Abstract
The community needed a reliable and user friendly tool to quickly produce robust phylogenetic trees which are crucial in evolutionary studies and genomes’ functional annotation. FIGENIX is software dedicated to this and was published in 2005. Several laboratories around the world use it in their research, but it was difficult to use for non-expert users, thus we developed a new graphical user interface for the benefit of all biologists.
Keywords: phylogeny, tree, phylogenetic, bioinformatics, evolution
Introduction
FIGENIX1 is a software platform dedicated to the automated execution of complex bioinformatics pipelines for reliable reconstruction of phylogenetic trees. The phylogeny pipeline’s originality comes from the fact that it mixes computational steps based on mathematical algorithms and expertize steps based on laboratory and state of the art knowledge in evolutionary biology. By expertize steps we mean that knowledge and reasoning in the evolutionary field are not always modeled by mathematical algorithms integrable in a bioinformatics pipeline. But it can be modeled, quite easily, by logical rules defined in the expert system integrated in FIGENIX. For example: detecting nonmonophyletic domain’s repeats, to eliminate them from the phylogeny pipeline (they distort the final phylogenetic tree), is done by automatic reading and decisions taken from a temporary domain’s repeats tree built during the process (see 2005 publication for more details). Note that other phylogeny online tools exist, like the most recent: phylogeny.fr,2 but their associated pipelines offer no improvement compared to our pipeline. And unlike ours they do not include all the essential bias correction steps, eg, dataset filtering by complex scopes definition, non-monophyletic domain repeats filtering, amino acid composition test, saturation test, domains congruence tests, multiple algorithm trees building, comparison and compilation, etc. Without these elements, phylogenetic trees building is not reliable. With FIGENIX we produced the phylogenetic trees (approximately 100000) for the Chordate History Database project (http://ioda.univ-provence.fr). To do this, FIGENIX was driven by another software system called DAGOBAH.3 DAGOBAH automatically reads phylogenetic trees built by FIGENIX by using PhyloPattern software library,4 with the aim to detect the evidence of genetic events patterns. (it tries to verify them at the genomic level as well as to correlate them). Several complex gene families were studied using phylogenetic reconstructions and functional information propagated through orthologous relationships have been carried out using FIGENIX5–10 thereby increasing demand of the software. In fact the system was initially built as an intranet system dedicated to experts from our laboratory. All parameters used in the phylogeny pipeline, from the simplest to the most sophisticated, was accessible in the same way and with no description. Moreover the parameters were dispatched on many steps of the pipeline and were difficult to locate. However, biologists who are not phylogeny specialists also often need to build trees to answer specific questions. Therefore, we have developed a new version of FIGENIX graphical user interface, because the older version was difficult to use with a steep learning curve. The new web interface (http://ioda.univ-provence.fr/IodaSite/Figenix.jsp) is freely accessible by all.
Graphical User Interface
FIGENIX new web interface can be used in two modes: identified mode or unidentified mode. A guest can create an account (“Create account” menu item) at any time to be an identified user. There is no difference in bioinformatic functionalities of the two modes, but identified users can follow task execution and look back more easily to the history of their tasks produced with FIGENIX. Note that the user accesses FIGENIX through the IODA (http://ioda.univ-provence.fr) web site. There is a FIGENIX menu, which allows the user direct access to the phylogeny creation tab. As one can see on Figure 1, we try to simplify phylogeny task creation by focusing on main parameters like protein Fasta data or file and the database (NR, Swissprot) used for homologs searching. More complex parameters are accessible by unfolding the “Expert parameters” panel. Each parameter has its own description and default value. As described in earlier version of FIGENIX publication, phylogenetic tree reconstruction is a very complex process, which consists of about fifty computation and expertize steps. When one chooses default parameters, we evaluate, quite precisely, that 66% of tasks are successful. That means that the final phylogenetic trees are built. For the other 33%, the biological parameters of the task must be modified. For example, the maximum number of kept homologs, the gaps threshold, etc. FIGENIX new online interface includes a user manual with a troubleshooting guide, accessible by selecting the dedicated menu item, that aids the user in understanding what expert parameters must be modified for the successful construction of the phylogeny. When a task is completed, the user receives an email with the potential problems and with a link to a results page. In identified mode, the user can follow the progression of its running task, and consult any time the partial or final results of its tasks, by using the “Tasks and Results” tab (see Fig. 1). With this tab the user can also see tasks for a variable time period, gain quick access to the parameters for a task, decide to clone an old task and to modify a specific parameter (for example: the species scope of phylogeny, or the maximum number of used sequences … For the task’s results pages, we choose to keep the continuity with the old interface, but with minor changes on displayed information, by eliminating the technical information from all the pages. On the first results page, now only the user parameters, with an added description, are displayed. Results hyperlinks are biologically labeled. Note that phylogenetic trees are directly drawn on web pages as shown on Figure 2.
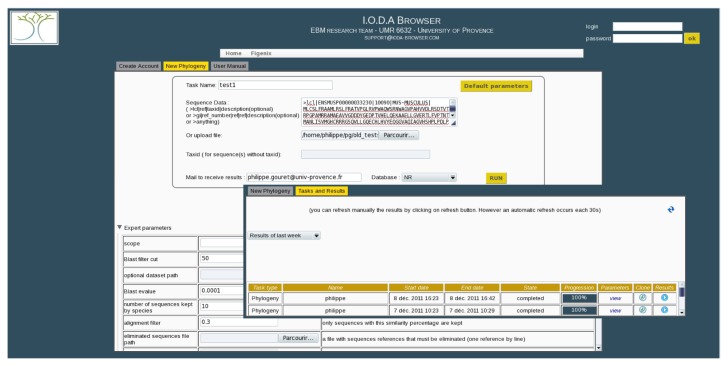
Figure 1.
At the top of the task creation window (upper left on the figure), one can see the most commonly used parameters, specially the protein sequence to be used as a query and the database to search homolog sequences.
Notes: At the bottom of the task creation window, expert parameters are displayed when the area is unfolded. Each parameter has a default value and a description. The tasks management window (lower right on the figure) is a table with the list of tasks started by the user during a chosen period of time. (Set to “week” on the figure). By reading one line of the table, one can see that the user can follow the progression and the state of a task, its parameters, its results (even if the task is not over). User can also clone tasks.
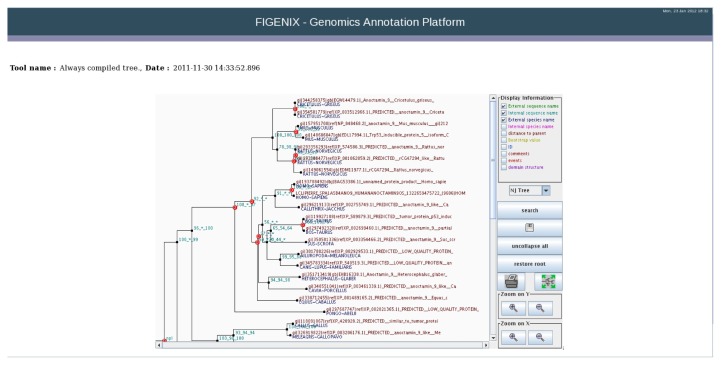
Figure 2.
An example of final tree produced by FIGENIX.
Notes: Visualization is assumed by a Java applet. On the tree area of the window, the user can fold or unfold nodes, edit node information, display chosen subtrees. With the control panel the user can choose a vizualisation mode (distance, clado, radial (hit R key to rotate tree)) and information displayed on nodes and branches. User can also zoom the tree, print and save the tree in NH format (http://evolution.genetics.washington.edu/phylip/newicktree.html) or NHX format (http://phylosoft.org/NHX/).
Conclusion
Phylogenetic reconstruction plays a significant role in evolution study and in the functional annotation of genes family through evolutionary approaches. This new version of FIGENIX graphical user interface can be used by biologists to infer phylogenies easily, and make the phylogenetic methodology available to non specialist.
Supplementary Data
FIGENIX phylogenetic trees creation interface is accessible at: http://ioda.univ-provence.fr/IodaSite/Figenix.jsp. It is implemented with GWT 2.4.0 (http://code.google.com/intl/fr/webtoolkit) and Java 1.6 (http://www.oracle.com/technetwork/java) technologies on a Tomcat 6 web server (http://tomcat.apache.org/).
Acknowledgements
We acknowledge Dr. Pontarotti and Dr. Hemalatha for biological discussions, tests of the tool and for manuscript reading and corrections.
Footnotes
Author Contributions
JP developed the GUI (Graphical User Interface) software components and produced tests. PG supervised all the software developments, realized the bridge between the web site and FIGENIX platform and produced tests. PG wrote the manuscript. All authors reviewed and approved of the final manuscript.
Funding
We acknowledge ANR (French National Agency for Research) grants: [grant numbers ANR-2010- GENM-017-05, ANR-08-GENM-033-04] for their partial support of the work.
Competing Interests
Author(s) disclose no potential conflicts of interest.
Disclosures and Ethics
As a requirement of publication author(s) have provided to the publisher signed confirmation of compliance with legal and ethical obligations including but not limited to the following: authorship and contributorship, conflicts of interest, privacy and confidentiality and (where applicable) protection of human and animal research subjects. The authors have read and confirmed their agreement with the ICMJE authorship and conflict of interest criteria. The authors have also confirmed that this article is unique and not under consideration or published in any other publication, and that they have permission from rights holders to reproduce any copyrighted material. Any disclosures are made in this section. The external blind peer reviewers report no conflicts of interest.
References
- 1.Gouret P, Vitiello V, Balandraud N, Gilles A, Pontarotti P, Danchin EG. FIGENIX: Intelligent automation of genomic annotation: expertise integration in a new software platform. BMC Bioinformatics. 2005;6:198. doi: 10.1186/1471-2105-6-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dereeper A, Guignon V, Blanc G, et al. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008:W465–9. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gouret P, Paganini J, Dainat J, et al., editors. Integration of evolutionary biology concepts for functional annotation and automation of complex research in evolution: the multi-agent software system DAGOBAH. Springer & Pontarotti Pierre (Ed); 2011. [Google Scholar]
- 4.Gouret P, Thompson JD, Pontarotti P. PhyloPattern: regular expressions to identify complex patterns in phylogenetic trees. BMC Bioinformatics. 2009;10:298. doi: 10.1186/1471-2105-10-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biesebeke R, Levasseur A, Boussier A, Record E, van den Hondel CA, Punt PJ. Phylogeny of fungal hemoglobins and expression analysis of the Aspergillus oryzae flavohemoglobin gene fhbA during hyphal growth. Fungal Biol. 2009;114(2–3):135–43. doi: 10.1016/j.mycres.2009.08.007. [DOI] [PubMed] [Google Scholar]
- 6.Bobe J, Guiguen Y, Fostier A. Diversity and biological significance of sex hormone-binding globulin in fish, an evolutionary perspective. Mol Cell Endocrinol. 2010;316(1):66–78. doi: 10.1016/j.mce.2009.09.017. [DOI] [PubMed] [Google Scholar]
- 7.Gohin M, Bobe J, Chesnel F. Comparative transcriptomic analysis of follicle-enclosed oocyte maturational and developmental competence acquisition in two non-mammalian vertebrates. BMC Genomics. 2010;11:18. doi: 10.1186/1471-2164-11-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ohm RA, de Jong JF, Lugones LG, et al. Genome sequence of the model mushroom Schizophyllum commune. Nat Biotechnol. 2010;28(9):957–63. doi: 10.1038/nbt.1643. [DOI] [PubMed] [Google Scholar]
- 9.Rescan P, Ralliere C. A Sox5 gene is expressed in the myogenic lineage during trout embryonic development. Int J Dev Biol. 2010;54(5):913–8. doi: 10.1387/ijdb.092893pr. [DOI] [PubMed] [Google Scholar]
- 10.Shen TH, Tarczy-Hornoch P, Detwiler LT, Cadag E, Carlson CS. Evaluation of probabilistic and logical inference for a SNP annotation system. J Biomed Inform. 2010;43(3):407–18. doi: 10.1016/j.jbi.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]