Abstract
This study created a database of pediatric age-specific MRI brain templates for normalization and segmentation. Participants included children from 4.5 through 19.5 years, totaling 823 scans from 494 subjects. Open-source processing programs (FSL, SPM, ANTS) constructed head, brain and segmentation templates in 6 month intervals. The tissue classification (WM, GM, CSF) showed changes over age similar to previous reports. A volumetric analysis of age-related changes in WM and GM based on these templates showed expected increase/decrease pattern in GM and an increase in WM over the sampled ages. This database is available for use for neuroimaging studies (blindedforreview).
Studies of pediatric brain development have used MRI processing steps with implicit or explicit use of reference data derived from adults. Advances in our understanding of pediatric brain development would be facilitated with the availability of a comprehensive series of age-specific brain templates constructed from magnetic resonance images (MRI) obtained from healthy children from birth through young adulthood. A recent publication produced a series of MRI brain templates covering anatomical brain development of healthy children at 2-weeks through 4-years, 5-months of age, i.e., a single age-specific template for each of the following 13 age groups: 2-weeks, 3, 4.5, 6, 7.5, 9, 12, 15, 18-months; and 2, 2.5, 3, 4-years of age (Sanchez, Richards, & Almli, 2012). In the present report, we describe the construction of a comprehensive series of 32 age-specific MRI brain templates spanning healthy children and young adults from 4.5 years through 24 years of age. The individual MR images used for construction of these averaged brain templates were obtained from four sources as described in the methods section.
Due to the historical lacking of a comprehensive series of pediatric MRI brain templates, many of the MRI processing steps in pediatric neuroimaging have required implicit or explicit use of reference data derived from adult MRI (e.g., Evans et al., 1993; Mazziotta et al., 2001; Talairach & Tournoux, 1988). However, numerous studies have reported some of the unavoidable problems associated with normalizing a child brain to an adult brain (Hoeksma et al., 2005; Muzik et al., 2000; Wilke et al., 2002; Wilke et al., 2003; Yoon et al., 2009). Notably, developmental age greatly affects brain morphology, and structural variation, in terms of global and local developmental changes, that can make the seemingly similar child brain very different from an adult brain (Gogtay et al., 2004; Lenroot & Giedd, 2006; Sowell et al., 2004). Further, although it has been argued (Muzik et al., 2000) that the MNI-305 adult template (Evans et al., 1993; Mazziotta et al., 2001) may be suitable for use with the spatial normalization of children over the age of six years, the avoidance of adult biases has become more critical in pediatric research with the increased availability and use of higher resolution imaging techniques (Wilke et al., 2008; Yoon et al., 2009). Specifically, differences in shape, size, composition and contrast pose problems when referencing pediatric brains to adult brains, especially when investigating a wide range of ages in children (Muzik et al., 2000; Wilke et al., 2002). Additionally, the use of adult reference templates with pediatric neuroimaging data has been reported to yield increased amounts of deformations in nonlinear transformations (Wilke et al., 2002; Yoon et al., 2009), more variable contours of the cortex (Hoeksma et al., 2005; Muzik et al., 2000), and misclassifications of brain tissue (Wilke et al., 2003). Also, pediatric neuroimaging studies have pointed to the strong bias that adult a priori information imposes and consequently, the necessity of age-appropriate segmentation maps (Murgasova et al., 2007; Wilke et al., 2003).
The above limitations associated with using adult brain reference data in the processing of pediatric images have underscored the necessity of age-specific, pediatric brain MRI templates for imaging research with children ages 4 years through young adulthood. However, pediatric research has been historically limited by the paucity of developmental MRI studies with large samples of healthy and demographically representative children (BDCG, 2006, Almli, Rivkin, McKinstry, et al, 2007, Waber et al., 2007; Sanchez, Richards, Almli, 2012), and the published pediatric brain templates for children at 4 years of age through young adulthood generally include only one or a few age-groups, low numbers of children per template, templates averaged over a wide age range, used inappropriate normalization and registration procedures, and so on. Thus, at the present time, there is no comprehensive series of fine-grained, age-specific, demographically representative MRI brain templates for healthy children from 4 years of age through young adulthood.
Examples of published studies creating MRI brain templates for children between the ages of 4 years to 18-plus years of age are organized in Table 1. The table includes published pediatric template studies and lists certain characteristics of those studies (i.e., the reference, sequences used, study age range for templates, template ages and number, source of MRI scans, MRI magnet strength, and averaging technique. The present report is also included in Table 1 for comparison purposes. For example, some of the studies created only a single template or limited set of templates averaged across many years of age (e.g., over 13 years: Wilke et al., 2003; Yoon et al., 2009), whereas other studies have multiple templates, but each template is averaged over a wide age range (e.g., over 4 plus years: Fonov et al., 2011). Fonov et al. (2011) constructed pediatric atlases that provided templates with multi-year widths that were based on estimated pubertal status.
Table 1.
Pediatric MRI template studies for children fro 4 to 24 years of age
| Reference | Sequences | Age Range | Specific ages | Source | Magnet Strength | Averaging Technique |
|---|---|---|---|---|---|---|
| Sanchez, Richards, & Almli, 2012 (this study) | T1W, T2W, N=1289 scans, | 4.5 years to 24 years | 6 mo increments, and 20–24Years | NIHPD Objective 1, Release 3, MCBI, IXI, OASIS | 1.5, 3.0 | Non-linear iterative |
| Wilke, Holland, Altaye, & Gaser, 2008 | T1W N=404 |
4.5 years to 24 years | “Template- O-Matic” | NIHPD Objective 1, Release 1 | 1.5 | Linear, based on segment modeling |
| Fonov, Evans, Botteron, Almli, McKinstry, Collins, 2011 | T1W, T2W, PDW, N=476 | 4.5 years to 24 years | 4.5–8.5, 7.0–11.0, 7.5–13.5, 10.0–14.0, 13.0–18.5 years | NIHPD Objective 1, Release 3, ICBM 152 | 1.5 | Non-linear iterative |
| Yoon, Fonov, Perusse, & Evans, 2009. | T1W, T2W, PDW N=205 |
8.2 years | One template | Quebec Normal Twin Study | 1.5 | Non-linear iterative |
| Wilke, Schmithorst, & Holland, 2003 | T1W, N=200 | 5 to 18 years | One template, avg 11.3 years | Cincinnati Children’s Hospital Med Center | 1.5 | Linear registration |
The Table 1 studies that covered same large age ranges (i.e., Fonov et al., 2011 [4.5–24 years of age]; Wilke et al., 2003 [5 to 18 years of age]) as the current report also differed from the current report for other attributes. Whereas Fonov et al. (2011) grouped template ages into multi-year groupings with partially or fully overlapping increments, both Wilke et al (2008) and the current report created more discrete age ranges. Further, Wilke et al. (2008) created a template-building platform based on linear registration techniques through which users could specify the participant’s age ranges and sex for template construction. The present report built 32 more restricted, age-specific templates (grouped in 6 month increments). Both Fonov et al. (2011) and the current report utilized also used nonlinear registration and iterative techniques to build the templates. Linear registration has been described to blur anatomical details and decreased overlap between subjects when compared to nonlinear registration (Ashburner & Friston, 1999). While iterative techniques, where subsequent iteration processing is based on the previous average, avoid biasing the templates to adult reference data. Finally, the current report and Fonov et al. (2011) created their brain templates from the same demographically representative and healthy participant database, which allowed averaging from a larger MRI scan pool than the other studies. Although the aforementioned MRI template studies organized in Table 1 show many differences, they may prove useful for certain types of research and clinical research.
Although the use of pediatric reference data improves normalization and segmentation of pediatric MRIs, the wide variation seen in the brains of developing children at different ages has advanced the notion of more fine-grained and explicit pediatric age-specific templates (i.e., templates averaged over a more restricted age-ranges, such as the 6-month age-range used in the present report. For example, age demonstrated a large effect on brain structure and correlated strongly with the amount of deformation during spatial normalization, despite the use of a single pediatric template that covered a wide age range (Wilke et al., 2008; Wilke et al., 2002). Wilke et al. (2003) divided their pediatric sample into three different pediatric age groups and reported notable differences in tissue distribution among age groups, suggesting that a narrower age range for age-specific templates may be more accurate and appropriate for pediatric data processing (Wilke et al., 2003). Yoon et al. (2009) utilized a single, age-specific template (i.e., based on 8 year old children) to compare with normalization using an adult template. The authors reported that the use of an age-matched template contributed to considerably different tissue distributions than those obtained from adult templates.
The main goal of the current report was to examine the ability of a developmental series of age-specific MRI brain templates to classify normal brain development. The availability of a large brain scan database for demographically representative and healthy children from 4.5 years of age through 18 plus years of age has afforded the opportunity to construct age-specific pediatric templates based on a large sample of participants with unified brain scan protocols across ages (BDCG, 2006; Almli et al., 2007; Waber et al., 2007; Sanchez, Richards, Almli, 2012). The current investigation developed average T1-Weighted and T2-Weighted brain templates based on age increments of 6 months from 4.5 to 19.5 years using a systematic, iterative approach. We used methods of template construction (e.g., nonlinear registration and transformation, iterative routines) used by others (Fonov et al., 2011; Sanchez, Richards, Almli, 2012) to guide our template construction. Importantly, Sanchez, Richards & Almli (2011) created infant and young child templates from birth to 4.3 years of age using the same MRI database and with similar methodology as in the current report. This allowed a uniform method for brain scan template construction for participants ranging in age from 2 weeks to 4.3 years (Sanchez, Richards, Almli, 2012) and 4.5 years through 20–24 years (present report). These standard neurodevelopmental templates provide for increase precision in the normalization and segmentation of pediatric brains from birth through young adulthood.
Material and methods
Participants
The MRI images for construction of the templates came from children, adolescents, and young adults, ranging in age from 4.5 to 24 years of age. The images originated from four data sources: (1) the NIH Study of Normal Brain Development, Pediatric MRI Data Repository (NIHPD) (BDCG, 2006; Fonov et al., 2011); (2) blindedforreview (3) the Open Access Series of Imaging Studies project (OASIS; Marcus et al 2007, 2009); (4) the Information Extraction from Medical Images (IXI; Heckemann et al. 2003; Ericsson et al 2008) project. The first two data sources collected mostly child and adolescent data, whereas the latter two sources contained adult data.
The majority of the child and adolescent participants of the present study were from the Pediatric MRI Data Repository created by the NIHPD Objective 1 (BDCG, 2006; Waber et al, 2007). The NIHPD participants ranged in age from 4.5 to 18 plus years (N = 431 participants; 207 M/224 F; completed Version 3 data release), and were recruited across six Pediatric Centers using community-based sampling techniques to reflect the gender, income, and race/ethnicity variation in the United States Census 2000. The NIHPD aimed to study the participants three times (at 2 year intervals) with both imaging and clinical/behavioral measures over the course of the study. Participants were screened on a number of factors The screening, inclusion and exclusion criteria, and behavioral and clinical assessments are described in detail by others (BDCG, 2006, Waber et al., 2007, Almli et al., 2007, for additional detail, see also https://nihpd.crbs.ucsd.edu/nihpd/info/index.html), and briefly described here. The screening criteria included behavior problems (e.g., Child Behavior Checklist, Achenbach, 2001; scores >= 70), academic problems (learning disorder), intelligence (Weschler Abbreviated Scale of Intelligence scores < 70; Weschler 1999), and other factors that might impact healthy brain development or prohibit the full completion of the study protocol (e.g., product of high-risk pregnancy, preterm birth, history of psychiatric disorder in first order family members, physical growth delay of the participant, failure of neurological or physical examination, no parent competent in reading or speaking English, participant with significant vision or hearing problems).
An additional set of participants from 10 to 19.9 years (N = 100; 55 M/45 F) came from studies performed at the blindedforreview The participants at the blindedforreview were healthy and were excluded if they had a history of previous neurological or psychiatric illness, head trauma with loss of consciousness, current or past psycho-stimulant medication, abnormal findings on brain MRIs and technical or clinical contraindications to an MRI exam.
Participants at 20–24 years came from the NIHPD data set (N = 28; 16 M/12 F), the BLINDEDFORREVIEW (N = 87, 40 M/47 F), the OASIS project (N = 82, 28 M/54 F), and the IXI project (N = 37, 20 M/16 F/1 Unknown). The participants for the 20–24 years average served to expand an adult comparison database. Institutional review board approval and informed consent were obtained for all participants.
Age groupings
Both NIHPD and BLINDEDFORREVIEW participants were grouped in 6-month increments (e.g. 4.5 to 4.99 years represented the 4.5 year group) through age 19.9 years. The age intervals were chosen to provide relatively fine-grained age intervals while achieving sufficient numbers of participants in each age group. Age groups based on 6 month increments served to minimize variability associated with age and maximize numbers in each group. The 20–24 year old average was constructed to expand an adult comparison template similar to the ages of the MNI-152 template. Most participants in the NIHPD study had more than one MRI scan at two year intervals. The repeated scans from participants were used in the construction of the templates for the different and appropriate age of the scan; we did not keep track of the repeated scans in any of the analyses. Overall 1289 scans from 742 participants were used for creation of the templates (see Table 2 for age group information).
Table 2.
Gender composition and frequency of alternate scans in each age group
| Age Range | Total n | Gender (# Female) | Percentage Female | Alternate T1 scan | Alternate T2 scan | BLINDEDFORREVIEW 3T |
|---|---|---|---|---|---|---|
| 4.5–4.9 | 9 | 7 | 77 | 7 | 8 | |
| 5.0–5.4 | 14 | 7 | 50 | 5 | 10 | |
| 5.5–5.9 | 17 | 8 | 47 | 7 | 9 | |
| 6.0–6.4 | 27 | 15 | 55 | 12 | 16 | |
| 6.5–6.9 | 36 | 20 | 55 | 8 | 11 | |
| 7.0–7.4 | 27 | 15 | 55 | 5 | 9 | |
| 7.5–7.9 | 44 | 19 | 43 | 9 | 10 | |
| 8.0–8.4 | 56 | 30 | 53 | 5 | 9 | 10 |
| 8.5–8.9 | 40 | 22 | 55 | 4 | 7 | |
| 9.0–9.4 | 46 | 23 | 50 | 5 | 9 | |
| 9.5–9.9 | 41 | 22 | 53 | 1 | 2 | |
| 10.0–10.4 | 72 | 32 | 44 | 2 | 6 | 10 |
| 10.5–10.9 | 52 | 34 | 65 | 3 | 5 | 1 |
| 11.0–11.4 | 31 | 18 | 58 | 0 | 0 | |
| 11.5–11.9 | 40 | 22 | 55 | 4 | 7 | |
| 12.0–12.4 | 47 | 23 | 48 | 0 | 4 | 10 |
| 12.5–12.9 | 30 | 18 | 60 | 1 | 2 | 1 |
| 13.0–13.4 | 34 | 16 | 47 | 1 | 1 | |
| 13.5–13.9 | 29 | 14 | 48 | 2 | 2 | |
| 14.0–14.4 | 42 | 24 | 57 | 1 | 1 | |
| 14.5–14.9 | 31 | 15 | 48 | 1 | 1 | |
| 15.0–15.4 | 32 | 23 | 71 | 1 | 2 | |
| 15.5–15.9 | 23 | 15 | 65 | 2 | 2 | |
| 16.0–16.4 | 44 | 21 | 47 | 0 | 1 | 10 |
| 16.5–16.9 | 29 | 10 | 34 | 0 | 1 | 1 |
| 17.0–17.4 | 25 | 17 | 68 | 0 | 1 | |
| 17.5–17.9 | 25 | 9 | 36 | 1 | 3 | |
| 18.0–18.4 | 28 | 9 | 32 | 0 | 1 | 10 |
| 18.5–18.9 | 29 | 4 | 13 | 0 | 0 | 17 |
| 19.0–19.4 | 23 | 6 | 26 | 0 | 0 | 13 |
| 19.5–19.9 | 22 | 6 | 27 | 0 | 0 | 17 |
| 20.0–24.9 | 244 | 129 | 52 | 0 | 0 | 87 |
|
| ||||||
| Total | 1289 | 653 | 50 | 87 | 140 | 187 |
MRI data acquisition
The procedures for the NIHPD Objective 1 are described in detail by others (BDCG, 2006; Fonov et al., 2011). Briefly, the brain scan data acquisition lasted 30–45 minutes on a 1.5 T with 1 mm in-plane resolution and 1–1.6 mm slice thickness for the entire head (1.0 mm N = 317; 1.0 – 1.4 mm N = 199; 1.5 mm 270; 1.6 mm N = 70). Variations in slice thickness were due to use of different scanner types at different data collection sites (i.e., Siemens Medical Systems or General Electric). The scans included multiple sequences which included T1-weighted, T2-weighted and proton density acquisition as described below: A 3D T1-weighted spoiled gradient recalled (SPGR) echo sequences (TR = 22 – 25 ms, TE = 10 – 11 ms, flip angle = 30°, FoV = 256 mm IS × 256 mm AP, matrix size = 256 × 256: 1–1.6 × 1 × 1 mm3 voxels, 160–180 slices of sagittal orientation). A dual contrast, proton density- and T2-weighted sequence used an optimized 2D multislice (2mm) Fast Turbo spin echo, ETL = 8 (TR = 3500 ms, flip angle = 90°, FoV = 256 mm AP × 224 mm LR, matrix size = 256 × 256: 1 × 1 × 2 mm3 voxels, axial orientation). Some children could not tolerate the standard imaging protocol and were scanned with “alternate 2D acquisitions” that minimized scan duration but provided adequate structural scans (BDCG, 2006). The alternate MRI acquisition protocol consisted of 2D T1-weighted spin echo sequences (TR = 500 ms, TE = 12 ms, flip angle = 90°, FoV = 256 mm AP × 192 mm LR, matrix size = 256 × 192: 1 × 1 × 3 mm3 voxels, axial orientation) and a T2-weighted 2D Fast Turbo spin echo sequence (TR = 3500 ms, flip angle = 90°, FoV = 256 mm AP × 192 mm LR, matrix size = 256 × 192: 1 × 1 × 3 mm3 voxels, axial orientation. Table 2 lists the number of participants in each age group who underwent the “Alternate” 2D T1- and T2- weighted scans. The actual pixel resolutions and dimensions found in the NIH online data for alternate scans varied more than originally formulated and are found in Tables 3 and 4 (e.g., the 3D T1W scans varied from 98 to 176 sagittal slices and the 2D alternate scans varied from 44 to 66 axial slices, with a FoV of 192 × 256 or 256 × 256 mm). The scans were conducted at different sites with Siemens Medical Systems (Sonata, Magnetom) and GE (Signa Excite) scanners. The scans were obtained from the NIHPD www site in compressed NIFTI format.
Table 3.
Frequency distribution for NIH alternate 2D scans of T1W dimensions and pixels.
| Dimensions (Number of slices, FoV) | Pixel Resolution | ||||||
|---|---|---|---|---|---|---|---|
| Sagittal | Coronal | Axial | n | Sagittal | Coronal | Axial | n |
| 192 | 256 | 44 | 2 | 0.938 | 0.938 | 2.907 | 2 |
| 192 | 256 | 52 | 1 | 0.938 | 0.938 | 2.915 | 2 |
| 192 | 256 | 56 | 1 | 0.938 | 0.938 | 2.926 | 1 |
| 192 | 256 | 66 | 21 | 0.938 | 0.938 | 2.927 | 2 |
| 192 | 256 | 84 | 1 | 0.938 | 0.938 | 3 | 23 |
| 256 | 256 | 42 | 1 | 0.977 | 0.977 | 2.907 | 1 |
| 256 | 256 | 45 | 1 | 0.977 | 0.977 | 3 | 11 |
| 256 | 256 | 46 | 2 | 0.977 | 0.977 | 3.001 | 1 |
| 256 | 256 | 48 | 6 | 0.977 | 0.977 | 3.002 | 1 |
| 256 | 256 | 50 | 4 | 1 | 1 | 3 | 36 |
| 256 | 256 | 51 | 4 | 1.016 | 1.016 | 2.943 | 1 |
| 256 | 256 | 52 | 18 | 1.016 | 1.016 | 3 | 6 |
|
|
|||||||
| 256 | 256 | 53 | 1 | 87 | |||
|
|
|||||||
| 256 | 256 | 54 | 3 | ||||
| 256 | 256 | 60 | 6 | ||||
| 256 | 256 | 62 | 1 | ||||
| 256 | 256 | 65 | 1 | ||||
| 256 | 256 | 66 | 13 | ||||
|
|
|||||||
| Total | 87 | ||||||
|
|
|||||||
Table 4.
Frequency distribution of T2W dimensions and pixels for NIH alternate scans
| Sagittal | Coronal | Axial | n |
|---|---|---|---|
| 0.938 | 0.938 | 2.907 | 2 |
| 0.977 | 0.977 | 2.907 | 2 |
| 0.977 | 0.977 | 2.914 | 1 |
| 0.938 | 0.938 | 2.915 | 2 |
| 0.977 | 0.977 | 2.915 | 1 |
| 0.938 | 0.938 | 2.926 | 2 |
| 0.938 | 0.938 | 2.927 | 2 |
| 1.016 | 1.016 | 2.928 | 1 |
| 1.016 | 1.016 | 2.943 | 1 |
| 0.938 | 0.938 | 3 | 34 |
| 0.977 | 0.977 | 3 | 19 |
| 1 | 1 | 3 | 65 |
| 1.016 | 1.016 | 3 | 7 |
| 0.977 | 0.977 | 3.006 | 1 |
|
| |||
| Total | 140 | ||
The data at the BLINDEDFORREVIEW were collected on a Siemens Medical Systems 3T Trio, and the three scans comprising this MRI protocol and had an overall duration of about 15 min. First, a localizer sequence allowed for subsequent approximate orientation to the anterior commissure (AC) and posterior commissure (PC) plane. Next, a 3D T1-weighted “MPRAGE” RF-spoiled rapid flash scan in the sagittal plane employed the following parameters: TR = 2250 ms, TE = 4.52 ms, flip angle = 9°, FoV = 256 mm × 256 mm, matrix size = 1 × 1 × 1 mm3 (the sagittal dimension of the T1W ranged from 160 to 212 slices). The third sequence, a T2-weighted multi-slice axial 2D dual Fast Turbo spin-echo scan (ETL = 5) utilized the following parameters: TR = 3200 ms, TE = 213, and matrix size = 1 × 1 × 1 mm3. The sagittal dimensions of the T2W ranged from 176 to 177 mm, with coronal and axial slices of 256 × 256 mm. DICOMM files from BLINDEDFORREVIEW were exported from the Siemens system and read into compressed NIFTI format (http://nifti.nimh.nih.gov/). All scans, whether from the NIHPD database (normal/alternate) or from USC-MCBI, were processed in the same manner using NIFTI compressed format (32 bit floating point).
Imaging data from the OASIS (Marcus et al 2007, 2009) and the IXI (Heckemann et al. 2003; Ericsson et al 2008) contributed to the adult template only (20–24 year old template). The OASIS study implemented a T1-weighted MPRAGE on a 1.5T Vision Scanner (TR = 9.7 ms, TE = 4.0 ms, flip angle = 10°, FoV = 256 mm × 256 mm, matrix size = 1 × 1 × 1 mm3). The IXI images, in total, consisted of 600 MR images from normal, healthy subjects ranging in age from 20 to 80 years of age (FoV = 256 mm × 256 mm, matrix size = .9375 × .9375 × 1.2 mm3). The multislice spin echo T1 images were collected at 3 sites with 1.5 and 3T scanners.
File preparation
MR images were prepared for pipeline processing in three steps. First, the brains were extracted from the whole-head MRI volume using the brain extraction tools of FSL. An automated bash script using the FSL tools (Smith, et al., 2004; Woolrich, et al., 2009) completed this task with the following actions: register the head to the MNI-152 head (Collins et al., 1995; Mazziotta et al., 2001); inverse-transform a MNI-brain-mask to the participant space; use the mask to get a preliminary brain; use betsurf to get a binary skull mask; use the skull mask to delineate a second preliminary brain; use bet2 to extract the brain from the second preliminary brain mask for the final brain (Jenkinson et al., 2005; Smith, 2002). The use of bet2 and betsurf together followed standard FSL script. We visually inspected each brain for accuracy, and adjusted some of the bet2 variables (e.g., fractional intensity threshold) to get a well-formed brain volume (Jenkinson et al., 2005).
Second, we adjusted the MRI intensity variations found in the datasets (NIHPD, USC-MCBI, OASIS, IXI) that stemmed from different machines, different recording sites and slight differences in protocol. Bias field inhomogeneity was corrected with a N4 bias field correction procedure (Avants et al., 2011; Tustison et al., 2010). Then, the FSL FAST procedure, “FMRIB’s Automated Segmentation Tool” (Zhang et al., 2001) segmented the T1W scan into GM, WM, and CSF. The MRI voxels with partial volume estimates of 1.0 in the GM segments were averaged to determine the average voxel intensity for GM. The scan was then renormed with this value to have a value of 100, which resulted in the peak of the GM intensity in the MRI histogram curve equaling 100. The T2W intensity values were then renormed. Cerebrospinal fluid is identified as voxels with the brightest intensities in the T2W MRI volume. The T2W voxel threshold was set as the average voxel quantity found in the lateral ventricles, which gave an approximate map of cerebrospinal fluid in the T2W. The T2W brain was transformed by relative voxel intensity into a probability map for T2W-CSF, and the T2W was renormed so that the CSF had an average intensity of 100.
Construction of age-specific templates
We constructed the age-specific pediatric with the iterative routine found in Sanchez et al. (2011; also see Fonov et al, 2011, Guimond, 2000, Yoon et al., 2009). The whole-head MRI volumes and brain-extracted MRI volumes were performed separately with the purpose of providing templates for MRI and EEG use. Figure 1 is a schematic representation of the steps used in the construction of the template for a specific age group for both whole-head and brain-extracted templates. The first step of the iterative procedure was to construct a tentative average (Figure 1, “A0”) based on a rigid rotation (FLIRT 6 parameter linear registration and transformation; (Jenkinson & Smith, 2001) to the MNI-152 adult template (ICBM-152 defined in Mazziotta et al, 2001; Joshi et al., 2004). The iterative procedure consisted of a non-linear registration (ANTS, “Advanced Normalization Tools”; Avants et al., 2008, 2011) to the current reference average (An-1), a transformation of each participant MRI into the template space (Vn), and then a averaging of the transformed MRIs.(An,). This average was then used as the reference model in the next iteration (An-1 on next step). The root mean square (RMS) difference between successive average reference models was calculated, and the iterative procedure was done until leveling of the successive RMS values was obtained. The final reference model is the “age-specific” template. More details of this procedure may be found in Sanchez et al. (2011).
Figure 1.
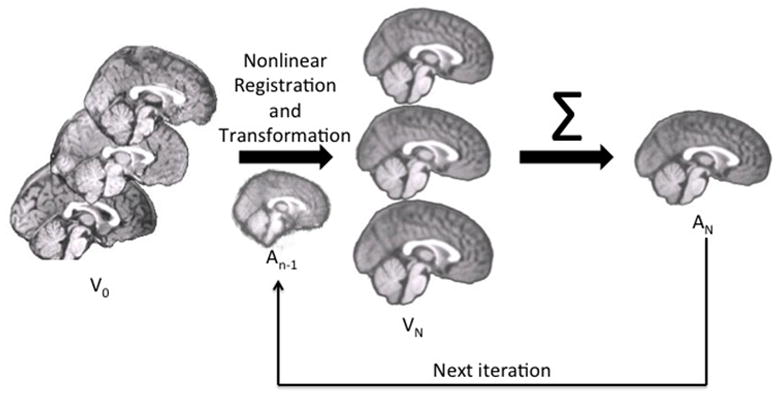
The pipeline for age-specific template creation. (A) In Step 0, the rigid registration occurred using FLIRT to the MNI brain with 6 DOF, with an output that was the same volume size as the original so as to keep pediatric sizes. Rigidly registered brains (V0) were averaged to create a rough template (A0). This template was used as the first guide in Step N. (B) With each iteration of Step N, the rigidly registered brains were nonlinearly registered to an iterative average (An-1) and transformed and then averaged to create a new average (An) for the next iteration.
We calculated T2W image templates from the dataset through use of the T2W volumes from the NIHPD and BLINDEDFORREVIEW MRI datasets. The T2W volumes were registered with rigid-rotation affine methods to the original MRI volume (FLIRT; both T2W whole head, and T2W brain). The T2W volumes were then averaged in the iteration procedure similar to the T1W volumes (e.g., rigid-rotation to MNI-152 head; initial averaging; successive ANTS registration, transformation, averaging; see Figure 1). We used one additional step in the procedure for the T2W volumes. The average template for the T2W volume on each step (AVGn in Figure 1) was registered with affine parameters (FLIRT) to the relevant T1W average template before being used as the AVGN-1 in the following step. This step retained the T2W average template in the same size and orientation as the T1W volume to allow for comparisons. The average brain template(s) are not precisely registered to the corresponding head template(s) since the whole head and extracted-brain constructions were separate. We therefore extracted the brain from the completed head template to provide a brain model corresponding to the space of the average head MRI template.
We used open-source and publicly available tools for these methods. An automated image processing pipeline was constructed with the Linux bash scripting language. The FSL FLIRT tool (Jenkinson & Smith, 2001) performed the rigid rotation to the MNI-152 volume (6-degree-of-freedom). The ANTS program, “Advanced Normalization Tools”, (Avants et al., 2008), performed an affine and diffeomorphic registration (symmetric normalization, SyN) of the source volumes to the reference volumes. The ANTS program uses symmetric diffeomorphic normalization to capture shape differences and enhance alignment of image features. The ANTS tool, AverageImages (non-normed) was used for the averaging step. The Sun Grid Engine (SGE; http://wikis.sun.com/display/gridengine62u2/Home) was used to compute the iterations on a SGE with 58 processors on the fastest Intel microprocessors (3.0+ GHz).
Transformation parameters
Each iteration step from the ANTS procedure resulted in a coefficient matrix for the affine transformation, three volumes with non-linear deformation values for the forward-transformation (Warp x, y, z) of the source volume to the reference volume, and three MRI volumes with non-linear deformation values for the inverse transformation (InverseWarp x, y, z) of the reference volume to the source volume. The spatial transformation coefficients and deformation-volumes on the final iteration represent the linear and non-linear registration of the individual participant MRI volumes to the average volumes. The transformation matrix and transformation volumes were retained for the tissue classification and volume-measurement steps.
Tissue classification
The individual participant MRI volumes were classified into GM, WM, and CSF. We used four procedures to do this in order to determine how different segmentation processes affect classification for age-specific templates. First, the FSL FAST procedure (Zhang et al., 2001) was used to segment the T1W scans without using any prior classification volumes (“Image”). Second, the FSL FAST procedure was used with the MNI-152 segmentation maps. The segmentation maps were computed by first using the ANTS linear/SyN registration of the MNI-152 brain to the average template for the age-appropriate template for a participant, transforming the MNI-152 segmentation maps to the average template space, then using the participant-average template transformation parameters to put the segmentation maps in the participant space. The resulting segmentation maps were then submitted to the FAST program with the individual participant MRI scan as segmentation maps and the identity matrix as warp coefficients (“A Priori” segmentation maps). Third, the same segmentation maps were used with the FAST -P option, which applies the segmentation maps to the results, emphasizing the segmentation maps in an a posteriori manner (“A Posteriori” segmentation maps). Fourth, the SPM8b segmenting routine was used with the same segmentation maps as in the second and third methods (options- no affine regularization since the segmentation maps were in the same size and orientation as the participant MRI volume; “SPM8” segmentation maps). All four methods resulted in a set of partial volume estimates (PVE) for GM, WM, and CSF, for each participant’s MRI volume. The individual participant PVE volumes were then forward-transformed by the participant-template transformation parameters into the average template size and orientation. An average of the transformed participant volumes was made separately for the GM, WM, and CSF volumes for each segmentation procedure in order to create “Image”, “A Priori”, “A Posteriori” and “SPM8” segmentation maps for each age-specific template. This resulted in tissue segmentation maps for each age-specific template, and separate segmentation maps for the four methods.
Results
The database consists of age-specific templates divided into half-year increments from 4.5 to 24 years of age, for a total of 32 age group templates (see Table 2 for N per age group). Templates for T1W head, T1W brain, T2W head and T2W brain exist for each age group. Additionally, partial volume estimates and binary segmented images were created for each age group from the four different segmentation methods: Image (no segmentation maps), A Priori, A Posteriori, and SPM8b. The T1W average templates represent gray matter values normalized to 100, so that variations in voxel intensity between MRI volumes did not affect the averages. All averaged templates result from an initial linear registration and transformation with the MNI-152 template. Subsequent steps utilized nonlinear transformations with age-specific templates created from prior steps, which minimized the influence of the adult templates. The resulting age specific head and brain templates represent the average size for the participants at that specific age. The templates are loosely oriented to the MNI-152 volume.
Figure 2 shows the change in age-specific template fit with each successive iteration. This was calculated as the RMS difference between the current iteration to the product from the previous iteration. The RMS difference measured the intensity difference between successive iterations at each voxel. Each line in Figure 2 represents the result from one age-template. The iterations for the T1W brain and head showed a general trend for the iterations to converge to a minimum level after about 4 or 5 iterations (Figure 2A, 2B). After 2 iterations with low resolution (50×0×0, 50×50×0, iterations 0 and 1) and one iteration at higher resolution (at 50×50×10, iteration 2), few additional iterations were needed to reach asymptotic RMS for all age groups. We show the iteration sequences for the normed T2W (Figure 2C). The same pattern of convergence occurred as with the T1W.
Figure 2.
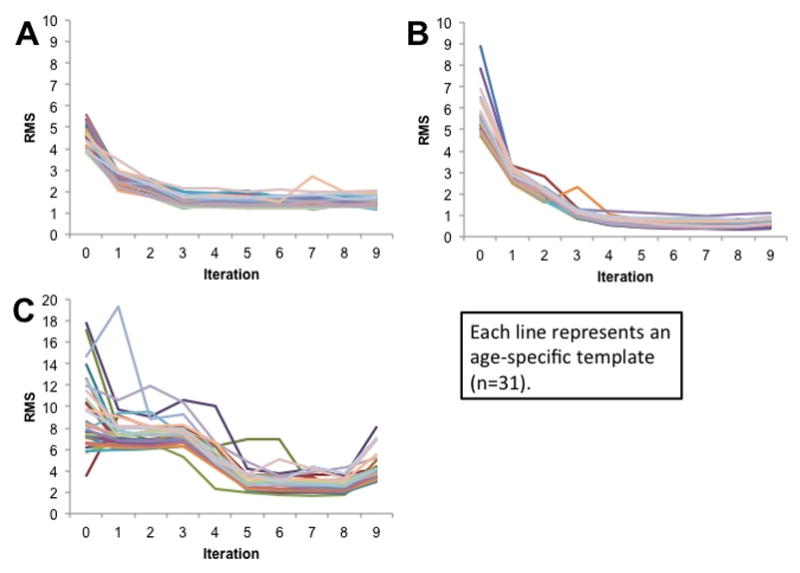
Degree of fit between successive iterations for (A) T1W Brain (B) T1W Head and (C) T2W Brain. Each line represents an age-specific template (n = 32) undergoing iterations. The dependent variable (RMS) represents the root-mean-squared difference between the intensity of voxels at the same location in the successive iterations.
Mid-sagittal slices for each age-specific T1W head template are demonstrated in Figure 3. The sagittal view of the averaged templates exhibited small morphological structures in fine detail, as can be seen with cortical and subcortical anatomy. The gradual change in head size was noticeable when comparing the 4.5 age group to the 20–24 age group. Despite the differential use of normal and alternate scans, all templates appeared to be relatively consistent in regards to level of detail and clarity and no gross errors or blurring was visually detected.
Figure 3.
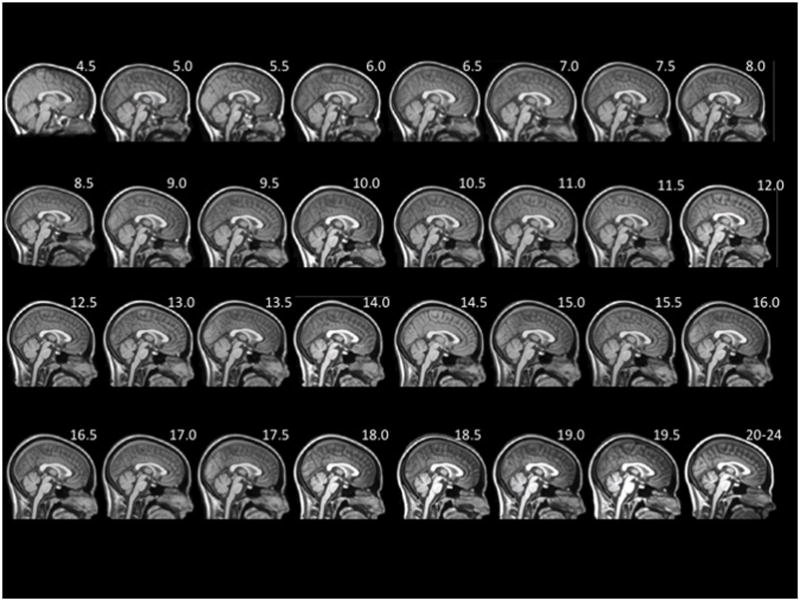
Mid-sagittal slice of whole-head average template T1W MRI volumes across ages of study.
An axial slice at the AC-PC level for selected ages is shown in Figure 4 for T1W head, T1W brain and T2W brain. The use of the same contrast parameters for each type of scan allows for the discernment of changes between ages. White matter intensity changed from hypointense to hyperintense relative to gray matter as the templates were followed vertically from younger to older ages. Additionally, the amount of gray matter appeared to decline when moving from the 5 year old template to the 20–24 year old template; at the same time, there was an increase in the amount of white matter. This can be seen in the bright sections of the T1W, and the dark sections of the T2W.
Figure 4.
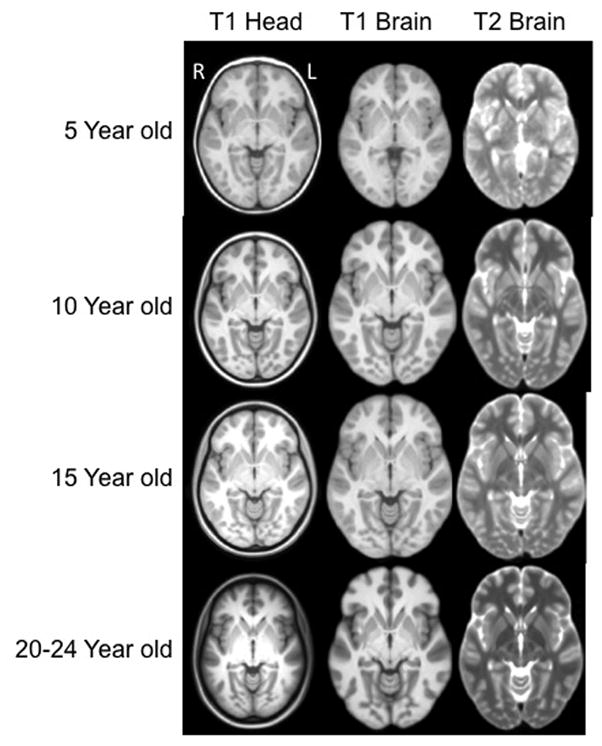
Head and brain templates with T1W and T2W weighted images, for 5, 10, 15 and 20–24 years of age. The displayed data is based on final averaged output as outlined in Figure 1. (axial slices at AC-PC level). Each figure is show approximately as the same size and not scaled according to the actual age head size. Right of head is on left of figure.
Tissue Classification
Figure 5 shows the results of gray matter tissue classification using four different approaches (Image, A Priori, A Posteriori, and SPM8) for selected age-specific templates. Segmentation procedures produced similar patterns of tissue distribution and tissue changes over development, such that GM became less thick and WM became more extensive as the ages of participants increased. Slight differences in tissue classification occurred, with the most notable differences manifesting in the segmented images created by SPM8. Segmented images produced by SPM8 appear to have thicker gray matter than the other segmentation procedures. Further segmentation analyses (see below) addresses the SPM8 discrepancies with other segmentation techniques.
Figure 5.
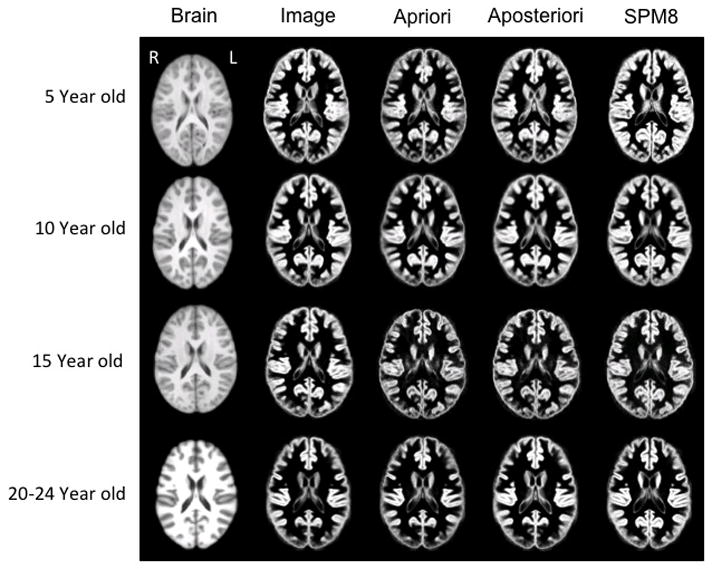
Brain template (left column) and associated gray matter tissue probability maps (in the four right columns) based on segmentation methods used for 5, 10, 15 and 20–24 year old templates (axial slices at AC-PC level). Figures are not to scale with actual head size; right of head is on left of figure.
Utility of brain templates for developmental differences in GM-WM-CSF
The neurodevelopmental database of MRI volumes were used to examine developmental changes in the composition of the brain. We segmented all MRI volumes, calculated partial volume estimates and summed across GM/WM/CSF for each participant. Figure 6 shows GM/WM materials for selected ages for the FSL A Posteriori image segmentation and the SPM8 segmentation. Examination across the select age-specific probability maps showed a gradual increase in WM volume over the entire age interval for both the A Posteriori image, F (32,1485) = 5.27, p < .001, Cohen’s f̂ = .295) and the SPM8 segmentation, F (32, 1485) = 2.85, p < .001, Cohen’s f̂ = .194). The GM showed an increase during childhood, a relatively stable level over most of adolescence, and a decline in GM after about age 16 years. Both the A Posteriori image segmentation and the SPM segmentation were significantly affected by recording age; F (32,1485) = 33.87, p < .001, Cohen’s f̂ = .808 and 15.97, p < .001, Cohen’s f̂ = .553. Figure 7 shows the quantitative differences in tissue voxel counts for every age-specific template for both segmentation procedures (A Posteriori, SPM) and CSF derived from the T2W. Note the discrepancies of WM voxel counts between the A Posteriori and SPM8 methods: segmentation with SPM8 identified more voxels as GM and fewer voxels as WM compared to A Posteriori methods, which created larger differences in volume between GM and WM. Both methods, however, showed the expected volume GM changes (decrease over age) and the WM changes (gradual increase). The T2W-derived CSF volume did not change over this age range. Figure 8 shows the changes in GM and WM over age only for the cortex. This was computed by transforming a cortex atlas from the 20–24 year old template to the younger ages. The changes in the cortex reflected the changes in overall brain size and both GM and WM changed over age similar to the whole head volume.
Figure 6.
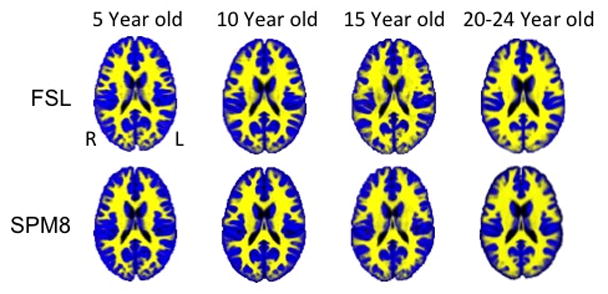
Tissue probability maps using “Image” segmentation procedure with FSL FAST and using SPM8b (with segmentation maps) across select age groups
Figure 7.
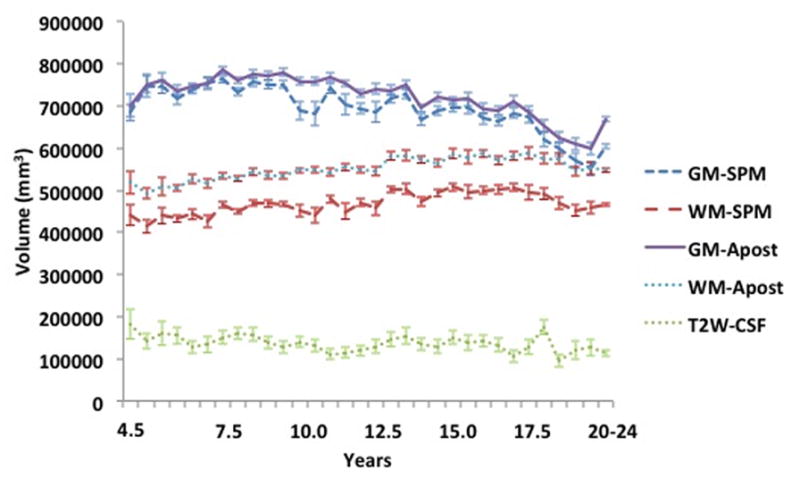
Whole brain voxel volume for gray matter, white matter and cerebral spinal fluid for each age group template. The volumes are based on segmentation with SPM and FSL-FAST (A Posteriori) processing for GM and WM. The CSF voxel volumes came from T2W-derived CSF volume.
Figure 8.
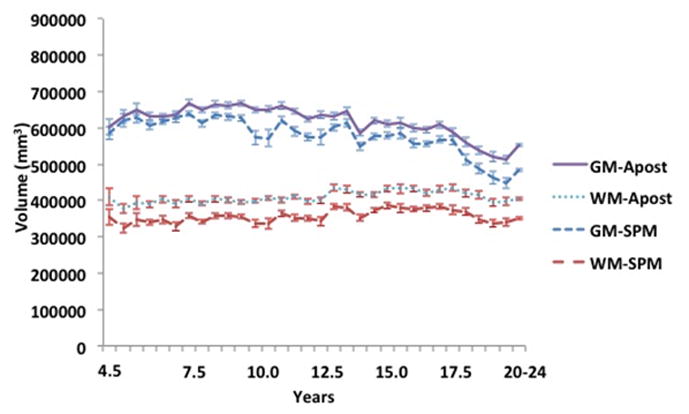
Cortex voxel volume for gray matter and white matter for each age group template. The volumes are based on segmentation with SPM and FSL-FAST (A Posteriori) processing.
Discussion
The main goal of the current study was to create a developmental series of relatively fine-grained, age-specific MRI brain templates to facilitate our ability to gain understanding of normal brain development and the diseased brain. Pediatric age specific templates were created that organize brain development into discrete 6-month periods for a healthy pediatric sample. The quality of the age-specific templates is validated through the replication of known trends in developmental neuroscience. The sizes of the heads agree with the established progression of brain development, such that there are broad peaks and maximal size between early school age in total cerebral volume (Giedd et al., 1996) and by adolescence (Lenroot et al., 2007). Additionally, the observed GM and WM changes confirm established trends for brain development (Casey et al., 2000; Courchesne et al., 2000), demonstrating GM thinning and WM growth with increasing age (Courchesne et al., 2000; Giedd et al., 1996). While the methods of segmentation produced similar patterns of brain development, they differed significantly in how they classified WM. These findings replicate previous research showing that brain tissue classification in pediatric samples depends largely on the segmentation scheme (Wilke et al., 2002)
The present study created age-specific segmentation maps to be used as a developmental reference tool for improved classification of pediatric brain tissue. These segmentation maps were created to decrease the influence of age and volume change on tissue probabilities. For all age groups, a priori segmentation maps were created in the form of tissue probability maps and three-class segmented volumes for the four segmentation methods: Image, A Priori, A Posteriori and SPM. We did not use a “gold standard” manual segmentation (e.g. the Internet Brain Segmentation Repository [http://www.cma.mgh.harvard.edu/ibsr/]) to test the tissue classification methods, so it cannot be stated how accurately and consistently tissues were labeled. A noticeable difference between SPM8b and other segmentation methods was identified. Specifically, the SPM8b methods appeared to code more tissue as GM globally throughout the brain. SPM8b-created segmented images showed markedly less overlap in GM than the other segmentation procedures (see Figure 7, Figure 8). This suggests that FSL-based segmentation techniques may be better than SPM8b for pediatric tissue classification. Future studies might directly compare the differences between segmentation outcomes to a “gold standard”.
Other groups have created MRI brain templates from the NIHPD database. Fonov et al. (2011) created templates based on ~ 4 year increments and Wilke et al. (2008) create templates grouped by user specification of gender and age. These two sets of templates make significant contributions to pediatric neuroimaging. The current study builds upon their work by providing a more fine-detailed picture of age-related changes in pediatric brain development. Age accounts for a significant amount of variability in the normalization and segmentation of pediatric images, despite the use of broad ranging pediatric templates (i.e., templates encompassing pediatric brains from 5 to 18 years of age; Wilke et al., 2002; Wilke et al., 2008). Furthermore, Yoon et al. (2009) showed that a pediatric-based template required 20% less deformation than an adult-based template. Based on these studies, we saw the next step to be the creation of more fine-grained, age-specific templates, and the NIHPD database allowed us to create 32 templates in very discrete 6 month age groupings across ages from 4.5 years through 18 plus years.
For the present report, the 6-month template intervals were chosen to provide a fine-grained series of developmental templates based on a workable number of participants contributing to each template. For now, the existence of this MRI template database with 6-month-old intervals should be useful since a sufficiently precise age-template is available for any age in the range covered in this study. Future studies might clarify the range of age-specific templates that may be most appropriate for a participant of a specific age. This could be done by comparing the results yielded from use of templates with varying age ranges (e.g., 6 months; 1.5 years; 4 years).
The present age-specific templates are intended for use on at least two levels: from a methodological perspective and from a database perspective. First, the method of template creation that we used here should prove useful to developmental neuroscience researchers. We utilized publically-available image post-processing software programs (FSL, SPM, ANTS) to produce our age-specific templates. Similar to others (Fonov et al. 2011; Sanchez et al., 2011), the age-specific templates were created through an iterative approach that minimized the influence of adult a priori data and maximized the preservation of the different sizes, shapes and tissue distribution of the pediatric data. The processing pipeline refined the images recursively, such that the optimization procedure was applied to the data at different resolutions, with successively higher resolution during the nonlinear registration. Our pipeline procedure should be useful to others who wish to create pediatric templates based on different parameters using state-of-the-art averaging programs. Second, the pediatric neuroimaging community will find that our templates can be used for neuroimaging studies involving structural image analysis, automated structural extraction, tissue classification, forward and reverse modeling for EEG/MEG, and functional image analysis. For example, the present age-specific templates provide brain templates and segmentation maps with skull intact to aid in the calculation of cortical source models of EEG and ERP (Reynolds & Richards, 2008).
One strength of the templates created from the NIHPD pertains to their potential to generalize to other subject populations. The majority of MR images that we used were collected as part of the NIHPD project which enrolled a diverse sample to characterize normal, healthy brain development. Thus, the pediatric templates largely represent the diversity present in the United States. Also, the MRI data used to construct the present templates were collected at multiple MRI centers, at differing volume resolutions, and with 1.5T and 3.0T scanners, so the current MRI data and templates reflect measurement error introduced by multiple variables (Jovicich, et al., 2009; Schnack, et al. 2004). The high number of 2D scans at early ages is a potential shortcoming of the NIHPD, but we prefer to preserve the younger age MRI categories due to the unique nature of brain imaging data capturing normal brain development in these young ages. Future studies might investigate the influence of scanner type on template creation. Potential users of this database should take caution in using 3D data with the younger age templates. Regardless, the inclusion of multiple scanner types and protocols should add a degree of robustness to the age-specific templates; a sample based on only data collected at a single site would limit generalization to other samples.
Our age-specific templates provide a framework with which to create pediatric templates and segmentation maps for use in developmental neuroimaging. Future investigations might create broader age ranges to classify different stages of childhood and adolescence, as well as creation of templates based on gender to investigate potential differences in developmental trajectories of males and females (Lenroot et al., 2007). A voxel by voxel registration of the T1W and T2W templates might aid investigations that examine the two types of scans. Also, future research might determine which segmentation procedure best represents pediatric tissue distribution. One question that could be answered with these methods is the age at which the pediatric brain may be represented by the adult brain, and for which brain regions these variations occur.
Discrepancies between adult and pediatric brains have continued to be an issue in pediatric neuroimaging and the need for age-specific templates is apparent. The NIHPD study with a large scale normative pediatric sample has provided the opportunity to address the need for age-specific brain templates. We implemented an automatic image processing pipeline for template creation that works well with pediatric and infant images (Sanchez et al., 2011) and created age-specific pediatric templates using state-of-the-art averaging techniques (Avants, et al, 2008; Avants, et al, 2011; Klein et al., 2009). The age-specific templates provide a priori information that reflects developmental trends to guide segmentation. Use of age-specific reference data will facilitate the generation of reliable conclusions about the normative ranges of brain growth and development for specific ages, as well as the ability to detect subtle differences in brain development. The regional brain variation present in early childhood and adolescence is acknowledged and preserved with this type of developmental neuroscience work, and understanding of brain development in health and disease will be facilitated.
The current work contributes to the pediatric clinical and research community by creating age-specific pediatric brain templates based on a narrow age range. The templates were created from a multi-center study, and despite the different imaging parameters, are collectively able to meaningfully represent anatomical changes in development. The templates, in 6-month increments, are of an unprecedented scale in terms of the narrow range of ages used to characterize a relatively long and dynamic period of brain development (e.g., Table 1). The age-specific templates created for this study, as well as other studies (Fonov et al., 2011; Wilke et al., 2008), stand to provide increased accuracy of neuroimaging procedures which will increase our understanding of brain development. The templates used in the current study are available online and will be updated as additional material becomes available (blindedforreview/).
Acknowledgments
This work was supported by the NIH grant to JER, #R37 HD18942, and by grants from the National Institute of Child Health and Human Development, the National Institute on Drug Abuse, the National Institute of Mental Health, and the National Institute of Neurological Disorders and Stroke (Contract #s N01-HD02-3343, N01-MH9-0002, and N01-NS-9-2314, -2315, -2316, -2317, -2319 and -2320) to CRA. A listing of the participating sites and a complete listing of the study investigators can be found at http://www.bic.mni.mcgill.ca/nihpd/info/participating_centers.html.
References
- Achenbach TM. Manual for the ASEBA School-Age Forms and Profiles (Child Behavior Checklist for Ages 6 – 18) ASBEA; Burlington, Vermont: 2001. [Google Scholar]
- Almli CR, Rivkin MJ, McKinstry RC. The nih mri study of normal brain development (objective-2): Newborns, infants, toddlers, and preschoolers. Neuroimage. 2007;35(1):308–325. doi: 10.1016/j.neuroimage.2006.08.058. [DOI] [PubMed] [Google Scholar]
- Altaye M, Holland SK, Wilke M, Gaser C. Infant brain probability templates for mri segmentation and normalization. Neuroimage. 2008;43:721–730. doi: 10.1016/j.neuroimage.2008.07.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner J, Friston KJ. Nonlinear spatial normalization using basis functions. Human Brain Mapping. 1999;7(4):254–266. doi: 10.1002/(SICI)1097-0193(1999)7:4<254::AID-HBM4>3.0.CO;2-G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avants BB, Epstein CL, Grossman M, Gee JC. Symmetric diffeomorphic image registration with cross-correlation: Evaluating automated labeling of elderly and neurodegenerative brain. Medical Image Analysis. 2008;12(1):26–41. doi: 10.1016/j.media.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avants BB, Tustison NJ, Song G, Cook PA, Klein A, Gee JC. A reproducible evaluation of ants similarity metric performance in brain registration. Neuroimage. 2011;54(3):2033–2044. doi: 10.1016/j.neuroimage.2010.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brain Development Cooperative Group (BDCG) The nih mri study of normal brain development. Neuroimage. 2006;30(1):184–202. doi: 10.1016/j.neuroimage.2005.09.068. [DOI] [PubMed] [Google Scholar]
- Casey BJ, Giedd JN, Thomas KM. Structural and functional brain development and its relation to cognitive development. Biological Psychology. 2000;54(1–3):241–257. doi: 10.1016/s0301-0511(00)00058-2. [DOI] [PubMed] [Google Scholar]
- Collins DL, Holmes CJ, Peters TM, Evans AC. Automatic 3-d model-based neuroanatomical segmentation. Human Brain Mapping. 1995;3(3):190–208. [Google Scholar]
- Courchesne E, Chisum HJ, Townsend J, Cowles A, Covington J, Egaas B, et al. Normal brain development and aging: Quantitative analysis at in vivo mr imaging in healthy volunteers. Radiology. 2000;216(3):672–682. doi: 10.1148/radiology.216.3.r00au37672. [DOI] [PubMed] [Google Scholar]
- Ericsson A, Alijabar P, Rueckert D. Construction of a patient-specific atlas of the brain: application to normal aging. ISBI IEEE. 2008:480–483. [Google Scholar]
- Evans AC, Collins DL, Mills SR, Brown ED, Kelly RL, Peters TM. 3d statistical neuroanatomical models from 305 mri volumes. Paper presented at the Proc IEEE Nucl Science Symp Medl Imaging Conf.1993. [Google Scholar]
- Fonov V, Evans AC, Botteron K, Almli CR, McKinstry RC, Collins DL. Unbiased average age-appropriate atlases for pediatric studies. Neuroimage. 2011;54(1):313–327. doi: 10.1016/j.neuroimage.2010.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giedd JN, Snell JW, Lange N, Rajapakse JC, Casey BJ, Kozuch PL, et al. Quantitative magnetic resonance imaging of human brain development: Ages 4–18. Cerebral Cortex. 1996;6(4):551–560. doi: 10.1093/cercor/6.4.551. [DOI] [PubMed] [Google Scholar]
- Gogtay N, Giedd JN, Lusk L, Hayashi KM, Greenstein D, Vaituzis AC, et al. Dynamic mapping of human cortical development during childhood through early adulthood. Proceedings Of The National Academy Of Sciences Of The United States Of America. 2004;101(21):8174–8179. doi: 10.1073/pnas.0402680101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimond A, Meunier J, et al. Average brain models: a convergence study. Computer Vision and Image Understanding. 2000;77:192–210. [Google Scholar]
- Heckemann RA, Hartkens T, Leung K, Hill DL, Hajnal JV, Rueckert D. Information Extraction from Medical Images (IXI): Developing an e-Science Application Based on the Globus Toolkit. 2nd UK e-Science All-hands Conference; Nottingham, UK. Sept 2003. [Google Scholar]
- Hoeksma MR, Kenemans JL, Kemner C, van Engeland H. Variability in spatial normalization of pediatric and adult brain images. Clinical Neurophysiology. 2005;116(5):1188–1194. doi: 10.1016/j.clinph.2004.12.021. [DOI] [PubMed] [Google Scholar]
- Jenkinson M, Pechaud M, Smith S. Bet2: Mr-based estimation of brain, skull and scalp surfaces. Paper presented at the Eleventh annual meeting of the Organization for Human Brain Mapping; Toronto, Canada. 2005. [Google Scholar]
- Jenkinson M, Smith S. A global optimisation method for robust affine registration of brain images. Medical Image Analysis. 2001;5(2):143–156. doi: 10.1016/s1361-8415(01)00036-6. [DOI] [PubMed] [Google Scholar]
- Joshi S, Davis B, Jomier M, Gerig G. Unbiased diffeomorphic atlas construction for computational anatomy. Neuroimage. 2004;23:S151–160. doi: 10.1016/j.neuroimage.2004.07.068. [DOI] [PubMed] [Google Scholar]
- Jovicich J, Czanner S, Han X, Salat D, van der Kouwe A, Quinn B, et al. MRI-derived measurements of human subcortical, ventricular and intracranial brain volumes: Reliability effects of scan session, acquisition sequences, data analyses, scanner upgrade, scanner vendors and field strengths. Neuroimage. 2009;46(1):177–192. doi: 10.1016/j.neuroimage.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenroot RK, Giedd JN. Brain development in children and adolescents: Insights from anatomical magnetic resonance imaging. Neuroscience And Biobehavioral Reviews. 2006;30(6):718–729. doi: 10.1016/j.neubiorev.2006.06.001. [DOI] [PubMed] [Google Scholar]
- Lenroot RK, Gogtay N, Greenstein DK, Wells EM, Wallace GL, Clasen LS, et al. Sexual dimorphism of brain developmental trajectories during childhood and adolescence. Neuroimage. 2007;36(4):1065–1073. doi: 10.1016/j.neuroimage.2007.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus DS, Wang TH, Parker J, Csernansky JG, Morris JC, Buckner RL. Open access series of imaging studies (OASIS): Cross-sectional MRI data in young, middle aged, nondemented, and demented older adults. Journal of Cognitive Neuroscience. 2007;19:1498–1507. doi: 10.1162/jocn.2007.19.9.1498. [DOI] [PubMed] [Google Scholar]
- Marcus DS, Fotenos AF, Csernansky JG, Morris JC, Buckner RL. Open acess series of imaging studies (OASIS): Longitudinal MRI data in nondemented and demented older adults. Journal of Cognitive Neuroscience. 2010;22(12):2677–2684. doi: 10.1162/jocn.2009.21407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazziotta J, Toga A, Evans A, Fox P, Lancaster J, Zilles K, et al. A probabilistic atlas and reference system for the human brain: International consortium for brain mapping (icbm) Philosophical Transactions Of The Royal Society B-Biological Sciences. 2001;356(1412):1293–1322. doi: 10.1098/rstb.2001.0915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murgasova M, Dyet L, Edwards D, Rutherford M, Hajnal J, Rueckert D. Segmentation of brain mri in young children. Academic Radiology. 2007;14(11):1350–1366. doi: 10.1016/j.acra.2007.07.020. [DOI] [PubMed] [Google Scholar]
- Muzik O, Chugani DC, Juhasz C, Shen CG, Chugani HT. Statistical parametric mapping: Assessment of application in children. Neuroimage. 2000;12(5):538–549. doi: 10.1006/nimg.2000.0651. [DOI] [PubMed] [Google Scholar]
- Reynolds GD, Richards JE. Cortical source localization of infant cognition. Developmental Neuropsychology. 2009;3:312–329. doi: 10.1080/87565640902801890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez CE, Richards JE, Almli CR. Neurodevelopmental MRI brain templates for children from 2 week to 4 years of age. Developmental Psychobiology. 2011 doi: 10.1002/dev.20579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnack HG, van Haren NEM, Pol HEH, Picchioni M, Weisbrod M, Sauer H, et al. Reliability of brain volumes from multicenter mri acquisition: A calibration study. Human Brain Mapping. 2004;22(4):312–320. doi: 10.1002/hbm.20040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi F, Yap PT, Wu G, Jia H, Gilmore JH, Lin W, Shen D. Infant brain atlases from neonates and 1-and 2-year-olds. PLoS ONE. 2011;6(4):e18746. doi: 10.1371/journal.pone.0018746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM. Fast robust automated brain extraction. Human Brain Mapping. 2002;17(3):143–155. doi: 10.1002/hbm.10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowell ER, Thompson PM, Leonard CM, Welcome SE, Kan E, Toga AW. Longitudinal mapping of cortical thickness and brain growth in normal children. Journal Of Neuroscience. 2004;24(38):8223–8231. doi: 10.1523/JNEUROSCI.1798-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talairach J, Tournoux P. Co-planar stereotaxic atlas of the human brain. New York: Thieme Medical Publishers; 1988. [Google Scholar]
- Tustison NJ, Avants BB, Cook PA, Zheng YJ, Egan A, Yushkevich PA, et al. N4itk: Improved n3 bias correction. IEEE Transactions on Imaging. 2011;29(6):13310–1320. doi: 10.1109/TMI.2010.2046908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waber DP, De Moor C, Forbes PW, Almli CR, Botteron KN, Leonard G, et al. The nih mri study of normal brain development: Performance of a population based sample of healthy children aged 6 to 18 years on a neuropsychological battery. Journal Of The International Neuropsychological Society. 2007;13(5):729–746. doi: 10.1017/S1355617707070841. [DOI] [PubMed] [Google Scholar]
- Wechsler D. Wechsler Abbreviated Scale of Intelligence. Psychological Corporation, Harcourt Brace and Company; San Antonio, TX: 1999. [Google Scholar]
- Wilke M, Holland SK, Altaye M, Gaser C. Template-o-matic: A toolbox for creating customized pediatric templates. Neuroimage. 2008;41(3):903–913. doi: 10.1016/j.neuroimage.2008.02.056. [DOI] [PubMed] [Google Scholar]
- Wilke M, Schmithorst VJ, Holland SK. Assessment of spatial normalization of whole-brain magnetic resonance images in children. Human Brain Mapping. 2002;17(1):48–60. doi: 10.1002/hbm.10053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilke M, Schmithorst VJ, Holland SK. Normative pediatric brain data for spatial normalization and segmentation differs from standard adult data. Magnetic Resonance In Medicine. 2003;50(4):749–757. doi: 10.1002/mrm.10606. [DOI] [PubMed] [Google Scholar]
- Woolrich MW, Jbbdi S, Patenaude B, Chappell M, Makni S, Behrens T, et al. Bayesian analysis of neuroimaging data in fsl. Neuroimage. 2009;45(1):S173–S186. doi: 10.1016/j.neuroimage.2008.10.055. [DOI] [PubMed] [Google Scholar]
- Yoon U, Fonov VS, Perusse D, Evans AC. The effect of template choice on morphometric analysis of pediatric brain data. Neuroimage. 2009;45(3):769–777. doi: 10.1016/j.neuroimage.2008.12.046. [DOI] [PubMed] [Google Scholar]
- Zhang YY, Brady M, Smith S. Segmentation of brain mr images through a hidden markove random field model and the expectation maximization algorithm. IEEE Transactions On Medical Imaging. 2001;20(1):45–57. doi: 10.1109/42.906424. [DOI] [PubMed] [Google Scholar]


