Abstract
The reduction of pathophysiologic levels of nitric oxide through inhibition of neuronal nitric oxide synthase (nNOS) has the potential to be therapeutically beneficial in various neurodegenerative diseases. We have developed a series of pyrrolidine-based nNOS inhibitors that exhibit excellent potencies and isoform selectivities (J. Am. Chem. Soc. 2010, 132, 5437). However, there are still important challenges, such as how to decrease the multiple positive charges derived from basic amino groups, which contribute to poor bioavailability, without losing potency and/or selectivity. Here we present an interdisciplinary study combining molecular docking, crystallography, molecular dynamics simulations, synthesis, and enzymology to explore potential pharmacophoric features of nNOS inhibitors and to design potent and selective monocationic nNOS inhibitors. The simulation results indicate that different hydrogen bond patterns, electrostatic interactions, hydrophobic interactions, and a water molecule bridge are key factors for stabilizing ligands and controlling ligand orientation. We find that a heteroatom in the aromatic head or linker chain of the ligand provides additional stability and blocks the substrate binding pocket. Finally, the computational insights are experimentally validated with double-headed pyridine analogs. The compounds reported here are among the most potent and selective monocationic pyrrolidine-based nNOS inhibitors reported to date, and 10 shows improved membrane permeability.
Introduction
The key physiologic mediator, nitric oxide (NO), which plays an important role in the regulation of various biological processes, is produced in mammalian cells by three distinct nitric oxide synthases (NOSs): neuronal NOS (nNOS), endothelial NOS (eNOS), and inducible NOS (iNOS).1,2 The three isoforms share significant sequence homology (~50%) and catalyze the oxidation of L-arginine to NO and citrulline with NADPH and O2 as cosubstrates.3,4 Each isoform consists of an N-terminal oxygenase domain that binds heme, substrate, and tetrahydrobiopterin (H4B); a central linker region that binds calmodulin; and a C-terminal reductase domain with binding sites for FAD, FMN, and NADPH.5,6
NO overproduction by nNOS has been associated with chronic neurodegenerative pathologies including Alzheimer’s, Parkinson’s, and Huntington’s diseases as well as neuronal damage resulting from stroke, cerebral palsy, and migraine headaches.1,7–10 The reduction in pathophysiologic levels of NO through inhibition of nNOS has the potential to be beneficial as an approach to develop new therapeutics for these diseases.11,12 However, the therapeutic control of NO synthesis is difficult because of the challenge to achieve highly selective inhibition of the specific isoforms.13 Each one of the three isozymes is associated with different functions: nNOS is devoted to neuronal signaling, iNOS to the immune response, and eNOS to smooth muscle relaxation and blood pressure regulation. 14 Therefore, selective inhibition of nNOS over the other isozymes is highly desirable for the treatment of neurodegenerative diseases to avoid undesirable effects related to iNOS and eNOS inhibition.15–19
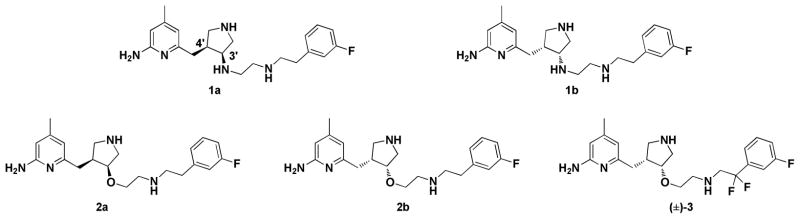
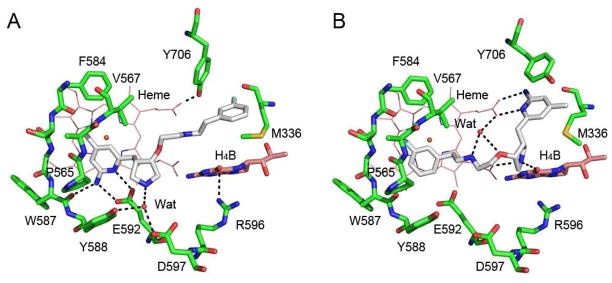
In our continuous efforts to develop nNOS selective inhibitors, we established a new approach for fragment-based de novo design and discovered a series of highly potent and nNOS-selective small-molecule inhibitors with a 2-aminopyridinomethyl pyrrolidine scaffold including 1a, 1b, 2a, and 2b (Figure 1).20–24 There are two chiral centers (the 3′ and 4′ carbons) in each structure. In vitro enzyme assays of the enantiopure compounds indicate that (3′R,4′R)-enantiomers 1b and 2b are much more potent and selective for nNOS than (3′S,4′S)-analogs 1a and 2a.20,21 These pyrrolidine-based inhibitors were designed from structure-activity studies based on a series of dipeptide inhibitors.25 The 2-aminopyridine moiety mimics the guanidine group in the dipeptide inhibitors for potential hydrogen bonding interactions with the NOS active site Glu592, which is conserved in all mammalian NOS isoforms. The pyrrolidine nitrogen mimics the α-amino group of dipeptide inhibitors and provides a positive charge next to the Glu592 in nNOS and close to another negatively charged residue, Asp597, which is Asn in eNOS. Therefore, nNOS has two negative charges in the active site compared to one in eNOS, and, as a result, nNOS provides better electrostatic stabilization to the positively charged inhibitors and is the primary reason why these inhibitors are selective for nNOS. An additional complication with the pyrrolidine inhibitors is that the binding orientation depends on the stereochemistry at the 3′- and 4′-positions of the pyrrolidine.20,21 The (3′S,4′S) inhibitors behave just like the dipeptide inhibitors, hydrogen bonding with their 2-aminopyridine to the NOS active side Glu residue, while the pyrrolidine nitrogen also interacts with this same Glu as does the α-amino group of dipeptide inhibitors. We refer to this binding mode as the “normal” binding mode. However, the (3′R,4′R) inhibitors bind in a 180° flipped over mode, so that the 2-aminopyridine makes bifurcated hydrogen bonds with heme propionate D, and the hydrophobic tail binds above the heme next to the active site Glu. We refer to this binding mode as the “flipped” binding mode. In the flipped binding mode there also is a new π-stacking interaction of the 2-aminopyridine with Tyr706. These two different binding modes are illustrated in Figure 2 for 2a and 2b bound to nNOS.
Figure 1.
Chemical structures of 1–3.
Figure 2.
Crystallographic binding conformation of 2a (A, PDB ID 3NLK) and 2b (B, PDB ID 3NLM) in rat nNOS.
Although the (3′R,4′R) inhibitors show great potency (Ki < 10 nM) and excellent selectivity for nNOS over eNOS (> 2500-fold) and iNOS (> 700-fold) (Table 1), the positive charges derived from the sidechain basic groups dramatically impair the ability of these inhibitors to penetrate blood brain barrier (BBB).26 To improve the bioavailability of these inhibitors, two fluorines were further introduced to the benzylic position of the m-fluorophenylethyl tail of 2b to lower the sidechain NH pKa (to about 5.5). The nNOS cell-based assay indicated that compound 3 (Figure 1, IC50 = 19 μM in cell-based assay) crossed the cell membrane 2.5-fold better than racemic 1.27,28 Computational studies indicate that the flipped mode binds more tightly as a result of better electrostatic interactions between the protein and inhibitor because in the “normal” binding mode the aminopyridine is only partially protonated as a result of unfavorable electrostatic interactions between the aminopyridine and pyrrolidine nitrogen atoms.20
Table 1.
Inhibition of NOS isozymes by the enantiomerically pure isomers of 1–3
| Compounds | Ki (μM)a | Selectivitya | |||
|---|---|---|---|---|---|
|
| |||||
| nNOS | eNOS | iNOS | n/e | n/i | |
| 1a | 0.052 | 26.4 | 3.8 | 506 | 74 |
| 1b | 0.005 | 20.3 | 3.9 | 3830 | 743 |
| 2a | 0.116 | 26.2 | 7.5 | 226 | 65 |
| 2b | 0.007 | 19.2 | 5.8 | 2667 | 806 |
| (±)-3 | 0.080 | 62.0 | 52.0 | 780 | 650 |
n/e and n/i are the selectivity ratio of Ki (eNOS or iNOS) to Ki (nNOS).
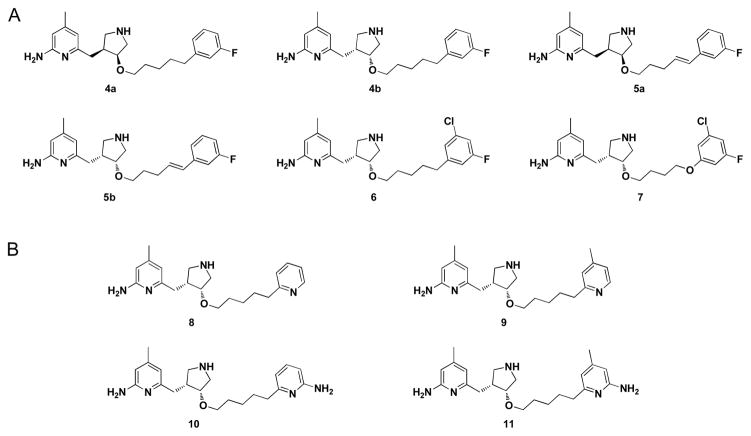
Removal of the sidechain NH group would be the most direct strategy to circumvent the sidechain positive charge and further improve BBB permeability. Here, we report the results of a series of monocationic pyrrolidine-based analogs as nNOS inhibitors (Figure 3A). In addition, on the basis of the crystal structures of these inhibitors with nNOS, we carried out steered molecular dynamics (SMD) simulations, which provide valuable information about the ligand-enzyme interactions and the pharmacophoric requirements needed for the design of potent inhibitors.29–31 Finally, the derivatives with doubly substituted pyridine heads (Figure 3B) were made to validate the predictions of our simulations.
Figure 3.
Target molecules synthesized and tested in this study.
Results and Discussion
Chemistry
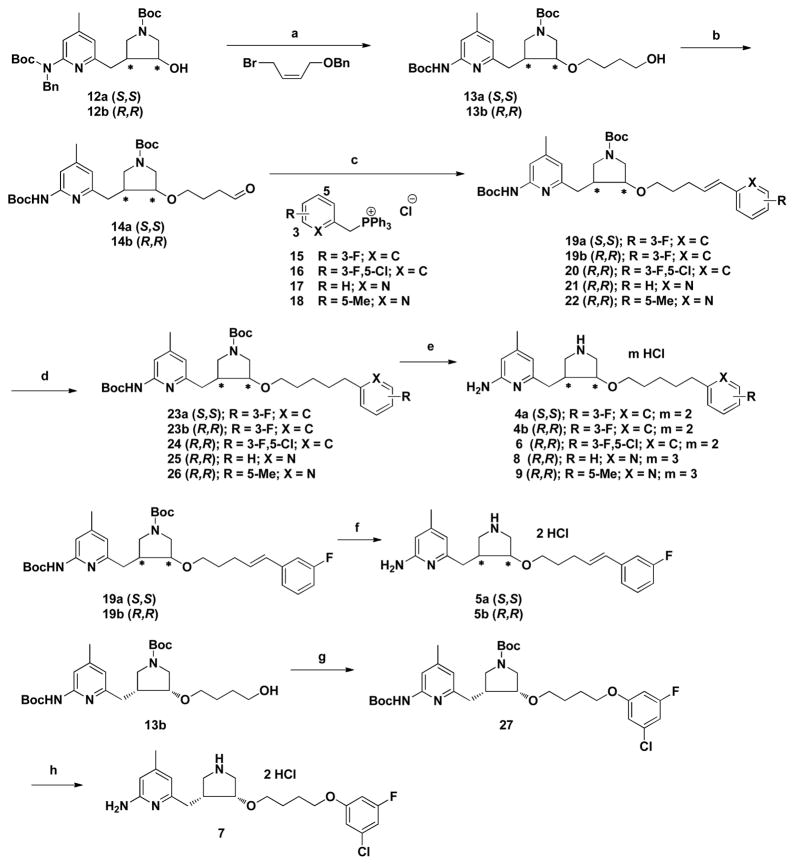
Scheme 1 shows the synthetic route for target compounds 4-9. The synthesis of 12a and 12b was described previously.22 Single enantiomer 12a or 12b was treated with NaH, and the resulting anion was allowed to react with (Z)-(((4-bromobut-2-en-1-yl)oxy)-methyl)benzene. Then catalytic hydrogenation of the crude product removed the benzyl-protecting group and also reduced the double bond, giving high yields of purified alcohols 13a or 13b. The oxidation of 13a and 13b gave aldehydes 14a and 14b, respectively. Next, a Wittig reaction of the aldehydes with the corresponding phosphorus ylides allowed the isolation of intermediates 19a, 19b, and 20–22 in moderate yields. Finally, the reduction of 19a, 19b, and 20–22 were performed to reduce the double bonds followed by Boc-deprotection to form corresponding target products 4a, 4b, 6, 8 and 9. The bromination of 13b, followed by nucleophilic substitution with 3-chloro-5-fluorophenol provided intermediate 27. The Boc-protecting groups of 19a, 19b, and 27 were removed to generate inhibitors 5a, 5b, and 7.
Scheme 1.
Synthesis of 4–9a
a Reagents and conditions: (a) (i) NaH, DMF, 0 °C, 3 h, (ii) 20 wt % Pd(OH)2/C, H2, EtOH, 60 °C, 36 h, 91–94%; (b) Dess-Martin periodinane, CH2Cl2, rt, 18 h, 89–92%; (c) (i) LHMDS, −78 °C to rt, THF, 8 h; (d) 10% Pd/C, H2, MeOH, rt, 12 h; (e) 3N HCl/MeOH, rt, 24 h, 47–69% for three steps; (f) 3N HCl/MeOH, rt, 24 h, 90–93%; (g) (i) PPh3, CBr4, CH2Cl2, 0 °C, 3 h, (ii) 3-chloro-5-fluorophenol, K2CO3, acetone, reflux, 12 h, quantitative; (h) 3N HCl/MeOH, rt, 24 h, quantitative.
The synthetic route for 10 and 11 is shown in Scheme 2. Boc-protected 2-aminopyridine analogs 28 and 29 were treated with n-butyllithium and allowed to react with (Z)-((4-bromobut-2-en-1-yl)oxy)(tert-butyl)dimethylsilane. The amino groups of the products were further protected with benzyl groups. Then the TBS protecting group was removed, followed by bromination to generate intermediates 30 and 31. Subsequently, a nucleophilic substitution of alcohol 12b with the allyl bromides gave the corresponding ethers. Debenzylation of the ethers with hydrogen gave 32 and 33 in which the double bond was also reduced. Finally, removal of the Boc-protecting group proceeded smoothly, giving high yields of target products 10 and 11.
Scheme 2.
Synthesis of 10 and 11a
a Reagents and conditions: (a) (i) n-BuLi, THF, −78 °C to rt, 2 h, (ii) NaH, DMF, 0 °C, 3 h, (iii) TBAF, DMF/THF, 0 °C, 2 h, (iv) PPh3, CBr4, CH2Cl2, 0 °C, 3 h, 26–43%; (b) (i) NaH, DMF, 0 °C, 3 h, (ii) 20 wt % Pd(OH)2/C, H2, EtOH, 60 °C, 36 h, 84–89%; (c) 3N HCl/MeOH, rt, 24 h, 89–94%.
In vitro NOS inhibition by 4-7
Table 2 shows the results of inhibition assays using purified NOS enzymes with 4-7. (3′R,4′R) Inhibitors 4b and 5b exhibit much better nNOS inhibitory potency and selectivity over the other two NOS isozymes than the corresponding (3′S,4′S)-enantiomers (4a and 5a), respectively. Introduction of a trans double bond into (3′S,4′S)-4a (compound 5a) results in a reduction in activity, whereas the potency increases a little by introduction of a trans double bond into (3′R,4′R)-4b (compound 5b). Compared to the 3-fluorophenyl analog (4b), the double halo-substituted phenyl compound (6) exhibits 4-fold better inhibitory potency and selectivity for nNOS over eNOS. Structural optimization of 6 leads to the generation of 3-fluoro-5-chlorophenyl ether 7, which exhibits two-fold better inhibitory potency but four-fold lower selectivity for nNOS over eNOS than 6.
Table 2.
Inhibition of NOS isozymes by synthetic compounds 4–7
| Compound |
Ki (μM)a
|
Selectivityb
|
|||
|---|---|---|---|---|---|
| nNOS | eNOS | iNOS | n/e | n/i | |
| 4a | 9.057 | 34.0 | 468.5 | 4 | 52 |
| 4b | 0.922 | 33.8 | 83.8 | 37 | 91 |
| 5a | 15.402 | 69.0 | 133.8 | 4 | 9 |
| 5b | 0.637 | 77.0 | 35.1 | 121 | 55 |
| 6 | 0.230 | 35.6 | 106.0 | 155 | 461 |
| 7 | 0.117 | 4.4 | 12.4 | 37 | 106 |
The apparent Ki values are represented as the mean of two or more independent experiments preformed in duplicate with five or six data points each.
n/e and n/i are the selectivity ratios of Ki (eNOS or iNOS) to Ki (nNOS).
Figure 4.
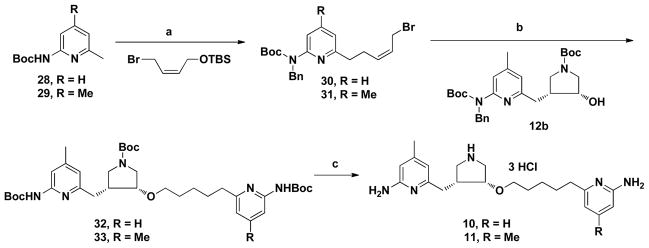
Crystallographic binding conformations of 4a (A), 4b (B), 5a (C), and 5b (D) with rat nNOS. The omit Fo – Fc electron density maps for the inhibitors are shown at 2.5 σ contour level. Major hydrogen bonds are depicted with dashed lines. The atom color schemes are: oxygen, red; nitrogen, blue; sulfur, yellow; fluorine, light cyan; chlorine, green.
Crystallographic Studies of 4-7
To better understand structure-activity relationships for 4-7, the crystal structures of nNOS complexed with these inhibitors were determined (Figure 4). Consistent with the binding preference of parental compound 2a, the (3′S,4′S)-isomers 4a and 5a are found to bind in the normal binding mode with their aminopyridine moiety hydrogen bonded to the side chain of Glu592 in nNOS (Figures 4A and 4C), and the pyrrolidine nitrogen also forms a hydrogen bond with Glu592. A water molecule provides a H-bonding bridge for the pyrrolidine nitrogen atom and the side chains of both Tyr588 and Asp597. The sidechain beyond the ether oxygen atom and the fluorophenyl end is disordered and only visible at low electron density contour levels. Surprisingly, (3′R,4′R)-isomers 4b and 5b break the rule of our previous pyrrolidine-based inhibitors. Despite their (3′R,4′R) stereochemistry, they exhibit the unexpected normal binding mode, similar to the conformation of the (3′S,4′S) enantiomers. The 2-aminopyridine moieties interact with Glu592 while the 3-fluorophenyl group extends out of the substrate-catalytic site and points toward the entrance of the binding pocket (Figures 4B and 4D). The locations of the pyrrolidine rings of 4b and 5b are similar to that of 4a. Although the pyrrolidine nitrogen can no longer directly hydrogen bond to Glu592, it can still form a hydrogen bond with the same water molecule found in the 4a-nNOS structure (Figure 4A) that bridges to a hydrogen bond network with both Tyr588 and Asp597. However, the rest of both inhibitor molecules beyond the ether oxygen is poorly defined and can only be modeled based on partial electron density at low contour levels. In chain B the position of the fluorophenyl tail of 4b is closer to heme propionate D with the Tyr706 side chain adopting the rotomer associated with the flipped binding mode (see Supporting Information Figure S1A). More interestingly, 5b shows alternate binding conformations in chain B and binds mostly (≈ 70%) in the normal mode, but with a small population (≈ 30%) in the flipped mode (Figure S1B). The crystallographic results indicate that both binding modes are almost isoenergetic, which means that small perturbations can apparently change the ligand binding conformation without significantly changing stability. Similar observations have been made for inhibitor binding to elastase.32
Despite their high structural similarity to 4b, both of the 3-fluoro-5-chlorophenyl analogs (6 and 7) adopt the flipped binding mode as expected from their (3′R,4′R) chirality (Figure 5). The aminopyridine makes bifurcated hydrogen bonds with heme propionate D. The pyrrolidine nitrogen atom hydrogen bonds to both heme propionate A and H4B. The flexible linker extending from the pyrrolidine brings the 3-fluoro-5-chlorophenyl ring to the substrate-catalytic site to form π-stacking interactions with the heme. However, the larger size of the 3-fluoro-5-chloro phenyl group forces Glu592 into an alternate rotamer position. This alternate Glu side chain position also was observed in the eNOS-2-bromo-7-nitroindazole complex.33 Overall, one extra ether oxygen in 7 does not change its binding conformation much from that seen for 6, which is consistent with the similar potency shared by the two inhibitors (Table 2).
Figure 5.
Crystallographic binding conformation of 6 (A) and 7 (B) with rat nNOS. The omit Fo – Fc electron density maps for inhibitors are shown at 2.5 σ contour level. Major hydrogen bonds are depicted with dashed lines. The same atom color schemes shown in Figure 4 are used.
Docking and Steered Molecular Dynamics Simulations
The normal binding mode of (3′R,4′R)-isomers 4b and 5b was completely unexpected because it breaks the precedent that had been established by many crystal structures, demonstrating that the (3′R,4′R)-pyrrolidine analogs adopt the flipped binding mode. Only one bioisosteric change (replacing the sidechain NH by CH2) from parent compound 2b both dramatically affects inhibitory potency and radically changes the binding mode. Clearly, the chemical nature of the sidechain and the tail end affects the inhibitor binding mode, but how? What kind of inhibitor-enzyme interactions, other than chirality, determine the binding mode? To study the molecular interactions involved in formation of the ligand-enzyme complex and to answer these questions, we carried out computational studies combining docking, equilibrium, and steered molecular dynamics simulations.
The two most populated clusters of docking conformations were obtained by using different nNOS receptors. One, in which 2a is bound, exhibits the normal binding mode and the other with 2b bound, which exhibits the flipped binding mode. After 2a and 2b were removed, the receptors were used to dock 4b, which gives 4b-nNOS complexes corresponding to the normal and flipped poses, respectively (Figure 6). Hereafter, the normal and flipped docking poses are referred to as 4b-N and 4b-F, respectively. Consistent with the binding preference of 4b in the crystal structure, the aminopyridine moiety of docking pose 4b-N establishes hydrogen bonding interactions with Glu592. Its pyrrolidine nitrogen also interacts with Glu592 and is part of a water-mediated hydrogen bonding network involving Tyr588 and Asp597. In contrast, the aminopyridine group in 4b-F makes bifurcated hydrogen bonds with the propionate of pyrrole ring D. Tyr706 rotates farther out, without making optimized π-stacking interaction with the aminopyridine ring. The pyrrolidine N atom in 4b-F makes favorable hydrogen bonds with propionate A, while the hydrogen bond with the O4 atom of H4B is broken.
Figure 6.
Docking conformation of 4b with different rat nNOS crystal structures. (A) 4b-N, in which 2a-nNOS complex (PDB ID 3NLK) was used as the receptor; (B) 4b-F, in which 2b-nNOS complex (PDB ID 3NLM) was used as the receptor.
The crystallographically flipped pose of 2b, crystallographically normal pose of 4b, and flipped docking pose 4b-F were subsequently submitted to 5 ns equilibrium molecular dynamics simulations. The root-mean-square derivation (RMSD) values of all atoms and the Cα atoms of nNOS as a function of time for the 5 ns MD simulation are shown in Figure S2 in the Supporting Information, suggesting the whole structure has been equilibrated after about 1.5 ns.
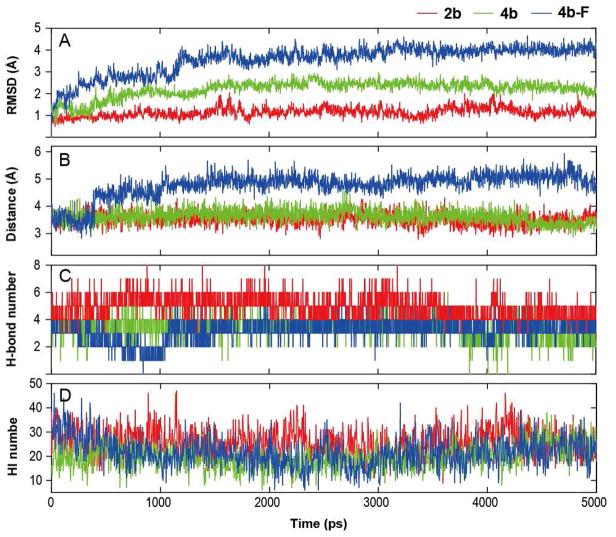
The RMSD values of ligands 2b, 4b, and 4b-F versus simulation time are shown in Figure 7A. These results indicate that both 4b and 4b-F are more flexible than 2b. The structure of 2b appears to have been stabilized with the RMSD value of ~1 Å after ~500 ps of simulation, while the structure of 4b and 4b-F are stabilized after ~1500 ps of simulation with the RMSD values of ~2 and ~3 Å, respectively. To further probe the differences among the moieties located in the substrate-catalytic pocket above the heme, center-center distances between the iron atom of the heme cofactor and the aromatic group of the ligands were monitored during the MD simulation, and the results are shown in Figure 7B. During the simulation, the aromatic heads (aminopyridine) of 2b and 4b vibrate slightly in the catalytic pocket, their distances to heme-Fe being relatively stable at ~3.5 Å. The profile of the center-center distance between the heme-Fe and the aromatic tail (fluorophenyl) of 4b-F forms the first platform from 0 to 380 ps at ~3.3 Å. Then the distance sharply increases to ~4.5 Å and forms a short platform from 380 to 1100 ps, followed by a long platform after 1000 ps at ~5 Å.
Figure 7.
(A) Time dependence of the RMSD; (B) distance between the heme Fe and the center of the aromatic head; (C) numbers for the direct hydrogen bonds; (D) numbers for hydrophobic interactions in the equilibrium MD simulations. All curves were obtained over 4 ps intervals.
To understand the interactions between nNOS and the ligands, the direct hydrogen bonds were analyzed. Compound 2b can potentially form more hydrogen bonds with the enzyme than 4b and 4b-F. Figure 7C shows the statistics of hydrogen bonds formed every 4 ps. Statistical analysis shows that the hydrogen bonds of 2b and 4b-F with the highest occurrence frequency involve heme propionate A, heme propionate D, and H4B. For the 4b-nNOS system, the dominant hydrogen bonds were formed by Glu592. In our simulation, Glu592 is a flexible residue, which can swing toward the sidechain N atom of 2b, forming a salt bridge or hydrogen bond and stabilize 2b (Figure 8A). Both the hydrogen bond numbers and the hydrophobic interaction numbers for 2b and 4b are relatively stable during the simulation, while those for 4b-F decrease gradually from 4 and 35 to 2 and 20 before 1000 ps, respectively, and then stabilize at 3–4 and ~20 after 1100 ps. This is in good agreement with the variation of the center-center distance between heme-Fe and the aromatic fluorophenyl tail of 4b-F.
Figure 8.
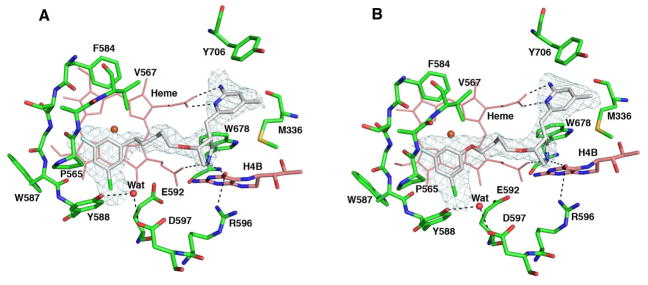
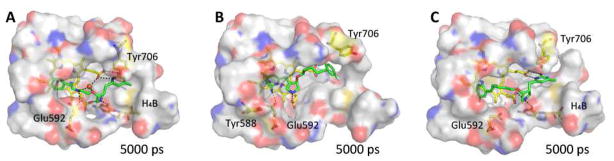
Snapshots of 2b (A), 4b (B), and 4b-F (C) isolated from the equilibrium MD at 5000 ps. The dashed lines represent hydrogen bonds and a water bridge. The water molecule in (A) is shown as a red sphere. Residues around the ligand within 8 Å were selected to show the surface.
Figure 8 displays snapshots of the structures of ligand-nNOS complexes at 5000 ps. Both the 2b-nNOS X-ray crystal structure and the dynamics simulation demonstrate the importance of the presence of water molecules in ligand binding to nNOS. A water molecule bridging the sidechain amine nitrogen in 2b with heme propionate D was seen during the entire simulation (Figure 8A). The water molecule and the atoms between the O atom and N atom in the sidechain of 2b form a rigid five-membered ring structure, which greatly stabilizes the flipped binding conformation of 2b. In contrast to 2b, no water bridge between the O atom of 4b-F (lack of the side chain N atom) and heme propionate D was detected in the simulation.
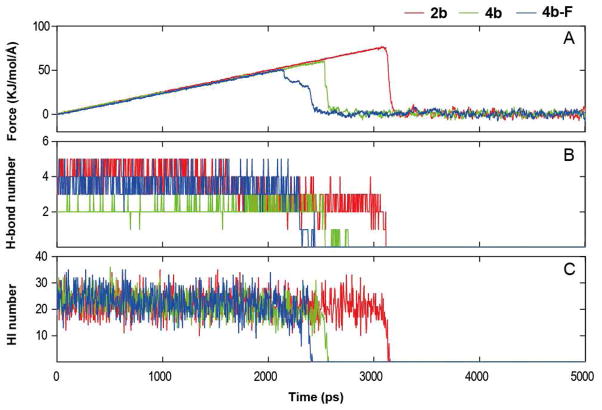
To investigate the molecular mechanism, at the atomic level, of a ligand leaving the binding pocket of nNOS, we performed SMD simulations, which can reveal features characteristic of the reverse process of binding. The profiles of the force exerted on the system to encourage the unbinding of the ligands along a carefully predefined reaction coordinate are shown in Figure 9A (see also the Experimental section and SI Figure S3 for further details). During the unbinding of 2b, the concerted rupture of the anchoring interactions (mainly electrostatic interactions, hydrogen bonds, and hydrophobic interactions) between nNOS and 2b defines the primary event with an unfolding force of ~77 kJ/mol/Å (Figure 9A), significantly higher than the 58.3 and 50.1 kJ/mol/Å measured for 4b and 4b-F, respectively, at the same pulling force constant. In particular, the unbinding is controlled by a breakup in the charge-reinforced hydrogen bonding network between the ligand pyrrolidine N atom and heme propionate A. The pulling force profile of 4b is similar to that of 2b; the forces increase in the beginning of ligand unbinding and reach a maximum around 2500 ps, followed by a return to zero. Notably, both the responses of 2b and 4b to the pulling force evolve in two distinct stages (Figure 9A): i) from 0 to 2600 ps (4b) or 3200 ps (2b), a buildup of force during which hydrogen bonds between the ligands with heme propionate A, heme propionate D, and Glu592 are ruptured; 2) after those times, the ligands are pulled out of the binding pocket.
Figure 9.
Plot of the rupture force (A), number of direct hydrogen bonds (B), and number of hydrophobic interactions (HI) (C) versus time in the equilibrium MD simulations. All curves were obtained over 4 ps intervals.
Interestingly, 4b-F suffers a three-state pathway for forced unbinding: an intermediate state is maintained from 2150 to 2350 ps between the maximum rupture force and breakdown of the primary unbinding barrier (Figure 9A). From 2140 to 2260 ps, the hydrophobic interactions between 4b-F and nNOS are broken, while the hydrogen bonding number remains relatively constant.
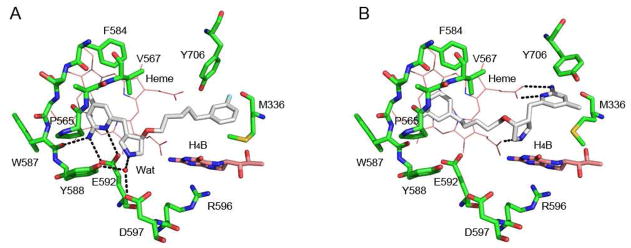
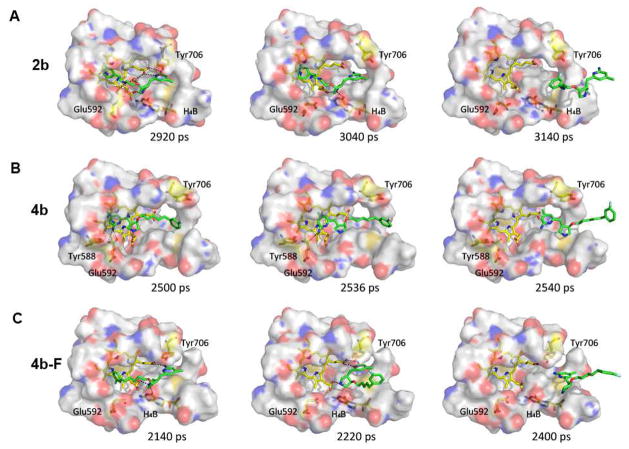
Ligands in nNOS are shown in their bound states before and after dissociation in Figure 10. A key observation during this time is that 4b-F curls up within the binding pocket. This curling, clearly discernible in Figure 10C, is the result of the rupture of hydrophobic interactions between the fluorobenzene moiety and the substrate binding pocket, while most hydrogen bonds between 4b-F and heme propionate D remain intact (Figures 9B and 9C). The curling then orients the fluorophenyl end of 4b-F into a position that permits it to easily dissociate from the binding pocket. These results clearly indicate the instability of the fluorophenyl end located in the catalytic pocket and favor 4b in a normal binding mode.
Figure 10.
Snapshots of 2b (A), 4b (B) and 4b-F (C) isolated from the SMD before and after dissociation. The dashed lines represent hydrogen bonds and the water bridge. The water molecule in (A) is shown as a red ball. Residues around the ligand within 8 Å were selected to show the surface.
What are the determining factors that result in a normal binding pose for the (3′R,4′R)-pyrrolidine analog 4b? The SMD simulation calculations provide some clues. The structural features of 4b have the potential to make it bind to nNOS with either the normal or flipped pose. We thought the hydrogen bonds from the aminopyridine and pyrrolidine to the surrounding protein would be the key factors that determine which way the inhibitors bind. However, the chemical nature of the sidechain and the aromatic end group also matters. SMD simulation results shown in Figure 10 indicate that 2b in its flipped binding mode utilizes its sidechain nitrogen to interact with a water molecule that bridges in between 2b and heme propionate D. This nitrogen atom also interacts with the Glu592 sidechain for another hydrogen bond. The sidechain and the fluorophenyl end of 4b-F are extremely unstable because of the lack of this sidechain N atom, resulting in a curling of the tail prior to its much easier dissociation from the substrate-catalytic pocket. Therefore, 4b prefers to adopt the normal binding mode. Comparisons of the force profiles for 4b and 2b (Figure 9A) show that the binding affinity of 4b is considerably weaker than that for 2b. This is understandable for the following reasons: (i) in the normal binding mode 4b has fewer hydrogen bonds, hydrophobic interactions, and electrostatic interactions than 2b in a flipped mode; (ii) the sidechain without the amino N atom causes the fluorophenyl moiety of 4b to vibrate more vigorously in the pocket surrounded by Met336, Leu337, and Tyr706 because of no hydrogen bonding possibilities; (iii) the orientation of the pyrrolidine ring of 4b in the normal mode makes it impossible to form a hydrogen bond to Glu592.
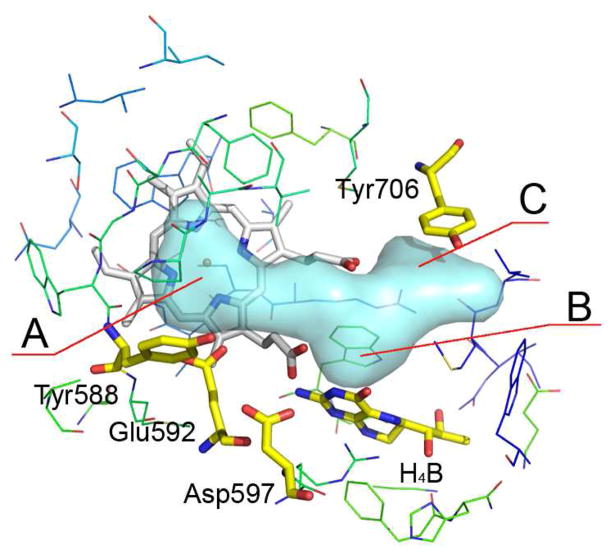
These results show that SMD simulations can be used to guide the development of better nNOS inhibitors. As shown in Figure 11, the substrate binding pocket (region A) should be occupied by an aromatic ring, which not only stack with the heme but should also interact with Glu592 for additional stability. This implies that introducing one or more heteroatoms embedded in the aromatic ring or linker chain should be favorable. In addition, an anchoring part that can interact with heme propionate A or Asp597 should be located in region B. This part functions to anchor the ligand in the substrate-catalytic pocket, and to position the tail aromatic ring to region A appropriately. Moreover, an aminopyridine fragment in region C is favorable to enhance the affinity through the bifurcated hydrogen bonds with heme propionate D and the π-stacking interaction with Tyr706.
Figure 11.
Pharmacophoric requirements needed for the design of potent nNOS inhibitors.
One final question to address is why 6 and 7 bind in the flipped mode, even though both 6 and 7 are close analogs of 4b, which binds in the normal orientation. The only difference between 4b and 6 is the additional Cl atom added to the fluorophenyl group of 4a. The bulkiness of the 2-substituted phenyl group forces the Glu592 side chain to adopt a different rotamer, resulting in less steric clashes between the chlorofluorophenyl group and Glu592. In addition, the second halogen atom increases the hydrophobic character of the protein-ligand interface within the active site pocket.34,35 It also reduces the electron density of the aromatic ring, allowing more favorable parallel π-stacking interactions between the chlorofluorophenyl tail and the heme.36,37 Both increased interactions enhance the binding affinity of the tail phenyl group to region A, thus tipping the balance in favor of the flipped orientation.
New Analog Design, NOS Inhibition, and Crystallographic Analysis of 8-11
As mentioned above, the simulation results imply that one or more heteroatoms embedded in the aromatic ring or linker chain should favorably stabilize the ligand by providing an additional hydrogen bond with the protein. A pyridine ring is a good candidate to replace the fluorophenyl moiety because it has the potential to form hydrogen bonding interactions with Glu592, and importantly, it would not be protonated under physiological conditions.
Both the activities and the crystal structures of double-headed pyridine analogs 8-11 correlate well with the insights gained from the above molecular dynamics simulations. The corresponding Ki values and nNOS selectivity ratios of 8-11 are shown in Table 3. The enzyme assay results indicate that the tested compounds are potent and selective inhibitors toward nNOS, with Ki values in the low nanomolar range. With the aim of investigating the effect of 4-methyl and 6-amino groups on the pyridine head, we synthesized compounds 9-11. Each of the substitutions increases the potency of the nNOS inhibitor relative to that of parent compound 8. Compounds 9 and 10 have the greatest combined potency for nNOS and selectivity over eNOS and iNOS of any of the monocationic pyrrolidine-based nNOS inhibitors reported to date. Compound 11, with both 4-methyl and 6-amino groups on the pyridine head, maintains a comparable nNOS inhibitory activity as 9 and 10, but the selectivities over eNOS (687-fold) and iNOS (172-fold) are decreased relative to those of 8-10.
Table 3.
Inhibition of NOS isozymes by synthetic compounds 8–11
| Compound |
Ki (μM)a
|
Selectivityb
|
|||
|---|---|---|---|---|---|
| nNOS | eNOS | iNOS | n/e | n/i | |
| 8 | 0.074 | 148.9 | 9.8 | 2012 | 132 |
| 9 | 0.031 | 45.2 | 17.3 | 1459 | 558 |
| 10 | 0.030 | 33.5 | 18.6 | 1117 | 619 |
| 11 | 0.038 | 26.1 | 6.5 | 687 | 172 |
The apparent Ki values are represented as the mean of two or more independent experiments preformed in duplicate with five or six data points each.
n/e and n/i are the selectivity ratio of Ki (eNOS or iNOS) to Ki (nNOS).
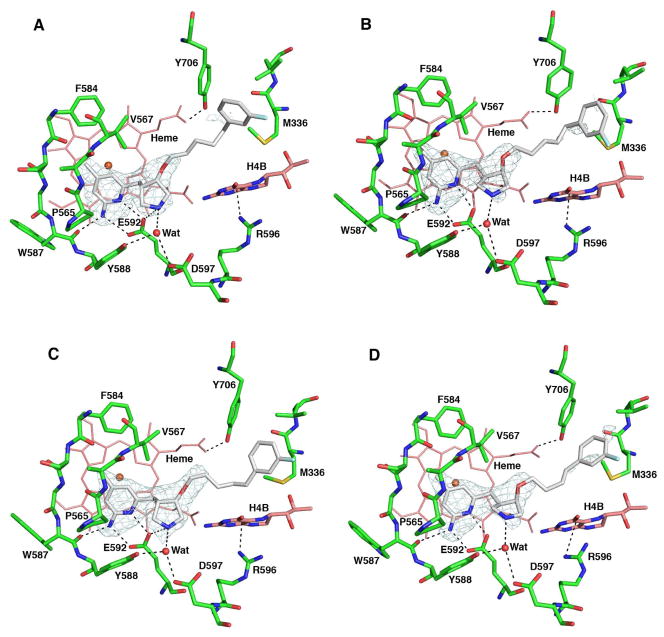
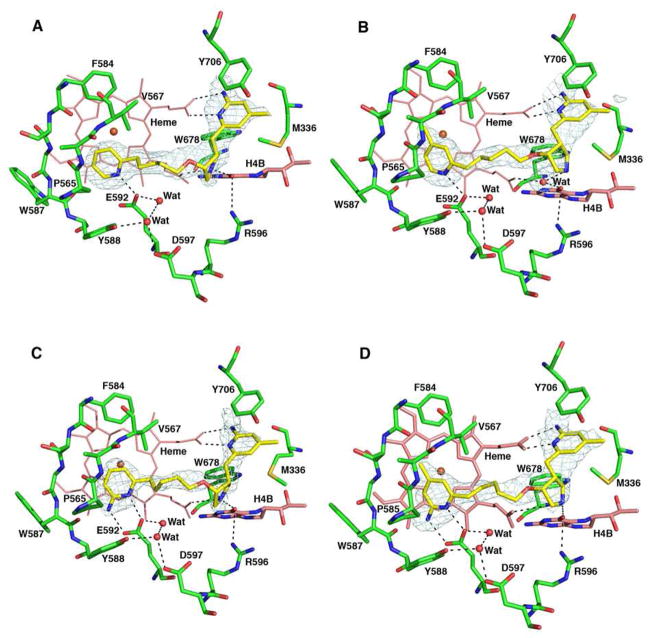
Crystal structures of nNOS complexed with 8 and 9 show that these inhibitors adopt the flipped binding mode again (Figures 12A and 12B). Their pyridine heads interact with active site Glu592, which stabilizes binding. The extra methyl group in the pyridine ring of 9 further helps to tightly position the ring by providing additional hydrophobic contacts with the protein. The positively charged nitrogen atoms in the pyrrolidine rings interact with heme propionate A, and the aminopyridine heads extend out of the active site, where they form bifurcated salt bridges with heme propionate D. The presence of the pyridine nitrogen atom strongly stabilizes inhibitor binding, which confirms the predictions from our molecular dynamics simulations.
Figure 12.
Crystallographic binding conformation of 8 (A), 9 (B), 10 (C), and 11 (D) with rat nNOS. The omit Fo – Fc electron density maps for inhibitors are shown at 2.5 σ contour level. Major hydrogen bonds are depicted with dashed lines. The same atom color schemes shown in Figure 4 are used.
Addition of the 6-amino group to the pyridine ring in 10 and 11 further enforces the hydrogen bonding interaction between the inhibitor and Glu592, as shown in Figures 12C and 12D, although it does not necessarily improve the potency (Table 3). These two compounds are double-headed aminopyridine inhibitors; however, they bind to the other two isoforms more tightly as well, thus lowering the selectivity.
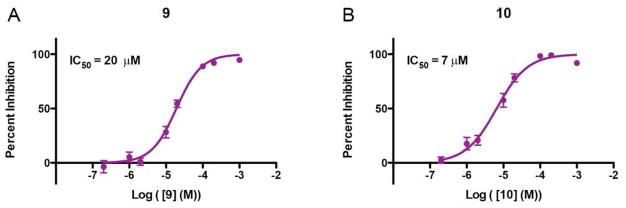
The two best inhibitors, 9 and 10, were further tested for their potency in a cell-based nNOS assay, which can provide information about their membrane permeability.38 As shown in Figure 13, the IC50 value for 9 is 20 μM, very similar to that of the difluorinated analog 3 (19 μM). Remarkably, compound 10 is 2.7 times more potent than 3, which demonstrates a successful improvement in membrane permeability for the pyrrolidine-based nNOS inhibitors and validates our approach toward eliminating the positively charged amines in the scaffold.
Figure 13.
Dose-response curves and IC50 values for 9 (A) and 10 (B) in a cell-based nNOS assay. Curves represent the average of three separate experiments.
Compared to their in vitro activities, the potencies of these pyrrolidine-based analogues in the cell-based assay are weaker, which may relate to both their membrane permeability and pharmacokinetic properties. Besides their potential affinity for one of the active efflux transporters (e.g., p-glycoprotein), metabolic properties also play important roles. 39 Metabolism may make the inhibitors ineffective or decompose them, thus weaken their cellular activities. The metabolic stability of 10 can be compared to that of (R,R)-3. The in vitro metabolic rate was 0.05 (nmol/min/mg protein) for (R,R)-3 and 0.02 (nmol/min/mg protein) for 10; 69% of (R,R)-3 and 85% of 10, respectively, remain after 60 minutes in the presence of NADPH. It is apparent that 10 has improved metabolic stability over (R,R)-3.
Conclusions
In summary, we have described an interdisciplinary study of a series of selective monocationic inhibitors of nNOS. Three stages of investigation allowed for the following conclusions:
The potencies and selectivities of a series of monocationic nNOS inhibitors decreased sharply, even with the small modification of removing the sidechain amino nitrogen compared to the potent and selective parent compound. The crystal structures revealed that they adopt an unexpected normal binding mode.
To investigate the underlying binding characteristics and reveal the important potential pharmacophoric features of the nNOS inhibitors, we carried out computational studies combining molecular docking, equilibrium, and steered molecular dynamics simulations. These computational results suggest that one or more heteroatoms embedded in the aromatic head or linker chain should be favorable to stabilize the ligand and allow it to tightly occupy the substrate-binding pocket.
The enzyme inhibition results of four double-headed pyridine analogs, in conjunction with their crystal structures, confirm the computational insights very well. Remarkably, 9 and 10 are the most combined potent and selective monocationic pyrrolidine-based nNOS inhibitors reported to date. Moreover, 10 has improved membrane permeability and serves as a lead molecule for further development.
Both computational and experimental insights from this study can be utilized to design even more effective nNOS inhibitors.
Experimental Section
Organic synthesis
All reagents were purchased from Sigma-Aldrich, Alfa Aesar, or TCI, and were used without further purification. Analytical thin layer chromatography was visualized by ultraviolet, ninhydrin, or phosphomolybdic acid (PMA). Flash column chromatography was carried out under a positive pressure of air. 1H NMR and 13C NMR spectra were recorded on 500 MHz and 125 MHz spectrometers, respectively. High-resolution mass spectra were measured on liquid chromatography/time-of-flight mass spectrometry (LC-TOF). For synthetic details, see the Supporting Information.
Molecular docking
AutoDock 4.2 was used to predict the potential binding between 4b and nNOS.40,41 The crystal structures of rat nNOS oxygenase domain in complex with 2a and 2b (PDB entry No.: 3NLK and 3NLM) were used as the receptors. The ligand and solvent molecules were removed from the crystal structures to obtain the docking grid, and the active site was defined using AutoGrid. The grid size was set to 60 × 60 × 60 points with grid spacing of 0.375Å. The grid box was centered on the center of the ligand from the corresponding crystal structure complexes. The Lamarckian genetic algorithm (LGA) was used for docking with the following settings: a maximum number of 2,500,000 energy evaluations, an initial population of 150 randomly placed individuals, a maximum number of 27,000 generations, a mutation rate of 0.02, a crossover rate of 0.80, and an elitism value (number of top individuals that automatically survive) of 1. The structure of 4b was built, and geometry optimized, using the molecular modeling suite of programs Sybyl-X 1.2 (Tripos International, 1699 South Hanley Rd., St. Louis, Missouri, 63144, USA). The ligand was fully optimized inside the binding site during the docking simulations. Finally, the conformation with the lowest predicted binding free energy of the most frequently occurring binding modes in the nNOS active pocket was selected.
Molecular dynamics simulation
The models used in the present study were built based on the X-ray crystal structures of the corresponding nNOS-ligand complexes. The coordinates of the missing residues (nNOS: 339–349 in chain A and 339–347 in chain B; eNOS: 109–121 in chain A and 110–120 in chain B) were added according to the intact X-ray crystal structures of nNOS and eNOS (PDB entry code 1ZVL and 1D0O for nNOS and eNOS, respectively) using the molecular modeling software Sybyl-X 1.2. The repaired residues were subjected to energy minimization in Sybyl-X 1.2 using the steepest descent method up to a gradient tolerance of 0.05 kcal/(mol·Å) to relieve possible steric clashes and overlaps of side chains with fixing the rest of the complex. The ionization states of the residues were determined by the web-based program H++ (http://biophysics.cs.vt.edu/H++).42,43 All of the simulations were carried out using the GROMACS 4.5.3 package with the identical protocol.44,45 The ligands were extracted from the corresponding complex crystal structures or docking results. The secondary amines were modeled as charged. Topology files and parameters for the ligands were generated using the AmberTools 1.5 package (http://ambermd.org/). A modification of the AMBER03 force field and the generalized amber force field were applied for the proteins and ligands, respectively.46,47 The reported AMBER force field parameters for the heme and Cys-Fe bond were adopted.33 TIP3P water molecules were added in cuboid periodic boxes, which were 120 Å ×110 Å ×120 Å.48 To ensure overall neutrality of the system, appropriate Na+ and Cl− were added at physiological concentration in the box.
All covalent bonds to hydrogen atoms were constrained using the linear constraint solver (LINCS) method. Electrostatic interactions were calculated using the particle-mesh Ewald (PME) algorithm.49,50 Periodic boundary conditions were applied to avoid edge effects in all calculations. Before the molecular dynamics run, the energy of these complexes was minimized to remove conflicting contacts. Then the systems were heated gradually from 0 to 310 K. After a 50 ps equilibration of water, two 250 ps equilibrations of the system were carried out with the restriction of main chain and carbon α–helix, respectively. Finally, 5 ns simulations were carried out, with coordinates saved every 2 ps during the entire process. Afterward, selective structures from the end of these trajectories were used as starting configurations for single molecule pulling simulations. Ligand in the active site of chain A was pulled out along the previously defined axis that extends from heme-CAB atom in chain A to the center of mass of atomic group of Trp711-CE2 in chain A and Trp306-CE2 in chain B. Chain A was used as an immobile reference for the Center-of-Mass (COM) pulling simulation. It should be emphasized that the pulling velocity is an important parameter in the COM pulling simulations.51,52 The simulations at higher pulling velocity may lead to substantial non-equilibrium effects. The low-velocity pulling simulations that were carried out on a millisecond time scale can overcome these disadvantages; however, the corresponding computational cost will be very expensive. To find an appropriate simulation velocity, several pulling simulations were carried out at different pulling velocities. The simulation results are listed in Figures S2 and S3 in the Supporting Information. Finally, the steered molecular dynamics simulations with a pulling rate of 0.01 Å·ps−1 and a force constant of 2.5 kJ·mol−1·Å−2 were selected. LIGPLOT 5.0.4 was used to analyze the hydrogen bonds and hydrophobic interactions between nNOS and the ligands in the molecular dynamics simulation trajectories. 53 Figures 2, 4–6, 8, and 10–12 were prepared with PyMol (http://www.pymol.org).
Enzyme Assays
The three isozymes, rat nNOS, murine macrophage iNOS, and bovine eNOS, were recombinant enzymes, overexpressed (in E. coli) and isolated as reported.54–56 IC50 values for inhibitors 4-11 were measured for the three different isoforms of NOS using L-arginine as a substrate. The formation of nitric oxide was measured using the hemoglobin capture assay described previously.57 All NOS isozymes were assayed using a Synergy H1 Hybrid Multi-Mode Microplate Reader (BioTek Instruments, Inc.) at 37 °C in a 100 mM Hepes buffer (pH 7.4) containing 10 μM L-arginine, 0.83 mM CaCl2, 320 units/mL calmodulin, 100 μM NADPH, 10 μM H4B, 3.0 μM oxyhemoglobin (for iNOS assays, no Ca2+ and calmodulin was added). The assay was initiated by the addition of enzyme, and the initial rates of the enzymatic reactions were determined by monitoring the formation of NO-hemoglobin complex at 401 nm for 60 sec. The apparent Ki values were obtained by measuring the percent enzyme inhibition in the presence of 10 μM L-arginine with at least five concentrations of inhibitor. The parameters of the following inhibition equation were fitted to the initial velocity data: % inhibition = 100[I]/{[I] + Ki(1+[S]/Km)}. Km values for L-arginine were 1.3 μM (nNOS), 8.2 μM (iNOS), and 1.7 μM (eNOS). The selectivity of an inhibitor was defined as the ratio of the respective Ki values.
Cell-based nNOS Inhibition Assay
HEK293t cells stably transfected with rat nNOS were cultured as previously described.38 The nNOS inhibition assay was performed as previously reported,38 with the following modifications: assays were performed in 96-well plates with a total volume of 100 μL, and 10 μM A23187 (Sigma Aldrich, St Louis, MO, USA) (added as 100x stock in 50% DMSO) was used in place of 5 μM. Eight concentrations of each inhibitor were tested, in at least triplicate wells. All inhibitors reported here were assayed within the same experiment to assure consistencies in cell concentration and passage number. The entire assay was repeated at least three times for each inhibitor, and the IC50 values averaged. Cells were plated in the 96-well plates 24 hours before activation at a concentration of approximately 8.5 × 106 cells/mL which resulted in 80–90 % confluency. After 6 hours of A23187 activation (in the presence or absence of inhibitor, which was added 30 minutes before activation) 50 μL aliquots of the media were removed from the wells. Nitrite production was quantified using Griess reagent and measuring absorbance at 540 nm.
Pharmacokinetic Data
All pharmacokinetic results were obtained by LC/MS/MS analysis in the DMPK group, Sai Advantium Pharma Ltd., Pune, India.
Inhibitor Complex Crystal Preparation
The nNOS heme domain proteins used for crystallographic studies were produced by limited trypsin digest from the corresponding full length enzymes and further purified through a Superdex 200 gel filtration column (GE Healthcare) as described previously. 58 The enzyme-inhibitor complex crystals were obtained by soaking, rather than co-crystallization, as reported in the earlier structural work.59 The nNOS heme domain at 7–9 mg/mL containing 20 mM histidine was used for the sitting drop vapor diffusion crystallization setup under the conditions reported before.58 Fresh crystals (1–2 day old) were first passed stepwise through cryo-protectant solutions as described58 and then soaked with 10 mM inhibitor for 4–6 h at 4 °C before being mounted on nylon loops and flash cooled by plunging into liquid nitrogen.
X-ray Diffraction Data Collection, Processing, and Structure Refinement
The cryogenic (100 K) X-ray diffraction data were collected remotely at various beamlines at Stanford Synchrotron Radiation Lightsource or Advanced Light Source through the data collection control software Blu-Ice60 and a crystal mounting robot. Raw data frames were indexed, integrated, and scaled using HKL2000.61 The binding of inhibitors was detected by the initial difference Fourier maps calculated with REFMAC.62 The inhibitor molecules were then modeled in COOT63 and refined using REFMAC. Water molecules were added in REFMAC and checked by COOT. The TLS64 protocol was implemented in the final stage of refinements with each subunit as one TLS group. The omit Fo – Fc density maps were calculated by repeating the last round of TLS refinement with inhibitor coordinates removed from the input PDB file to generate the coefficients DELFWT and SIGDELFWT. The refined structures were validated in COOT before deposition in the RCSB protein data bank. The crystallographic data collection and structure refinement statistics are summarized in Table 4 with PDB accession codes included.
Table 4.
Crystallographic data collection and refinement statistics
| Data set | nNOS-4a | nNOS-4b | nNOS-5a | nNOS-5b | nNOS-6 | nNOS-7 | nNOS-8 | nNOS-9 | nNOS-10 | nNOS-11 |
|---|---|---|---|---|---|---|---|---|---|---|
| Data collection | ||||||||||
| PDB code | 3UFO | 3UFP | 3UFQ | 3UFR | 3UFS | 3UFT | 3UFU | 3UFV | 3UFW | 4EUX |
| Space group | P212121 | P212121 | P212121 | P212121 | P212121 | P212121 | P212121 | P212121 | P212121 | P212121 |
| Cell dimensions | ||||||||||
| a, b, c (Å) | 52.2,111.6,165.0 | 52.0,110.9,164.5 | 52.1,111.0,164.5 | 51.9,111.3,164.5 | 51.8,111.3,164.2 | 51.8,111.2,164.1 | 52.1,110.8,164.4 | 51.7,110.2,164.3 | 51.9,111.2,164.1 | 52.0,111.2,164.3 |
| Resolution (Å) | 2.18 (2.22–2.18) | 2.10 (2.21–2.10) | 2.05 (2.09–2.05) | 2.10 (2.14–2.10) | 1.97 (2.00–1.97) | 2.08 (2.12–2.08) | 1.89 (1.92–1.89) | 2.08 (2.12–2.08) | 2.00 (2.03–2.00) | 2.14 (2.18–2.14) |
| Rsym or Rmerge | 0.079 (0.570) | 0.095 (0.489) | 0.067 (0.536) | 0.070/0.656 | 0.053 (0.460) | 0.084 (0.656) | 0.056 (0.546) | 0.055 (0.662) | 0.077 (0.654) | 0.096 (0.630) |
| I/σI | 22.5 (2.0) | 5.4 (1.6)b | 24.5 (1.9) | 24.1 (2.1) | 31.9 (2.4) | 29.8 (2.1) | 22.7 (1.9) | 24.3 (2.2) | 24.5 (1.6) | 20.4 (1.8) |
| No. unique reflections | 50,641 | 56,388 | 59,092 | 55,153 | 67,604 | 57,805 | 75,429 | 55,597 | 64,761 | 52,877 |
| Completeness (%) | 97.8 (94.6) | 99.7 (100.0) | 98.4 (94.5) | 97.2 (100.0) | 98.6 (83.0) | 99.6 (99.7) | 97.2 (96.8) | 96.7 (96.2) | 99.4 (99.9) | 99.3 (100.0) |
| Redundancy | 4.1 (4.1) | 3.9 (3.9) | 3.7 (3.7) | 4.1 (4.1) | 4.0 (3.7) | 4.8 (3.9) | 4.1 (4.2) | 3.9 (3.9) | 4.0 (3.9) | 3.9 (4.0) |
| Refinement | ||||||||||
| Resolution (Å) | 2.18 | 2.10 | 2.06 | 2.10 | 1.97 | 2.08 | 1.89 | 2.08 | 2.00 | 2.14 |
| No. reflections used | 47,872 | 53,442 | 55,921 | 52,229 | 64,012 | 54,727 | 71,429 | 52,658 | 61,504 | 50,181 |
| Rwork/Rfreea | 0.198/0.254 | 0.208/0.262 | 0.190/0.231 | 0.199/0.249 | 0.193/0.233 | 0.206/0.260 | 0.175/0.209 | 0.175/0.218 | 0.196/0.244 | 0.180/0.228 |
| No. atoms | ||||||||||
| Protein | 6678 | 6660 | 6660 | 6669 | 6671 | 6660 | 6681 | 6690 | 6665 | 6671 |
| Ligand/ion | 196 | 185 | 185 | 209 | 187 | 187 | 183 | 185 | 185 | 185 |
| Water | 195 | 259 | 270 | 220 | 284 | 167 | 367 | 325 | 252 | 268 |
| R.m.s. deviations | ||||||||||
| Bond lengths (Å) | 0.016 | 0.015 | 0.015 | 0.016 | 0.015 | 0.019 | 0.014 | 0.015 | 0.015 | 0.011 |
| Bond angles (deg) | 1.589 | 1.565 | 1.317 | 1.640 | 1.485 | 1.865 | 1.421 | 1.485 | 1.513 | 1.981 |
Rfree was calculated with the 5% of reflections set aside throughout the refinement. The set of reflections for the Rfree calculation were kept the same for all data sets according to those used in the data of the starting model (1OM4).
This data set was processed with MOSFLM. All others were processed with HKL2000.
Supplementary Material
Acknowledgments
The authors are grateful for financial support from the National Institutes of Health (GM049725 to RBS and GM057353 to TLP). We thank Dr. Bettie Sue Siler Masters (NIH grant GM52419, with whose laboratory P.M. and L.J.R. are affiliated). B.S.S.M. also acknowledges the Welch Foundation for a Robert A. Welch Distinguished Professorship in Chemistry (AQ0012). P.M. is supported by grant 0021620849 from MSMT of the Czech Republic. We also thank the beamline staff at SSRL and ALS for their assistance during the remote X-ray diffraction data collections.
Footnotes
Supporting Information Available: Details of chemical synthesis, alternate crystallographic binding conformation of 4b and 5b in chain B of rat nNOS, the rupture force versus time in the process of 2b leaving the nNOS binding gorge at different pulling force constant and pulling rates, time dependence of the RMSD of the ligand-nNOS complex for the all atoms and Cα in the equilibrium MD simulation, and copies of complete spectroscopic data of compounds 4–11. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Vallance P, Leiper J. Nat Rev Drug Discovery. 2002;1:939–950. doi: 10.1038/nrd960. [DOI] [PubMed] [Google Scholar]
- 2.Stuehr DJ. J Nutr. 2004;134:2748s–2751s. doi: 10.1093/jn/134.10.2748S. [DOI] [PubMed] [Google Scholar]
- 3.Rosen GM, Tsai P, Pou S. Chem Rev. 2002;102:1191–1199. doi: 10.1021/cr010187s. [DOI] [PubMed] [Google Scholar]
- 4.Granik VG, Grigor’ev NA. Russ Chem Bull. 2002;51:1973–1995. [Google Scholar]
- 5.Alderton WK, Cooper CE, Knowles RG. Biochem J. 2001;357:593–615. doi: 10.1042/0264-6021:3570593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li HY, Poulos TL. J Inorg Biochem. 2005;99:293–305. doi: 10.1016/j.jinorgbio.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 7.Olesen J. Neurotherapeutics. 2010;7:183–190. doi: 10.1016/j.nurt.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reif DW, McCarthy DJ, Cregan E, MacDonald JE. Free Radic Biol Med. 2000;28:1470–1477. doi: 10.1016/s0891-5849(00)00250-1. [DOI] [PubMed] [Google Scholar]
- 9.Villanueva C, Giulivi C. Free Radic Biol Med. 2010;49:307–316. doi: 10.1016/j.freeradbiomed.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aquilano K, Baldelli S, Rotilio G, Ciriolo MR. Neurochem Res. 2008;33:2416–2426. doi: 10.1007/s11064-008-9697-6. [DOI] [PubMed] [Google Scholar]
- 11.Sobolewski P, Gramaglia I, Frangos J, Intaglietta M, van der Heyde HC. Trends Parasitol. 2005;21:415–422. doi: 10.1016/j.pt.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 12.Matter H, Kotsonis P. Med Res Rev. 2004;24:662–684. doi: 10.1002/med.20005. [DOI] [PubMed] [Google Scholar]
- 13.Masic LP. Curr Med Chem. 2006;13:3627–3648. doi: 10.2174/092986706779026101. [DOI] [PubMed] [Google Scholar]
- 14.Oess S, Icking A, Fulton D, Govers R, Muller-Esterl W. Biochem J. 2006;396:401–409. doi: 10.1042/BJ20060321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Patman J, Bhardwaj N, Ramnauth J, Annedi SC, Renton P, Maddaford SP, Rakhit S, Andrews JS. Bioorg Med Chem Lett. 2007;17:2540–2544. doi: 10.1016/j.bmcl.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 16.Hesslinger C, Strub A, Boer R, Ulrich WR, Lehner MD, Braun C. Biochem Soc Trans. 2009;37:886–891. doi: 10.1042/BST0370886. [DOI] [PubMed] [Google Scholar]
- 17.Tsutsui M, Shimokawa H, Otsuji Y, Ueta Y, Sasaguri Y, Yanagihara N. Circ J. 2009;73:986–993. doi: 10.1253/circj.cj-09-0208. [DOI] [PubMed] [Google Scholar]
- 18.Silverman RB. Acc Chem Res. 2009;42:439–451. doi: 10.1021/ar800201v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maddaford S, Annedi SC, Ramnauth J, Rakhit S. Annu Rep Med Chem. 2009;44:27–50. [Google Scholar]
- 20.Delker SL, Ji HT, Li HY, Jamal J, Fang JG, Xue FT, Silverman RB, Poulos TL. J Am Chem Soc. 2010;132:5437–5442. doi: 10.1021/ja910228a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ji HT, Delker SL, Li HY, Martasek P, Roman LJ, Poulos TL, Silverman RB. J Med Chem. 2010;53:7804–7824. doi: 10.1021/jm100947x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ji H, Stanton BZ, Igarashi J, Li H, Martasek P, Roman LJ, Poulos TL, Silverman RB. J Am Chem Soc. 2008;130:3900–3914. doi: 10.1021/ja0772041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ji H, Li H, Martasek P, Roman LJ, Poulos TL, Silverman RB. J Med Chem. 2009;52:779–797. doi: 10.1021/jm801220a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ji H, Delker SL, Li H, Martasek P, Roman LJ, Poulos TL, Silverman RB. J Med Chem. 2010;53:7804–7824. doi: 10.1021/jm100947x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Flinspach ML, Li HY, Jamal J, Yang WP, Huang H, Hah JM, Gomez-Vidal JA, Litzinger EA, Silverman RB, Poulos TL. Nat Struct Mol Biol. 2004;11:54–59. doi: 10.1038/nsmb704. [DOI] [PubMed] [Google Scholar]
- 26.Lawton GR, Ranaivo HR, Chico LK, Ji HT, Xue FT, Martasek P, Roman LJ, Watterson DM, Silverman RB. Bioorg Med Chem. 2009;17:2371–2380. doi: 10.1016/j.bmc.2009.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xue F, Fang J, Lewis WW, Martasek P, Roman LJ, Silverman RB. Bioorg Med Chem Lett. 2010;20:554–557. doi: 10.1016/j.bmcl.2009.11.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xue F, Li H, Delker SL, Fang J, Martasek P, Roman LJ, Poulos TL, Silverman RB. J Am Chem Soc. 2010;132:14229–14238. doi: 10.1021/ja106175q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cui Y. J Pharm Sci. 2011;100:2000–2019. doi: 10.1002/jps.22392. [DOI] [PubMed] [Google Scholar]
- 30.Genchev GZ, Kallberg M, Gursoy G, Mittal A, Dubey L, Perisic O, Feng G, Langlois R, Lu H. Cell Biochem Biophys. 2009;55:141–152. doi: 10.1007/s12013-009-9064-5. [DOI] [PubMed] [Google Scholar]
- 31.Sotomayor M, Schulten K. Science. 2007;316:1144–1148. doi: 10.1126/science.1137591. [DOI] [PubMed] [Google Scholar]
- 32.Mattos C, Rasmussen B, Ding X, Petsko GA, Ringe D. Nat Struct Biol. 1994;1:55–58. doi: 10.1038/nsb0194-55. [DOI] [PubMed] [Google Scholar]
- 33.Raman CS, Li H, Martasek P, Southan G, Masters BS, Poulos TL. Biochemistry. 2001;40:13448–13455. doi: 10.1021/bi010957u. [DOI] [PubMed] [Google Scholar]
- 34.Ippolito JA, Christianson DW. Int J Biol Macromol. 1992;14:193–197. doi: 10.1016/s0141-8130(05)80026-1. [DOI] [PubMed] [Google Scholar]
- 35.Lu YX, Shi T, Wang Y, Yang HY, Yan XH, Luo XM, Jiang HL, Zhu WL. J Med Chem. 2009;52:2854–2862. doi: 10.1021/jm9000133. [DOI] [PubMed] [Google Scholar]
- 36.Hunter CA, Sanders JKM. J Am Chem Soc. 1990;112:5525–5534. [Google Scholar]
- 37.Cozzi F, Cinquini M, Annuziata R, Siegel JS. J Am Chem Soc. 1993;115:5330–5331. [Google Scholar]
- 38.Fang JG, Silverman RB. Anal Biochem. 2009;390:74–78. doi: 10.1016/j.ab.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Linnet K, Ejsing TB. Eur Neuropsychopharmacol. 2008;18:157–169. doi: 10.1016/j.euroneuro.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 40.Sanner MF. J Mol Graph Model. 1999;17:57–61. [PubMed] [Google Scholar]
- 41.Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. J Comput Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gordon JC, Myers JB, Folta T, Shoja V, Heath LS, Onufriev A. Nucleic Acids Res. 2005;33:W368–371. doi: 10.1093/nar/gki464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Myers J, Grothaus G, Narayanan S, Onufriev A. Proteins: Struct, Funct, Bioinf. 2006;63:928–938. doi: 10.1002/prot.20922. [DOI] [PubMed] [Google Scholar]
- 44.Hess B, Kutzner C, van der Spoel D, Lindahl E. J Chem Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 45.Van der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJC. J Comput Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 46.Oda A, Yamaotsu N, Hirono S. J Comput Chem. 2005;26:818–826. doi: 10.1002/jcc.20221. [DOI] [PubMed] [Google Scholar]
- 47.Duan Y, Wu C, Chowdhury S, Lee MC, Xiong GM, Zhang W, Yang R, Cieplak P, Luo R, Lee T, Caldwell J, Wang JM, Kollman P. J Comput Chem. 2003;24:1999–2012. doi: 10.1002/jcc.10349. [DOI] [PubMed] [Google Scholar]
- 48.Mark P, Nilsson L. J Phys Chem A. 2001;105:9954–9960. [Google Scholar]
- 49.Darden T, York D, Pedersen L. J Chem Phys. 1993;98:10089–10092. [Google Scholar]
- 50.Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG. J Chem Phys. 1995;103:8577–8593. [Google Scholar]
- 51.Xu YC, Shen JH, Luo XM, Silman I, Sussman JL, Chen KX, Jiang HL. J Am Chem Soc. 2003;125:11340–11349. doi: 10.1021/ja029775t. [DOI] [PubMed] [Google Scholar]
- 52.Lemkul JA, Bevan DR. J Phys Chem B. 2010;114:1652–1660. doi: 10.1021/jp9110794. [DOI] [PubMed] [Google Scholar]
- 53.Wallace AC, Laskowski RA, Thornton JM. Protein Eng. 1995;8:127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
- 54.Hevel JM, White KA, Marletta MA. J Biol Chem. 1991;266:22789–22791. [PubMed] [Google Scholar]
- 55.Roman LJ, Sheta EA, Martasek P, Gross SS, Liu Q, Masters BSS. Proc Natl Acad Sci U S A. 1995;92:8428–8432. doi: 10.1073/pnas.92.18.8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Martasek P, Liu Q, Liu JW, Roman LJ, Gross SS, Sessa WC, Masters BSS. Biochem Biophys Res Commun. 1996;219:359–365. doi: 10.1006/bbrc.1996.0238. [DOI] [PubMed] [Google Scholar]
- 57.Hevel JM, Marletta MA. Oxygen Radicals in Biological Systems, Pt C. 1994;233:250–258. [Google Scholar]
- 58.Li H, Shimizu H, Flinspach M, Jamal J, Yang W, Xian M, Cai T, Wen EZ, Jia Q, Wang PG, Poulos TL. Biochemistry. 2002;41:13868–13875. doi: 10.1021/bi020417c. [DOI] [PubMed] [Google Scholar]
- 59.Flinspach ML, Li H, Jamal J, Yang W, Huang H, Hah JM, Gomez-Vidal JA, Litzinger EA, Silverman RB, Poulos TL. Nat Struct Mol Biol. 2004;11:54–59. doi: 10.1038/nsmb704. [DOI] [PubMed] [Google Scholar]
- 60.McPhillips TM, McPhillips SE, Chiu HJ, Cohen AE, Deacon AM, Ellis PJ, Garman E, Gonzalez A, Sauter NK, Phizackerley RP, Soltis SM, Kuhn P. J Synchrotron Radiat. 2002;9:401–406. doi: 10.1107/s0909049502015170. [DOI] [PubMed] [Google Scholar]
- 61.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 62.Murshudov GN, Vagin AA, Dodson EJ. Acta Crystallogr. 1997;D53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 63.Emsley P, Cowtan K. Acta Crystallogr. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 64.Winn MD, Isupov MN, Murshudov GN. Acta Crystallogr. 2001;D57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.