Abstract
Background
Smad ubiquitination regulatory factor (Smurf) 1 and 2 are E3 ubiquitin ligases originally identified as inhibitors of transforming growth factor beta signaling and are shown to modulate multiple cellular activities. The roles of Smurfs in vertebrate embryogenesis, however, are not completely understood.
Results
Here we investigate the function of Smurf2 during early Xenopus development. We show that distinctly from Smurf1, overexpression of Smurf2 in presumptive mesoderm interfered with mesoderm induction and caused axial defects, whereas knockdown of Smurf2 with antisense morpholino oligonucleotides resulted in expansion of the mesoderm. These results imply that Smurf2 may modulate nodal-mediated mesodermal induction. Consistently, ventral expression of Smurf2 induced a partial secondary axis with head structures. In the ectoderm, Smurf2 resembled Smurf1 in controlling neural and epidermal marker expression and influencing head formation. Smurf1, but not Smurf2, additionally affected neural tube closure. Interestingly, both Smurfs could enhance as well as repress neural crest markers, implying that they modulate their targets dynamically during neural plate border specification.
Conclusion
Our data demonstrate that Smurf1 and Smurf2 have overlapping and distinct functionalities during early frog embryogenesis; collectively, they regulate ectodermal and mesodermal induction and patterning to ensure normal development of Xenopus embryos.
Keywords: Smurf1/2 ubiquitin ligases, activin/nodal/BMP signals, mesodermal induction and patterning, neural development, Xenopus
Introduction
Secreted growth factors belonging to the transforming growth factor beta (TGF-β) superfamily of cytokines play essential roles during vertebrate development and regulate multiple processes. In Xenopus, two branches of the TGF-β signaling pathway modulate different aspects of early embryogenesis. While activin/nodal-like ligands control mesodermal and endodermal induction and patterning, proteins in the bone morphogenetic protein (BMP) subfamily are critical for ventral cell fate determination in all three germ layers. In the ectoderm, BMPs promote epidermal development and inhibit neural specification in Xenopus gastrulae (Harland and Gerhart, 1997; Chang and Hemmati-Brivanlou, 1998a; Harland 2000; Whitman 2001; Schier 2003; De Robertis and Kuroda, 2004; Vonica and Brivanlou 2006). Stringent control of both activin/nodal and BMP signals is imperative to ensure normal development of Xenopus embryos, and is achieved by the actions of myriad of regulators of TGF-β signals that modulate signaling components at the extracellular, membrane, cytoplasmic and nuclear levels (Massague and Chen, 2000; Shi and Massague, 2003).
Canonical signal transduction of TGF-β growth factors involves activation of cytoplasmic receptor-regulated Smads (R-Smads) by transmembrane serine/threonine kinase receptors. Upon ligand binding, type I and type II TGF-β receptors form a complex and phosphorylate Smad2/3 downstream of TGF-β/activin/nodal signals and Smad1/5/8 downstream of BMP factors. These phosphorylated R-Smads then bind to a common partner Smad (co-Smad), Smad4, and are translocated into the nucleus to collaborate with other transcription factors to regulate target gene expression (Massague 1998). This signaling pathway is subjected to modulation by various positive and negative factors, including, for example, the inhibitory Smads (I-Smads) and the nuclear transcriptional co-repressors. The I-Smads, Smad6 and Smad7, inhibit phosphorylation of R-Smads by TGF-β receptors, compete with Smad4 for activated Smad1, and/or promote degradation of TGF-β receptors by ubiquitin-proteasome mediated pathway (Hayashi et al., 1997; Imamura et al., 1997; Nakao et al., 1997; Hata et al., 1998; Kavsak et al., 2000; Ebisawa et al., 2001; Murakami et al., 2003). The transcriptional co-repressors, such as Sloan-Kettering Institute proto oncogene (Ski), Ski-related novel gene, non-Alu-containing (SnoN), and TG-interacting factor (TGIF), are recruited to the R-Smad/Smad4 complex at the promoter region of TGF-β target genes to repress their expression (Liu et al., 2001; Luo 2004; Massague et al., 2005). The protein stability of TGF-β signaling molecules as well as their regulators can be controlled by ubiquitin-dependent degradation system, which influences the outcome of TGF-β signaling (Inoue and Imamura, 2011; Soond and Chantry, 2011).
Smurfs, Smad ubiquitination regulatory factors, were first characterized as HECT domain-containing E3 ubiquitin ligases that negatively regulated BMP signaling (Zhu et al., 1999; Lin et al., 2000; Zhang et al., 2001). Two Smurf members were identified, both interacted directly with Smad1/5 and down-regulated their protein levels. Subsequent studies reveal that the Smurfs target many additional proteins that can both enhance and reduce TGF-β signals, and the two Smurfs seem to have overlapping as well as distinct substrate specificities. Besides Smad1 and 5, both Smurfs can be recruited to TGF-β and BMP receptors via an intermediary protein, such as I-Smads or Fused, a cytoplasmic kinase previously shown to act in the Hedgehog pathway. These adaptor proteins facilitate the formation of a ternary complex with Smurfs and the receptors and promote receptor degradation (Kavsak et al., 2000; Ebisawa et al., 2001; Murakami et al., 2003; Xia et al., 2010). Similarly, via the intermediary proteins R-Smads and I-Smads, both Smurfs can be recruited to the Smad4 complex and reduce Smad4 levels (Moren et al., 2005). Smurf1 also down-regulates the protein level of TRAF4 (tumor necrosis factor-receptor-associated factor-4), a protein that potentiates both nodal and BMP signals (Kalkan et al., 2009). Smurf2, but not Smurf1, is shown to additionally enhance degradation of Smad2 in certain cells and promote multiple mono-ubiquitination of Smad3 to attenuate Smad3 signaling (Lin et al., 2000; Zhang et al., 2001; Tan et al., 2008; Tang et al., 2011). These activities of Smurfs decrease signaling capacity of TGF-β signal transducers, resulting in inhibition of TGF-β signals. However, Smurfs, especially Smurf2, can also decrease the levels of negative regulators of the pathway to augment TGF-β signals. Smurf2 is reported to facilitate degradation of the transcriptional co-repressors SnoN, Ski and TGIF to allow activation of the pathway (Bonni et al., 2001; Tan et al., 2008). Smurf1 and Smurf2 also promote degradation of Smad7 in the ternary complex with the TGF-β receptors (Kavsak et al., 2000; Ebisawa et al., 2001). Furthermore, Smurf2 can reduce the level of Smurf1 in cancer cells to modulate cell migration (Fukunaga et al., 2008). Thus, the effect of Smurfs on TGF-β signaling may depend on specific cell types, and degradation of distinct Smurf targets can influence spatial and temporal control of initiation, strength, and/or duration of the TGF-β signal.
The function of Smurfs during early vertebrate development has been analyzed in mice that are deficient for Smurf genes. Targeted inactivation of Smurf1 or 2 individually resulted in viable and fertile mice (Yamashita et al., 2005; Tang et al., 2011), suggesting redundant activities of the two Smurf genes. Disruption of both Smurf1 and 2 genes in mice led to embryonic lethality, with embryos displaying two distinct phenotypes (Narimatsu et al., 2009). One group of the mutants had planar cell polarity (PCP) defects that affected tissue convergent extension, so that the mice showed failure in neural tube closure. The defects were attributed to impairment in localized degradation of the PCP protein Prickle 1. The second group of the mutants displayed gastrulation defects. Though this could be caused by deregulated TGF-β signals, no detailed analyses of these mutants were reported. In Xenopus, antisense morpholino oligonucleotides (MO) – mediated knockdown approach was combined with the usage of a dominant negative mutant to assess the function of Smurf1 during early frog embryogenesis (Alexandrova and Thomsen, 2006). Smurf1 was found to regulate neural patterning and neurulation, but knockdown of the gene did not affect mesendodermal development. It is unclear whether this is due to redundant activities of Smurf2 present in the embryos that compensate for Smurf1 deficiency, and whether Smurf1 and Smurf2 regulate similar or distinct processes during early Xenopus embryogenesis. In this study, we used gain- and loss-of-function approaches to address the roles of Smurf2 in early frog development. We report that Smurf1 and Smurf2 have some functional overlaps, but they also harbor certain distinct activities, so that the two Smurfs work in concert to properly regulate the development of Xenopus embryos.
Results
Expression pattern of Smurf2 during early Xenopus embryogenesis
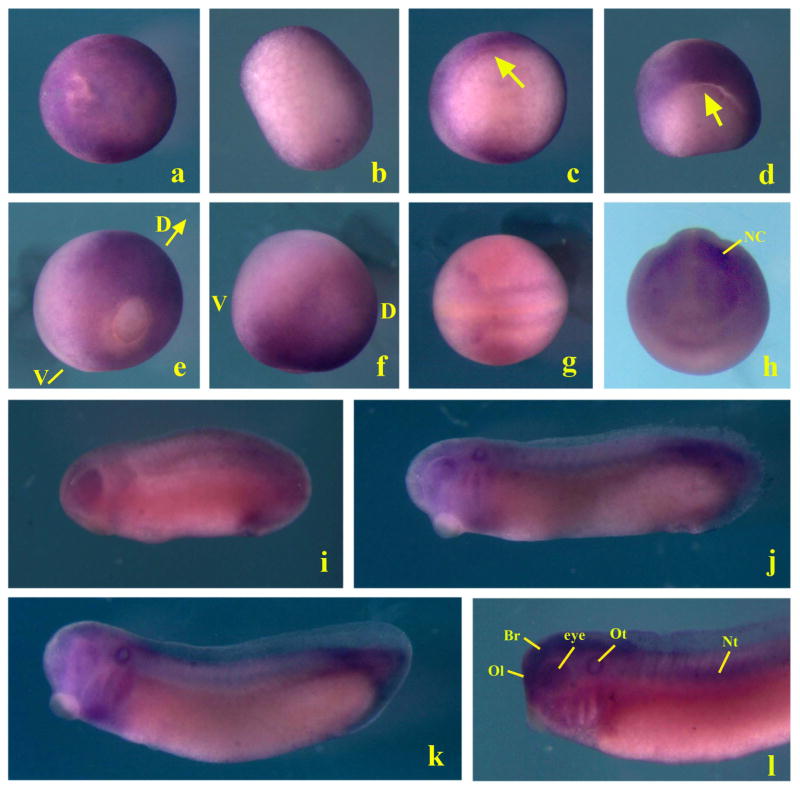
To understand the function of Smurf2 in early Xenopus development, we first analyzed its expression at different developmental stages by whole-mount in situ hybridization. At late blastula and early gastrula stages, Smurf2 transcripts were evenly distributed in the animal and the marginal regions without obvious dorsal-ventral differences (Fig. 1, panels a-c). By mid- to late gastrula stages, Smurf2 transcripts were reduced in the ventral region, so that high expression was observed only in the dorsal ectodermal and mesodermal domains (Fig. 1, panels d-f). At neurula stages, Smurf2 was predominately expressed in the neural and the neural crest cells (Fig. 1, panels g and h). As development progressed, Smurf2 continued to express in the neural tissues and the migrating neural crest, and was seen also in the sensory organs, such as the eyes, the olfactory placodes, and the otic vesicles. Expression in the notochord and the tail was observed in late stage embryos (Fig. 1, panels i-l). The wide expression of Smurf2 indicated that the gene might be involved in regulation of neural, neural crest as well as mesendodermal development.
Figure 1. Expression pattern of Smurf2 during early Xenopus development.
Whole mount in situ hybridization shows that Smurf2 transcripts are uniformly localized to the ectodermal and mesodermal regions in blastula (panels a and b, stage 9 embryos with animal and vegetal views, respectively) and early gastrula (c, stage 10 vegetal view with the arrow pointing to the blastopore lip) embryos. At mid- to late gastrula stages, its expression is enhanced in the dorsal region (panels d and e, stage 11 and 12 embryos, posterior view with dorsal side on top; and panel f, stage 12 embryo lateral view; D: dorsal; V, ventral). During neurulation, Smurf2 is seen in the neural plate and the migration neural crest cells (panels g and h, dorsal and anterior views respectively; NC, neural crest). At the tailbud to tadpole stages, Smurf2 transcripts are detected in the neural tissues, the olfactory and otic vesicles, eyes, migrating neural crest, notochord, and tail (panels i to l, lateral view with the head on the left side; Br: brain; Nt: notochord; Ol, Olfactory placode; Ot: otic vesicle).
Ectopic expression of Smurf2 in the ectoderm affects neural and neural crest development
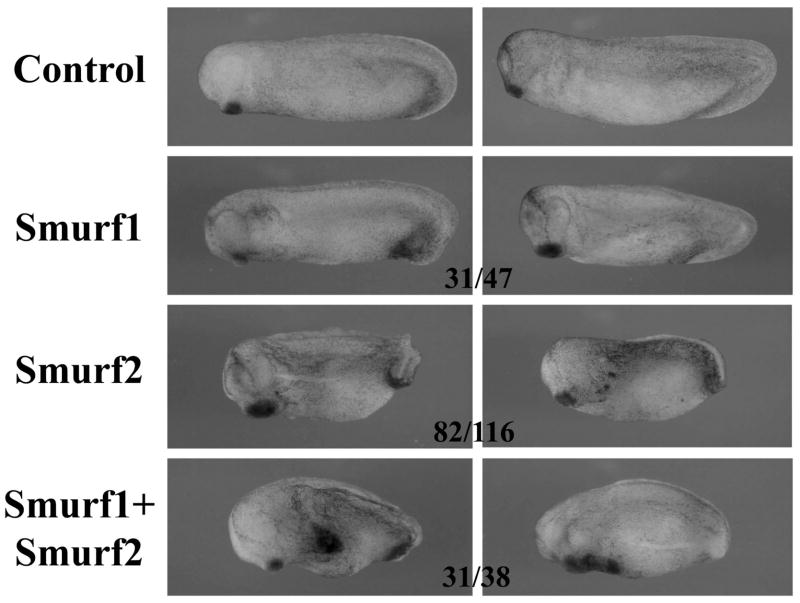
To address the role of Smurf2 in early frog embryogenesis, we first used the gain-of-function approach to examine the effect of increased levels of Smurf2 on early frog development. We thus injected the RNA encoding Smurf2 into the animal region of one- to two-cell stage embryos and analyzed the morphology of the resulting embryos at tailbud to tadpole stages. As shown in Fig. 2, overexpression of Smurf2 resulted in embryos with enlarged or ectopic cement gland, enlarged head, malformation of the eyes, and reduced body axis. When compared with the activities of Smurf1 that was injected into the animal region at the same dose and the same time, we found that elevated expression of Smurf1 also caused enlargement of the cement gland and the head, but the effect was milder than that induced by Smurf2 in that it rarely generated ectopic cement gland. When the RNAs encoding the two Smurfs were co-injected, the embryos displayed a more severe phenotype, with the majority of them having ectopic cement gland and reduced or bent body axis (Fig. 2 and data not shown). The results indicate that Smurf1 and Smurf2 collaborate in the ectoderm to control early embryogenesis.
Figure 2. Overexpression of Smurfs in the ectoderm induces enlarged or ectopic cement gland and enlarged head.
RNAs encoding Smurf1 or 2 were injected into the animal region of early frog embryos and the resulting tailbud embryos are shown here. The numbers indicate the embryos displaying the phenotype.
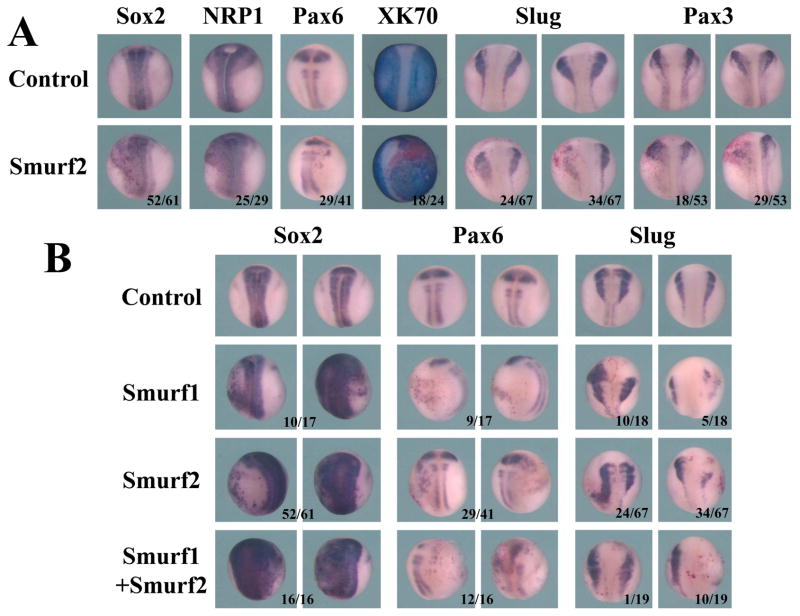
Since cement gland and head formation is often associated with early neural specification at low BMP signaling in the ectoderm, we next examined whether overexpression of Smurfs influenced the ectodermal cell differentiation. We thus performed whole-mount in situ hybridization to assess the expression of the neural, neural crest, and epidermal cell markers. To facilitate direct comparison of Smurf-expressing versus control tissues, we injected Smurf RNAs with a lineage tracer, the RNA encoding nuclear beta-galactosidase, into the animal region of one blastomere of 2-cell stage embryos. The embryos were then collected at the neurula stages, stained with the beta-galactosidase substrate Red-Gal to mark the injected side, and subjected to in situ hybridization. The side of the embryos that received Smurf2 injection consistently showed expansion or ectopic expression of the neural markers Sox2, NRP1 and Pax6 in the lateral epidermal regions, and this was accompanied by a reduction in the expression of the epidermal marker epidermal keratin XK70 (Fig. 3A). Interestingly, when we examined the markers expressed at the neural plate border, including the neural crest marker Slug and the neural crest/hatching gland marker Pax3, we found that the embryos exhibited two different expression patterns. One group of the embryos showed expansion of Slug and Pax3 domains, while the other group displayed reduced marker expression (Fig. 3A). Our data thus imply that Smurf2 dynamically modulates neural plate border genes.
Figure 3. Elevation of Smurf levels affects neural and neural crest development.
A) Increased levels of Smurf2 led to expansion or ectopic expression of the neural genes Sox2, NRP1 and Pax6 and reduction of the epidermal marker XK70, but could both up- and down-regulate the neural plate border genes Slug and Pax3. B) Smurf1 and Smurf2 function similarly to control neural and neural crest development; and together they facilitated expansion of the neural genes but reduction of the neural crest marker Slug. The numbers of the embryos displaying the shown changes in marker expression are indicated in the figure.
To investigate whether Smurf1 resembles Smurf2 in control of neural and neural crest differentiation, we repeated the experiment using Smurf1 RNA. Like Smurf2, Smurf1 induced the expression of the neural markers Sox2 and Pax6 in the epidermal region, and it could also both enhance and repress the expression of the neural crest marker Slug (Fig. 3B). When we coinjected Smurf1 and 2 RNAs, we discovered that the neural markers were expanded and the neural crest marker Slug was predominately inhibited (Fig. 3B). These results suggest that Smurf1 and Smurf2 act in a similar manner to regulate neural and neural crest genes during early frog development, and their control of the neural plate border genes requires a delicate balance of the protein levels of their targets.
Smurf2 overexpression impairs mesodermal development
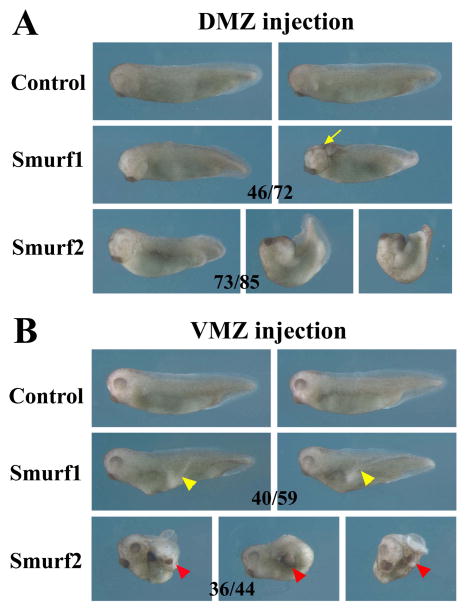
Smurf2, but not Smurf1, is shown to facilitate Smad2 degradation and Smad3 mono-ubiquitination to block activin/nodal branch of the TGF-β signals. However, this effect may be cell type- and dose-dependent, as Smad2 and Smad3 do not always appear to be the preferred targets of Smurf2 (Bonni et al., 2001; Zhang et al., 2001). To see whether Smurf2 can regulate activin/nodal as well as BMP signaling during early Xenopus development, we performed targeted injection of Smurf2 RNA into the dorsal or the ventral marginal zone (D/VMZ) of 4-cell stage embryos. Tissues from these regions give rise to dorsal-anterior and ventral-posterior mesendoderm, respectively, in activin/nodal- and BMP-dependent manners. When Smurf2 was ectopically expressed in the DMZ, embryos developed bent and reduced body axes, and many also had split dorsal axes indicative of gastrulation defects (Fig. 4A). On the other hand, overexpression of Smurf2 in the VMZ induced an incomplete secondary axis in many embryos. Intriguingly, the ectopic axis often contained tissues derived from the head, such as the cement gland and the hatching gland, and eyes were also present in a small percentage of the embryos (Fig. 4B and data not shown). The morphology of the embryos differed from those with elevated levels of Smurf1. Ectopic expression of Smurf1 in the DMZ resulted in reduction of the tail and malformation of the head in the tadpoles, with frequent occurrence of anterior neural tube closure defects; whereas enhanced Smurf1 in the VMZ led to the formation of a partial secondary axis that lacked any head tissues (Fig. 4). Co-injection of RNAs encoding the two Smurfs in the DMZ induced severe gastrulation defects in embryos (18/19 surviving tadpoles with gastrulation defects), whereas co-injection of these RNAs in the VMZ induced partial ectopic axes that often contained head structures in the resulting tadpoles (22/35 embryos with a partial secondary axis; among these, 14 had ectopic cement gland; not shown). The phenotypes of Smurf-overexpressing embryos reveal that though both Smurfs can alter mesodermal patterning and development, they have differential functions in this process.
Figure 4. Smurf1 and Smurf2 play different roles in mesodermal development.
A) Increased Smurf1 expression in dorsal mesoderm results in dorsal-anteriorization of the embryos (46/72 embryos), which often also show anterior neural tube closure defects (in 18 embryos; arrow points to the open brain). In contrast, increased Smurf2 expression in the dorsal mesoderm leads to disruption of axial structures, with embryos frequently displaying gastrulation defects (73/85 embryos with gastrulation defects; among these, 32 embryos had split dorsal structures characterized as the open back phenotype). B) Ectopic expression of Smurf1 in the VMZ induces a partial secondary dorsal axis with the trunk tissues (40/59 embryos; yellow arrowheads); whereas ectopic expression of Smurf2 in the VMZ results in induction of a partial secondary axis that often contains cement gland and hatching gland (36/44 embryos with an ectopic axis, and 25 of these contained cement gland; red arrowheads).
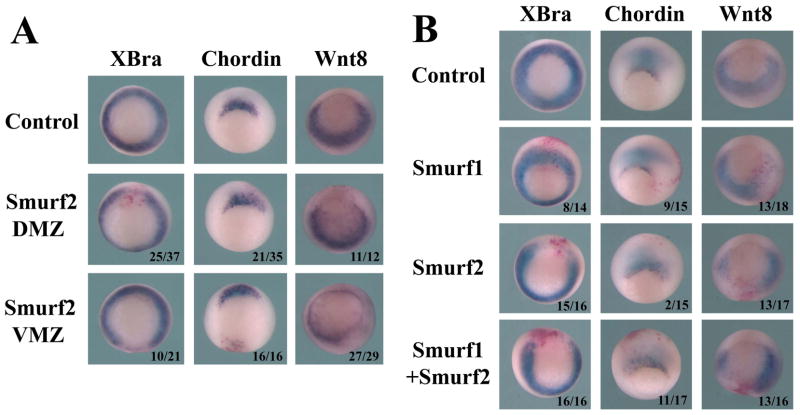
To understand how mesodermal genes were affected by Smurf2 overexpression, we next performed in situ hybridization to evaluate mesodermal marker expression. Elevation of Smurf2 levels in the mesoderm, either in the dorsal or the ventral region, disrupted the expression of the pan-mesodermal gene Brachyury (XBra). Smurf2 also weakly down-regulated the dorsal mesodermal marker chordin and inhibited the ventral-lateral mesodermal gene wnt8 when it was overexpressed in these regions (Fig. 5A). Interestingly, though ventral injection of Smurf2 induced a partial secondary axis, chordin expression was not robustly turned on in these embryos (Fig. 5A).
Figure 5. Smurf1 and Smurf2 differentially regulate mesodermal markers at gastrula stages.
A) Overexpression of Smurf2 in either DMZ or VMZ disrupts expression of the pan-mesodermal marker Brachyury (XBra). Dorsal expression of Smurf2 slightly reduces the dorsal mesoderm marker chordin, whereas ventral expression of Smurf2 inhibits the ventral-lateral mesodermal marker wnt8. B) Unlike Smurf2, Smurf1 does not block, but instead mildly expands, the expression of XBra or chordin; though like Smurf2, it also inhibits wnt8 expression. Co-expression of Smurf1 and 2 leads to reduction of all three mesodermal markers. The numbers indicate the embryos displaying the shown changes in marker expression.
To compare the activities of Smurf1 and Smurf2, we also examined the embryos injected with Smurf1 RNA. In this experiment, Smurf1 or Smurf2 RNA was injected into the marginal zone region of one blastomere of 2-cell stage embryos, and mesodermal marker expression was analyzed at the gastrula stages by in situ hybridization. Unlike Smurf2, enhanced Smurf1 levels did not block XBra or chordin expression, but instead mildly expanded the expression of these genes. Similarly to Smurf2, Smurf1 also inhibited wnt8 expression (Fig. 5B). When the two Smurfs were co-expressed, all three mesodermal markers were down-regulated, though the effect on chordin expression was weak (Fig. 5B). The data illustrate that in contrast to cell fate regulation in the ectoderm where Smurf1 and Smurf2 act similarly, regulation of cell differentiation in the mesoderm by Smurf1 and Smurf2 seems to differ, and this is likely caused by differential modulation of different targets by the two Smurfs.
Reduction of endogenous Smurf2 levels interferes with early embryonic development
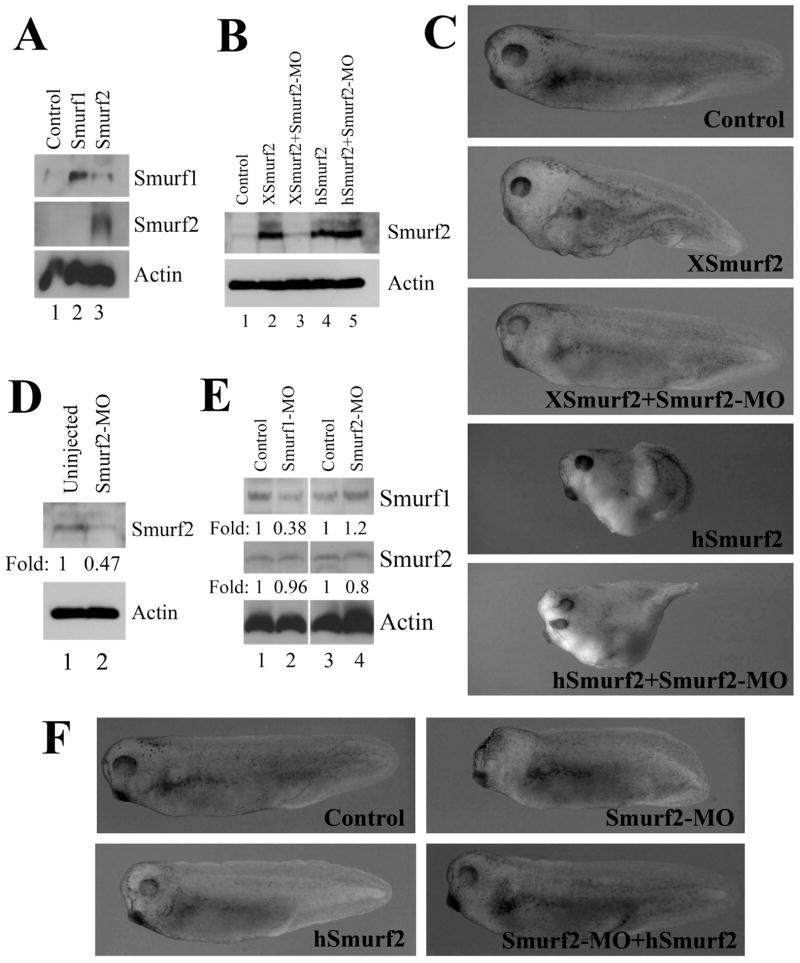
To complement the gain-of-function studies, we next adopted the loss-of-function approach to reduce the endogenous levels of Smurf2 and analyze its effect on early Xenopus embryogenesis. We used the antisense morpholino oligonucleotide (MO) – mediated knockdown method to decrease endogenous Smurf2 protein production. Two Xenopus laevis Smurf2 transcript sequences were found in the database, and they differ in their upstream 5’-untranslated regions. We therefore designed a Smurf2-MO that targeted the conserved sequence around the translational start site of both alleles, so that it could block translation of Smurf2 from both transcripts. When injected into early frog embryos, Smurf2-MO efficiently prevented protein translation from the introduced RNA encoding Xenopus Smurf2, as detected by a Smurf2-specific antibody; but it had no effect on protein production from the RNA that encoded human Smurf2 (Fig. 6A, B). In agreement with this, Smurf2-MO inhibited the induction of a secondary axis by XSmurf2, but it did not block secondary axis induction by hSmurf2 (Fig. 6C). Smurf2-MO also reduced the endogenous Smurf2 protein up to about 50% of its original level at the gastrula stages, and there was no appreciable degree of cross-regulation between Smurf1 and Smurf2 at the protein level (Fig. 6D, E). When injected into one-cell stage embryos, Smurf2-MO caused reduction of the head structures with malformation of the eyes in the morphants, and the defects were mainly rescued by co-expression of low doses (280pg) of hSmurf2 (Fig. 6F). Taken together, these results demonstrate that Smurf2-MO specifically blocks translation of Xenopus Smurf2 to influence early frog embryogenesis.
Figure 6. Smurf2 antisense morpholino oligonucleotides (Smurf2-MO) acts specifically to block translation and function of Xenopus Smurf2.
A) Smurf2 antibody recognizes Xenopus Smurf2, but not Smurf1. B) Smurf2-MO inhibits translation from the RNA encoding Xenopus Smurf2 (XSmurf2), but has no effect on translation of human Smurf2 (hSmurf2). C) Smurf2-MO blocks induction of a partial secondary axis by XSmurf2, but not by hSmurf2. D) Smurf2-MO reduces endogenous Smurf2 protein levels at the gastrula stages. E) Smurf1-MO does not regulate the protein level of Smurf2. F) Ectodermal expression of Smurf2-MO induces head defects, which are partially rescued by low doses of co-expressed hSmurf2.
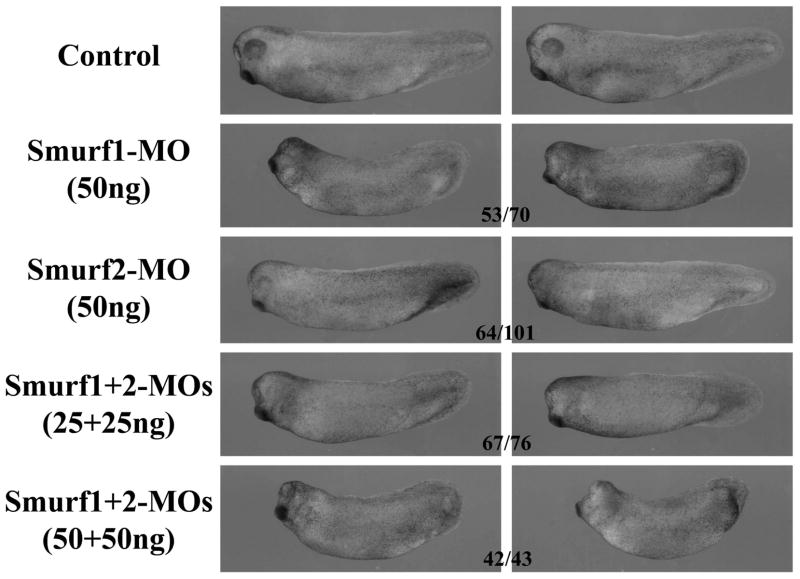
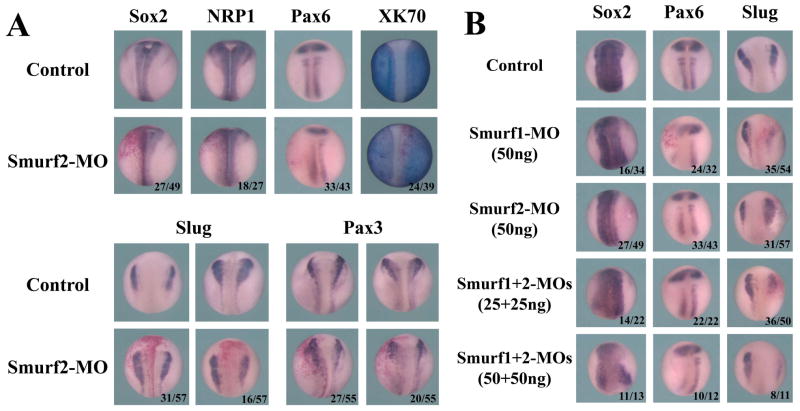
Since ectopic expression of Smurf2 expanded neural tissues in early embryos, we next asked whether knockdown of Smurf2 could disrupt neural development. We thus injected Smurf2-MO into the animal region of early embryos. Smurf2 morphants displayed head and eye malformation, a phenotype often associated with reduced anterior neural tissues (Fig. 7). When injected at the same dose, Smurf1-MO, which has previously been shown to be specific (Alexandrova and Thomsen, 2006), induced more severe defects in the head (Fig. 7), a result consistent with the previous reports that Smurf1 regulated neural development in early Xenopus embryos (Zhu et al., 1999; Alexandrova and Thomsen, 2006). Double knockdown of both Smurf1 and Smurf2 also resulted in embryos with reduced head and malformed eyes, and the effects seemed to be dose-dependent (Fig. 7). Whole-mount in situ hybridization of marker expression showed that knockdown of Smurf2 weakly decreased the expression of the neural genes Sox2, NRP1 and Pax6, and slightly expanded the domain of the epidermal gene XK70. Similar to the expression patterns observed in Smurf2-overexpressing embryos, the expression of the neural plate border genes Slug and Pax3 could also be enhanced or reduced slightly in Smurf2 morphants, though most morphants showed increased expression of these markers (Fig. 8A). This contrasted with the patterns observed in Smurf1 knockdown embryos where the expression of the neural crest markers was predominately decreased (Fig. 8B). Depletion of Smurf1 also resulted in somewhat wider and fainter domain of Sox2 and reduced Pax6 staining (Fig. 8B), and this might underlie the delay in neural fold closure and the eye defects in Smurf1 morphants (Fig. 7 and data not shown; also see Alexandrova and Thomsen, 2006). In Smurf1 and Smurf2 double knockdown embryos, Sox2 expression domain was disrupted, Pax6 was decreased, and Slug was preferentially inhibited (Fig. 8B). Data from these experiments suggest that both Smurfs participate in modulation of neural development and head formation during early Xenopus embryogenesis.
Figure 7. Smurf1 and Smurf2 are both required for normal head development in early Xenopus embryos.
Knockdown of either Smurf led to malformation of the head in the resulting tadpoles, with Smurf1 morphants displaying a more severe phenotype. Depletion of both Smurfs exacerbated the head defects, suggesting that the two Smurfs have redundant activities in regulation of head development. The numbers indicate the embryos showing the phenotype.
Figure 8. Smurfs regulate ectodermal marker expression.
A) Knockdown of Smurf2 slightly reduced the neural marker expression and expanded the epidermal marker XK70, but the neural plate border genes Slug and Pax3 could be both up- and down-regulated in the morphants. B) Knockdown of Smurf1 resulted in slightly reduced Sox2 in a wider domain and decreased Pax6 expression; Slug was also predominantly reduced (10/54 and 35/54 embryos with enhanced and decreased Slug expression, respectively). Depletion of both Smurf1 and Smurf2 caused reduction and disorganization of the Sox2 domain, and decreased expression of both Pax6 and Slug (8/50 and 36/50 embryos, respectively, with enhanced and reduced Slug in 25ng combination of both MOs, and no embryos showed higher expression of Slug in double morphants with 50ng combination of the MOs).
Smurf2 regulates mesendodermal development in early Xenopus embryos
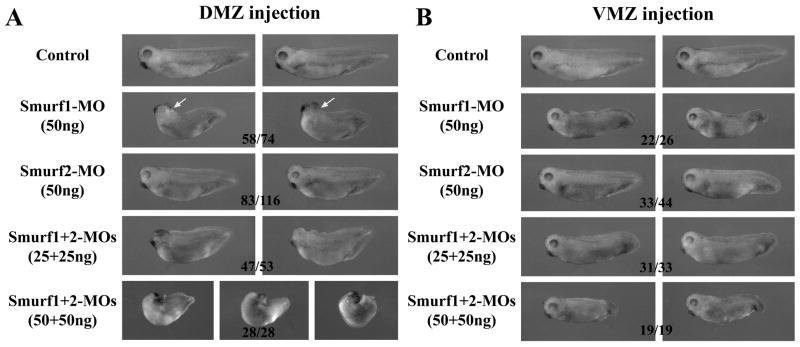
As Smurf2 transcripts were detected in both the ectoderm and the mesoderm in early frog embryos, we addressed whether Smurf2 played a role in regulation of mesoderm development. We thus performed targeted injection of Smurf2-MO into either dorsal or ventral marginal zone (DMZ and VMZ) region of 4-cell stage embryos. Knockdown of Smurf2 in the dorsal mesoderm led to consistent enlargement of endodermal mass in the resulting tadpoles. In contrast, knockdown of Smurf1 in DMZ resulted in embryos with a delay or failure in neural tube closure that was more prominent than ectodermal depletion of Smurf1. The resulting tadpoles displayed small head and bent body axis (Fig. 9A). When Smurf1 and Smurf2 were simultaneously knocked down, the embryos invariably developed microcephaly and axial defects. At high doses of the MOs, the embryos failed to complete gastrulation and had bent and split axial structures (Fig. 9A). Ventral depletion of Smurf2 did not affect head or endodermal tissues, but the resulting tadpoles had truncated tails compared with their uninjected siblings. Similar but more severe defects in tail extension were also observed in Smurf1-ventrally-depleted embryos. When Smurf1 and Smurf2 were both knocked down in the VMZ, the tadpoles failed to develop a tail (Fig. 9B).
Figure 9. Smurf1 and Smurf2 control mesendodermal development in early Xenopus embryos.
A) Knockdown of Smurf1 in dorsal tissues resulted in reduction of axial structures and malformation of the head, which was often accompanied by a delay or failure in neural tube closure (32 out of the 58 embryos with the shown phenotype had neural tube closure defects, arrow). In contrast, knockdown of Smurf2 led to enlargement of the endodermal mass in the embryos. When both Smurfs were knocked down, the embryos developed with severe axial defects, microcephaly, and failure in gastrulation. B) Knockdown of either Smurf1 or Smurf2 in the VMZ region induced tail truncation, though the phenotype was more pronounced in the Smurf1 morphants. Double knockdown of both Smurfs aggravated the tail defects. The numbers indicate the embryos showing the depicted phenotype.
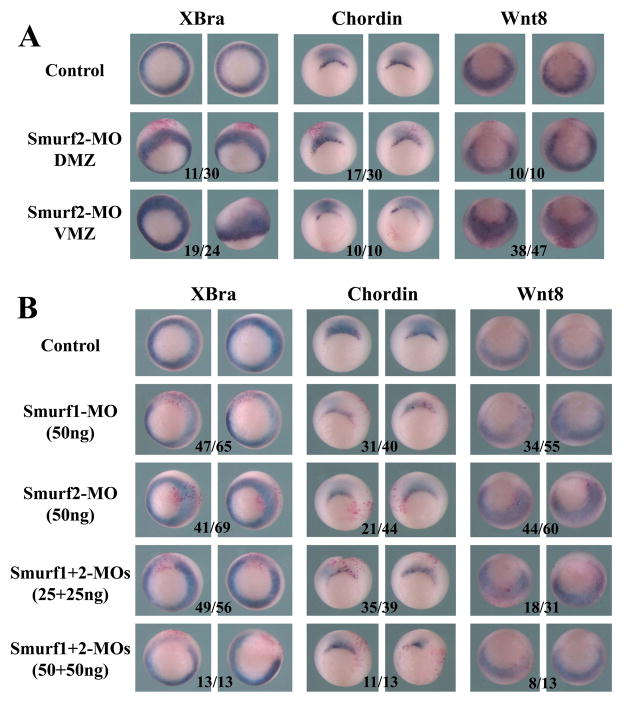
To gain insight into the defects of Smurf morphants at the molecular level, we performed in situ hybridization to analyze mesodermal marker expression at the gastrula stages. Depletion of Smurf2 in either DMZ or VMZ resulted in expansion of the pan-mesodermal marker XBra towards the animal pole, and this effect was more obvious when the Smurf2 level was reduced in the VMZ (Fig. 10A). In addition, knockdown of Smurf2 in the DMZ weakly decreased the expression of the dorsal marker chordin, but knockdown of Smurf2 in the VMZ expanded the domain of the ventral-lateral mesoderm marker wnt8 (Fig. 10A). To compare the function of Smurf1 and Smurf2, we next examined the embryos injected with Smurf1-MO or Smurf2-MO into the MZ of one blastomere of 2-cell stage embryos. Interestingly, unlike Smurf2 knockdown, Smurf1 knockdown reduced the expression of Xbra; but similarly to Smurf2 knockdown, Smurf1 knockdown also resulted in decreased expression of chordin (Fig. 10B). When both Smurf1 and Smurf2 were depleted, expression of Xbra and chordin were both down-regulated (Fig. 10B). The results indicate that both Smurf1 and Smurf2 regulate mesodermal development, but they have distinct as well as redundant functions in this process.
Figure 10. Smurf1 and Smurf2 differentially regulate mesodermal marker expression.
A) Knockdown of Smurf2 in the marginal zone region caused expansion of the pan-mesodermal marker XBra towards the ectodermal region. Dorsal knockdown of Smurf2 reduced expression of chordin, whereas ventral knockdown of Smurf2 led to expansion of wnt8 into the ectodermal domain. B) Unlike Smurf2 knockdown, Smurf1 knockdown reduced the expression of both XBra and chordin, and double knockdown of both Smurfs resulted in more efficient reduction of these markers. The numbers indicate the embryos displaying the shown changes of marker expression.
Discussion
Smurf1 and Smurf2 are E3 ubiquitin ligases that were originally identified as negative regulators of the TGF-β signaling. However, subsequent studies reveal that they can not only reduce, but also enhance, TGF-β signals, depending on the preferred proteins they target in a particular cellular environment (Inoue and Imamura, 2008; Soond and Chantry, 2011). Moreover, Smurfs have additional substrates that are not involved in the TGF-β pathway. By regulating stability and/or activities of their substrate proteins, Smurfs participate in a variety of biological processes, such as cell proliferation, senescence, migration, differentiation, and polarity (Zhao et al., 2003, 2010; Yamashita et al., 2005; Sahai et al., 2007; Schwamborn et al., 2007; Cheng et al., 2011; Kong et al., 2011). The roles of Smurfs during early vertebrate embryogenesis, however, are not completely understood.
In this study, we used the animal model of African clawed frog, Xenopus laevis, and both gain- and loss-of-function approaches to address how Smurf2 regulates early development of frog embryos and whether Smurf1 and Smurf2 have similar or distinct activities. We describe that similarly to that reported for Smurf1, Smurf2 is expressed in the ectoderm and the mesoderm in early frog embryos. However, the expression patterns of the two genes diverge at later stages. Transcripts of both genes are detected in the central nervous system, eyes, otic vesicles, neural crest derivatives, and the notochord, but Smurf1 alone is also detected in the kidney and the somites (Zhu et al., 1999; Alexandrova and Thomsen, 2006). Functionally, both Smurfs modulate mesodermal and neural development during Xenopus embryogenesis, but they do not simply have the same and redundant activities. As discussed below, our results help to shed light on the following issues.
1. Smurf2 likely modulates both activin/nodal and BMP branches of the TGF-β signals during early vertebrate development
One issue that remained unresolved is whether Smurf2 regulates TGF-β/activin/nodal branch of the signal in vivo. Though it has been reported that Smurf2 can facilitate degradation of Smad1 and Smad2 by ubiquitin-dependent machinery to terminate all TGF-β signaling, some groups suggest that Smurf2 is ineffective towards Smad2 but promotes Smad1 clearance efficiently (Lin et al., 2000; Bonni et al., 2001; Zhang et al., 2001; Tan et al., 2008). Subsequently, it is shown that Smurf2 can induce multiple mono-ubiquitination of Smad3 to block the TGF-β pathway without down-regulation of Smad3 protein level (Tang et al., 2011). In addition, though both Smurf1 and Smurf2 have been shown to facilitate degradation of TGF-β receptors in mammalian cells (Kavsak et al., 2000; Ebisawa et al., 2001), knockdown or knockout of Smurf1 does not affect TGF-β/nodal signaling (Yamashita et al., 2005; Alexandrova and Thomsen, 2006). It is thus unclear whether during vertebrate embryogenesis Smurf2 modulates both activin/nodal and BMP signals. Data from our studies imply that Smurf2 likely regulates both branches of the TGF-β pathway during early Xenopus development. Elevation of Smurf2 levels leads to disruption of mesodermal induction, as evident from the inhibition of the pan-mesodermal marker Brachyury and the development of the axial defects in the resulting embryos. Conversely, knockdown of Smurf2 results in expansion of the Brachyury domain toward the ectodermal region in gastrula embryos and enlargement of the endoderm mass in tadpoles of the morphants. As induction of the mesoderm and the endoderm in Xenopus relies on activin/nodal signaling (Whitman 2001; Schier 2003), our results indicate that Smurf2 likely modulates this pathway to control mesoderm development. This conclusion is consistent with our observation that overexpression of Smurf2 in the VMZ induces a partial secondary axis with the head structures, and this is achieved without robust induction of ectopic dorsal mesoderm as indicated by the lack of ectopic chordin expression. Induction of a secondary neural axis with the head tissues by simultaneous inhibition of both nodal and BMP signals has been documented previously (Piccolo et al., 1999), and the similar phenotypes we observe here are in agreement with the notion that Smurf2 blocks both Smad1- and Smad2-mediated signaling pathways. Examination of Smad2 protein levels in embryos overexpressing Smurf2 revealed no difference from that in the control embryos (data not shown), suggesting that Smurf2 may utilize a mechanism other than Smad2 degradation to modulate activin/nodal signals.
2. Smurf1 and Smurf2 have overlapping and distinct activities during early frog embryogenesis
Another issue that deserves attention is whether Smurf1 and Smurf2 act similarly to affect vertebrate embryogenesis. Many proteins have been identified as common substrates for both Smurfs (e.g. Smad1/5 or TGF-β type I receptor), while others have not been characterized in parallel for the two Smurfs (e.g. TRAF4). Though a limited number of unique targets for each Smurf have also been found (e.g. Smad2/3 for Smurf2), it is unclear whether during early vertebrate development the two Smurfs have redundant or distinct functions in particular tissues. In mice, targeted deletion of either Smurf gene does not instigate developmental defects in early embryos, suggesting that the functions of the two Smurfs are redundant and can compensate for each other (Yamashita et al., 2005; Tang et al., 2011). However, our data suggest that in Xenopus Smurf1 and Smurf2 do not always exert similar effects, and both are required for normal development of Xenopus embryos. Overexpression of Smurf1 does not disrupt mesodermal induction; instead it alters mesodermal patterning, so that the ventral-posterior marker wnt8 is inhibited and a secondary dorsal axis containing the trunk tissue is induced. Knockdown of Smurf1 in the DMZ leads to reduction of the head and the axial structures, a phenotype also seen in embryos with enhanced BMP signaling (Harland and Gerhart, 1997; De Robertis and Kuroda, 2004; Alexandrova and Thomsen, 2006) and is different from that resulted from Smurf2 knockdown in the DMZ. These results are compatible with the idea that Smurf1 promotes degradation of Smad1 but not Smad2 and suggest that Smurf1 can function differently from Smurf2 in regulation of early frog embryogenesis.
3. Smurf1 and Smurf2 likely regulate multiple substrates to influence cell fate determination in the ectoderm and the mesoderm
A third important issue pertains to the substrates that Smurfs regulate during early Xenopus development. We propose that Smurfs control both positive and negative regulators of the TGF-β signals in a tissue-dependent manner, and Smurfs also modulate additional protein substrates that are not in the TGF-β pathway to influence developmental processes in Xenopus. These points are relevant in both ectodermal and mesodermal development. In the ectoderm, overexpression or knockdown of either Smurf can both enhance and inhibit the expression of the neural crest marker Slug, and this is achieved without overt alteration in expression of the somitic mesodermal marker myoD (not shown). This phenotype has not been reported for other regulators of the neural crest and suggests a critical requirement for fine balances of signals that control neural crest development. The mechanism underlying such delicate balance of signals is likely through dynamic modulation of Smurf targets temporally and spatially. It has been reported that a BMP signaling gradient in the ectoderm determines the status of cell differentiation, with the formation of the epidermis at high BMP levels, the induction of the neural fate at low BMP levels, and the generation of the neural crest at immediate BMP levels (Baker and Bronner-Fraser, 1997; Aybar and Mayor, 2002; Barembaum and Bronner-Fraser, 2005). Smurfs can modulate both positive and negative factors in the BMP pathway, thus it is possible to imagine that Smurfs may simultaneously regulate R-Smads/TRAF4 to reduce, and I-Smads/Ski/SnoN to enhance, BMP signaling in the neural plate border region. As Smurfs likely have differential capacity to modulate the stability/activity of these substrates that may also be engaged in feedback regulation among themselves, the doses and the timing of Smurfs expressed in the region will determine the ratio of positive versus negative factors of the BMP pathway present in this domain, hence the expansion or the reduction of the neural crest markers. Besides modulation of the BMP signaling, we can envision an alternative scenario in which Smurfs regulate proteins that do not act in the TGF-β/BMP pathway. For example, Wnt signaling has been implicated in neural crest induction in Xenopus (Saint-Jeannet et al., 1997; Chang and Hemmati-Brivanlou, 1998b; LaBonne and Bronner-Fraser, 1998), and a crucial inhibitory component of the Wnt signal transduction, GSK-3β, is a target of Smurf2 in chondrocyte. By promoting GSK-3β degradation, Smurf2 helps to stabilize β-catenin and stimulate canonical Wnt signal transduction (Wu et al., 2008, 2009). It is possible that Smurfs may also promote degradation of GSK-3β in the neural crest cells to up-regulate Wnt signaling, while at the same time facilitate clearance of R-Smads/TRAF4 to inhibit BMP signals. As the affinity and/or kinetics of Smurfs towards these substrates likely differ, the doses of Smurfs present at the neural plate border region will influence the balance between the Wnt and the BMP signals, which in turn affects the expression of the neural crest markers. In addition to regulating cell fate in the ectoderm, Smurf1, but not Smurf2, seems to control neural morphogenesis. Dorsal expression of Smurf1 or Smurf1-MO induces a delay or a failure in neural tube closure. Since this phenotype has not been associated with altered levels of BMP signaling, it is likely that Smurf1 acts on other proteins to modulate this process. The most probable target is Prickle1, a component in the planar cell polarity pathway that is shown to be regulated by Smurfs in mouse to control neural tube closure (Narimatsu et al., 2009). The other possible target is RhoA, a Smurf1 substrate that regulates neuronal cell polarity and cancer cell migration (Sahai et al., 2007; Tian et al., 2011). Smurfs therefore seem to modulate both TGF-β/BMP signaling components and other proteins to control ectodermal cell differentiation and morphogenesis.
In the mesoderm, Smurfs also likely modulate multiple proteins to influence induction and patterning of this germ layer. Knockdown of either Smurf1 or Smurf2 in the VMZ leads to tail truncation of the morphants, and this phenotype has not been reported when nodal or BMP signaling is enhanced in this region. The results imply that in addition to inhibition of nodal and BMP pathways in the VMZ, as indicated by expansion of XBra and/or wnt8 domain in Smurf VMZ morphants, Smurfs may also regulate other substrates to control ventral-posterior tissue formation. Similarly, when Smurf1 is depleted in the mesoderm, the pan-mesodermal marker Brachyury is reduced. This result has not been seen with enhanced BMP signaling and again suggests that substrates other than R-Smads and TRAF4 are involved. Interestingly, though Brachyury is expanded into the ectodermal region in Smurf2 morphants, this marker expression is reduced in the mesoderm of Smurf1 and Smurf2 double knockdown embryos. This indicates that a mesodermal inhibitory molecule is stabilized when both Smurfs are depleted. It is possible that I-Smads (Smad6 and 7) or Ski/SnoN, all of which are present during the gastrula stages in the mesoderm (Amaravadi et al., 1997; Bhushan et al., 1998; Casellas and Hemmati-Brivanlou, 1998; Nakayama et al., 1998; Seufert et al., 2005), are the candidate molecules that regulate mesoderm formation in these embryos. However, since the expression of Wnt8 is expanded rather than inhibited in Smurf morphants, it is likely that the BMP signaling is not dampened in these embryos. Other proteins may be affected by altered Smurf levels to regulate mesodermal induction and patterning.
Taken together, our studies reveal that Smurf1 and Smurf2 likely modulate both positive and negative factors in the TGF-β/BMP pathways as well as many other protein substrates, some of them may yet to be discovered, to regulate ectodermal and mesodermal development during early Xenopus embryogenesis. Smurf1 and Smurf2 have both redundant functions in regulation of neural and tail development as well as unique activities, with Smurf1 playing more important roles in neural tube closure and Smurf2 having essential function in controlling mesodermal induction. The totality of the biological roles of Smurfs in early vertebrate embryogenesis may be conserved across species, though the involvement of individual Smurfs in particular processes may differ. Future investigation is needed to tease out the exact substrate(s) involved in mediating the effects of Smurf overexpression and knockdown in particular tissue contexts during early Xenopus development.
Experimental Procedures
Plasmids construction and RNA synthesis
The full length Xenopus laevis Smurf2 gene was cloned by RT-PCR into the pCS105 vector, using the primers Smurf2-N: 5’-GGAATTCACCATGTCTAATCAGGGATCCCGGC-3’ and Smurf2-C: 5’-GCTCTAGATCATTCCACAGCAAATCCACAAGT-3’. For mRNA injection, the plasmid was linearized with AscI and transcribed using SP6 in vitro transcription kit (mMessage mMachine kit, Ambion). Unless otherwise stated, the results shown in the figures were from embryos injected with 2ng of RNAs. For whole-mount in situ hybridization of Smurf2, the plasmid was cut with EcoRI and the digoxygenin-UTP-labeled antisense RNA probe was synthesized using T3 RNA polymerase. Xenopus Smurf1 and human Smurf2 genes were used as described previously (Zhu et al., 1999; Zhang et al., 2001).
Embryo injection and processing for whole mount in situ hybridization
The X. laevis embryos were obtained and injected as described (Nie and Chang, 2007). The sequence of the antisense Smurf2 MO is: 5’-TCCCTGATTAGACATGGCACCGGGC-3’ (Gene Tools, LLC), which hybridizes to the -10 to +15 nucleotide position relative to translational start site of Smurf2. Injected embryos were collected at gastrula or neurula stages for whole mount in situ hybridization analyses of marker expression, or at tailbud to tadpole stages for phenotype examination. In situ hybridization was performed as described by Harland (1991), using the neural or the mesodermal probes described previously (Chang and Harland, 2007; Burn et al., 2011). Xenopus Smurf1-MO was used as previously reported (Alexandrova and Thomsen, 2006). All the experiments were repeated at least three times.
Western blot analyses
For Western blot analyses, injected or control embryos were collected at early gastrula stages for protein extraction. Western blot was performed using the anti-Smurf2 antibody (Santa Cruz Biotechnology) at the 1:200 dilution or anti-Smurf1 antibody (Abcam) at the 1:500 dilution.
Key findings.
Smurf2 regulates mesodermal induction and patterning, suggesting that it may control both nodal and BMP signals in the mesoderm in vivo
Smurf2 modulates neural development in collaboration with Smurf1, a result consistent with their roles in inhibition of BMP signals
Smurf2 can both up- and down-regulate neural crest markers, implying that it dynamically modulates its substrates temporally and/or spatially at the neural plate border
Smurf1 and Smurf2 have overlapping and distinct functions during early Xenopus embryogenesis
Acknowledgments
This work is supported by NIH grant R01 GM083029.
References
- Alexandrova EM, Thomsen GH. Smurf1 regulates neural patterning and folding in Xenopus embryos by antagonizing the BMP/Smad1 pathway. Dev Biol. 2006;299:398–410. doi: 10.1016/j.ydbio.2006.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaravadi LS, Neff AW, Sleeman JP, Smith RC. Autonomous neural axis formation by ectopic expression of the protooncogene c-ski. Dev Biol. 1997;192:392–404. doi: 10.1006/dbio.1997.8780. [DOI] [PubMed] [Google Scholar]
- Aybar MJ, Mayor R. Early induction of neural crest cells: lessons learned from frog, fish and chick. Curr Opin Genet Dev. 2002;12:452–458. doi: 10.1016/s0959-437x(02)00325-8. [DOI] [PubMed] [Google Scholar]
- Baker CV, Bronner-Fraser M. The origins of the neural crest. Part I: embryonic induction. Mech Dev. 1997;69:3–11. doi: 10.1016/s0925-4773(97)00132-9. [DOI] [PubMed] [Google Scholar]
- Barembaum M, Bronner-Fraser M. Early steps in neural crest specification. Semin Cell Dev Biol. 2005;16:642–646. doi: 10.1016/j.semcdb.2005.06.006. [DOI] [PubMed] [Google Scholar]
- Bhushan A, Chen Y, Vale W. Smad7 inhibits mesoderm formation and promotes neural cell fate in Xenopus embryos. Dev Biol. 1998;200:260–268. doi: 10.1006/dbio.1998.8965. [DOI] [PubMed] [Google Scholar]
- Bonni S, Wang HR, Causing CG, Kavsak P, Stroschein SL, Luo K, Wrana JL. TGF-beta induces assembly of a Smad2-Smurf2 ubiquitin ligase complex that targets SnoN for degradation. Nat Cell Biol. 2001;3:587–595. doi: 10.1038/35078562. [DOI] [PubMed] [Google Scholar]
- Burn B, Brown S, Chang C. Regulation of early Xenopus development by the PIAS genes. Dev Dyn. 240:2120–2126. doi: 10.1002/dvdy.22701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casellas R, Brivanlou AH. Xenopus Smad7 inhibits both the activin and BMP pathways and acts as a neural inducer. Dev Biol. 1998;198:1–12. doi: 10.1006/dbio.1998.8893. [DOI] [PubMed] [Google Scholar]
- Chang C, Harland RM. Neural induction requires continued suppression of both Smad1 and Smad2 signals during gastrulation. Development. 2007;134:3861–3872. doi: 10.1242/dev.007179. [DOI] [PubMed] [Google Scholar]
- Chang C, Hemmati-Brivanlou A. Cell fate determination in embryonic ectoderm. J Neurobiol. 1998a;36:128–151. [PubMed] [Google Scholar]
- Chang C, Hemmati-Brivanlou A. Neural crest induction by Xwnt7B in Xenopus. Dev Biol. 1998b;194:129–134. doi: 10.1006/dbio.1997.8820. [DOI] [PubMed] [Google Scholar]
- Cheng PL, Lu H, Shelly M, Gao H, Poo MM. Phosphorylation of E3 ligase Smurf1 switches its substrate preference in support of axon development. Neuron. 69:231–243. doi: 10.1016/j.neuron.2010.12.021. [DOI] [PubMed] [Google Scholar]
- De Robertis EM, Kuroda H. Dorsal-ventral patterning and neural induction in Xenopus embryos. Annu Rev Cell Dev Biol. 2004;20:285–308. doi: 10.1146/annurev.cellbio.20.011403.154124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebisawa T, Fukuchi M, Murakami G, Chiba T, Tanaka K, Imamura T, Miyazono K. Smurf1 interacts with transforming growth factor-beta type I receptor through Smad7 and induces receptor degradation. J Biol Chem. 2001;276:12477–12480. doi: 10.1074/jbc.C100008200. [DOI] [PubMed] [Google Scholar]
- Fukunaga E, Inoue Y, Komiya S, Horiguchi K, Goto K, Saitoh M, Miyazawa K, Koinuma D, Hanyu A, Imamura T. Smurf2 induces ubiquitin-dependent degradation of Smurf1 to prevent migration of breast cancer cells. J Biol Chem. 2008;283:35660–35667. doi: 10.1074/jbc.M710496200. [DOI] [PubMed] [Google Scholar]
- Harland R. Neural induction. Curr Opin Genet Dev. 2000;10:357–362. doi: 10.1016/s0959-437x(00)00096-4. [DOI] [PubMed] [Google Scholar]
- Harland R, Gerhart J. Formation and function of Spemann's organizer. Annu Rev Cell Dev Biol. 1997;13:611–667. doi: 10.1146/annurev.cellbio.13.1.611. [DOI] [PubMed] [Google Scholar]
- Harland RM. In situ hybridization: an improved whole-mount method for Xenopus embryos. Methods Cell Biol. 1991;36:685–695. doi: 10.1016/s0091-679x(08)60307-6. [DOI] [PubMed] [Google Scholar]
- Hata A, Lagna G, Massague J, Hemmati-Brivanlou A. Smad6 inhibits BMP/Smad1 signaling by specifically competing with the Smad4 tumor suppressor. Genes Dev. 1998;12:186–197. doi: 10.1101/gad.12.2.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi H, Abdollah S, Qiu Y, Cai J, Xu YY, Grinnell BW, Richardson MA, Topper JN, Gimbrone MA, Jr, Wrana JL, Falb D. The MAD-related protein Smad7 associates with the TGFbeta receptor and functions as an antagonist of TGFbeta signaling. Cell. 1997;89:1165–1173. doi: 10.1016/s0092-8674(00)80303-7. [DOI] [PubMed] [Google Scholar]
- Imamura T, Takase M, Nishihara A, Oeda E, Hanai J, Kawabata M, Miyazono K. Smad6 inhibits signalling by the TGF-beta superfamily. Nature. 1997;389:622–626. doi: 10.1038/39355. [DOI] [PubMed] [Google Scholar]
- Inoue Y, Imamura T. Regulation of TGF-beta family signaling by E3 ubiquitin ligases. Cancer Sci. 2008;99:2107–2112. doi: 10.1111/j.1349-7006.2008.00925.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalkan T, Iwasaki Y, Park CY, Thomsen GH. Tumor necrosis factor-receptor-associated factor-4 is a positive regulator of transforming growth factor-beta signaling that affects neural crest formation. Mol Biol Cell. 2009;20:3436–3450. doi: 10.1091/mbc.E08-03-0325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavsak P, Rasmussen RK, Causing CG, Bonni S, Zhu H, Thomsen GH, Wrana JL. Smad7 binds to Smurf2 to form an E3 ubiquitin ligase that targets the TGF beta receptor for degradation. Mol Cell. 2000;6:1365–1375. doi: 10.1016/s1097-2765(00)00134-9. [DOI] [PubMed] [Google Scholar]
- Kong Y, Cui H, Zhang H. Smurf2-mediated ubiquitination and degradation of Id1 regulates p16 expression during senescence. Aging Cell. 10:1038–1046. doi: 10.1111/j.1474-9726.2011.00746.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaBonne C, Bronner-Fraser M. Neural crest induction in Xenopus: evidence for a two-signal model. Development. 1998;125:2403–2414. doi: 10.1242/dev.125.13.2403. [DOI] [PubMed] [Google Scholar]
- Lin X, Liang M, Feng XH. Smurf2 is a ubiquitin E3 ligase mediating proteasome-dependent degradation of Smad2 in transforming growth factor-beta signaling. J Biol Chem. 2000;275:36818–36822. doi: 10.1074/jbc.C000580200. [DOI] [PubMed] [Google Scholar]
- Liu X, Sun Y, Weinberg RA, Lodish HF. Ski/Sno and TGF-beta signaling. Cytokine Growth Factor Rev. 2001;12:1–8. doi: 10.1016/s1359-6101(00)00031-9. [DOI] [PubMed] [Google Scholar]
- Luo K. Ski and SnoN: negative regulators of TGF-beta signaling. Curr Opin Genet Dev. 2004;14:65–70. doi: 10.1016/j.gde.2003.11.003. [DOI] [PubMed] [Google Scholar]
- Massague J. TGF-beta signal transduction. Annu Rev Biochem. 1998;67:753–791. doi: 10.1146/annurev.biochem.67.1.753. [DOI] [PubMed] [Google Scholar]
- Massague J, Chen YG. Controlling TGF-beta signaling. Genes Dev. 2000;14:627–644. [PubMed] [Google Scholar]
- Massague J, Seoane J, Wotton D. Smad transcription factors. Genes Dev. 2005;19:2783–2810. doi: 10.1101/gad.1350705. [DOI] [PubMed] [Google Scholar]
- Moren A, Imamura T, Miyazono K, Heldin CH, Moustakas A. Degradation of the tumor suppressor Smad4 by WW and HECT domain ubiquitin ligases. J Biol Chem. 2005;280:22115–22123. doi: 10.1074/jbc.M414027200. [DOI] [PubMed] [Google Scholar]
- Murakami G, Watabe T, Takaoka K, Miyazono K, Imamura T. Cooperative inhibition of bone morphogenetic protein signaling by Smurf1 and inhibitory Smads. Mol Biol Cell. 2003;14:2809–2817. doi: 10.1091/mbc.E02-07-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakao A, Afrakhte M, Moren A, Nakayama T, Christian JL, Heuchel R, Itoh S, Kawabata M, Heldin NE, Heldin CH, ten Dijke P. Identification of Smad7, a TGFbeta-inducible antagonist of TGF-beta signalling. Nature. 1997;389:631–635. doi: 10.1038/39369. [DOI] [PubMed] [Google Scholar]
- Nakayama T, Snyder MA, Grewal SS, Tsuneizumi K, Tabata T, Christian JL. Xenopus Smad8 acts downstream of BMP-4 to modulate its activity during vertebrate embryonic patterning. Development. 1998;125:857–867. doi: 10.1242/dev.125.5.857. [DOI] [PubMed] [Google Scholar]
- Narimatsu M, Bose R, Pye M, Zhang L, Miller B, Ching P, Sakuma R, Luga V, Roncari L, Attisano L, Wrana JL. Regulation of planar cell polarity by Smurf ubiquitin ligases. Cell. 2009;137:295–307. doi: 10.1016/j.cell.2009.02.025. [DOI] [PubMed] [Google Scholar]
- Nie S, Chang C. Regulation of Xenopus gastrulation by ErbB signaling. Dev Biol. 2007;303:93–107. doi: 10.1016/j.ydbio.2006.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccolo S, Agius E, Leyns L, Bhattacharyya S, Grunz H, Bouwmeester T, De Robertis EM. The head inducer Cerberus is a multifunctional antagonist of Nodal, BMP and Wnt signals. Nature. 1999;397:707–710. doi: 10.1038/17820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahai E, Garcia-Medina R, Pouyssegur J, Vial E. Smurf1 regulates tumor cell plasticity and motility through degradation of RhoA leading to localized inhibition of contractility. J Cell Biol. 2007;176:35–42. doi: 10.1083/jcb.200605135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint-Jeannet JP, He X, Varmus HE, Dawid IB. Regulation of dorsal fate in the neuraxis by Wnt-1 and Wnt-3a. Proc Natl Acad Sci U S A. 1997;94:13713–13718. doi: 10.1073/pnas.94.25.13713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schier AF. Nodal signaling in vertebrate development. Annu Rev Cell Dev Biol. 2003;19:589–621. doi: 10.1146/annurev.cellbio.19.041603.094522. [DOI] [PubMed] [Google Scholar]
- Schwamborn JC, Muller M, Becker AH, Puschel AW. Ubiquitination of the GTPase Rap1B by the ubiquitin ligase Smurf2 is required for the establishment of neuronal polarity. EMBO J. 2007;26:1410–1422. doi: 10.1038/sj.emboj.7601580. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Seufert DW, Hegde RS, Nekkalapudi S, Kelly LE, El-Hodiri HM. Expression of a novel Ski-like gene in Xenopus development. Gene Expr Patterns. 2005;6:22–28. doi: 10.1016/j.modgep.2005.05.004. [DOI] [PubMed] [Google Scholar]
- Shi Y, Massague J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113:685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- Soond SM, Chantry A. How ubiquitination regulates the TGF-beta signalling pathway: new insights and new players: new isoforms of ubiquitin-activating enzymes in the E1-E3 families join the game. Bioessays. 33:749–758. doi: 10.1002/bies.201100057. [DOI] [PubMed] [Google Scholar]
- Tan R, He W, Lin X, Kiss LP, Liu Y. Smad ubiquitination regulatory factor-2 in the fibrotic kidney: regulation, target specificity, and functional implication. Am J Physiol Renal Physiol. 2008;294:F1076–1083. doi: 10.1152/ajprenal.00323.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang LY, Yamashita M, Coussens NP, Tang Y, Wang X, Li C, Deng CX, Cheng SY, Zhang YE. Ablation of Smurf2 reveals an inhibition in TGF-beta signalling through multiple mono-ubiquitination of Smad3. EMBO J. 30:4777–4789. doi: 10.1038/emboj.2011.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian M, Bai C, Lin Q, Lin H, Liu M, Ding F, Wang HR. Binding of RhoA by the C2 domain of E3 ligase Smurf1 is essential for Smurf1-regulated RhoA ubiquitination and cell protrusive activity. FEBS Lett. 585:2199–2204. doi: 10.1016/j.febslet.2011.06.016. [DOI] [PubMed] [Google Scholar]
- Vonica A, Brivanlou AH. An obligatory caravanserai stop on the silk road to neural induction: inhibition of BMP/GDF signaling. Semin Cell Dev Biol. 2006;17:117–132. doi: 10.1016/j.semcdb.2005.11.013. [DOI] [PubMed] [Google Scholar]
- Whitman M. Nodal signaling in early vertebrate embryos: themes and variations. Dev Cell. 2001;1:605–617. doi: 10.1016/s1534-5807(01)00076-4. [DOI] [PubMed] [Google Scholar]
- Wu Q, Chen D, Zuscik MJ, O'Keefe RJ, Rosier RN. Overexpression of Smurf2 stimulates endochondral ossification through upregulation of beta-catenin. J Bone Miner Res. 2008;23:552–563. doi: 10.1359/JBMR.071115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Huang JH, Sampson ER, Kim KO, Zuscik MJ, O'Keefe RJ, Chen D, Rosier RN. Smurf2 induces degradation of GSK-3beta and upregulates beta-catenin in chondrocytes: a potential mechanism for Smurf2-induced degeneration of articular cartilage. Exp Cell Res. 2009;315:2386–2398. doi: 10.1016/j.yexcr.2009.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia L, Jia S, Huang S, Wang H, Zhu Y, Mu Y, Kan L, Zheng W, Wu D, Li X, Sun Q, Meng A, Chen D. The Fused/Smurf complex controls the fate of Drosophila germline stem cells by generating a gradient BMP response. Cell. 143:978–990. doi: 10.1016/j.cell.2010.11.022. [DOI] [PubMed] [Google Scholar]
- Yamashita M, Ying SX, Zhang GM, Li C, Cheng SY, Deng CX, Zhang YE. Ubiquitin ligase Smurf1 controls osteoblast activity and bone homeostasis by targeting MEKK2 for degradation. Cell. 2005;121:101–113. doi: 10.1016/j.cell.2005.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Chang C, Gehling DJ, Hemmati-Brivanlou A, Derynck R. Regulation of Smad degradation and activity by Smurf2, an E3 ubiquitin ligase. Proc Natl Acad Sci U S A. 2001;98:974–979. doi: 10.1073/pnas.98.3.974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, Qiao M, Oyajobi BO, Mundy GR, Chen D. E3 ubiquitin ligase Smurf1 mediates core-binding factor alpha1/Runx2 degradation and plays a specific role in osteoblast differentiation. J Biol Chem. 2003;278:27939–27944. doi: 10.1074/jbc.M304132200. [DOI] [PubMed] [Google Scholar]
- Zhu H, Kavsak P, Abdollah S, Wrana JL, Thomsen GH. A SMAD ubiquitin ligase targets the BMP pathway and affects embryonic pattern formation. Nature. 1999;400:687–693. doi: 10.1038/23293. [DOI] [PubMed] [Google Scholar]