Abstract
Chronic ethanol exposure impairs insulin signaling in the liver. Peroxisome-proliferator activated receptor (PPAR) agonists function as insulin sensitizers and are used to treat type 2 diabetes mellitus. We examined the therapeutic effectiveness of PPAR agonists in reducing alcoholic hepatitis and hepatic insulin resistance in a model of chronic ethanol feeding. Adult male Long Evans rats were pair fed with isocaloric liquid diets containing 0% (control) or 37% ethanol (caloric content; 9.2% v/v) for 8 weeks. After 3 weeks on the diets, the rats were treated with vehicle, or a PPAR-α, PPAR-δ, or PPAR-γ agonist twice weekly by i.p. injection. Livers were harvested for histopathological, gene expression (RT-PCR), protein (Western and ELISA), and receptor binding studies. Ethanol-fed rats developed steatohepatitis with disordered hepatic chord architecture, increased hepatocellular apoptosis, reduced binding to the insulin, IGF-1, and IGF-2 receptors, and decreased expression of glyceraldehyde-3-phosphate dehydrogenase and aspartyl-(asparaginyl)-β-hydroxylase (mediates remodeling), which are regulated by insulin/IGF signaling. PPAR-α, PPAR-δ, or PPAR-γ agonist treatments reduced the severity of ethanol-mediated liver injury, including hepatic architectural disarray and steatosis. In addition, PPAR-δ and PPAR-γ agonists reduced insulin/IGF resistance and increased insulin/IGF-responsive gene expression. In conclusion, PPAR agonists may help reduce the severity of chronic ethanol-induced liver injury and insulin/IGF resistance, even in the context of continued high-level ethanol consumption.
Key terms: Alcoholic liver disease, insulin resistance, receptor binding, aspartyl-asparaginyl-β-hydroxylase, insulin sensitizers, PPAR-agonist
Introduction
Chronic high-level ethanol consumption inhibits DNA synthesis and compromises the regenerative and reparative capacities of the liver (1), which in part are due to inhibition of insulin signaling (2). Insulin mediates its pro-growth and pro-metabolic effects by binding to its cell surface receptors and activating signal transduction pathways through the insulin receptor substrate, type 1 (IRS-1). IRS-1 activates downstream pathways through SH2 and SH3 domain-containing proteins, phosphatidylinositol-3-kinase (PI3 Kinase), and c-Jun N-terminal kinase (JNK) (2). IRS-1 phosphorylation of Syp, an SH2 domain containing protein tyrosine phosphatase, and Grb2, which recruits Sos1, results in activation of the Ras-MAPK growth-promoting pathway. IRS-1 activation of PI3 kinase catalyzes the formation of phosphatidylinositol-3,4,5-triphosphate, a lipid second messenger, which activates Akt and atypical protein kinase C. Akt mediates pro-growth, pro-survival, and pro-metabolic signaling through PRAS40, mammalian target of rapamycin (mTOR), and S6 kinase (3, 4). Inhibitory effects of ethanol on insulin signaling are due to reduced ligand binding to the insulin receptor (2, 5), reduced tyrosine phosphorylation and activation of insulin receptor tyrosine kinase (6), and inhibition of signaling through IRS-1 (7), PI3 Kinase-Akt (8, 9), PRAS40-mTOR-S6Kinase (10), and Erk MAPK (11).
Peroxisome proliferator-activated receptors (PPARs) are nuclear hormone receptors that bind to DNA and regulate gene transcription in a broad range of cells and tissues (12-15). PPARs are regulated by ligand binding, and they mediate their effects by heterodimerizing with the retinoid × receptor (13). Three distinct isoforms of PPARs exist, PPAR-α, PPAR-δ (also referred to as PPAR-β), and PPAR-γ. PPAR-α is most abundantly expressed in brown adipose tissue and liver, followed by kidney, heart and skeletal muscle. PPAR-α is activated by polyunsaturated fatty acids and fibrates, and it regulates adipocyte growth and differentiation, lipid metabolism, lipoprotein synthesis, and tissue inflammatory responses. PPAR-δ is widely expressed, but most abundant in gut, kidney and heart. PPAR-δ regulates expression of acyl-CoA synthetase 2 in brain, and may also participate in placental implantation and decidualization. In addition, PPAR-δ has a functional role in adaptive responses to the environment. PPAR-γ is primarily expressed in adipose tissue, followed by colon, immune cells, and retina (12, 14). PPAR-γ influences storage of fatty acids in adipose tissue by regulating lipogenic metabolic and transport pathways. The enhanced insulin sensitivity imparted by PPAR-agonists led to their common use for treating type 2 diabetes mellitus (16-19).
Since our previous studies demonstrated that chronic ethanol-induced liver injury was partly mediated by hepatic insulin resistance (2, 11, 20), we utilized a robust experimental animal model of chronic ethanol feeding to evaluate the effectiveness of PPAR agonists in reversing hepatic steatosis and improving insulin receptor binding and insulin-responsive gene expression. Insulin-like growth factor (IGF) types 1 and 2 polypeptide and receptor expression and ligand-receptor binding were also evaluated because cross-talk and functional overlap occur among the corresponding signal transduction pathways, and previous studies demonstrated that PPAR agonists can affect IGF receptor binding and IGF-responsive gene expression (21, 22).
Methods
Chronic ethanol exposure model
Adult male (∼200-250 g) Long Evans rats (Harlan Sprague Dawley, Inc., Indianapolis, Indiana) were pair-fed isocaloric liquid diets (BioServ, Frenchtown, NJ) containing 0% (control) or 37% ethanol by caloric content (9.2% v/v) for 8 weeks (2, 5, 11). Rats were monitored daily to ensure equivalent food consumption and maintenance of body weight. During the last 3 weeks, rats were administered twice weekly (Mondays and Thursdays) intra-peritoneal (i.p.) injections of vehicle (saline), a PPAR-α (GW7647; 25 μg/Kg), PPAR-δ (L-160,043; 2 μg/Kg), or PPAR-γ (F-L-Leu; 20 μg/Kg) agonist (CalBiochem, Carlsbad, CA). The PPAR doses, routes of administration, and frequency were based on empirical in vivo and in vitro studies demonstrating effectiveness of this approach for restoring insulin responsiveness following ethanol exposure. The i.p. rather than p.o. route of drug delivery ensured that treatment was the same for all rats. At the conclusion of the experiment, rats were anesthetized with vaporized isofluorane (SurgiVet, Inc. Waukesha, WI), and liver and blood were harvested for analysis. Liver samples were fixed in Histochoice (Amresco Corp., Solon, OH) and embedded in paraffin. Histological sections were stained with Hematoxylin and Eosin, periodic acid-Schiff (PAS), or Gomori Trichrome, and examined under code. Liver tissue was also snap-frozen and stored at -80°C for mRNA and protein studies. Rats were housed under humane conditions and kept on a 12-hour light/dark cycle with free access to food. All experiments and protocols conformed to guidelines established by the National Institutes of Health and were approved by the Institutional Animal Care and Use Committee at the Lifespan-Rhode Island Hospital.
Analysis of mRNA
Total RNA was extracted from liver using TRIzol® (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol. RNA concentrations and purity were determined from the absorbances measured at 260 nm and 280 nm. RNA (2 μg) was reverse transcribed using the AMV First Strand cDNA synthesis kit (Roche Diagnostics Corporation, Indianapolis, IN) and random oligodeoxynucleotide primers. Quantitative reverse transcriptase polymerase chain reaction (qRT-PCR) assays were used to measure specific mRNA transcripts as previously described (2, 5, 11) using gene-specific primer pairs as published previously (2, 5).
Receptor Binding Assays
Competitive saturation binding studies were used to determine if the PPAR agonist treatments improved ethanol-impaired insulin and IGF receptor binding in liver. Fresh frozen liver tissue was homogenized in buffer containing 50 mM Tris-HCl, pH 7.5, 1% NP-40, 150 mM NaCl, 1 mM EDTA, 2 mM EGTA), plus protease (1 mM PMSF, 0.1 mM TPCK, 1 μg/ml aprotinin, 1 μg/ml pepstatin A, 0.5 μg/ml leupeptin, 1 mM NaF, 1 mM Na4P2O7) and phosphatase (2 mM Na3VO4) inhibitors. Protein concentration was measured with the bicinchoninic acid (BCA) assay (Pierce, Rockford, IL). Exploratory studies determined that to achieve 20% specific binding, insulin receptor binding assays required 100 μg of sample protein, while IGF-1 receptor binding required 25 μg protein, and IGF-2 receptor binding required 10 μg protein per reaction. Binding curves were generated using pooled samples from 8 rats per group. Total binding and non-specific binding were measured in duplicate reactions containing binding buffer (100 mM HEPES, pH 8.0, 118 mM NaCl, 1.2 mM MgSO4, 8.8 mM dextrose, 5 mM KCl, 1% bovine serum albumin), and 0.0031 to 1 μCi/ml of [125I] (2000 Ci/mmol) insulin, IGF-1, or IGF-2, in the absence or presence of 0.1 μM unlabeled ligand (5, 23). After 16-hours incubation at 4°C, reactions were vacuum harvested (Corning, Lowell, MA) onto GF/C filters (5, 23), and bound [125I] insulin, IGF-1, or IGF-2 was measured in a TopCount machine (Packard Instrument Company, Meriden, CT). Specific binding (fmol/mg protein) was calculated by subtracting non-specifically bound isotope from the total bound isotope. The data were analyzed using GraphPad Prism 5 software (GraphPad Software, Inc., San Diego, CA).
Protein Studies
Immunoreactivity to aspartyl-(asparaginyl)-β-hydroxylase (AAH), glyceraldehydes-3-phosphate dehydrogenase (GAPDH), p85 subunit of PI3 kinase (control) and β–actin (control) was examined by Western blot analysis or enzyme-linked immunosorbant assay (ELISA) (2, 5, 11). Tissues were homogenized in radio-immunoprecipitation assay (RIPA) buffer containing protease and phosphatase inhibitors (2, 5, 11). Protein concentrations were determined using the BCA assay (Pierce, Rockford, IL). Western blot membranes were incubated with primary antibody (0.5-1 μg/ml) over night at 4°C. Immunoreactivity was detected with horseradish peroxidase (HRP) conjugated secondary antibody, enhanced chemiluminescence (ECL) reagents (Pierce, Rockford, IL), and the Kodak Digital Science Imaging Station (NEN Life Sciences, Boston, MA). ELISAs were performed as previously described (2) and used to confirm results obtained by Western blot analysis. ELISA immunoreactivity was detected with HRP-conjugated secondary antibody and the Amplex Red soluble fluorophore (Invitrogen, Carlsbad, CA). Fluorescence was measured (Ex 530/Em 595) in a SpectraMax M5 micro-plate reader (Molecular Devices Corp., Sunnyvale, CA).
Source of reagents
The PPAR agonists, GW7647 (PPAR-α), L165, 041 (PPAR-δ), and Fmoc-Leu (PPAR-γ) were purchased from Calbiochem (EMD Chemicals, Gibbstown, NJ). Human recombinant [125I] Insulin, IGF-1, and IGF-2 were purchased from Amersham Biosciences (Boston, MA). Unlabeled human insulin, recombinant IGF-1, and recombinant IGF-2 were purchased from Bachem (Torrance, CA). QuantiTect SYBR Green PCR Mix was obtained from (Qiagen Inc, Valencia, CA). Monoclonal antibodies to GAPDH and β-actin were purchased from Chemicon (Temecula, CA). The A85G6 mouse monoclonal antibody used to detect AAH was generated with purified recombinant human protein (24). All other fine chemicals were purchased from CalBiochem (EMD Chemicals, Gibbstown, NJ) or Sigma-Aldrich (St. Louis, MO).
Statistical Analysis
Data depicted in graphs represent the mean ± S.E.M. Inter-group comparisons were made using repeated measures two-way analysis of variance (ANOVA) and the post-hoc Bonferroni test of significance. Statistical analyses were performed using GraphPad Prism 5 software (GraphPad Software, Inc., San Diego, CA).
Results
PPAR Agonists Reverse Ethanol-Induced Liver Pathology
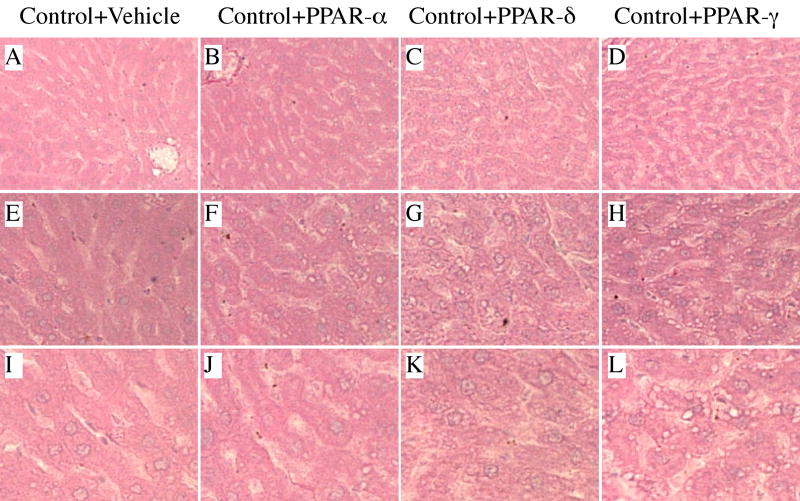
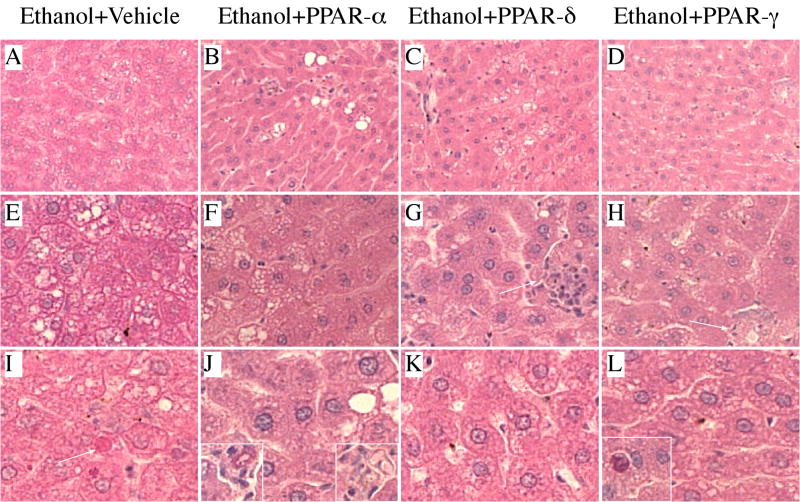
Livers from control rats (liquid diets and chow) exhibited the expected well-organized lobular architecture with minimal evidence of steatosis, variation in nuclear size, or hepatocyte drop-out (Figures 1A, 1E, 1I). In contrast, ethanol exposed livers had microvesicular and macrovesicular steatosis with multiple foci of intralobular lympho-mononuclear inflammatory cell infiltrates, scattered areas of apoptosis and/or necrosis (Figures 2A, 2E, 2I), and hepatic architectural disarray with loss of regular chords and increased variability in size of hepatocyte nuclei. There was no evidence of increased fibrosis, regenerating nodule formation, or cirrhosis in ethanol-exposed livers (Gomori trichrome stain). Control rats treated with the PPAR-α agonist had no detectable histological changes in liver (Figures 1B, 1F, 1J) relative to vehicle-treated controls. In contrast, control rats treated with the PPAR-δ (Figures 1C, 1G, 1K) or PPAR-γ (Figures 1D, 1H, 1L) agonist had less well-organized hepatic architecture due to sinusoidal widening and apparently increased hepatocyte crowding. In addition, PPAR-δ or PPAR-γ agonist treatments resulted in increased nuclear prominence and micro-vacuolation of hepatocyte cytoplasm. The micro-vacuolation was associated with increased PAS staining, corresponding to glycogen accumulation (data not shown). Among ethanol-fed rats, treatment with PPAR agonists had variable effects on liver histology in terms of reducing the architectural disarray, steatosis, and cell death (Figure 2). Treatment with the PPAR-α, PPAR-δ, or PPAR-γ agonist reduced the disordered architecture and resulted in more of a chord-like arrangement of hepatocytes, and both micro- and macrosteatosis were reduced. Nonetheless, small foci of necrosis (Figure 2J, insets), inflammation (Figure 2G, inset and Figure 2H, arrow), and apoptosis (Figure 2L, inset) were still readily detected, although these lesions were less conspicuous than in vehicle-treated, ethanol-exposed livers. The most striking improvements in liver histology occurred in ethanol-fed rats that had been treated with the PPAR-δ (Figures 2C and 2K) or PPAR-γ (Figures 2D and 2L) agonist.
Figure 1.
Peroxisome-proliferator activated receptor (PPAR) agonists produce subtle changes in normal liver histology: Adult male Long Evans rats were pair-fed for 8 weeks with isocaloric liquid diets containing 0% or 37% ethanol by caloric content. After 3 weeks on liquid diets, control rats were subdivided into groups and treated with (a, e, i) vehicle, or a (b, f, j) PPAR-α, (c, g, k) PPAR-δ, or (d, h, l) PPAR-γ agonist, twice weekly by i.p. injection for the duration of the study (see Methods). Liver samples were immersion fixed, embedded in paraffin, and histological sections were stained with H&E. Note regular chord-like architecture in (a, e, i) vehicle-treated and (b, f, j) PPAR-α agonist control livers, and slight architectural disorganization with increased crowding and cytoplasmic vacuolation in livers from (c, g, k) PPAR-δ and (d, h, l) PPAR-γ agonist treated rats. (Original magnifications: a–d, ×200; e–h, ×400; i–l, ×600).
Figure 2.
Peroxisome-proliferator activated receptor (PPAR) agonists partially reverse ethanol-induced liver histopathology: Adult male Long Evans rats were pair-fed for 8 weeks with isocaloric liquid diets containing 0% or 37% ethanol by caloric content. After 3 weeks on liquid diets, ethanol-fed rats were subdivided into groups and treated with (a, e, i) vehicle, or a (b, f, j) PPAR-α, (c, g, k) PPAR-δ, or (d, h, l) PPAR-γ agonist, twice weekly by i.p. injection for the duration of the study (see Methods). Livers were immersion fixed, embedded in paraffin, and histological sections were stained with H&E. (a, e, i) Chronic ethanol feeding resulted in (a) hepatic architectural disorganization with (e) increased steatosis, inflammation (not shown), and (i) apoptosis (arrow). (b, f, j) PPAR-α agonist treatment (b) improved hepatic chord architecture, (f) reduced the steatosis, but (j) was still associated with foci of chronic inflammation (insets). (c, g, k) PPAR-δ agonist treatment also (c) improved hepatic chord architecture and reduced (g) steatosis and (k) apoptosis, but was associated with (g) small foci of inflammation. (d, h, l) PPAR-γ agonist treatment (d) had little effect on hepatic architecture, and although it reduced both (h) steatosis and inflammation, (l) foci of apoptosis were still readily detected. (Original magnifications: a–d, ×200; e–h, ×400; i–l, ×600).
PPAR agonist treatments alter cell population profiles in liver
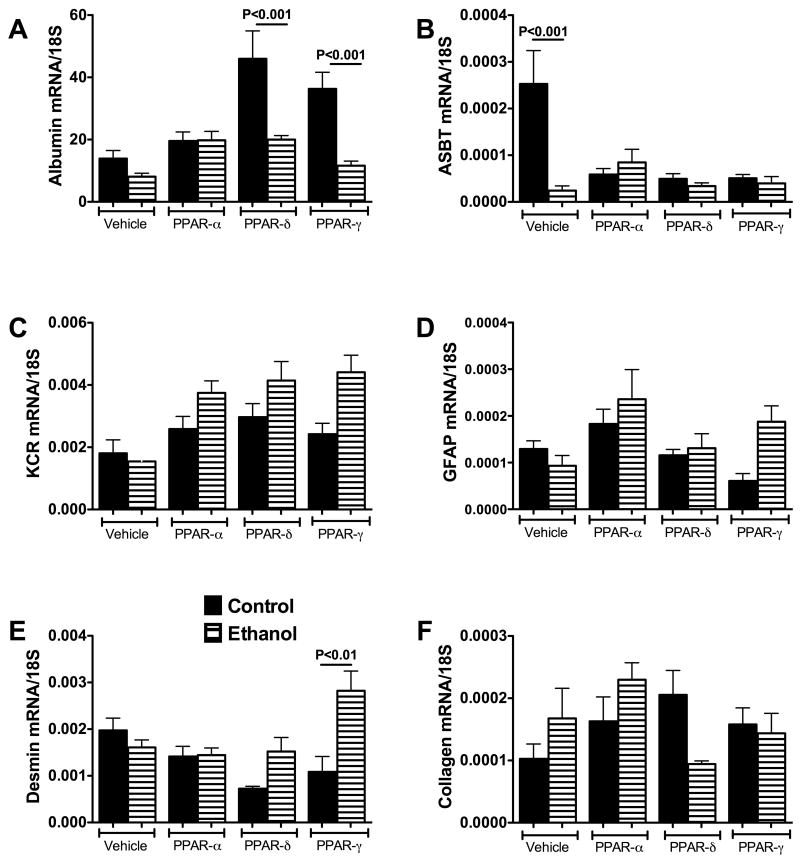
Liver mRNA levels of albumin, apical sodium-dependent bile transporter protein (ASBT), glial fibrillary acidic protein (GFAP), Kupffer cell receptor (KCR), desmin, and collagen were measured by qRT-PCR analysis. Albumin expression was used as an index of hepatocyte abundance/function. ASBT reflects bile duct epithelium. GFAP is an early marker of stellate cell activation and desmin marks transdifferentiation of hepatic stellate cells into myofibroblasts. KCR is a maker of Kupffer cells and increased expression could reflect an intra-hepatic response to injury. Collagen gene expression corresponds to fibrogenic potential or active fibrogenesis. Altogether, these assessments of gene expression, which we have termed “cell profiling”, enabled us to quantify ethanol and PPAR agonist associated shifts in liver cell type and function. In previous studies, we used this approach to characterize differential in vivo effects of chronic ethanol exposure or gene delivery on survival and proliferation of specific cell populations in relation to disease (5).
Albumin expression was similar in livers of control and ethanol fed vehicle- or PPAR-α agonist-treated rats (Figure 3A). Treatment with the PPAR-δ or PPAR-γ agonist increased albumin expression in control livers, resulting in significantly higher levels of albumin mRNA relative to the corresponding ethanol-exposed livers (both P<0.001).
Figure 3.
Peroxisome-proliferator activated receptor (PPAR) agonist treatments alter cell population profiles in liver: Control and chronic ethanol-fed rats were treated twice weekly by i.p. injection of vehicle, or a PPAR-α, PPAR-δ, or PPAR-γ agonist (see Methods). RNA extracted from liver was used to measure gene expression corresponding to (a) albumin, (b) apical sodium-dependent bile transporter protein (ASBT), (c) Kupffer cell receptor (KCR), (d) glial fibrillary acidic protein (GFAP), (e) desmin, and (f) collagen by qRT-PCR (See Methods). Gene expression levels were normalized to 18S rRNA. Graphs depict the mean ± S.E.M. of each specific mRNA/18S ratio. Inter-group comparisons were made using two-way repeated measures analysis of variance (ANOVA) with the post hoc Tukey-Kramer test for significance. Significant P-values reflecting pairwise differences are indicated over the bars. Significant differences relative to the corresponding vehicle-treated, control or ethanol diet fed groups are indicated by asterisks (*P < 0.05; **P < 0.001). Black bars represent control, while hatched bars represent ethanol.
ASBT expression was significantly higher in vehicle-treated control livers relative to all other groups (P<0.001). Among rats treated with a PPAR-agonist, ASBT expression was similar for corresponding control and ethanol-exposed groups (Figure 3B).
KCR expression was lowest in vehicle-treated control and ethanol-exposed livers. In the control group, treatment with a PPAR agonist did not significantly alter KCR expression. In ethanol exposed livers, KCR expression was significantly increased by treatment with the PPAR-α (P<0.05), PPAR-δ (P<0.01), or PPAR-γ (P<0.01) agonist. However, there were no significant differences in KCR expression between control and ethanol-fed rats within corresponding treatment groups (Figure 3C).
GFAP is an early marker of stellate cell activation. Hepatic GFAP mRNA levels were not significantly different between control and ethanol-fed rats, and the levels were not significantly altered by PPAR agonist treatments (Figure 3D).
Desmin is an intermediate filament expressed in stellate cells during transdifferentiation into myofibroblast-like cells. Desmin expression was similar in vehicle-treated control and ethanol-exposed livers. Among control rats, desmin expression was significantly reduced by PPAR-δ agonist treatment relative to vehicle (P<0.05). In addition, PPAR-γ agonist treatment significantly increased desmin expression in ethanol-exposed relative to corresponding control livers (P<0.01) (Figure 3E).
Collagen gene expression reflects fibrogenesis. Collagen mRNA levels were similar for each of the corresponding control and ethanol groups (Figure 3F). However, collagen expression was reduced by PPAR-δ agonist treatment in ethanol-fed rats, although the difference resulting from that response did not reach statistical significance (P=0.08).
Effects of ethanol and PPAR agonists on hepatic expression of insulin and IGF polypeptides, their receptors, and IRS molecules
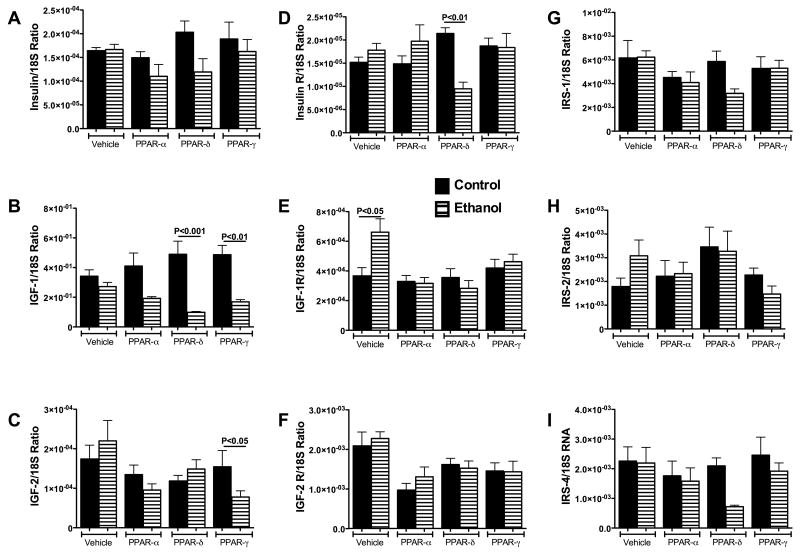
QRT-PCR studies demonstrated expression of insulin, IGF-1, IGF-2 polypeptide genes, their corresponding receptors, and IRS-1, IRS-2, and IRS-4 in livers of both control and ethanol-fed rats (Figure 4), indicating that the upstream genes required for insulin and IGF signaling are all expressed in adult rat livers. Among the polypeptide genes, insulin was least abundant, followed by IGF-2; IGF-1 was most abundantly expressed (Figures 4A-4C).
Figure 4.
Ethanol and Peroxisome-proliferator activated receptor (PPAR) agonists minimally alter expression of genes required for insulin and IGF signaling. Effects of ethanol and PPAR agonist treatments on hepatic mRNA expression of (a) insulin, (b) insulin-like growth factor, type 1 (IGF-I), (c) IGF-II, (d) insulin receptor (R), (e) IGF-I receptor, (f) IGF-II receptor, (g) insulin receptor substrate, type 1 (IRS-1), (h) IRS-2, and (i) IRS-4 were demonstrated using qRT-PCR (See Methods). mRNA levels were normalized to 18S rRNA. Graphs depict the mean ± S.E.M. of each specific mRNA/18S ratio. Inter-group comparisons were made using two-way repeated measures analysis of variance (ANOVA) with the post-hoc Tukey-Kramer test for significance. Significant P-values reflecting pair-wise differences are indicated over the bars. Significant differences relative to the corresponding vehicle-treated control are indicated by asterisks over the bars (*P < 0.05).
Insulin gene expression was similar in vehicle-treated control and ethanol-exposed livers. Although PPAR-δ and PPAR-γ agonists increased insulin expression in control rats, and PPAR-α and PPAR-δ agonists reduced insulin expression in ethanol-fed rats, the inter-group differences did not reach statistical significance (Figure 4A).
IGF-1 mRNA levels were similar in vehicle-treated control and ethanol-exposed livers (Figure 4B). PPAR agonist treatments did not significantly change IGF-1 expression relative to vehicle among control or ethanol-fed rats. However, the mean hepatic levels of IGF-1 were significantly lowered by PPAR-δ (P<0.001) or PPAR-γ (P<0.01) treatment of ethanol-fed relative to corresponding control rats.
Hepatic IGF-2 mRNA levels were similar for corresponding vehicle- or PPAR-agonist treated control and ethanol-exposed rats (Figure 4C). However, PPAR-γ treatment significantly reduced IGF-2 expression relative to vehicle (P<0.05) among ethanol-fed rats, whereas among controls, PPAR agonist treatments did not significantly alter hepatic IGF-2 expression.
Insulin receptor expression was similar among controls treated with vehicle or a PPAR agonist, whereas among ethanol-fed rats, insulin receptor was significantly reduced following PPAR-δ agonist treatment relative to vehicle, PPAR-α, and PPAR-γ treatment (P<0.05) (Figure 4D). In addition, insulin receptor expression was significantly reduced in ethanol-fed, PPAR-δ agonist treated relative corresponding controls (P<0.01), whereas for the other treatment groups, there were no significant differences between control and ethanol associated levels of hepatic insulin receptor expression.
IGF-1 receptor expression was higher in vehicle-treated, ethanol-fed versus control rats (P<0.05) (Figure 4E). PPAR agonist treatments had no significant effect on IGF-1R expression among control rats. In contrast, PPAR-α (P<0.01) and PPAR-δ (P<0.001) agonist treatments significantly down-regulated IGF-1R expression in livers of ethanol-fed rats. In addition, IGF-1R expression was also lower in ethanol+PPAR-γ treated rats, but the difference from vehicle treatment did not reach statistical significance. Consequently, the mean hepatic IGF-1R mRNA levels were similar in control and ethanol-fed rats that received the same treatments.
IGF-2 receptor expression was highest in vehicle-treated control and ethanol-fed rats. Treatment with PPAR agonists reduced hepatic IGF-2R expression in both control and ethanol-fed rats, but the differences relative to vehicle treatment were significant only with respect to PPAR-α treatment (Figure 4F). There were no significant differences in the mean levels of IGF-2R between similarly treated control and ethanol-exposed livers (Figure 4F).
In livers of both control and ethanol-exposed rats, IRS-1 mRNA levels were highest (Figure 4G), followed by IRS-2 (Figure 4H), and then IRS-4 (Figure 4I). The mean levels of IRS-1 were similar for control and ethanol-fed rats (Figure 4G). There were no significant differences in the mean levels of IRS-1, IRS-2, or IRS-4 between control and ethanol exposed rats that had similar treatments. However, modest reductions (not statistically significant) in IRS-1, IRS-2, and IRS-4 expression occurred in ethanol-fed rats that were treated with the PPAR-δ or PPAR-γ agonist (Figures 4G-4I).
Effects of ethanol and PPAR agonist treatments on insulin and IGF receptor binding
Competitive saturation binding assays were used to demonstrate the effects of ethanol and PPAR agonist treatments on insulin, IGF-1 and IGF-2 receptor binding. Binding curves ± 95% C.I.L. (data not shown), computations of Kd (dissociation constant; affinity) and BMAX (top-level binding), and inter-group statistical comparisons (Table 1) were generated with Prism Graphics 5 software. In all experimental conditions, a single-site model produced the highest R2, i.e. best fit. Among controls, the BMAX and Kd for insulin receptor binding were similar following vehicle, PPAR-δ, or PPAR-γ treatment, whereas both were significantly reduced after PPAR-α treatment relative to vehicle (Table 1A). Among ethanol fed rats, the BMAX and Kd for insulin receptor binding were similar, irrespective of treatment. However, the BMAX and Kd were significantly reduced relative to correspondingly treated controls, except for PPAR-α treatment, which significantly increased the BMAX for insulin receptor binding in ethanol fed rats (Table 1A).
TABLE 1.
| TABLE 1A: Insulin Receptor Binding | |||||
|---|---|---|---|---|---|
| Group | BMAX ± S.E.M. | 95% C.I.-BMAX | Kd ± S.E.M | 95% C.I.-KD | R2 |
| Control + Vehicle | 7.23 ± 2.66 | 1.83-12.63 | 165.5 ± 73 | 16.6-314.5 | 0.929 |
| Control + PPAR-α | 1.03 ± 0.16 ξξξ | 0.69-1.36 | 34.0 ± 9.4 ξξξ | 14.93-53.14 | 0.884 |
| Control + PPAR-δ | 4.93 ± 1.99 | 0.89-8.97 | 165.7 ± 80.7 | 2.23-329.1 | 0.920 |
| Control + PPAR-γ | 8.06 ± 10.73 | -13.78-29.91 | 300.6 ± 448.5 | -612.4-1214 | 0.778 |
| Ethanol + Vehicle | 2.59 ± 0.61 ** | 1.36-3.82 | 51.38 ± 18.63*** | 13.62-89.15 | 0.84 |
| Ethanol + PPAR-α | 2.04 ± 0.35 ** | 1.33-2.75 | 8.45 ± 4.06*** | 0.21-16.69 | 0.57 |
| Ethanol + PPAR-δ | 3.21 ±0.45 ** | 2.29-4.12 | 43.85 ± 9.91** | 23.76-63.94 | 0.92 |
| Ethanol + PPAR-γ | 2.08 ± 0.42 *** | 1.22-2.93 | 23.26 ± 9.33** | 4.3-42.22 | 0.746 |
| TABLE 1B: IGF-1 RECEPTOR BINDING | |||||
|---|---|---|---|---|---|
| Group | BMAX ± S.E.M. | 95% C.I.-BMAX | Kd ± S.E.M | 95% C.I.-KD | R2 |
| Control + Vehicle | 3.62 ± 0.77 | 2.04-5.21 | 131.9 ± 49.7 | 30.64-233.1 | 0.79 |
| Control + PPAR-α | 2.52 ± 0.31 ξξ | 1.87-3.16 | 63.3 ± 16.2 ξξ | 30.2-95.4 | 0.84 |
| Control + PPAR-δ | 1.64 ± 0.37 ξξ | 0.88-2.39 | 57.2 ± 27.1 | 0.88-2.4 | 0.48 |
| Control + PPAR-γ | 6.43 ± 2.00 ξ | 2.32-10.63 | 425.7 ± 178.1 ξξ | 80.9-790.4 | 0.91 |
| Ethanol + Vehicle | 2.66 ± 0.38* | 1.88-3.45 | 43.57 ± 13.71** | 15.49-71.68 | 0.78 |
| Ethanol + PPAR-α | 4.21 ± 0.88 | 2.42-6.00 | 127.3 ± 45.86 | 32.2-222.4 | 0.76 |
| Ethanol + PPAR-δ | 3.81 ±0.89 | 1.99-5.62 | 120.6 ± 52.1 | 14.4-226.8 | 0.71 |
| Ethanol + PPAR-γ | 2.74 ± 0.91 | 0.87-4.60 | 100.1 ± 63.2 | -28.9-229.0 | 0.48 |
| TABLE 1C: IGF-2 RECEPTOR BINDING | |||||
|---|---|---|---|---|---|
| Group | BMAX ± S.E.M. | 95% C.I.-BMAX | Kd ± S.E.M | 95% C.I.-KD | R2 |
| Control + Vehicle | 40.12 ± 3.04 | 33.96-46.27 | 29.3 ± 5.8 | 17.6-40.9 | 0.92 |
| Control + PPAR-α | 40.27 ± 3.28 | 33.62-46.9 | 31.4 ± 6.4 | 18.3-44.5 | 0.91 |
| Control + PPAR-δ | 29.01 ± 1.99ξξξ | 24.99-33.04 | 21.5 ± 4.3 | 12.6-29.9 | 0.91 |
| Control + PPAR-γ | 25.20 ± 2.04ξξξ | 21.06-29.34 | 14.6 ± 3.9ξξ | 6.6-22.6 | 0.83 |
| Ethanol + Vehicle | 22.63 ± 5.25* | 12.00-33.25 | 32.4 ± 18.7 | -5.5-70.4 | 0.53 |
| Ethanol + PPAR-α | 28.88 ± 6.20 | 16.31-41.44 | 40.5 ± 20.0 | 16.3-41.4 | 0.59 |
| Ethanol + PPAR-δ | 51.50 ± 13.53 | 24.11-78.90 | 75.6 ± 37.1 | 0.52-150.8 | 0.60 |
| Ethanol + PPAR-γ | 22.56 ± 4.43 | 13.60-31.53 | 32.3 ± 15.8 | 0.26-64.38 | 0.60 |
P<0.05;
P<0.01;
P<0.001 relative to corresponding vehicle-treated group
P<0.05;
P<0.001;
P<0.0001 relative to corresponding control diet group
Among controls, PPAR-α and PPAR-δ agonist treatments reduced the BMAX for IGF-1R binding, the PPAR-α agonist also reduced the Kd, whereas the PPAR-γ agonist increased both the BMAX and Kd. Chronic ethanol feeding (vehicle treated) significantly reduced the BMAX (P<0.05) and Kd (P<0.01) for IGF-1R binding relative to control (+vehicle), but those adverse effects of ethanol were rescued by PPAR agonist treatments (Table 1B).
For IGF-2R binding, the BMAX was similar in livers from vehicle and PPAR-α-treated controls, but significantly reduced in control rats that had been treated with the PPAR-δ or PPAR-γ agonist (both P<0.001). The Kd's were similar for all but PPAR-γ treated group, in which it was significantly reduced relative to control (P<0.01). Chronic ethanol feeding reduced the BMAX of IGF-2R binding relative to vehicle-treated control (P<0.05). Treatment with PPAR agonists restored IGF-2R binding, and resulted in similar BMAX levels in livers of ethanol-fed and corresponding controls. In contrast, the Kd's of IGF-2R binding were not significantly altered by ethanol feeding, irrespective of treatment (Table 1C).
Effects of ethanol and PPAR agonist treatments on insulin/IGF responsive gene expression related to energy metabolism and tissue remodeling
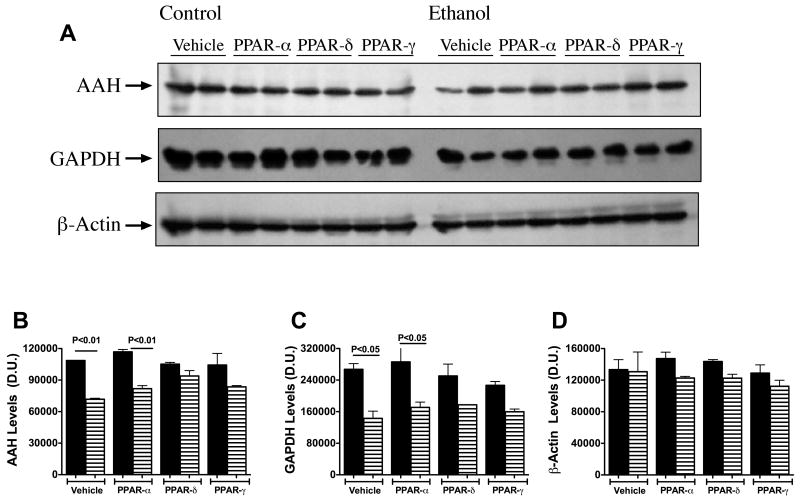
AAH expression increases with insulin, IGF-1, or IGF-2 stimulation (25), and has positive effects on hepatocellular growth and motility (25, 26). GAPDH is an insulin-responsive gene that has an important role in glucose metabolism. Western blot analysis detected AAH, GAPDH, and β-actin expression in all samples (Figure 5A). Re-probing the blots with monoclonal antibody to β-actin demonstrated approximately equal protein loading in all lanes. Digital image quantification of the Western blot signals revealed similar levels of AAH (Figure 5B), GAPDH (Figure 5C), and β-actin (Figure 5D) expression across all control groups, irrespective of PPAR agonist treatment. Chronic ethanol feeding (+ vehicle treatment) reduced AAH (P<0.05) and GAPDH (P<0.05), but not β-Actin expression relative to control. The PPAR agonist treatments slightly increased AAH immunoreactivity; this effect abolished the statistical significance of the inter-group differences. In contrast, GAPDH expression remained similarly reduced in livers of ethanol-fed rats, although the differences from corresponding controls were not significant for the PPAR-δ and PPAR-γ treated rats. β-actin expression was similar in control and ethanol-fed rats that received similar treatments. These results were confirmed by ELISA studies (data not shown).
Figure 5.
Peroxisome-proliferator activated receptor (PPAR) agonists increase insulin and insulin-like growth factor (IGF) responsive gene expression in ethanol-exposed livers. Effects of ethanol and PPAR agonist treatments on insulin/IGF responsive gene expression related to tissue remodeling and energy metabolism were examined by (a) Western blot analysis with (b–d) digital image quantification. Control and chronic ethanol-fed rats were treated twice weekly by i.p. injection of vehicle, or a PPAR-α, PPAR-δ, or PPAR-γ agonist (see Methods). Western blot analysis, performed with specific monoclonal antibodies (0.5–1 μg/ml), horseradish peroxidase (HRP)-conjugated secondary antibody, and enhanced chemiluminescence reagents, was used to detect aspartyl-asparaginyl-β-hydroxylase (AAH), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and β-actin. Representative immunoblot results are shown in Panel (a). The Western blot signals corresponding to (b) AAH, (c) GAPDH, and (d) β-actin were quantified (arbitrary densitometry units; D.U.) with the Kodak Digital Science Imaging Station. Results (mean ± S.E.M) are depicted graphically. Inter-group comparisons were made using two-way repeated measures analysis of variance (ANOVA) with the post-hoc Tukey-Kramer test for significance. Significant P values reflecting pair-wise differences are shown over the bars.
Discussion
Previous studies demonstrated that Long Evans rats were highly susceptible to the hepatotoxic effects of ethanol (5, 11, 27) since, within a period of 5 or 6 weeks of ethanol feeding, Long Evans rats develop prominent macrosteatosis, inflammation, DNA damage, and hepatocellular apoptosis (5, 11, 27). Therefore, the histopathological abnormalities produced in the Long Evans rat model resemble chronic alcoholic hepatitis seen in humans. Given the known inhibitory effects of ethanol on insulin signaling and insulin-responsive gene expression in liver, and the adverse consequences of these abnormalities in terms of liver regeneration (1, 28, 29), we sought to determine if chronic alcoholic liver disease (ALD) produced in the Long Evans rat model was associated with hepatic insulin resistance, and at the same time, explored the hypothesis that insulin sensitizer agents could be used to restore liver histology in the clinical setting of continued ethanol consumption. To conduct these studies, we treated the rats with a PPAR-α, PPAR-δ, or PPAR-γ agonist, since all three receptors are expressed in liver, and it was not certain which class of PPAR agonist would be most effective for treating ALD.
The chronic ethanol feeding produced histopathological changes that resemble ALD in humans, including prominent architectural disarray with variation in nuclear size and hepatocyte drop-out, yet there was no evidence of interlobular or bridging fibrosis, cirrhosis, or regenerative nodule formation, or neoplastic transformation. Treatment with a PPAR-α, PPAR-δ, or PPAR-γ agonist produced minimal changes in control liver histology, but they strikingly reduced ethanol-associated architectural disarray and steatosis, despite continued ethanol exposure. The PPAR-δ and PPAR-γ agonists were more effective than the PPAR-α agonist in restoring hepatic architecture and reducing foci of inflammation and necrosis or apoptosis. These effects of the PPAR agonists correspond with their known anti-inflammatory actions, in addition to their insulin sensitizer properties (19).
To further examine effects of PPAR agonist treatments on liver function, we utilized a qRT-PCR-based method of measuring gene expression corresponding to different cell types in liver. Although chronic ethanol feeding did not impair albumin expression, it did inhibit PPAR-δ and PPAR-γ agonist stimulated increases in albumin, which is a marker of hepatocyte function. Ethanol inhibition of ASBT expression, further suggests that bile duct epithelial cell function is impaired in ALD. The studies also demonstrated increased KCR expression in PPAR agonist treated, ethanol-exposed livers. One possible interpretation is that, in correlation with the improvements in liver histology, increased Kupffer cell function is needed to manage the repair process (30, 31). Corresponding with the absence of collagen deposition, none of the biomarkers of stellate cell activation (GFAP and desmin) (32, 33) or fibrogenesis (collagen) were significantly increased in ethanol-exposed livers. On the other hand, PPAR-δ agonist treatment reduced desmin expression in control livers, and collagen expression in ethanol-exposed livers. These trends suggest that progression of ALD might be reduced by PPAR agonist treatments.
We used qRT-PCR analysis to assess the integrity of “machinery” required for insulin and IGF signaling. The studies demonstrated that the expression levels of the insulin, IGF-1 and IGF-2 polypeptides, their corresponding receptors, and IRS molecules were relatively preserved in chronic ethanol-exposed livers, indicating that impairments in insulin or IGF signaling caused by chronic ethanol exposure could not be attributed to local growth factor deficiencies, down-regulation or loss of receptors, or impaired expression of major docking molecules that transmit downstream signals. Of note was that treatment with the PPAR-δ or PPAR-γ agonist significantly increased IGF-1 expression in control livers, but either inhibited or had no significant effect on trophic factor expression in ethanol-exposed livers. Similarly, insulin, IGF-1, and IGF-2 receptors, and IRS-1, IRS-2, and IRS-4 expression levels were generally not significantly modulated by PPAR agonist treatments. Therefore, any improvements in liver histology associated with PPAR agonist treatment in ethanol-fed rats were not likely due to enhanced expression of local growth factors, growth factor receptors, or IRS genes.
Effective ligand binding is critical to the signaling cascade, and many of the downstream effects of impaired insulin signaling that have been reported in ethanol-exposed livers, including reduced cell survival, could be mediated by inhibition of insulin binding to its receptor (2, 11, 25). Given the fact that signaling through IGF-1 or IGF-2 activates IRS pathways either directly or via cross-talk (22, 25), it was of interest to also measure IGF-1 and IGF-2 receptor binding in our model. The studies using competitive saturation binding assays demonstrated that chronic ethanol exposure significantly impairs ligand binding to the insulin, IGF-1, and IGF-2 receptors as manifested by the reduced BMAX (top-level binding). The PPAR-δ, and in some instances PPAR-γ agonist treatments significantly increased insulin, IGF-1, and/or IGF-2 receptor binding, resulting in higher, i.e. normalized BMAX values in ethanol-exposed livers. Although the mechanism of increased receptor binding has not yet been determined, this effect could have been mediated by corrections in membrane lipid composition, since in previous studies, we and others demonstrated that ligand binding to the insulin and IGF receptors is impaired by membrane cholesterol depletion, and restored by membrane cholesterol repletion (34-36). Regardless of the mechanism, it is likely that the PPAR agonist-associated increases in insulin and IGF receptor binding had critical roles in restoring liver structure and function, including insulin/IGF responsive gene expression, despite continued ethanol exposure.
To examine the consequences of PPAR agonist-mediated increases in insulin and IGF receptor binding in ethanol-exposed livers, we assessed insulin and IGF responsive gene expression by Western blot analysis and ELISA. As expected, the ethanol-exposed livers of vehicle-treated rats had significantly reduced levels of GAPDH and AAH, which respectively mediate energy metabolism and cell motility required for regeneration and remodeling of tissue (26, 37). Treatment with a PPAR agonist increased AAH expression to levels that were no longer significantly reduced relative to control. PPAR agonist treatment was less effective with respect to ethanol's inhibitory effects on GAPDH expression, although the slight increased resulting from PPAR-δ or PPAR-γ agonist treatments rendered the differences from corresponding controls not statistically significant. Therefore, the downstream consequences of increased ligand-receptor binding included increased insulin/IGF responsive gene expression and/or improved liver histology, despite continued ethanol exposure. Increased GAPDH expression could have helped reduce hepatic steatosis due to improved energy metabolism and ATP production, whereas increased AAH expression may have aided in liver remodeling and repair, and thereby helped to restore the normal liver architecture. Together, the results suggest that PPAR agonist treatments may help reverse some of the adverse effects of chronic ALD, particularly with respect to restoring liver structure and function. However, the findings also suggest that different subtypes PPAR agonists may differ in their therapeutic effects on ALD.
Acknowledgments
Research Supported by AA02666, AA02169, AA11431, AA12908, and AA-16126 from the National Institutes of Health
References
- 1.Wands JR, Carter EA, Bucher NL, Isselbacher KJ. Inhibition of hepatic regeneration in rats by acute and chronic ethanol intoxication. Gastroenterology. 1979 Sep;77(3):528–31. [PubMed] [Google Scholar]
- 2.de la Monte SM, Yeon JE, Tong M, Longato L, Chaudhry R, Pang MY, et al. Insulin resistance in experimental alcohol-induced liver disease. J Gastroenterol Hepatol. 2008 Aug;23(8 Pt 2):e477–86. doi: 10.1111/j.1440-1746.2008.05339.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu M, Yang X, Chan C. A dynamic analysis of IRS-PKR signaling in liver cells: a discrete modeling approach. PLoS One. 2009;4(12):e8040. doi: 10.1371/journal.pone.0008040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vander Haar E, Lee SI, Bandhakavi S, Griffin TJ, Kim DH. Insulin signalling to mTOR mediated by the Akt/PKB substrate PRAS40. Nat Cell Biol. 2007 Mar;9(3):316–23. doi: 10.1038/ncb1547. [DOI] [PubMed] [Google Scholar]
- 5.Denucci SM, Tong M, Longato L, Lawton M, Setshedi M, Carlson RI, et al. Rat strain differences in susceptibility to alcohol-induced chronic liver injury and hepatic insulin resistance. Gastroenterol Res Pract. 2010;2010 doi: 10.1155/2010/312790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mohr L, Tanaka S, Wands JR. Ethanol inhibits hepatocyte proliferation in insulin receptor substrate 1 transgenic mice. Gastroenterology. 1998 Dec;115(6):1558–65. doi: 10.1016/s0016-5085(98)70036-8. [DOI] [PubMed] [Google Scholar]
- 7.Sasaki Y, Wands JR. Ethanol impairs insulin receptor substrate-1 mediated signal transduction during rat liver regeneration. Biochem Biophys Res Commun. 1994 Feb 28;199(1):403–9. doi: 10.1006/bbrc.1994.1243. [DOI] [PubMed] [Google Scholar]
- 8.Pang M, de la Monte SM, Longato L, Tong M, He J, Chaudhry R, et al. PPARdelta agonist attenuates alcohol-induced hepatic insulin resistance and improves liver injury and repair. J Hepatol. 2009 Jun;50(6):1192–201. doi: 10.1016/j.jhep.2009.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He J, de la Monte S, Wands JR. Acute ethanol exposure inhibits insulin signaling in the liver. Hepatology. 2007 Dec;46(6):1791–800. doi: 10.1002/hep.21904. [DOI] [PubMed] [Google Scholar]
- 10.Hong-Brown LQ, Brown CR, Kazi AA, Huber DS, Pruznak AM, Lang CH. Alcohol and PRAS40 knockdown decrease mTOR activity and protein synthesis via AMPK signaling and changes in mTORC1 interaction. J Cell Biochem. 2010 Apr 15;109(6):1172–84. doi: 10.1002/jcb.22496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yeon JE, Califano S, Xu J, Wands JR, De La Monte SM. Potential role of PTEN phosphatase in ethanol-impaired survival signaling in the liver. Hepatology. 2003 Sep;38(3):703–14. doi: 10.1053/jhep.2003.50368. [DOI] [PubMed] [Google Scholar]
- 12.Dreyer C, Krey G, Keller H, Givel F, Helftenbein G, Wahli W. Control of the peroxisomal beta-oxidation pathway by a novel family of nuclear hormone receptors. Cell. 1992 Mar 6;68(5):879–87. doi: 10.1016/0092-8674(92)90031-7. [DOI] [PubMed] [Google Scholar]
- 13.Issemann I, Green S. Activation of a member of the steroid hormone receptor superfamily by peroxisome proliferators. Nature. 1990 Oct 18;347(6294):645–50. doi: 10.1038/347645a0. [DOI] [PubMed] [Google Scholar]
- 14.Kliewer SA, Forman BM, Blumberg B, Ong ES, Borgmeyer U, Mangelsdorf DJ, et al. Differential expression and activation of a family of murine peroxisome proliferator-activated receptors. Proc Natl Acad Sci U S A. 1994 Jul 19;91(15):7355–9. doi: 10.1073/pnas.91.15.7355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kramer DK, Al-Khalili L, Guigas B, Leng Y, Garcia-Roves PM, Krook A. Role of AMP kinase and PPARdelta in the regulation of lipid and glucose metabolism in human skeletal muscle. J Biol Chem. 2007 Jul 6;282(27):19313–20. doi: 10.1074/jbc.M702329200. [DOI] [PubMed] [Google Scholar]
- 16.Folli F, Saad MJ, Backer JM, Kahn CR. Regulation of phosphatidylinositol 3-kinase activity in liver and muscle of animal models of insulin-resistant and insulin-deficient diabetes mellitus. J Clin Invest. 1993 Oct;92(4):1787–94. doi: 10.1172/JCI116768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goldstein BJ, Rosenstock J, Anzalone D, Tou C, Ohman KP. Effect of tesaglitazar, a dual PPAR alpha/gamma agonist, on glucose and lipid abnormalities in patients with type 2 diabetes: a 12-week dose-ranging trial. Curr Med Res Opin. 2006 Dec;22(12):2575–90. doi: 10.1185/030079906x154169. [DOI] [PubMed] [Google Scholar]
- 18.Luquet S, Gaudel C, Holst D, Lopez-Soriano J, Jehl-Pietri C, Fredenrich A, et al. Roles of PPAR delta in lipid absorption and metabolism: a new target for the treatment of type 2 diabetes. Biochim Biophys Acta. 2005 May 30;1740(2):313–7. doi: 10.1016/j.bbadis.2004.11.011. [DOI] [PubMed] [Google Scholar]
- 19.Odegaard JI, Ricardo-Gonzalez RR, Goforth MH, Morel CR, Subramanian V, Mukundan L, et al. Macrophage-specific PPARgamma controls alternative activation and improves insulin resistance. Nature. 2007 Jun 28;447(7148):1116–20. doi: 10.1038/nature05894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ronis MJ, Wands JR, Badger TM, de la Monte SM, Lang CH, Calissendorff J. Alcohol-induced disruption of endocrine signaling. Alcohol Clin Exp Res. 2007 Jul;31(8):1269–85. doi: 10.1111/j.1530-0277.2007.00436.x. [DOI] [PubMed] [Google Scholar]
- 21.Degenhardt T, Matilainen M, Herzig KH, Dunlop TW, Carlberg C. The insulin-like growth factor-binding protein 1 gene is a primary target of peroxisome proliferator-activated receptors. J Biol Chem. 2006 Dec 22;281(51):39607–19. doi: 10.1074/jbc.M605623200. [DOI] [PubMed] [Google Scholar]
- 22.Denley A, Carroll JM, Brierley GV, Cosgrove L, Wallace J, Forbes B, et al. Differential activation of insulin receptor substrates 1 and 2 by insulin-like growth factor-activated insulin receptors. Mol Cell Biol. 2007 May;27(10):3569–77. doi: 10.1128/MCB.01447-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de la Monte SM, Tong M, Lester-Coll N, Plater M, Jr, Wands JR. Therapeutic rescue of neurodegeneration in experimental type 3 diabetes: relevance to Alzheimer's disease. J Alzheimers Dis. 2006 Sep;10(1):89–109. doi: 10.3233/jad-2006-10113. [DOI] [PubMed] [Google Scholar]
- 24.de la Monte SM, Tong M, Carlson RI, Carter JJ, Longato L, Silbermann E, et al. Ethanol inhibition of aspartyl-asparaginyl-beta-hydroxylase in fetal alcohol spectrum disorder: potential link to the impairments in central nervous system neuronal migration. Alcohol. 2009 May;43(3):225–40. doi: 10.1016/j.alcohol.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cantarini MC, de la Monte SM, Pang M, Tong M, D'Errico A, Trevisani F, et al. Aspartyl-asparagyl beta hydroxylase over-expression in human hepatoma is linked to activation of insulin-like growth factor and notch signaling mechanisms. Hepatology. 2006 Aug;44(2):446–57. doi: 10.1002/hep.21272. [DOI] [PubMed] [Google Scholar]
- 26.de la Monte SM, Tamaki S, Cantarini MC, Ince N, Wiedmann M, Carter JJ, et al. Aspartyl-(asparaginyl)-beta-hydroxylase regulates hepatocellular carcinoma invasiveness. J Hepatol. 2006 May;44(5):971–83. doi: 10.1016/j.jhep.2006.01.038. [DOI] [PubMed] [Google Scholar]
- 27.Derdak Z, Lang CH, Villegas KA, Tong M, Mark NM, de la Monte SM, et al. Activation of p53 enhances apoptosis and insulin resistance in a rat model of alcoholic liver disease. J Hepatol. 2010 doi: 10.1016/j.jhep.2010.08.007. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hsu MK, Qiao L, Ho V, Zhang BH, Zhang H, Teoh N, et al. Ethanol reduces p38 kinase activation and cyclin D1 protein expression after partial hepatectomy in rats. J Hepatol. 2006 Feb;44(2):375–82. doi: 10.1016/j.jhep.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 29.Sasaki Y, Hayashi N, Ito T, Fusamoto H, Kamada T, Wands JR. Influence of ethanol on insulin receptor substrate-1-mediated signal transduction during rat liver regeneration. Alcohol Alcohol Suppl. 1994;29(1):99–106. [PubMed] [Google Scholar]
- 30.Meijer C, Wiezer MJ, Diehl AM, Schouten HJ, Schouten HJ, Meijer S, et al. Kupffer cell depletion by CI2MDP-liposomes alters hepatic cytokine expression and delays liver regeneration after partial hepatectomy. Liver. 2000 Feb;20(1):66–77. doi: 10.1034/j.1600-0676.2000.020001066.x. [DOI] [PubMed] [Google Scholar]
- 31.Prins HA, Meijer C, Boelens PG, Diks J, Holtz R, Masson S, et al. Kupffer cell-depleted rats have a diminished acute-phase response following major liver resection. Shock. 2004 Jun;21(6):561–5. doi: 10.1097/01.shk.0000126649.96850.36. [DOI] [PubMed] [Google Scholar]
- 32.Morini S, Carotti S, Carpino G, Franchitto A, Corradini SG, Merli M, et al. GFAP expression in the liver as an early marker of stellate cells activation. Ital J Anat Embryol. 2005 Oct-Dec;110(4):193–207. [PubMed] [Google Scholar]
- 33.Nitou M, Ishikawa K, Shiojiri N. Immunohistochemical analysis of development of desmin-positive hepatic stellate cells in mouse liver. J Anat. 2000 Nov;197(Pt 4):635–46. doi: 10.1046/j.1469-7580.2000.19740635.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huo H, Guo X, Hong S, Jiang M, Liu X, Liao K. Lipid rafts/caveolae are essential for insulin-like growth factor-1 receptor signaling during 3T3-L1 preadipocyte differentiation induction. J Biol Chem. 2003 Mar 28;278(13):11561–9. doi: 10.1074/jbc.M211785200. [DOI] [PubMed] [Google Scholar]
- 35.Matthews LC, Taggart MJ, Westwood M. Effect of cholesterol depletion on mitogenesis and survival: the role of caveolar and noncaveolar domains in insulin-like growth factor-mediated cellular function. Endocrinology. 2005 Dec;146(12):5463–73. doi: 10.1210/en.2005-0236. [DOI] [PubMed] [Google Scholar]
- 36.Soscia SJ, Tong M, Xu XJ, Cohen AC, Chu J, Wands JR, et al. Chronic gestational exposure to ethanol causes insulin and IGF resistance and impairs acetylcholine homeostasis in the brain. Cell Mol Life Sci. 2006 Sep;63(17):2039–56. doi: 10.1007/s00018-006-6208-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dhar-Chowdhury P, Harrell MD, Han SY, Jankowska D, Parachuru L, Morrissey A, et al. The glycolytic enzymes, glyceraldehyde-3-phosphate dehydrogenase, triose-phosphate isomerase, and pyruvate kinase are components of the K(ATP) channel macromolecular complex and regulate its function. J Biol Chem. 2005 Nov 18;280(46):38464–70. doi: 10.1074/jbc.M508744200. [DOI] [PMC free article] [PubMed] [Google Scholar]