Abstract
Pharmacological responses of G protein-coupled receptors (GPCRs) can be fine-tuned by allosteric modulators. Structural studies of such effects have been limited due to the medium resolution of GPCR structures. We re-engineered the human A2A adenosine receptor by replacing its third intracellular loop with apo-cytochrome b562RIL and solved the structure at 1.8 angstrom resolution. The high-resolution structure identified about 57 ordered waters inside the receptor comprising three major clusters. The central cluster harbors a putative sodium ion bound to the highly conserved Asp2.50. Additionally, two cholesterols stabilize the conformation of helix VI, and one of 23 ordered lipids intercalates inside the ligand-binding pocket. These high-resolution details shed light on the potential role of structured waters, sodium ions and lipids/cholesterol in GPCR stabilization and function.
G protein-coupled receptors (GPCRs) comprise the largest and most diverse family of membrane proteins in eukaryotes, sharing a common architecture of seven transmembrane (7TM) α-helices. GPCRs transduce a variety of signals across the cell membrane that regulate diverse biological and (patho)physiological processes, and, hence, are favored drug targets. Ligand-dependent GPCR activation triggers dramatic conformational changes in the 7TM helical bundle that are coupled to activation of G proteins and other downstream effectors. Over the last four years, breakthroughs in protein engineering and crystallography have yielded structures of fourteen different GPCRs responding to diffusible ligands in different functional states (1–9), providing the basic structural framework for understanding ligand binding and activation mechanisms. However, despite the progress in GPCR stabilization for crystallographic studies, the resolution of GPCR structures has remained in the 2.2–3.5 Å range (1). The function of GPCRs relies on a specific lipid environment and often strongly depends on the presence of cholesterol and ions (10–12). Understanding the structural mechanism of such effects has been limited due to the absence of higher resolution GPCR structures.
Here we replaced the third intracellular loop (ICL3) of the human A2A adenosine receptor (A2AAR) with a thermostabilized apocytochrome b562RIL (BRIL) (13, 14) and determined the crystal structure of this chimeric protein (referred to as A2AAR-BRIL-ΔC (15)) in complex with a high-affinity, subtype-selective antagonist, ZM241385, at 1.8 Å resolution (table S1). The high resolution enabled us to identify a comprehensive network of 57 internal waters, a highly conserved binding site for a sodium ion, three cholesterol molecules, and 23 lipid acyl chains bound to the receptor.
The ligand binding and functional characteristics of the A2AAR-BRIL-ΔC fusion protein were extensively characterized and compared to the A2AAR-WT (wild type) and the A2AAR-ΔC (C-terminus truncation) constructs. Antagonist [3H]ZM241385 radioligand binding and agonist displacement assays confirmed that the ligand recognition site of the A2AAR-BRIL-ΔC fusion protein is very similar to that of both the A2AAR-WT and the A2AAR-ΔC constructs (fig. S1). The BRIL insertion did not have an impact on receptor expression and trafficking to the cell plasma membrane (fig. S2), but, expectedly, prevented the receptor construct from activating available Gs proteins (fig. S3).
The high-resolution receptor structure is nearly identical to the original 2.6 Å resolution crystal structure of A2AAR-T4L-ΔC/ZM241385 (PDB ID 3EML) (16), with an all-atom RMSD = 0.45 Å over 81% of A2AAR. The conformations of the cytoplasmic ends of helices V and VI near the BRIL junction sites closely resemble the conformations in the A2AAR structure with unmodified ICL3 (17), in contrast to a distorted conformation caused by the T4L fusion in A2AAR-T4L-ΔC/ZM241385 (16). A distal part of the extracellular loop 2 (ECL2), which was missing in all inactive state structures of A2AAR, is fully resolved (fig. S4). Most importantly, the higher resolution revealed atomic details for the structurally conserved regions including intricate networks of water molecules, sodium ion, lipids and cholesterols.
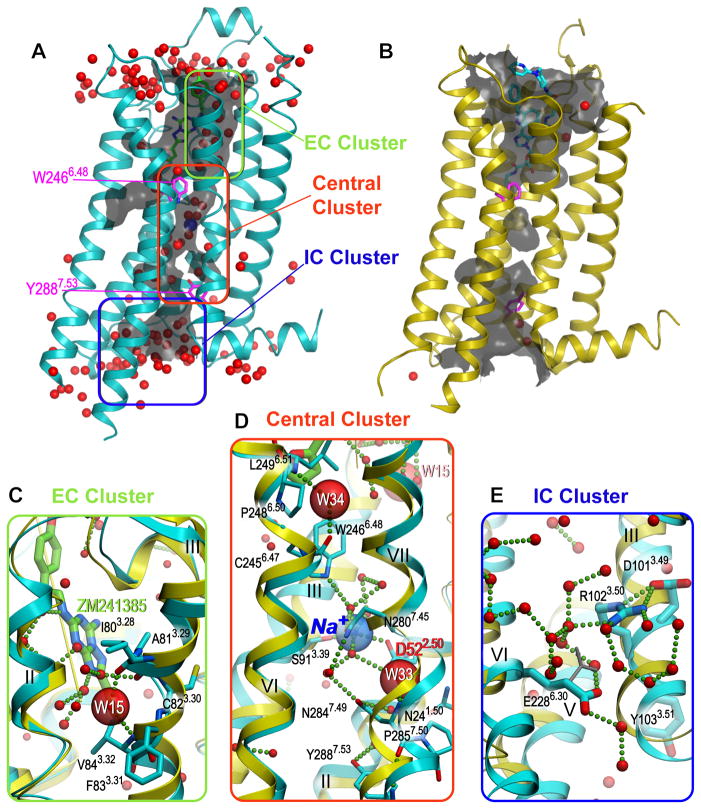
The 1.8 Å resolution structure of A2AAR-BRIL-ΔC/ZM241385 contains 177 structured waters, 57 of which occupy the interior of the 7TM bundle. The interior waters form an almost continuous channel extending from the ligand-binding site to the site of G protein interaction (Fig. 1A), which is comprised of three bulky water molecule clusters, as well as several scattered waters. The channel has two narrow “bottlenecks” restricted by Trp2466.48 and Tyr2887.53 (18) to slightly less than the diameter of one water molecule (2.4 and 2.0 Å, respectively). The rearrangements of the receptor backbone and side chains upon activation disrupt the channel continuity in these sites (Fig. 1B), substantially reducing its volume.
Fig. 1. Distribution of ordered waters in A2AAR.
In all panels, the antagonist-bound high-resolution structure is shown in cyan and the agonist-bound, active-like state structure (PDB ID 3QAK) is shown in yellow, waters are represented as red spheres, and salt bridges and hydrogen bonds as small green spheres. (A) Interior watersin A2AAR-BRIL-ΔC/ZM241385 structure form an almost continuous water channel (grey; calculated using the program Hollow (37)) containing three major water clusters. (B) The channel is disrupted in the active-like state structure (PDB ID 3QAK). (C) Close-up of the extracellular (EC) water cluster deep in the ligand binding pocket. The water molecule W15 shown as a large red semitransparent sphere stabilizes the kink in helix III. (D) Close-up of the central cluster, which includes waters and a sodium ion (blue transparent sphere). Water molecules W34 and W33 stabilizing the Pro-induced kinks in helices VI and VII are shown as large red semitransparent spheres. (E) Close-up of the intracellular (IC) cluster around the D[E]RY motif in helix III. Despite of close proximity Arg1023.50 and Glu2286.30 do not form an ionic interaction, instead both amino acids form hydrogen bonds with neighboring waters. An alternative rotamer of Glu2286.30, making a potential 3.0 Å contact with Arg1023.50 side chain, is shown in a grey stick representation.
The first of the three water clusters, the EC cluster, located inside the orthosteric ligand-binding pocket, plays a role in ligand binding and selectivity (19) (Fig. 1C and fig. S5). One of the waters in this cluster stabilizes the conformation of a non-proline kink in helix III by forming hydrogen bonds with both the main-chain carboxyl of Ile803.28 and the main-chain nitrogen of Val843.32. In the active-like state agonist-bound A2AAR structures (20, 21), the kink in helix III straightens, thereby precluding water binding, which points to a role of water rearrangement at this location in A2AAR activation. This ligand-induced and water-mediated conformational change is accompanied by an overall activation-related shift of helix III (20).
The second, central water cluster includes a sodium ion and 10 waters that completely fill a cavity in the middle of the 7TM bundle (Fig. 1D). The central cluster spans a distance of more than 13 Å between two functionally important and conserved residues, Trp2466.48 and Tyr2887.53. Three water molecules from this cluster were previously observed in the A2AAR-T4L-ΔC/ZM241385 complex (16) (fig. S6A), and positions of four of the ten water molecules are similar to those found in bovine rhodopsin (PDB ID 1U19) (22) (fig. S6B), suggesting that such an internal network is common in Class A GPCRs and could be important for their function (23, 24). Finally, the IC cluster containing approximately 20 water molecules is found near the conserved D[E]RY motif (Fig. 1E), which is inferred in the stabilization of different functional receptor states and in interactions with G proteins and other effectors.
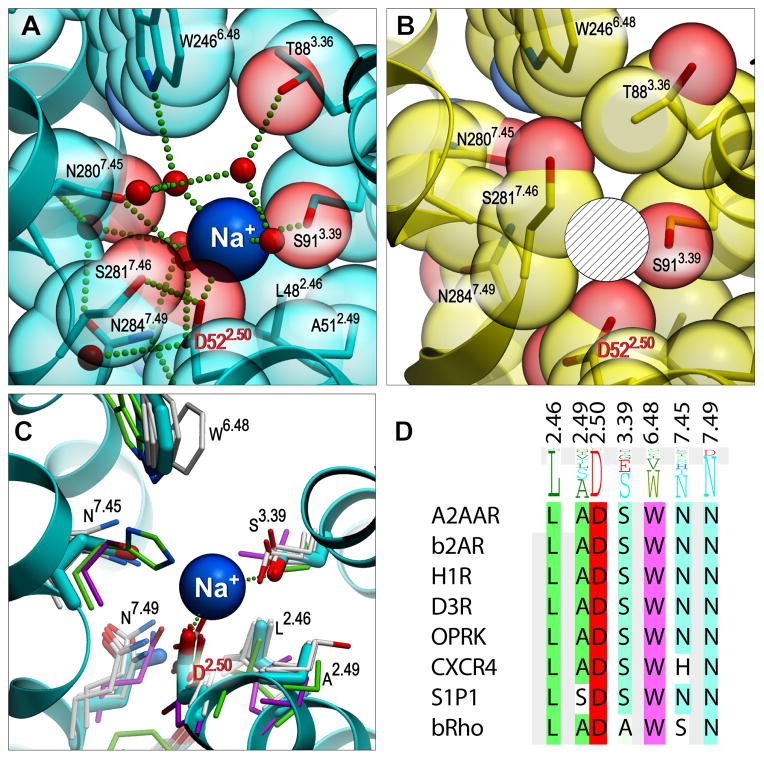
Evidence for the presence of a sodium ion in the central water cluster came from a strong spherical electron density with a distorted octahedral coordination and short distances to coordinating atoms in its neighborhood. Such geometry was poorly compatible with the presence of a water molecule. However, Na+ modeled in this density is coordinated by five oxygen atoms including OD1 of Asp522.50 (2.45 Å), OG of Ser913.39 (2.54 Å) and three waters (2.21, 2.44 and 2.59 Å) (Fig. 2A, fig. S7), which is in excellent agreement with coordination and distances for Na+ in protein structures (25). Moreover, the presence of Na+ in the structure is consistent with conditions employed during purification (0.8 M Na+) and crystallization (~0.15 M Na+). Allosteric effects of Na+ on ligand binding, stability and crystallization of A2AAR have been documented in previous studies (11, 16, 26). We confirmed the allosteric effects of physiological concentrations of Na+ on agonist and antagonist binding and thermal stability of A2AAR (Fig. 3, table S2).
Fig. 2. Structural details of the Na+ allosteric site in the inactive and active-like state A2AAR.
(A) Sodium ion (blue sphere) in the middle of the 7TM bundle coordinated by highly conserved Asp522.50, Ser913.39 and 3 water molecules. Receptor is shown as a ribbon, with residues lining the Na+ cavity shown as sticks and transparent spheres with carbon atoms colored cyan and oxygen atoms red. Water molecules in the cluster are shown as small red spheres, while the salt bridge between Na+ and Asp522.50 and hydrogen bonds are shown as green dotted lines. (B) The pocket collapses in the active-like state A2AAR-T4L-ΔC/UK432,097 structure, precluding Na+ binding at this site (hatched sphere designates the position of Na+ in the inactive structure). (C) Structural conservation of the allosteric pocket among solved GPCR structures. (A2AAR - cyan, CXCR4 - green, rhodopsin - magenta, all other - grey). (D) Sequence conservation of the pocket residues among all class A GPCRs (shown as a residue profile in the top row), and among the solved GPCR structures.
Figure 3. Modulation of A2AAR by sodium ions, amiloride and cholesterol.
(A) [3H]ZM241385 or (B) [3H]NECA equilibrium binding to A2AAR-WT and A2AAR–BRIL-ΔC constructs transiently expressed on HEK293 cell membranes in the presence of buffer (control) or buffer supplemented with 150 mM NaCl, 100 μM amiloride, combinations of 100 μM amiloride and 150 mM NaCl, and 150 mM choline chloride. The figures represent data combined from three separate experiments performed in duplicate. Differences in specific binding were analyzed by a Student’s t-test. Significant differences were observed for the effect of modulators on control binding and are noted as follows: ** p < 0.01, *** p < 0.001. Significant differences were observed for the effect of NaCl on amiloride modulation and noted as follows: # p < 0.05, ## p < 0.01. There was no significant effect of choline chloride on [3H]ZM241385 or [3H]NECA binding, further proof that Na+ rather than Cl− ions caused the effect of NaCl. (C) Shifts in thermostability of A2AAR-BRIL-ΔC construct purified in detergent micelles upon addition of 150 mM NaCl, 100 μM amiloride, combinations of 100 μM amiloride and 150 mM NaCl, 1 μM ZM241385, 1 μM ZM241385 and 150 mM NaCl, and 0.01% CHS. Experiments with ZM241385 have been repeated 6 times, with a standard deviation of less than 1 °C. The composition of control buffer was 25 mM Hepes pH 7.5, 0.05% DDM, 0.01% CHS for all samples except for the study of the effect of CHS, in which the control buffer was 25 mM Hepes pH 7.5, 0.05% DDM, 800 mM NaCl.
During the last 40 years allosteric modulation by Na+ has been observed for many GPCRs, and was linked to motifs in helix II, including the highly conserved Asp2.50 (27). Mutation of this residue has been the subject of many studies on a multitude of receptors; in the GPCRDB database (28), it is mentioned >100 times for dozens of receptors of human origin and from other species. The residue has not been mutated in the A2AAR, but in the closely homologous A1 and A3 adenosine receptors, the Na+ effect was largely abrogated when the residue was mutated to alanine or asparagine, respectively (29, 30). Despite this indirect evidence, the nature of Na+ interactions with GPCRs remained hypothetical (31, 32) and the sodium ion remained undetected in crystal structures of GPCRs solved at medium resolution.
In this high-resolution A2AAR structure, we were not only able to determine the precise location of Na+, but also resolved the complete network of water molecules around it and conformations of all residues involved in direct or water-mediated coordination. The central water cluster harboring the sodium ion is surrounded by and engaged in hydrogen bonding with several highly conserved residues including Asn241.50, Asp522.50, Ser913.39, Trp2466.48, Asn2807.45, Asn2847.49 and Tyr2887.53 (Fig. 2A). Structural alignment of these residues from known GPCR structures reveals a well preserved site capable of binding Na+ along with several water molecules (Fig. 2C,D). Small deviations were observed in bovine rhodopsin, in which Asn7.45 is substituted with a serine, and Asn7.49 adopts a different rotamer, and in CXCR4 where Asn7.45 is replaced with a histidine. Interestingly, in many olfactory receptors, Ser3.39 is substituted with a glutamate, resulting in two acidic side chains pointing inside the central water cluster, which may favor binding of a divalent cation in this site.
The central water cluster with Na+ likely plays an important role in receptor activation. Comparison between the inactive and active-like states of A2AAR (PDB ID 3QAK) suggests that the inward movement of helix VII in this region upon activation collapses this pocket from 200 to 70 Å3 and shifts the pocket towards helix VI (Fig. 2B). The resulting pocket in the active-like state can accommodate a maximum of three water molecules and does not provide sufficient coordination for Na+. This comparison suggests that high-affinity agonist binding and the presence of Na+ are indeed mutually exclusive, which is consistent with the observed negative allosteric effects of Na+ on binding of agonists (Fig 3 and table S2) (16, 20).
The high resolution structure also allows us to investigate off-target interactions of the diuretic and sodium channel blocker amiloride with GPCRs that were first documented by Howard et al. (33) and Nunnari et al. (34). They demonstrated that amiloride and derivatives non-competitively displaced a radiolabeled antagonist from the α2-adrenergic receptor by enhancing the radioligand’s dissociation rate. Moreover, Howard et al. (33) provided evidence that NaCl competed with amiloride, suggesting that Na+ and amiloride share an allosteric binding site. We docked amiloride into the central cluster cavity (fig. S8), with only minor changes in the conformations of the surrounding side chains, i.e. Trp2466.48. Multiple docking runs consistently led to an amiloride orientation, in which its charged guanidinium group interacts with Asp522.50, while other polar groups form a network of hydrogen bonds with residues in the pocket, mimicking polar interactions in the Na+/water cluster. Successful docking of bulkier amiloride derivatives, such as 5-(N,N-hexamethylene)amiloride (HMA), required more substantial rearrangements of several side chains and possibly the main-chain (not shown), which would potentially perturb the conformation of the deepest part of the orthosteric ligand-binding pocket. This result is consistent with the higher potencies of such derivatives in accelerating orthosteric antagonists’ off-rates (11). This docking study also corroborates the results of our experiments (Fig. 3), in which amiloride reduced [3H]ZM241385 binding (probably due to the conformational changes in Trp2466.48), inhibited binding of the agonist [3H]NECA, and competed with Na+ ions for the same binding site.
The 1.8 Å structure includes 23 ordered lipid chains and 3 cholesterols per receptor. Together, they form an almost complete lipid bilayer around each protein molecule, mediating crystal contacts (Fig. 4A,B and fig. S9). Lipids on the extracellular side have stronger electron densities and appear to be more ordered. All three cholesterols in this structure are bound to the extracellular half of the receptor and have low B-factors (25 to 27 Å2) in comparison to other lipids (41 to 61 Å2). Two of these cholesterols (CLR1 and CLR3) are bound to symmetry-related receptors and mediate crystal lattice packing by forming face-to-face interactions. The third cholesterol molecule (CLR2) does not participate in crystal contacts. Interestingly, CLR2 and CLR3 occupy hydrophobic grooves along helix VI and form extensive contacts with the aromatic ring of Phe2556.57, which is sandwiched between them (Fig. 4C, fig. S10). In the adenosine family of receptors, position 6.57 is conserved as Ile, Val, or Phe: hydrophobic residues that could all support the type of stacking interaction observed in this structure. In addition, CLR2 forms a hydrogen bond (2.7 Å) with the main-chain carboxyl of Ser263, and the hydroxyl of CLR3 has a polar interaction with sulfur of Cys259 (3.8 Å) in ECL3, the loop that is stabilized by the Cys259-Cys262 disulfide bond. Specific binding and conformational stabilization of this region of helix VI by cholesterols may play a functional role in A2AAR by fixing the position of Asn2536.55 in the ligand binding pocket of the receptor (Fig. 4C). This key residue exists in all adenosine receptors and anchors the exocyclic amine of the ligand’s central core in both agonist and antagonist complexes (20). Such direct cholesterol binding to A2AAR is also consistent with observations that addition of cholesterol hemisuccinate (CHS) increases the thermostability of the receptor purified in detergent micelles (Fig. 3).
Fig. 4. Lipid-receptor interactions.
(A) Top view of A2AAR (cyan ribbon) including crystallographic neighbors (green: translational symmetry, magenta: rotational, yellow: antiparallel arrangement). Cholesterol molecules are shown as balls with yellow carbons, lipid molecules are shown as sticks with grey carbons. (B) Side view of A2AAR. (C) Potential stabilizing effect of two cholesterols, CLR2 and CLR3, on the conformation of helix VI. Side chains Phe2556.57 and Asn2536.55 of the A2AAR-BRIL-ΔC/ZM241385 complex are shown as sticks with cyan carbons. The superimposed active-like state A2AAR-T4L-ΔC/UK432,097 is shown as orange ribbon (helix VI only) and sticks (Asn2536.55 side chain and NECA scaffold of the UK432,097 agonist only). (D) Lipid molecule (OLA11, grey balls) is inserted in between helices I and VII inside the ligand-binding pocket.
While most lipid chains form non-specific hydrophobic contacts filling grooves on the protein surface, several interactions of lipids with A2AAR may be specific: for example, interactions between the lipids’ polar groups and the main chain and side chain polar groups of ECL1 and ECL3. In addition, one lipid (OLA11) intercalates inside the TM bundle between helices I and VII, thereby protruding into the ligand-binding pocket (Fig. 4D). This lipid molecule apparently stabilizes the conformation of the first eight N-terminal residues of helix I, which do not make any direct contacts with the rest of the helical bundle.
It is intriguing to speculate that GPCRs are allosteric machines (35–38), controlled in part by ions like sodium, membrane components such as lipids and cholesterol, as well as water molecules. Although this concept of allostery is different, with more and more high resolution eukaryotic membrane protein structures and complementary biophysical studies, it seems likely we will start to observe the control of membrane proteins by more than just the pharmacological ligands or the traditional definition of allostery being a singly defined binding site that directly perturbs the orthosteric binding site. The small molecules mentioned can dramatically affect the stability of a protein as well as the function, and this effect can have a pronounced effect on the physiological signaling of GPCRs in very important ways.
Supplementary Material
Acknowledgments
This work was supported by the NIH Common Fund in Structural Biology grant P50 GM073197 for technology development, the NIH/NIGMS PSI:Biology grant U54 GM094618 for biological studies and structure production, the NIGMS grant R01 GM089857 to VC, the Dutch Research Council NWO TOP grant (#714.011.001: “the crystal structure of the adenosine A2A receptor: the follow-up”) to APIJ, and the NWO Veni grant (#11188) to LHH. We thank J. Velasquez for help on molecular biology; T. Trinh, K. Allin and M. Chu for help on baculovirus expression; T. Mulder-Krieger and H. de Vries for their technical expertise in the biochemical characterization, G. van Westen for his educated comments on the crystal structure; M. Mileni for reviewing the final structure; J.P. Changeux for discussions on GPCR allostery and the potential for allosteric stabilizers; K. Kadyshevskaya for assistance with figure preparation; A. Walker for assistance with manuscript preparation; J. Smith, R. Fischetti, and N. Sanishvili for assistance in development and use of the minibeam and beamtime at GM/CA-CAT beamline 23-ID at the Advanced Photon Source, which is supported by National Cancer Institute grant Y1-CO-1020 and National Institute of General Medical Sciences grant Y1-GM-1104. The coordinates and the structure factors have been deposited in the Protein Data Bank under the accession code (4EIY).
Footnotes
This manuscript has been accepted for publication in Science. This version has not undergone final editing. Please refer to the complete version of record at http://www.sciencemag.org/. The manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior, written permission of AAAS.”
Materials and Methods
References (39–56)
References and Notes
- 1.Katritch V, Cherezov V, Stevens RC. Diversity and modularity of G protein-coupled receptor structures. Trends Pharmacol Sci. 2012;33:17. doi: 10.1016/j.tips.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rasmussen SG, et al. Crystal structure of the beta2 adrenergic receptor-Gs protein complex. Nature. 2011;477:549. doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hanson MA, et al. Crystal structure of a lipid G protein-coupled receptor. Science. 2012;335:851. doi: 10.1126/science.1215904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haga K, et al. Structure of the human M2 muscarinic acetylcholine receptor bound to an antagonist. Nature. 2012;482:547. doi: 10.1038/nature10753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kruse AC, et al. Structure and dynamics of the M3 muscarinic acetylcholine receptor. Nature. 2012;482:552. doi: 10.1038/nature10867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu H, et al. Structure of the human κ-opioid receptor in complex with JDTic. Nature. 2012 doi: 10.1038/nature10939. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Manglik A, et al. Crystal structure of the micro-opioid receptor bound to a morphinan antagonist. Nature. 2012 doi: 10.1038/nature10954. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thompson AA, et al. Structure of the Nociceptin/Orphanin FQ Receptor in Complex with a Peptide Mimetic. Nature. 2012 doi: 10.1038/nature11085. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Granier S, et al. Structure of the delta opioid receptor bound to naltrindole. Nature. 2012 doi: 10.1038/nature11111. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burger K, Gimpl G, Fahrenholz F. Regulation of receptor function by cholesterol. Cell Mol Life Sci. 2000;57:1577. doi: 10.1007/PL00000643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao ZG, IJzerman AP. Allosteric modulation of A(2A) adenosine receptors by amiloride analogues and sodium ions. Biochem Pharmacol. 2000;60:669. doi: 10.1016/s0006-2952(00)00360-9. [DOI] [PubMed] [Google Scholar]
- 12.Goblyos A, IJzerman AP. Allosteric modulation of adenosine receptors. Biochim Biophys Acta. 2010;1808:1309. doi: 10.1016/j.bbamem.2010.06.013. [DOI] [PubMed] [Google Scholar]
- 13.Chun E, et al. Fusion partner toolchest for the stabilization and crystallization of G protein-coupled receptors. Structure. 2012 doi: 10.1016/j.str.2012.04.010. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alexandrov AI, Mileni M, Chien EY, Hanson MA, Stevens RC. Microscale fluorescent thermal stability assay for membrane proteins. Structure. 2008;16:351. doi: 10.1016/j.str.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 15.Schapira M, Abagyan R, Totrov M. Nuclear hormone receptor targeted virtual screening. J Med Chem. 2003;46:3045. doi: 10.1021/jm0300173. [DOI] [PubMed] [Google Scholar]
- 16.Jaakola VP, et al. The 2.6 angstrom crystal structure of a human A2A adenosine receptor bound to an antagonist. Science. 2008;322:1211. doi: 10.1126/science.1164772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dore AS, et al. Structure of the adenosine A(2A) receptor in complex with ZM241385 and the xanthines XAC and caffeine. Structure. 2011;19:1283. doi: 10.1016/j.str.2011.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Palczewski K, et al. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 2000;289:739. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 19.Katritch V, Kufareva I, Abagyan R. Structure based prediction of subtype-selectivity for adenosine receptor antagonists. Neuropharmacology. 2010;60:108. doi: 10.1016/j.neuropharm.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu F, et al. Structure of an agonist-bound human A2A adenosine receptor. Science. 2011;332:322. doi: 10.1126/science.1202793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lebon G, et al. Agonist-bound adenosine A2A receptor structures reveal common features of GPCR activation. Nature. 2011;474:521. doi: 10.1038/nature10136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okada T, et al. The retinal conformation and its environment in rhodopsin in light of a new 2.2 A crystal structure. J Mol Biol. 2004;342:571. doi: 10.1016/j.jmb.2004.07.044. [DOI] [PubMed] [Google Scholar]
- 23.Nygaard R, Frimurer TM, Holst B, Rosenkilde MM, Schwartz TW. Ligand binding and micro-switches in 7TM receptor structures. Trends Pharmacol Sci. 2009;30:249. doi: 10.1016/j.tips.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 24.Angel TE, Chance MR, Palczewski K. Conserved waters mediate structural and functional activation of family A (rhodopsin-like) G protein-coupled receptors. Proc Natl Acad Sci U S A. 2009;106:8555. doi: 10.1073/pnas.0903545106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harding MM. Metal-ligand geometry relevant to proteins and in proteins: sodium and potassium. Acta Crystallogr. 2002;D58:872. doi: 10.1107/s0907444902003712. [DOI] [PubMed] [Google Scholar]
- 26.Magnani F, Shibata Y, Serrano-Vega MJ, Tate CG. Co-evolving stability and conformational homogeneity of the human adenosine A2a receptor. Proc Natl Acad Sci U S A. 2008;105:10744. doi: 10.1073/pnas.0804396105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parker MS, Wong YY, Parker SL. An ion-responsive motif in the second transmembrane segment of rhodopsin-like receptors. Amino acids. 2008;35:1. doi: 10.1007/s00726-008-0637-6. [DOI] [PubMed] [Google Scholar]
- 28.Vroling B, et al. GPCRDB: information system for G protein-coupled receptors. Nucl Acids Res. 2010;39:D309. doi: 10.1093/nar/gkq1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barbhaiya H, McClain R, IJzerman A, Rivkees SA. Site-directed mutagenesis of the human A1 adenosine receptor: influences of acidic and hydroxy residues in the first four transmembrane domains on ligand binding. Mol Pharmacol. 1996;50:1635. [PubMed] [Google Scholar]
- 30.Gao ZG, et al. Identification of essential residues involved in the allosteric modulation of the human A(3) adenosine receptor. Mol Pharmacol. 2003;63:1021. doi: 10.1124/mol.63.5.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhorov BS, Ananthanarayanan VS. Signal transduction within G-protein coupled receptors via an ion tunnel: a hypothesis. J Biomolec Struct Dyn. 1998;15:631. doi: 10.1080/07391102.1998.10508980. [DOI] [PubMed] [Google Scholar]
- 32.Selent J, Sanz F, Pastor M, De Fabritiis G. Induced effects of sodium ions on dopaminergic G-protein coupled receptors. PLoS Comp Biol. 2010;6 doi: 10.1371/journal.pcbi.1000884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Howard MJ, Hughes RJ, Motulsky HJ, Mullen MD, Insel PA. Interactions of amiloride with alpha- and beta-adrenergic receptors: amiloride reveals an allosteric site on alpha 2-adrenergic receptors. Mol Pharmacol. 1987;32:53. [PubMed] [Google Scholar]
- 34.Nunnari JM, Repaske MG, Brandon S, Cragoe EJ, Jr, Limbird LE. Regulation of porcine brain alpha 2-adrenergic receptors by Na+,H+ and inhibitors of Na+/H+ exchange. J Biol Chem. 1987;262:12387. [PubMed] [Google Scholar]
- 35.Canals M, Sexton PM, Christopoulos A. Allostery in GPCRs: ‘MWC’ revisited. Trends Biochem Sci. 2011;36:663. doi: 10.1016/j.tibs.2011.08.005. [DOI] [PubMed] [Google Scholar]
- 36.Changeux JP. Allostery and the Monod-Wyman-Changeux Model After 50 Years. Annu Rev Biophys. 2012;41:103. doi: 10.1146/annurev-biophys-050511-102222. [DOI] [PubMed] [Google Scholar]
- 37.Ho BK, Gruswitz F. HOLLOW: generating accurate representations of channel and interior surfaces in molecular structures. BMC Struct Biol. 2008;8:49. doi: 10.1186/1472-6807-8-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kenakin TP. Biased Signaling and Allosteric Machines; New Vistas and Challenges for Drug Discovery. Br J Pharmacol. 2012;165:1659. doi: 10.1111/j.1476-5381.2011.01749.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.