regulation of Anthocyanin Gene Expression.
(A) Light perceived by chloroplasts and photoreceptors in mature leaves generates a systemic signal that is crucial for proper morphological development of young leaves.
(B) Transfer of plants to an altered photoperiod modifies the redox homeostasis in chloroplasts. Signal directly from PET or mediated by GUN1 is transferred to cytosol by an unknown mechanism. This chloroplast-derived signal may control the expression of anthocyanin genes independently or via the components of the light receptor signaling pathway.
Disturbed energy balance may be a major cause of the developmental disorders in mutants with dysfunctional chloroplasts. For example, mutant alleles of the SCABRAS3 gene encoding the nuclear-encoded plastid RNA polymerase showed roundish vegetative leaves with lateral teeth and protruding leaf laminae and severely impaired differentiation of mesophyll cells (Hricova et al., 2006). The authors suggested that proliferation of mesophyll cells and chloroplast biogenesis are coordinated during leaf development, which may be controlled by the energy signaling network. Recently, a central integrator of transcription networks linking the plant stress, energy, and developmental signaling was identified (Baena-Gonzalez and Sheen, 2008). The KIN10/11 protein kinases were shown to have a pivotal role in controlling energy balance, growth, and survival of Arabidopsis. Since chloroplasts are essential for plant energy homeostasis, the chloroplast-generated signals are obvious factors contributing to the transcription networks controlled by KIN10/11.
Interference of chloroplast-generated signals with other signaling networks in plants raises a question about the homogeneity of the signals coming from the chloroplasts. In plant cells, all chloroplasts are autonomous with regard to biogenesis and function and they communicate with the nucleus independently from each other (Yu et al., 2007). Besides the variegated-type mutants, in which each cell line has either functional or undifferentiated chloroplasts, mutations in the nuclear-encoded chloroplast proteins can generate photosynthetic cells with heterogeneous plastids (Aseeva et al., 2007; Nakanishi et al., 2009). For example, both wild-type chloroplasts and irregularly differentiated plastids were detected in a single cell of the knockout lines of a regulatory protein, CHLOROPLAST NADPH-THIOREDOXIN REDUCTASE (NTRC) (Figure 1; Lepistö et al., 2009). The presence of heterogeneous plastids in mesophyll cells of the ntrc knockout mutants was accompanied by the irregularly shaped palisade mesophyll cells (Figure 1). Heterogeneous chloroplast population in an ntrc cell may send contradictory signals to the nucleus, thereby confusing the nuclear-controlled developmental processes. The ultrastructure of chloroplasts as well as the leaf phenotype of the moderate vipp1 knock-down mutant (VESICLE INDUCING PLASTID PROTEIN 1; see Aseeva et al., 2007) substantially resembles that of the ntrc line. VIPP1 has been suggested to be essential to the formation of thylakoid membrane lipid bilayers (Kroll et al., 2001; Westphal et al., 2001). Interestingly, the dose of VIPP1 protein in leaves affects the differentiation of chloroplasts; vipp1 knock-down mutants with only 20% VIPP1 left in the leaves had undifferentiated chloroplasts, whereas, in the mutants with 40% VIPP1, both functional chloroplasts and undifferentiated plastids are present in a single cell (Aseeva et al., 2007). The variation in chloroplast differentiation stage in a single cell of the vipp1 knock-down and ntrc knockout mutants suggests that (1) a threshold amount of the certain activity missing in these mutants is needed for the proper differentiation of chloroplasts and (2) the nuclear-encoded resources are not equally distributed to every chloroplast in a single cell. Deterioration of the morphological development and acclimation capacity of leaf cells detected in ntrc (Lepistö et al., 2009) and other pale green mutants of chloroplast proteins (Yu et al., 2007) suggests that the contradictory signals from chloroplasts with different functional status interfere with the nuclear-controlled developmental processes.
Case Studies
Below, we describe two case studies indicating how light intensity-dependent, photoperiodic, and chloroplast signaling pathways act in the developmental process (stomatal development) or control a biosynthesis of cellular component (anthocyanin biosynthesis).
Case Study 1: Stomatal Development Controlled by Environmental Cues
Operation of stomata is closely associated with the photosynthetic performance of leaves. Stomata restrict excess loss of water from plants but simultaneously they allow sufficient supply of CO2 to photosynthesis. This trade-off situation is controlled by short-term regulation of stomatal aperture in leaves and by long-term regulation of the number of stomata in leaf epidermis. A complex regulatory network consisting of basic-helix-loop-helix (bHLH) transcription factors and negative regulators controls the differentiation and distribution of stomata in Arabidopsis epidermis (Bergmann and Sack, 2007; Casson and Gray, 2008; Serna, 2009). The negative regulators (e.g. TOO MANY MOUTHS, YODA) control the density of stomata in leaf epidermis by preventing the development of adjacent protodermal cells to guard cells (Bergmann and Sack, 2007). The number of stomata in leaf blade is modulated by environmental variables, such as light intensity, photoperiod, and CO2 level. High light, long photoperiod, and low CO2 increase the stomatal index in leaves, while low light, short photoperiod, and high CO2 have an opposing effect (Lake et al., 2001; Casson et al., 2009; Lepistö et al., 2009). As in the acclimation of leaf anatomy to light intensity, the environmental signal to stomatal development is perceived by the mature leaves and transported to developing leaves by an unknown mechanism (Lake et al., 2001). Photoreceptors control the stomatal development by COP1-dependent signaling (Kang et al., 2009). Casson et al. (2009) showed that light intensity-dependent distribution of stomata in epidermis relied on PhyB and PIF4 transcription factor. The question is, what are the downstream components controlled by a systemic signal? The membrane-bound negative regulators, SUBTILISIN-LIKE PROTEASE 1 (SDD1) and EPIDERMAL PATTERING FACTORs (EPF) 1 and 2, control the stomatal index in leaves by repressing the differentiation of guard cells in epidermis (Bergmann and Sack, 2007; Casson and Gray, 2008). Accordingly, mutations in SDD1 or EPF1 increased the stomatal index in leaf epidermis in comparison to wild-type plants (Berger and Altmann, 2000; Hara et al., 2007). Coupe et al. (2006) also reported that shading of the mature leaves induced the expression of SDD1 in non-shaded young leaves, in which the differentiation of guard cells is reduced. Thereby, the expression of SDD1 may be a potential target of light-intensity-dependent systemic signal in Arabidopsis (Figure 3A).
Besides photoreceptors, chloroplasts in mature leaves likely mediate the light-dependent signal to expression of the negative regulators of stomatal development, SDD1, EPF1, and EPF2. Mutations in PhaNGs frequently modify the ability of a leaf to correctly respond to environmental changes, which likely is due to the misleading signals from malfunctional chloroplast to nuclear gene expression. Accordingly, a slightly lower stomatal index and substantially increased stomatal density were detected in the ntrc lines than in wild-type Arabidopsis acclimated to short photoperiod that may be due to the detected repression of SDD1 and EPF1 expression in the ntrc line (Lepistö et al., 2009). We hypothesize that, as in low light, acclimation of plant to short photoperiod enhances the expression of the negative regulators SDD1, EPF1, and EPF2 in developing leaves, resulting in the reduced differentiation of guard cells in epidermis and lower value of stomatal index in SD-leaves (Lepistö et al., 2009). In ntrc lines, the signals from malfunctional chloroplasts restrain the full activation of negative regulators of stomatal differentiation in SD-grown plants that consequently allows more meristemoid mother cells to develop to guard cells in leaf primordium (Figure 3A).
Case Study 2: Biosynthesis of Anthocyanins in Arabidopsis Leaves
Anthocyanins are pigmented flavonoids that are synthesized as a response to various environmental cues including light, temperature, water deficiency, herbivores, and pathogens. They are proposed to protect plant leaves from photodamage induced by high light or altered light conditions (Jaakola and Hohtola, 2010). Accordingly, different qualities of light (white, red, blue, far red, UVA, and UVB) and high light activate the expression of the anthocyanin genes (Vanderauwera et al., 2005; Shin et al., 2007; Cominelli et al., 2008; Jaakola and Hohtola, 2010). Production of anthocyanins is frequently used as a visible marker in the studies of light-induced signaling pathways. Expression of anthocyanin biosynthetic genes is controlled by MYB- and basic helix-loop-helix-related (bHLH) transcription factors (Vom Endt et al., 2002). These interacting transcription factors form a complex regulatory network that both positively and negatively controls the anthocyanin biosynthesis genes (Allan et al., 2008; Cominelli et al., 2008; Dubos et al., 2008).
Light-dependent environmental factors diversely modulate the balance and the number of the regulatory complexes induced by MYB and bHLH transcription factors (Dubos et al., 2008). The photoreceptor-dependent signaling components PIF3 and HY5 have been reported to act as positive regulators of anthocyanin biosynthesis (Shin et al., 2007). Furthermore, high light strongly activates the expression of anthocyanin biosynthesis genes (Page et al., 2011). Accordingly, a high increase in the anthocyanin accumulation was observed also in sweet potato grown under LD photoperiod as compared to growth under SD photoperiod (Carvalho et al., 2010), supporting the conclusion that long photoperiod mimics the high-light conditions in plant acclimation. Accordingly, a massive repression of the biosynthetic and regulatory anthocyanin genes is detected in SD-grown Arabidopsis if compared to LD-grown plants (Table 3).
Table 3.
Stimulation/Repression of Genes Encoding Anthocyanin Biosynthetic Enzymes in Col-0 and Mutant Lines Treated with Different Light Quantity, Quality and Photoperiod, and with Hydrogen Peroxide.
| Experimental set–upa | Controla | Expression of anthocyanin genesb | Materialc | Accession number in Genevestigatord |
| Col-0 L/white light | Col-0 Dark | ++ | Seedlings grown in light/dark | AT-00002 |
| Col-0/blue light | Col-0 Dark | +++ | 7–day-old seedlings, GL | AT-00246 |
| Col-0/HL/3 h | Col-0 GL | +++ | 7–day-old seedlings, GL | AT-00246 |
| cry1 /HL/3 h | Col-0 HL/3 h | –– | 7–day-old seedlings, GL | AT-00246 |
| hy5/HL/3 h | Col-0 HL/3 h | ––– | 7–day-old seedlings, GL | AT-00246 |
| cry1/HL/3 h | cry1 GL | ++ | 7–day-old seedlings, GL | AT-00246 |
| hy5/HL/3 h | hy5 GL | ++ | 7–day-old seedlings, GL | AT-00246 |
| gun1gun5/white light | Col-0/white light | –– | Seedlings with cotyledons fully open | AT-00083 |
| Col-0/SD | Col-0/LD | –––– | Rosettes with eight leaves | AT-00214 |
| flu/light | Col-0 | NC | Adult rosette leaves | AT-00287 |
| Col-0/10 mM H2O2 | Col-0 Water | –– | Hypocotyl and cotyledon emergence | AT-00185 |
Expression of 23 genes encoding enzymes in flavonoid biosynthetic pathway (Vanderauwera et al., 2005) was analyzed using Genevestigator database of Arabidopsis ATH1 22k mircoarray experiments. The experiments tested are indicated by the accession number in Genevestigator
Description of treated and control plants used in the experiments. GL, growth light 100 μmol photons m−2 s−1; HL, high light 1000 μmol photons m−2 s−1; SD and LD, plants grown under short and long photoperiods, respectively.
Stimulation or repression of the expression of the gene cluster is indicated as follows: the majority of the tested genes are up-regulated under experimental set-up: ++, the transcript ratio of the treated sample to the control sample is on average 1.5–2.5; +++, the transcript ratio of the treated sample to the control sample is on average >2.5. The majority of the tested genes are repressed under experimental set-up: --, the transcript ratio of the treated sample to the control sample is on average 0.5–0.8; ---, the transcript ratio of the treated sample to the control sample is on average 0.2–0.5; ----, the transcript ratio of the treated sample to the control sample is on average < 0.3. NC, no changes.
Plant growth condition and age of plants used in the experiments.
Publications or contributors indicated in data depository: AT-00002, M. Alvarez; AT-00246, Kleine et al., 2007; AT-00083, A. McCormac; AT-00214, Wigge et al., 2005; AT-00287, Lee et al., 2007; AT-00185, R. Mittler, R. Mittler, H. Townsend, Z. Emmerson, B. Schildknecht.
In photoreceptor mutants and in mutants deficient in the light signaling component, anthocyanin genes are not induced with high-light treatment (Table 3), indicating that the photoreceptors mediate the high-light signals to anthocyanin genes. However, if the expression of anthocyanin genes in high-light-exposed cry1 and hy5 mutants is compared with the growth-light-illuminated cry1 and hy5 mutants, a substantial activation of the anthocyanin genes is still detected (Table 3). Thereby, other signaling pathways also likely contribute to the regulation of anthocyanin genes. Manipulation of the activity of the photosynthetic electron transfer chain (PET) has demonstrated that the reduction of PET induced and the oxidation of PET reduced the accumulation of anthocyanins in Lemna gibba (Akhtar et al., 2010). Furthermore, the ntrc mutants with heterogeneous chloroplasts (Figure 1) accumulate a significantly lower amount of anthocyanin than wild-type Arabidopsis (Lepistö et al., 2009), suggesting a chloroplast-originated signal in the regulation of anthocyanin biosynthesis. Accordingly, the genes of the anthocyanin biosynthesis are strongly repressed in illuminated gun1 gun5 double mutant (Table 3). Furthermore, anthocyanins were nearly absent in gun1 cry1 double mutants illuminated with high light, whereas about 40% of anthocyanins were present in the single cry1 mutant (Ruckle and Larkin, 2009). Low accumulation of anthocyanin was also detected in gun1 mutants under conditions that stimulated anthocyanin synthesis in wild-type plants (Ruckle and Larkin, 2009; Cottage et al., 2010). Thereby, GUN1 likely mediates a positive chloroplast signal to nuclear anthocyanin genes (Ruckle and Larkin, 2009; Figure 3B).
Chloroplast signal has been suggested to rely on the reactive oxygen species, but, in the case of light-dependent regulation of anthocyanin biosynthesis, ROS likely plays a minor role. The expression of anthocyanin genes did not change significantly in flu mutants producing 1O2 in chloroplast (Table 3). The external treatment of plants with H2O2 induced a slight repression of anthocyanin gene expression (Table 3), whereas the induction of anthocyanin genes by high light was delayed in H2O2-accumulating cat2 mutant deficient in peroxisomal catalase activity (Vanderauwera et al., 2005). These experiments indicate that the accumulation of ROS in cells has an opposite effect on the expression of anthocyanin genes than high light. Thereby, a response of anthocyanin biosynthesis to high light is mediated by the signaling pathway not related to ROS signaling.
We hypothesize that photoperiodic signal to anthocyanin biosynthesis mimics the high-light signaling and can be mediated by chloroplast components (Figure 3B). Transfer to shorter/longer photoperiod than a plant has experienced previously modifies the redox homeostasis of chloroplasts probably by modification of PET. GUN1 acts downstream of PET and mediates the signal to cytoplasm by an unknown mechanism. The chloroplast-derived signal may control the expression of anthocyanin genes independently or interfere with the components of the light signaling pathway.
ACCESSION NUMBERS
Array design and data from this article have been deposited at ArrayExpress under accession number E-MEXP-3331.
FUNDING
This work was supported by the Academy of Finland (grant numbers 127819 and 130192) and Turku University Foundation.
Acknowledgments
We thank Ulla-Maija Suoranta and Mika Keränen for excellent technical assistance and Eva-Mari Aro for critical reading of the manuscript. We also thank the personnel of the Laboratory of Electron Microscopy at the Medical Faculty of the University of Turku, Leena Salminen for sample preparation, and Markus Peurla for assistance with the microscope. No conflict of interest declared.
References
- Akhtar TA, Lees HA, Lampi MA, Enstone D, Brain RA, Greenberg BM. Photosynthetic redox imbalance influences flavonoid biosynthesis in Lemna gibba . Plant Cell Environ. 2010;33:1205–1219. doi: 10.1111/j.1365-3040.2010.02140.x. [DOI] [PubMed] [Google Scholar]
- Allan AC, Hellens RP, Laing WA. MYB transcription factors that colour our fruit. Trends Plant Sci. 2008;13:99–102. doi: 10.1016/j.tplants.2007.11.012. [DOI] [PubMed] [Google Scholar]
- Aseeva E, et al. Vipp1 is required for basic thylakoid membrane formation but not for the assembly of thylakoid protein complexes. Plant Physiol. Biochem. 2007;45:119–128. doi: 10.1016/j.plaphy.2007.01.005. [DOI] [PubMed] [Google Scholar]
- Bae G, Choi G. Decoding of light signals by plant phytochromes and their interacting proteins. Annu. Rev. Plant Biol. 2008;59:281–311. doi: 10.1146/annurev.arplant.59.032607.092859. [DOI] [PubMed] [Google Scholar]
- Baena-Gonzalez E, Sheen J. Convergent energy and stress signaling. Trends Plant Sci. 2008;13:474–482. doi: 10.1016/j.tplants.2008.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian S, Sureshkumar S, Lempe J, Weigel D. Potent induction of Arabidopsis thaliana flowering by elevated growth temperature. PLoS Genet. 2006;2:e106. doi: 10.1371/journal.pgen.0020106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartoli CG, Yu J, Gomez F, Fernandez L, McIntosh L, Foyer CH. Inter-relationships between light and respiration in the control of ascorbic acid synthesis and accumulation in Arabidopsis thaliana leaves. J. Exp. Bot. 2006;57:1621–1631. doi: 10.1093/jxb/erl005. [DOI] [PubMed] [Google Scholar]
- Berger D, Altmann T. A subtilisin-like serine protease involved in the regulation of stomatal density and distribution in Arabidopsis thaliana . Genes Dev. 2000;14:1119–1131. [PMC free article] [PubMed] [Google Scholar]
- Bergmann DC, Sack FD. Stomatal development. Annu. Rev. Plant Biol. 2007;58:163–181. doi: 10.1146/annurev.arplant.58.032806.104023. [DOI] [PubMed] [Google Scholar]
- Bohlenius H, Huang T, Charbonnel-Campaa L, Brunner AM, Jansson S, Strauss SH, Nilsson O. CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees. Science. 2006;312:1040–1043. doi: 10.1126/science.1126038. [DOI] [PubMed] [Google Scholar]
- Bu Q, Castillon A, Chen F, Zhu L, Huq E. Dimerization and blue light regulation of PIF1 interacting bHLH proteins in Arabidopsis . Plant Mol. Biol. 2011;77:501–511. doi: 10.1007/s11103-011-9827-4. [DOI] [PubMed] [Google Scholar]
- Carvalho IS, Cavaco T, Carvalho LM, Duque P. Effect of photoperiod on flavonoid pathway activity in sweet potato (Ipomoea batatas (L.) Lam.) leaves. Food Chem. 2010;118:384–390. [Google Scholar]
- Casson S, Gray JE. Influence of environmental factors on stomatal development. New Phytol. 2008;178:9–23. doi: 10.1111/j.1469-8137.2007.02351.x. [DOI] [PubMed] [Google Scholar]
- Casson SA, Franklin KA, Gray JE, Grierson CS, Whitelam GC, Hetherington AM. Phytochrome B and PIF4 regulate stomatal development in response to light quantity. Curr. Biol. 2009;19:229–234. doi: 10.1016/j.cub.2008.12.046. [DOI] [PubMed] [Google Scholar]
- Cominelli E, Gusmaroli G, Allegra D, Galbiati M, Wade HK, Jenkins GI, Tonelli C. Expression analysis of anthocyanin regulatory genes in response to different light qualities in Arabidopsis thaliana . J. Plant Physiol. 2008;165:886–894. doi: 10.1016/j.jplph.2007.06.010. [DOI] [PubMed] [Google Scholar]
- Cookson SJ, Chenu K, Granier C. Day length affects the dynamics of leaf expansion and cellular development in Arabidopsis thaliana partially through floral transition timing. Ann. Bot. 2007;99:703–711. doi: 10.1093/aob/mcm005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbesier L, et al. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis . Science. 2007;316:1030–1033. doi: 10.1126/science.1141752. [DOI] [PubMed] [Google Scholar]
- Cottage A, Mott EK, Kempster JA, Gray JC. The Arabidopsis plastid-signalling mutant gun1 (genomes uncoupled1) shows altered sensitivity to sucrose and abscisic acid and alterations in early seedling development. J. Exp. Bot. 2010;61:3773–3786. doi: 10.1093/jxb/erq186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coupe SA, Palmer BG, Lake JA, Overy SA, Oxborough K, Woodward FI, Gray JE, Quick WP. Systemic signalling of environmental cues in Arabidopsis leaves. J. Exp. Bot. 2006;57:329–341. doi: 10.1093/jxb/erj033. [DOI] [PubMed] [Google Scholar]
- Covington MF, Maloof JN, Straume M, Kay SA, Harmer SL. Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biol. 2008;9:R130. doi: 10.1186/gb-2008-9-8-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd AN, Salathia N, Hall A, Kevei E, Toth R, Nagy F, Hibberd JM, Millar AJ, Webb AA. Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science. 2005;309:630–633. doi: 10.1126/science.1115581. [DOI] [PubMed] [Google Scholar]
- Dubos C, Le Gourrierec J, Baudry A, Huep G, Lanet E, Debeaujon I, Routaboul JM, Alboresi A, Weisshaar B, Lepiniec L. MYBL2 is a new regulator of flavonoid biosynthesis in Arabidopsis thaliana . Plant J. 2008;55:940–953. doi: 10.1111/j.1365-313X.2008.03564.x. [DOI] [PubMed] [Google Scholar]
- Foyer CH, Noctor G. Redox regulation in photosynthetic organisms: signaling, acclimation, and practical implications. Antioxid. Redox Signal. 2009;11:861–905. doi: 10.1089/ars.2008.2177. [DOI] [PubMed] [Google Scholar]
- Gibon Y, Pyl ET, Sulpice R, Lunn JE, Hohne M, Gunther M, Stitt M. Adjustment of growth, starch turnover, protein content and central metabolism to a decrease of the carbon supply when Arabidopsis is grown in very short photoperiods. Plant Cell Environ. 2009;32:859–874. doi: 10.1111/j.1365-3040.2009.01965.x. [DOI] [PubMed] [Google Scholar]
- Hara K, Kajita R, Torii KU, Bergmann DC, Kakimoto T. The secretory peptide gene EPF1 enforces the stomatal one-cell-spacing rule. Genes Dev. 2007;21:1720–1725. doi: 10.1101/gad.1550707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer SL. The circadian system in higher plants. Annu. Rev. Plant Biol. 2009;60:357–377. doi: 10.1146/annurev.arplant.043008.092054. [DOI] [PubMed] [Google Scholar]
- He Y. Control of the transition to flowering by chromatin modifications. Mol. Plant. 2009;2:554–564. doi: 10.1093/mp/ssp005. [DOI] [PubMed] [Google Scholar]
- Hendriks JH, Kolbe A, Gibon Y, Stitt M, Geigenberger P. ADP-glucose pyrophosphorylase is activated by posttranslational redox-modification in response to light and to sugars in leaves of Arabidopsis and other plant species. Plant Physiol. 2003;133:838–849. doi: 10.1104/pp.103.024513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hricova A, Quesada V, Micol JL. The SCABRA3 nuclear gene encodes the plastid RpoTp RNA polymerase, which is required for chloroplast biogenesis and mesophyll cell proliferation in Arabidopsis . Plant Physiol. 2006;141:942–956. doi: 10.1104/pp.106.080069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P. Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Adv. Bioinformatics. 2008;2008:420747. doi: 10.1155/2008/420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi T. Arabidopsis circadian clock and photoperiodism: time to think about location. Curr. Opin. Plant Biol. 2010;13:83–89. doi: 10.1016/j.pbi.2009.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaba T. Bilateral communication between plastid and the nucleus: plastid protein import and plastid-to-nucleus retrograde signaling. Biosci. Biotechnol. Biochem. 2010;74:471–476. doi: 10.1271/bbb.90842. [DOI] [PubMed] [Google Scholar]
- Jaakola L, Hohtola A. Effect of latitude on flavonoid biosynthesis in plants. Plant Cell Environ. 2010;33:1239–1247. doi: 10.1111/j.1365-3040.2010.02154.x. [DOI] [PubMed] [Google Scholar]
- Jackson SD. Plant responses to photoperiod. New Phytol. 2009;181:517–531. doi: 10.1111/j.1469-8137.2008.02681.x. [DOI] [PubMed] [Google Scholar]
- Jang S, Marchal V, Panigrahi KC, Wenkel S, Soppe W, Deng XW, Valverde F, Coupland G. Arabidopsis COP1 shapes the temporal pattern of CO accumulation conferring a photoperiodic flowering response. EMBO J. 2008;27:1277–1288. doi: 10.1038/emboj.2008.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang D, Wang Y, Wang Y, He Y. Repression of FLOWERING LOCUS C and FLOWERING LOCUS T by the Arabidopsis Polycomb repressive complex 2 components. PLoS One. 2008;3:e3404. doi: 10.1371/journal.pone.0003404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez S, Li Z, Reighard GL, Bielenberg DG. Identification of genes associated with growth cessation and bud dormancy entrance using a dormancy-incapable tree mutant. BMC Plant Biol. 2010;10:25. doi: 10.1186/1471-2229-10-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung HS, Chory J. Signaling between chloroplasts and the nucleus: can a systems biology approach bring clarity to a complex and highly regulated pathway? Plant Physiol. 2010;152:453–459. doi: 10.1104/pp.109.149070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanervo E, Singh M, Suorsa M, Paakkarinen V, Aro E, Battchikova N, Aro EM. Expression of protein complexes and individual proteins upon transition of etioplasts to chloroplasts in pea (Pisum sativum) Plant Cell Physiol. 2008;49:396–410. doi: 10.1093/pcp/pcn016. [DOI] [PubMed] [Google Scholar]
- Kang CY, Lian HL, Wang FF, Huang JR, Yang HQ. Cryptochromes, phytochromes, and COP1 regulate light-controlled stomatal development in Arabidopsis . Plant Cell. 2009;21:2624–2641. doi: 10.1105/tpc.109.069765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kangasjärvi S, Nurmi M, Tikkanen M, Aro EM. Cell-specific mechanisms and systemic signalling as emerging themes in light acclimation of C3 plants. Plant Cell Environ. 2009;32:1230–1240. doi: 10.1111/j.1365-3040.2009.01982.x. [DOI] [PubMed] [Google Scholar]
- Kim C, Meskauskiene R, Apel K, Laloi C. No single way to understand singlet oxygen signalling in plants. EMBO Rep. 2008;9:435–439. doi: 10.1038/embor.2008.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine T, Kindgren P, Benedict C, Hendrickson L, Strand A. Genome-wide gene expression analysis reveals a critical role for CRYPTOCHROME1 in the response of Arabidopsis to high irradiance. Plant Physiol. 2007;144:1391–1406. doi: 10.1104/pp.107.098293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine T, Voigt C, Leister D. Plastid signalling to the nucleus: messengers still lost in the mists? Trends Genet. 2009;25:185–192. doi: 10.1016/j.tig.2009.02.004. [DOI] [PubMed] [Google Scholar]
- Knappe S, Lottgert T, Schneider A, Voll L, Flugge UI, Fischer K. Characterization of two functional phosphoenolpyruvate/phosphate translocator (PPT) genes in Arabidopsis–AtPPT1 may be involved in the provision of signals for correct mesophyll development. Plant J. 2003;36:411–420. doi: 10.1046/j.1365-313x.2003.01888.x. [DOI] [PubMed] [Google Scholar]
- Kobayashi Y, Imamura S, Hanaoka M, Tanaka K. A tetrapyrrole-regulated ubiquitin ligase controls algal nuclear DNA replication. Nat. Cell Biol. 2011;13:483–487. doi: 10.1038/ncb2203. [DOI] [PubMed] [Google Scholar]
- Kobayashi Y, Kanesaki Y, Tanaka A, Kuroiwa H, Kuroiwa T, Tanaka K. Tetrapyrrole signal as a cell-cycle coordinator from organelle to nuclear DNA replication in plant cells. Proc. Natl Acad. Sci. U S A. 2009;106:803–807. doi: 10.1073/pnas.0804270105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koussevitzky S, Nott A, Mockler TC, Hong F, Sachetto-Martins G, Surpin M, Lim J, Mittler R, Chory J. Signals from chloroplasts converge to regulate nuclear gene expression. Science. 2007;316:715–719. [PubMed] [Google Scholar]
- Kozarewa I, Ibanez C, Johansson M, Ogren E, Mozley D, Nylander E, Chono M, Moritz T, Eriksson ME. Alteration of PHYA expression change circadian rhythms and timing of bud set in Populus. Plant Mol. Biol. 2010;73:143–156. doi: 10.1007/s11103-010-9619-2. [DOI] [PubMed] [Google Scholar]
- Kroll D, Meierhoff K, Bechtold N, Kinoshita M, Westphal S, Vothknecht UC, Soll J, Westhoff P. VIPP1, a nuclear gene of Arabidopsis thaliana essential for thylakoid membrane formation. Proc. Natl Acad. Sci. U S A. 2001;98:4238–4242. doi: 10.1073/pnas.061500998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagercrantz U. At the end of the day: a common molecular mechanism for photoperiod responses in plants? J. Exp. Bot. 2009;60:2501–2515. doi: 10.1093/jxb/erp139. [DOI] [PubMed] [Google Scholar]
- Lake JA, Quick WP, Beerling DJ, Woodward FI. Plant development: signals from mature to new leaves. Nature. 2001;411:154. doi: 10.1038/35075660. [DOI] [PubMed] [Google Scholar]
- Larkin RM, Ruckle ME. Integration of light and plastid signals. Curr. Opin. Plant Biol. 2008;11:593–599. doi: 10.1016/j.pbi.2008.10.004. [DOI] [PubMed] [Google Scholar]
- Lee KP, Kim C, Landgraf F, Apel K. EXECUTER1- and EXECUTER2-dependent transfer of stress-related signals from the plastid to the nucleus of Arabidopsis thaliana . Proc. Natl Acad. Sci. U S A. 2007;104:10270–10275. doi: 10.1073/pnas.0702061104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepistö A, Kangasjärvi S, Luomala EM, Brader G, Sipari N, Keränen M, Keinänen M, Rintamäki E. Chloroplast NADPH-thioredoxin reductase interacts with photoperiodic development in Arabidopsis . Plant Physiol. 2009;149:1261–1276. doi: 10.1104/pp.108.133777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Wakao S, Fischer BB, Niyogi KK. Sensing and responding to excess light. Annu. Rev. Plant Biol. 2009;60:239–260. doi: 10.1146/annurev.arplant.58.032806.103844. [DOI] [PubMed] [Google Scholar]
- Liu L, Zhang Y, Li Q, Sang Y, Mao J, Lian H, Wang L, Yang H. COP1-mediated ubiquitination of CONSTANS is implicated in cryptochrome regulation of flowering in Arabidopsis . Plant Cell. 2008;20:292–306. doi: 10.1105/tpc.107.057281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Gehan JP, Sharkey TD. Daylength and circadian effects on starch degradation and maltose metabolism. Plant Physiol. 2005;138:2280–2291. doi: 10.1104/pp.105.061903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto F, Obayashi T, Sasaki-Sekimoto Y, Ohta H, Takamiya K, Masuda T. Gene expression profiling of the tetrapyrrole metabolic pathway in Arabidopsis with a mini-array system. Plant Physiol. 2004;135:2379–2391. doi: 10.1104/pp.104.042408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meskauskiene R, Nater M, Goslings D, Kessler F, op den Camp R, Apel K. FLU: a negative regulator of chlorophyll biosynthesis in Arabidopsis thaliana . Proc. Natl Acad. Sci. U S A. 2001;98:12826–12831. doi: 10.1073/pnas.221252798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelet L, Krieger-Liszkay A. Reactive oxygen intermediates produced by photosynthetic electron transport are enhanced in short-day grown plants. Biochimica et Biophysica Acta (BBA)—Bioenergetics. 2011 doi: 10.1016/j.bbabio.2011.11.014. in press. [DOI] [PubMed] [Google Scholar]
- Mikkelsen R, Mutenda KE, Mant A, Schurmann P, Blennow A. Alpha-glucan, water dikinase (GWD): a plastidic enzyme with redox-regulated and coordinated catalytic activity and binding affinity. Proc. Natl Acad. Sci. U S A. 2005;102:1785–1790. doi: 10.1073/pnas.0406674102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R, Vanderauwera S, Gollery M, Van Breusegem F. Reactive oxygen gene network of plants. Trends Plant Sci. 2004;9:490–498. doi: 10.1016/j.tplants.2004.08.009. [DOI] [PubMed] [Google Scholar]
- Mochizuki N, Brusslan JA, Larkin R, Nagatani A, Chory J. Arabidopsis genomes uncoupled 5 (GUN5) mutant reveals the involvement of Mg-chelatase H subunit in plastid-to-nucleus signal transduction. Proc. Natl Acad. Sci. U S A. 2001;98:2053–2058. doi: 10.1073/pnas.98.4.2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki N, Tanaka R, Tanaka A, Masuda T, Nagatani A. The steady-state level of Mg-protoporphyrin IX is not a determinant of plastid-to-nucleus signaling in Arabidopsis . Proc. Natl Acad. Sci. U S A. 2008;105:15184–15189. doi: 10.1073/pnas.0803245105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulin M, McCormac AC, Terry MJ, Smith AG. Tetrapyrrole profiling in Arabidopsis seedlings reveals that retrograde plastid nuclear signaling is not due to Mg-protoporphyrin IX accumulation. Proc. Natl Acad. Sci. U S A. 2008;105:15178–15183. doi: 10.1073/pnas.0803054105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlenbock P, Szechynska-Hebda M, Plaszczyca M, Baudo M, Mateo A, Mullineaux PM, Parker JE, Karpinska B, Karpinski S. Chloroplast signaling and LESION SIMULATING DISEASE1 regulate crosstalk between light acclimation and immunity in Arabidopsis . Plant Cell. 2008;20:2339–2356. doi: 10.1105/tpc.108.059618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy F, Schafer E. Phytochromes control photomorphogenesis by differentially regulated, interacting signaling pathways in higher plants. Annu. Rev. Plant Biol. 2002;53:329–355. doi: 10.1146/annurev.arplant.53.100301.135302. [DOI] [PubMed] [Google Scholar]
- Nakanishi H, Suzuki K, Kabeya Y, Miyagishima SY. Plant-specific protein MCD1 determines the site of chloroplast division in concert with bacteria-derived MinD. Curr. Biol. 2009;19:151–156. doi: 10.1016/j.cub.2008.12.018. [DOI] [PubMed] [Google Scholar]
- Nott A, Jung HS, Koussevitzky S, Chory J. Plastid-to-nucleus retrograde signaling. Annu. Rev. Plant Biol. 2006;57:739–759. doi: 10.1146/annurev.arplant.57.032905.105310. [DOI] [PubMed] [Google Scholar]
- Olsen JE. Light and temperature sensing and signaling in induction of bud dormancy in woody plants. Plant Mol. Biol. 2010;73:37–47. doi: 10.1007/s11103-010-9620-9. [DOI] [PubMed] [Google Scholar]
- Page M, Sultana N, Paszkiewicz K, Florance H, Smirnoff N. The influence of ascorbate on anthocyanin accumulation during high light acclimation in Arabidopsis thaliana: further evidence for redox control of anthocyanin synthesis. Plant Cell Environ. 2011 doi: 10.1111/j.1365-3040.2011.02369.x. [DOI] [PubMed] [Google Scholar]
- Pesaresi P, Schneider A, Kleine T, Leister D. Interorganellar communication. Curr. Opin. Plant Biol. 2007;10:600–606. doi: 10.1016/j.pbi.2007.07.007. [DOI] [PubMed] [Google Scholar]
- Pfannschmidt T, Brautigam K, Wagner R, Dietzel L, Schroter Y, Steiner S, Nykytenko A. Potential regulation of gene expression in photosynthetic cells by redox and energy state: approaches towards better understanding. Ann. Bot. 2009;103:599–607. doi: 10.1093/aob/mcn081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfannschmidt T, Nilsson A, Tullberg A, Link G, Allen JF. Direct transcriptional control of the chloroplast genes psbA and psaAB adjusts photosynthesis to light energy distribution in plants. IUBMB Life. 1999;48:271–276. doi: 10.1080/713803507. [DOI] [PubMed] [Google Scholar]
- Piippo M, Allahverdiyeva Y, Paakkarinen V, Suoranta UM, Battchikova N, Aro EM. Chloroplast-mediated regulation of nuclear genes in Arabidopsis thaliana in the absence of light stress. Physiol. Genomics. 2006;25:142–152. doi: 10.1152/physiolgenomics.00256.2005. [DOI] [PubMed] [Google Scholar]
- Pogson BJ, Woo NS, Forster B, Small ID. Plastid signalling to the nucleus and beyond. Trends Plant Sci. 2008;13:602–609. doi: 10.1016/j.tplants.2008.08.008. [DOI] [PubMed] [Google Scholar]
- Pursiheimo S, Mulo P, Rintamäki E, Aro EM. Coregulation of light-harvesting complex II phosphorylation and lhcb mRNA accumulation in winter rye. Plant J. 2001;26:317–327. doi: 10.1046/j.1365-313x.2001.01033.x. [DOI] [PubMed] [Google Scholar]
- Queval G, Issakidis-Bourguet E, Hoeberichts FA, Vandorpe M, Gakiere B, Vanacker H, Miginiac-Maslow M, Van Breusegem F, Noctor G. Conditional oxidative stress responses in the Arabidopsis photorespiratory mutant cat2 demonstrate that redox state is a key modulator of daylength-dependent gene expression, and define photoperiod as a crucial factor in the regulation of H2O2-induced cell death. Plant J. 2007;52:640–657. doi: 10.1111/j.1365-313X.2007.03263.x. [DOI] [PubMed] [Google Scholar]
- Reinbothe S, Reinbothe C, Lebedev N, Apel K. PORA and PORB, two light-dependent protochlorophyllide-reducing enzymes of angiosperm chlorophyll biosynthesis. Plant Cell. 1996;8:763–769. doi: 10.1105/tpc.8.5.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rintamäki E, Martinsuo P, Pursiheimo S, Aro EM. Cooperative regulation of light-harvesting complex II phosphorylation via the plastoquinol and ferredoxin-thioredoxin system in chloroplasts. Proc. Natl Acad. Sci. U S A. 2000;97:11644–11649. doi: 10.1073/pnas.180054297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritte G, Lloyd JR, Eckermann N, Rottmann A, Kossmann J, Steup M. The starch-related R1 protein is an alpha -glucan, water dikinase. Proc. Natl Acad. Sci. U S A. 2002;99:7166–7171. doi: 10.1073/pnas.062053099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso D, Bode R, Li W, Krol M, Saccon D, Wang S, Schillaci LA, Rodermel SR, Maxwell DP, Huner NP. Photosynthetic redox imbalance governs leaf sectoring in the Arabidopsis thaliana variegation mutants immutans, spotty, var1, and var2. Plant Cell. 2009;21:3473–3492. doi: 10.1105/tpc.108.062752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruckle ME, Larkin RM. Plastid signals that affect photomorphogenesis in Arabidopsis thaliana are dependent on GENOMES UNCOUPLED 1 and cryptochrome 1. New Phytol. 2009;182:367–379. doi: 10.1111/j.1469-8137.2008.02729.x. [DOI] [PubMed] [Google Scholar]
- Ruckle ME, DeMarco SM, Larkin RM. Plastid signals remodel light signaling networks and are essential for efficient chloroplast biogenesis in Arabidopsis . Plant Cell. 2007;19:3944–3960. doi: 10.1105/tpc.107.054312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serna L. Cell fate transitions during stomatal development. Bioessays. 2009;31:865–873. doi: 10.1002/bies.200800231. [DOI] [PubMed] [Google Scholar]
- Shin J, Park E, Choi G. PIF3 regulates anthocyanin biosynthesis in an HY5-dependent manner with both factors directly binding anthocyanin biosynthetic gene promoters in Arabidopsis . Plant J. 2007;49:981–994. doi: 10.1111/j.1365-313X.2006.03021.x. [DOI] [PubMed] [Google Scholar]
- Sirpiö S, Khrouchtchova A, Allahverdiyeva Y, Hansson M, Fristedt R, Vener AV, Scheller HV, Jensen PE, Haldrup A, Aro EM. AtCYP38 ensures early biogenesis, correct assembly and sustenance of photosystem II. Plant J. 2008;55:639–651. doi: 10.1111/j.1365-313X.2008.03532.x. [DOI] [PubMed] [Google Scholar]
- Sokolov LN, Dominguez-Solis JR, Allary AL, Buchanan BB, Luan S. A redox-regulated chloroplast protein phosphatase binds to starch diurnally and functions in its accumulation. Proc. Natl Acad. Sci. U S A. 2006;103:9732–9737. doi: 10.1073/pnas.0603329103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song YH, Ito S, Imaizumi T. Similarities in the circadian clock and photoperiodism in plants. Curr. Opin. Plant Biol. 2010;13:594–603. doi: 10.1016/j.pbi.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparla F, Costa A, Lo Schiavo F, Pupillo P, Trost P. Redox regulation of a novel plastid-targeted beta-amylase of Arabidopsis . Plant Physiol. 2006;141:840–850. doi: 10.1104/pp.106.079186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staiger D, Zecca L, Wieczorek Kirk DA, Apel K, Eckstein L. The circadian clock regulated RNA-binding protein AtGRP7 autoregulates its expression by influencing alternative splicing of its own pre-mRNA. Plant J. 2003;33:361–371. doi: 10.1046/j.1365-313x.2003.01629.x. [DOI] [PubMed] [Google Scholar]
- Stettler M, Eicke S, Mettler T, Messerli G, Hortensteiner S, Zeeman SC. Blocking the metabolism of starch breakdown products in Arabidopsis leaves triggers chloroplast degradation. Mol. Plant. 2009;2:1233–1246. doi: 10.1093/mp/ssp093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strand A, Asami T, Alonso J, Ecker JR, Chory J. Chloroplast to nucleus communication triggered by accumulation of Mg-protoporphyrinIX. Nature. 2003;421:79–83. doi: 10.1038/nature01204. [DOI] [PubMed] [Google Scholar]
- Streatfield SJ, Weber A, Kinsman EA, Hausler RE, Li J, Post-Beittenmiller D, Kaiser WM, Pyke KA, Flugge UI, Chory J. The phosphoenolpyruvate/phosphate translocator is required for phenolic metabolism, palisade cell development, and plastid-dependent nuclear gene expression. Plant Cell. 1999;11:1609–1622. doi: 10.1105/tpc.11.9.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan JA, Deng XW. From seed to seed: the role of photoreceptors in Arabidopsis development. Dev. Biol. 2003;260:289–297. doi: 10.1016/s0012-1606(03)00212-4. [DOI] [PubMed] [Google Scholar]
- Sun X, Feng P, Xu X, Guo H, Ma J, Chi W, Lin R, Lu C, Zhang L. A chloroplast envelope-bound PHD transcription factor mediates chloroplast signals to the nucleus. Nature Communications. 2011;2:477. doi: 10.1038/ncomms1486. [DOI] [PubMed] [Google Scholar]
- Tepperman JM, Hwang YS, Quail PH. phyA dominates in transduction of red-light signals to rapidly responding genes at the initiation of Arabidopsis seedling de-etiolation. Plant J. 2006;48:728–742. doi: 10.1111/j.1365-313X.2006.02914.x. [DOI] [PubMed] [Google Scholar]
- Tikkanen M, Grieco M, Kangasjarvi S, Aro EM. Thylakoid protein phosphorylation in higher plant chloroplasts optimizes electron transfer under fluctuating light. Plant Physiol. 2010;152:723–735. doi: 10.1104/pp.109.150250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turck F, Fornara F, Coupland G. Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu. Rev. Plant Biol. 2008;59:573–594. doi: 10.1146/annurev.arplant.59.032607.092755. [DOI] [PubMed] [Google Scholar]
- Valverde F, Mouradov A, Soppe W, Ravenscroft D, Samach A, Coupland G. Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science. 2004;303:1003–1006. doi: 10.1126/science.1091761. [DOI] [PubMed] [Google Scholar]
- Vanderauwera S, Zimmermann P, Rombauts S, Vandenabeele S, Langebartels C, Gruissem W, Inze D, Van Breusegem F. Genome-wide analysis of hydrogen peroxide-regulated gene expression in Arabidopsis reveals a high light-induced transcriptional cluster involved in anthocyanin biosynthesis. Plant Physiol. 2005;139:806–821. doi: 10.1104/pp.105.065896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasileuskaya Z, Oster U, Beck CF. Involvement of tetrapyrroles in inter-organellar signaling in plants and algae. Photosynth. Res. 2004;82:289–299. doi: 10.1007/s11120-004-2160-x. [DOI] [PubMed] [Google Scholar]
- Vener AV, van Kan PJ, Rich PR, Ohad I, Andersson B. Plastoquinol at the quinol oxidation site of reduced cytochrome bf mediates signal transduction between light and protein phosphorylation: thylakoid protein kinase deactivation by a single-turnover flash. Proc. Natl Acad. Sci. U S A. 1997;94:1585–1590. doi: 10.1073/pnas.94.4.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victor KJ, Fennell AY, Grimplet J. Proteomic analysis of shoot tissue during photoperiod induced growth cessation in V. riparia Michx. grapevines. Proteome Sci. 2010;8:44. doi: 10.1186/1477-5956-8-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vom Endt D, Kijne JW, Memelink J. Transcription factors controlling plant secondary metabolism: what regulates the regulators? Phytochemistry. 2002;61:107–114. doi: 10.1016/s0031-9422(02)00185-1. [DOI] [PubMed] [Google Scholar]
- von Gromoff ED, Alawady A, Meinecke L, Grimm B, Beck CF. Heme, a plastid-derived regulator of nuclear gene expression in Chlamydomonas. Plant Cell. 2008;20:552–567. doi: 10.1105/tpc.107.054650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Gromoff ED, Schroda M, Oster U, Beck CF. Identification of a plastid response element that acts as an enhancer within the Chlamydomonas HSP70A promoter. Nucleic Acids Res. 2006;34:4767–4779. doi: 10.1093/nar/gkl602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner D, et al. The genetic basis of singlet oxygen-induced stress responses of Arabidopsis thaliana . Science. 2004;306:1183–1185. doi: 10.1126/science.1103178. [DOI] [PubMed] [Google Scholar]
- Walters RG, Horton P. Acclimation of Arabidopsis thaliana to the light environment: changes in composition of the photosynthetic apparatus. Planta. 1995;195:248–256. [Google Scholar]
- Walters RG, Rogers JJ, Shephard F, Horton P. Acclimation of Arabidopsis thaliana to the light environment: the role of photoreceptors. Planta. 1999;209:517–527. doi: 10.1007/s004250050756. [DOI] [PubMed] [Google Scholar]
- Waters MT, Wang P, Korkaric M, Capper RG, Saunders NJ, Langdale JA. GLK transcription factors coordinate expression of the photosynthetic apparatus in Arabidopsis . Plant Cell. 2009;21:1109–1128. doi: 10.1105/tpc.108.065250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westphal S, Heins L, Soll J, Vothknecht UC. Vipp1 deletion mutant of Synechocystis: a connection between bacterial phage shock and thylakoid biogenesis? Proc. Natl Acad. Sci. U S A. 2001;98:4243–4248. doi: 10.1073/pnas.061501198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigge PA, Kim MC, Jaeger KE, Busch W, Schmid M, Lohmann JU, Weigel D. Integration of spatial and temporal information during floral induction in Arabidopsis . Science. 2005;309:1056–1059. doi: 10.1126/science.1114358. [DOI] [PubMed] [Google Scholar]
- Woodson JD, Chory J. Coordination of gene expression between organellar and nuclear genomes. Nat. Rev. Genet. 2008;9:383–395. doi: 10.1038/nrg2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson JD, Perez-Ruiz JM, Chory J. Heme synthesis by plastid ferrochelatase I regulates nuclear gene expression in plants. Curr. Biol. 2011;21:897–903. doi: 10.1016/j.cub.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano S, Terashima I. Separate localization of light signal perception for sun or shade type chloroplast and palisade tissue differentiation in Chenopodium album . Plant Cell Physiol. 2001;42:1303–1310. doi: 10.1093/pcp/pce183. [DOI] [PubMed] [Google Scholar]
- Yu F, Fu A, Aluru M, Park S, Xu Y, Liu H, Liu X, Foudree A, Nambogga M, Rodermel S. Variegation mutants and mechanisms of chloroplast biogenesis. Plant Cell Environ. 2007;30:350–365. doi: 10.1111/j.1365-3040.2006.01630.x. [DOI] [PubMed] [Google Scholar]
- Yu TS, et al. The Arabidopsis sex1 mutant is defective in the R1 protein, a general regulator of starch degradation in plants, and not in the chloroplast hexose transporter. Plant Cell. 2001;13:1907–1918. doi: 10.1105/TPC.010091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeeman SC, Kossmann J, Smith AM. Starch: its metabolism, evolution, and biotechnological modification in plants. Annu. Rev. Plant Biol. 2010;61:209–234. doi: 10.1146/annurev-arplant-042809-112301. [DOI] [PubMed] [Google Scholar]
- Zeeman SC, Smith SM, Smith AM. The diurnal metabolism of leaf starch. Biochem. J. 2007;401:13–28. doi: 10.1042/BJ20061393. [DOI] [PubMed] [Google Scholar]
- Zhao H, Li Y, Duan B, Korpelainen H, Li C. Sex-related adaptive responses of Populus cathayana to photoperiod transitions. Plant Cell Environ. 2009;32:1401–1411. doi: 10.1111/j.1365-3040.2009.02007.x. [DOI] [PubMed] [Google Scholar]