Abstract
Renal cell carcinomas (RCC) are emerging as a complex set of diseases with major socioeconomic impact and a continued rise in incidence throughout the world. As the field of urologic oncology faces these trends, several major genomic and mechanistic discoveries have altered our core understanding of this multitude of cancers, including several new rare subtypes of renal cancers. This review will examine these new findings, and place them in the context of the well-established association of clear cell RCC (ccRCC) with mutations in the von Hippel-Lindau (VHL) gene and resultant aberrant hypoxia inducible factor (HIF) signaling. The impact of novel ccRCC-associated genetic lesions on chromatin remodeling and epigenetic regulation is explored. The effects of VHL mutation on primary ciliary function, extracellular matrix homeostasis, and tumor metabolism are discussed. VHL proteostasis is reviewed, with the goal of harnessing the proteostatic machinery to refunctionalize mutant VHL. Translational efforts using molecular tools to understand discriminating features of ccRCC tumors and develop improved prognostic and predictive algorithms are presented and new therapeutics arising from the earliest molecular discoveries in ccRCC are summarized. By creating an integrated review of the key genomic and molecular biological disease characteristics of ccRCC and placing these data in the context of the evolving therapeutic landscape, we intend to facilitate interaction between basic, translational and clinical researchers involved in the treatment of this devastating disease, and accelerate progress towards its ultimate eradication.
Keywords: Renal cell carcinoma, VHL, HIF, Epigenetics, Genetics, Tumor subtypes, Biomarkers
Introduction
A rapid series of discoveries in ccRCC, bolstered by advances in genomic biology and the embrace of targeted therapy have ushered in a new era of biological investigation and therapeutic opportunity for this challenging disease. ccRCC is unresponsive to traditional chemotherapies, highly radiation resistant, and lacks the hallmark genetic features of solid tumors such as KRAS or TP53 mutations. The unique tight association between ccRCC and mutations in the VHL gene, and the resulting constitutive stabilization of hypoxia inducible factors (HIF-1α and HIF-2α), have been a source of intense study over the past nearly two decades. Stemming directly from the studies of VHL insufficiency is an enhanced understanding of the intricate relationship between this tumor type and the tumor endothelial vascular network, and the result has been the development of therapies which not only reduce tumor burden, but also have extended the natural life expectancy of patients with metastatic disease.
This review will examine recent developments poised yet again to produce a paradigm shift in our understanding of the biology of ccRCC and other tumors, as well as to generate a landscape ripe for development of new therapeutics. An international panel of experts provides a concise description of the most relevant developments in their field for ccRCC. Andy Futreal summarizes the discoveries arising out of the deep sequencing studies performed over the last few years, and Ian Davis and Cheryl Walker describe the impact of these mutations on cellular behaviour. The potential consequences of this finding are enormous, and provide an explanation for the source of tumor heterogeneity as well as a target for therapeutic intervention. Understanding of the VHL gene and HIF signaling continues to evolve as well. Pathways are never as simple as they initially appear, and the intense focus on HIF-1a signaling associated with VHL mutation has gradually shifted to a focus on HIF-2α as the offending culprit in this disease, with definitive evidence now available. Sean Bailey and William Kim describe these findings in more detail.
RCC is increasingly being recognized as a metabolic disease, and key lesions in nutrient sensing and processing have been detected. These metabolic abnormalities provide protection for the tumor but also may provide a source of vulnerability and therapeutic opportunity. James Brugarolas and Amato Giaccia describe this important network. The same is true for the recently described abnormalities in extracellular matrix engendered by loss of VHL function, which are elucidated by Ghada Kurban and Arnim Pause. VHL is increasingly being recognized as an important regulator of the primary cilium, and by extension, of the cilia centrosome cycle. A better understanding of the role VHL plays in this pathway can potentially lead to insights in RCC carcinogenesis. Cheryl Walker provides a summary of this complex and intriguing function.
It has been well established that VHL mutations lead to malfolded, and poorly functioning VHL protein. A better understanding of VHL proteostasis may allow us to develop strategies to refold or otherwise refunctionalize point mutated, full length VHL. Eric Jonasch and Judith Frydman report on recent developments in this emerging field. Numerous biomarkers have emerged to clarify the presence of heterogeneity among tumors that can be exploited for prognostic value or intervention. Kimryn Rathmell reviews the emergence of molecular classification for RCC, and Amado Zurita describes prognostic and predictive biomarkers under development. Finally, the goal of all of this outstanding science is to allow us to offer patients better chances for survival and more healthy lives. The therapeutic options for ccRCC have evolved rapidly in the last six years, and continue unabated. Both targeted therapies directed at features uncovered in molecular and genetic studies, and improved opportunities to re-direct the immune system have significant potential to continue to improve the outlook for ccRCC. Brian Rini describes current and emerging molecularly targeted agents, and Pam Sharma and Michael Atkins review the exciting new developments in immunotherapy for RCC.
GENETICS
Renal cell carcinoma (RCC) is a collective term applied to a set of cancers arising in the epithelium of the renal tubules comprised of three main histopathological entities. Clear cell RCC (ccRCC) is the dominant histology, accounting for approximately 65% of reported cases, followed by papillary and chromophobe RCC, accounting for approximately 15–20 and 5%, respectively. Other more rare subtypes make up the remainder of RCC cases including collecting duct, mucinous tubular, spindle cell, renal medullary, and MiTF-TFE translocation carcinomas.
Hereditary RCC, which accounts for around 4% of cases, has been a relatively dominant area of RCC genetics. Causative genes have been identified in several familial cancer syndromes that predispose to RCC including VHL mutations in von Hippel-Lindau disease that predispose to ccRCC[1], MET mutations in familial papillary renal cancer[2], FH (fumarate hydratase) mutations in hereditary leiomyomatosis and renal cell cancer that predispose to papillary RCC[3] and FLCN (folliculin) mutations in Birt-Hogg-Dubé syndrome that predispose to primarily chromophobe RCC[4]. In addition, germline mutations in the TSC1/2 genes predispose to tuberous sclerosis complex where approximately 3% of cases develop ccRCC[5] and SDHB (succinate dehydrogenase type B) germline mutations in patients with paraganglioma syndrome give rise to increased risk of developing multiple types of RCC[6]. Moving away from rare monogenic disease to population-based RCC susceptibility, genome wide association study results from a recent study of almost 6000 RCC cases has implicated loci on 2p21 and 11q13.3 in RCC susceptibility[7]. 2p21 contains the EPAS1 gene encoding a transcription factor operative in hypoxia-regulated responses while the other region has no known coding genes.
There has been, however, comparatively less progress in the elaboration of the somatic genetics of sporadic RCC. By far, the most studied somatically mutated gene is VHL, which follows the classic tumor suppressor gene paradigm of a germline cancer susceptibility gene also manifesting as being somatically mutated in the sporadic form of cancer type[8]. VHL is somatically mutated in up to 80% of ccRCC[9]. The majority of these mutations are protein-terminating mutations with loss of the wild-type allele via large-scale loss of heterozygosity of chromosome 3p. There is a further small proportion (5–10%) of cases with no apparent somatic mutations that apparently methylate the locus and thus are functionally VHL null[9–11]. Along a similar theme of congruence of germline and somatic genetics, albeit with a diminished magnitude of effect, there are dominantly activating kinase domain MET mutation reported in 4–10% of sporadic papillary RCC[2]. Conversely, somatic mutations in FLCN in chromophobe RCC are rare[12] and somatic FH mutations in sporadic papillary renal cancers were not found [12–14]. Similarly, somatic mutations of TSC12 and SDHB were not identified in sporadic RCC [13, 14]. Recently however, somatic mutations in TSC1 have been found in sporadic ccRCC [15]. TSC1 mutations occur in 5% of ccRCCs and may predict for extraordinary sensitivity to mTORC1 inhibitors clinically [15].
Further investigation of RCC somatic genetics has included evaluation of cancer genes important in other adult epithelial cancers. Taking all histologies combined, the COSMIC database reports somatic point mutations in TP53 in 10% of cases, KRAS/HRAS/NRAS combined ≤1%, CDKN2A 10%, PTEN 3%, RB1 3%, STK11/LKB1 ≤1%, PIK3Ca ≤1%, EGFR 1% and BRAF ≤1% (http://www.sanger.ac.uk/ genetics/CGP/cosmic/). MYC has been reported to be amplified in papillary RCC [16] and rare cases of RCC have been reported with EGFR amplification [17]. Focusing on the most prevalent histology, ccRCC, the contribution of cancer genes commonly mutated in other tumor types provides limited insight into what additional somatic genetic events are contributing to pathogenesis.
With this as a background, systematic approaches have been undertaken to elaborate the somatic genetics of ccRCC. A screen of 3,544 protein coding genes via PCR-based exon re-sequencing in 101 cases of ccRCC identified several new cancer genes in RCC [18, 19]. Remarkably, four out of five genes with robust statistical support for being new cancer genes encode proteins involved in histone methylation/demethylation. Truncating mutations were identified in KDM6A/UTX, SETD2 and KDM5C/JARID1C which encode an histone 3 lysine 27 (H3K27) demethylase, an histone 3 lysine 36 (H3K36) methyltransferase and an histone 3 lysine 4 (H3K4) demethylase, respectively. MLL2, an H3K4 methyltransferase, was also mutated at a significant rate. These data implicate deregulation of histone H3, known to be a major regulator of euchromatin/transcription, as a new area of RCC biology for exploration. Of note and further confirming the utility of large-scale systematic approaches, NF2 truncating mutations were unexpectedly identified in a significant proportion of the small subset of ccRCC that are VHL wildtype. Taken together, however, these genes are mutated in less than 15% of ccRCC suggesting the existence of additional cancer genes.
A subsequent study has moved to solution capture and sequencing of the coding exons of 20,000 protein coding genes utilizing next-generation sequencing technologies to more comprehensively investigate ccRCC somatic genetics. This work identified a second major somatically mutated cancer gene in ccRCC and thus substantially reshaped the field of RCC genetics. Truncating mutations in the PBRM1 gene were identified in a remarkable 41% (92/227) of ccRCC [20]. PBRM1 encodes the Baf180 protein, a chromatin targeting subunit of the SWI/SNF chromatin remodeling complex implicated in multiple chromatin/transcriptionally mediated processes through interaction with histone H3 [21, 22], reinforcing the striking theme of deregulated chromatin in ccRCC biology. Of note, VHL, SETD2 and PBRM1 are all located on chromosome 3p, thus providing a likely explanation for the near pathognomonic loss of 3p seen in ccRCC. Indeed, half of all cases with a demonstrable VHL point mutation in this series have a PBRM1 truncating mutation and 9/9 cases with a SETD2 mutation also have concurrent VHL and PBRM1 mutations. This work has framed important new areas for ccRCC basic and clinical research.
Recent work performing deep sequencing on samples from a variety of locations from individual large tumors and metastatic lesions shows that considerable heterogeneity exists within these tumors suggesting a branched pattern of evolution [23]. Mutational events, such as the VHL mutation among others were ubiquitous to all samples, however, certain mutations were present only in either the primary tumor or the metastatic lesions, and many mutations were private. Of particular interest, different phylogenetic branches demonstrated distinct SETD2 mutations, indicating a convergent pattern of selection for certain genotypic events. More work to understand this process, and the implications for biomarker development is needed.
Given the findings of these recent studies, it is a certainty that other RCC cancer genes and driver mutations remain to be identified. To this end, there are international efforts underway (ICGC www.icgc.org and TCGA cancergenome.nih.gov) to sequence large numbers of RCC at the whole genome level, coupled with transcriptomic and epigenomic analyses. This work is proceeding at pace, and thus the comprehensive structure of RCC somatic architecture will be revealed in the coming few years.
EPiGENETICS
Together with long-standing insights into HIF deregulation through VHL loss, these findings suggest that RCC development may represent a nexus of epigenetic and transcriptional deregulation, and exploration of epigenetic modification could reveal critical biological properties and offer clues to novel therapeutic approaches.
Genetic alterations in epigenetic regulators
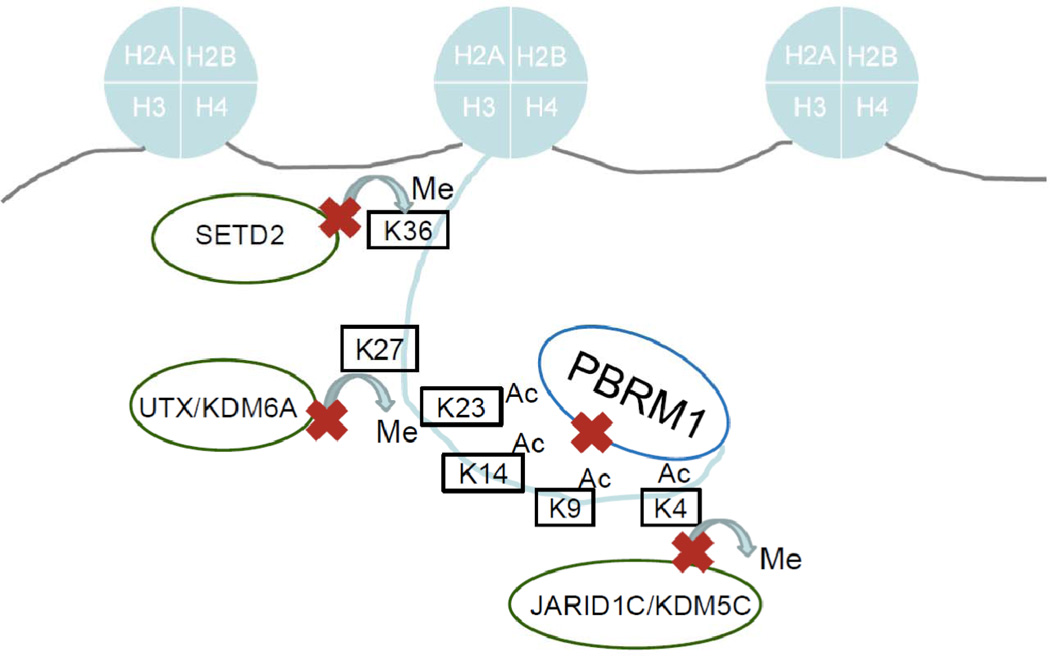
As described above, high throughput genetic studies of RCC have identified recurrent mutations in genes encoding several epigenetic regulators. Mutated genes have been implicated in chromatin regulation through nucleosome repositioning and histone tail modification. PBRM1, which was found to be mutated in nearly 40% of human RCC [20, 24], is a component of the Polybromo BRG1-associated factor complex (PBAF, SWI/SNF-B). PBAF, like SWI/SNF, functions as a nucleosome remodeler and has been shown to be involved in transcriptional regulation [25–27]. Less common mutations were also identified in two methyltransferases, SETD2 and MLL2, and two demethylases, UTX (KDM6A) and JARID1C (KDM5C) [18] (Figure 1). Deletion of 3p, a common finding in ccRCC associated with the loss of VHL at 3p25 can also affect SETD2 and PBRM1, both at 3p21 [28]. SETD2 mediates the trimethylation of H3K36 [29], a histone mark that is placed during transcription and may be important for maintaining faithful transcription [30, 31] whereas MLL2 mediates H3K4me3, a mark associated with active transcription. UTX demethylates H3K27me3 [32–34], a histone mark associated with repressed chromatin. Interestingly, UTX associates with MLL2 [32, 35], suggesting that demethylation of repressive marks is linked to placement of marks associated with transcriptional activation. JARID1C demethylates H3K4 [36]. The finding of mutations in MLL2 and JARID1C, which act oppositely on the same residue, suggests that the genomic effects of mutations in these genes are likely to be complex (Figure 1). Although some mutations may result in widespread epigenetic variation, the effect of others may demonstrate preference for specific regions of the genome [37].
Figure 1. Overview of Histone 3 modifications implicated in RCC genetics.
A number of histone modifying genes are mutated in renal cell carcinoma. These include the H3K36 trimethylase SETD2, the H3K27 demethylase UTX/KDM6A, the H3K4 demethylase JARID1C/KDM5C and the SWI/SNF complex compenent PBRM1, shown in this cartoon to represent their relative activities on Histone H3.
HIF- and hypoxia-mediated epigenetic regulation
The hypoxia response pathway has previously been shown to have a direct effect on histone modification. HIF been shown to activate several chromatin demethylases, including JMJD1A (KDM3A), JMJD2B (KDM4B), JMJD2C (KDM4C) and JARID1B (KDM5B), all of which are directly targeted by HIF [38–41]. Re-expression of pVHL in VHL deficient cell lines increased H3K4me3 levels associated with decreasing levels of JARID1C, a target of HIF2α [24]. Silencing JARID1C in VHL-deficient tumor cells augmented tumor growth in a xenografted mouse model suggesting that JARID1C acts as a tumor suppressor. In contrast, hypoxia may increase methylation through HIF independent mechanisms. Like HIF prolyl hydroxylase (EGLN3), histone demethylases are members of the dioxygenase superfamily, which require oxygen as well as iron and 2-oxoglutarate for activity [42, 43]. In a manner analogous to stabilization of HIF via decreased hydroxylation, hypoxia was shown to suppress JARID1A (KDM5A) activity resulting in increased H3K4me3 levels [44]. This suggests the hypothesis that loss of demethylases (and by analogy increased histone methylation) is part of a “hypoxia phenotype” which is selected for in RCC. This “hypoxia phenotype”, which is mimicked by VHL loss, would also be mimicked by loss of histone demethylase activity, which as noted above, is a high-frequency event in RCC.
Chromatin organization also influences HIF function. Studies of HIF, induced under conditions of hypoxia, demonstrated preferential targeting of HIF to previously nucleosome-depleted chromatin regions [27]. Moreover, the co-expression of SWI/SNF components BRG1, BAF170 and BAF57 augmented HIF activity from a HIF responsive reporter [26]. This study demonstrated that BRG1, but not BRM silencing, decreased HIF responsiveness, suggesting that PBAF may be more critical for HIF function than SWI/SNF.
To what extent mutations of epigenetic regulators influence chromatin or HIF targeting remains unknown. Because of the direct influence of hypoxia on demethylase activity, it is likely that the relationship between epigenetic variation and HIF targeting will differ under conditions of hypoxia in primary cells and in the context of specific epigenetic alterations in tumor cells. Altering the activity of an individual epigenetic regulator that functions as part of a complex may result in pleiotropic effects resulting from alterations in the stoichiometry of active complexes.
In addition to epigenetic regulation through histone tail modification, DNA methylation in RCC has been well recognized. Through studies of tumors, urine and RCC-derived cell lines, hypermethylation of several tumor suppressor genes have been demonstrated. RASSF1 may be hypermethylated in over half of RCC with less common hypermethylation of VHL and CDKN2A [10, 45–48]. Additional studies have identified methylation and silencing of other genes include TIMP3 and secreted frizzled-related protein 2 ([49, 50] [51]). The use of genome-wide assays of methylation and studies of differential methylation will likely identify many more loci that are methylated in ccRCC [52, 53], however, the relationship between DNA hypermethylation and histone modification in the context of RCC remains unclear.
Therapeutic implications
Epigenetic differences may predict variation in patient outcome. Global decreases in H3K4 methylation and H3K18 acetylation have associated with decreased patient survival [37, 54]. Since epigenetic alterations and transcriptional deregulation are central to RCC, employing agents with predicted epigenetic influences may have an effect on disease outcomes. In preclinical studies, treatment with the histone deacetylase inhibitor vorinostat augmented the activity of the mTOR inhibitor temsirolimus to induce apoptosis in xenografted RCC cell lines [55]. However, a phase II trial of a different HDAC inhibitor, panobinostat, in patients with refractory metastatic RCC failed to demonstrate an objective response [56]. A more precise understanding of the role of epigenetic alterations could indicate other targetable strategies.
HIFs AND HIF TARGET GENES
The hypoxia-inducible factors (HIF) are a family of transcription factors that contain a basic helix-loop-helix domain and function in a heterodimeric complex [57]. There are three known HIFα subunits (HIF-1α, HIF-2α, HIF-3α) which heterodimerize with their binding partner ARNT (HIF-1β) to transcriptionally regulate target genes containing hypoxia response elements (HREs). HIF-1α and HIF-2α, are best characterized and are known to regulate transcriptional programs associated with cellular and physiological adaptation to hypoxia such as erythropoietin (EPO), vascular endothelial growth factor (VEGF), and carbonic anhydrase 9 (CA9), amongst others [58]. While there is significant overlap in genes that are transcriptionally activated by HIF-1α and HIF-2α, each HIF family member is thought to also transactivate unique target genes [59]. For example, HIF-1α has been linked to regulating genes in pathways associated with glycolytic metabolism such as, SLC2A1 (GLUT1), LDHA and autophagy BNIP3 while HIF-2α is uniquely responsible for transcriptionally activating genes associated with proliferation and de-differentiation, TGFα, CCND1 (Cyclin D1), and Oct4, respectively.
VHL regulation of HIF
An important realization in understanding the molecular pathogenesis of VHL deficient RCC was that under conditions of normoxia, the pVHL complex binds to and polyubiquitinates HIFα subunits, resulting in their targeting and destruction by the proteasome[57]. The interaction between HIF and pVHL is mediated by an enzymatic, post-translational hydroxylation of HIF on conserved prolyl residues by a family of HIF prolyl hydroxylases (PHDs or EGLNs). In keeping with the notion that regulation of HIF is an important function of pVHL, the majority of disease associated VHL mutations are predicted to abolish the interaction between pVHL and HIF[60]. Moreover, studies in mice suggest that HIF activation (in particular HIF-2α) mediate the majority of the phenotypes seen in the setting of VHL loss [61–63].
Role of HIF in RCC
Early in vitro and cell line xenograft studies suggested that while HIF-2α is both necessary and sufficient for the growth of transformed RCC cell lines [64–66] HIF-1α is not[67], indicating that HIF-1α is expendable for RCC growth. However, it appears that HIF-1α is not merely dispensable in the context of RCC but actually functions as a tumor suppressor gene. Several lines of evidence support this hypothesis. First, targeted exon sequencing of RCC has demonstrated (albeit rarely) inactivating mutations in HIF-1α, while copy number analysis RCC cell lines and primary tumors suggest that the HIF-1α locus is frequently lost along with the long arm of chromosome 14 (14q) [18, 68]. Secondly, while all VHL defective clear cell renal cell carcinomas appear to overexpress HIF-2α, about one third of these tumors appear to lack HIF-1α expression as well [69]. Finally, functional studies in vitro and in vivo suggest that over expression of HIF-1α in VHL wild type cells restrains tumor growth while suppression of HIF-1α in VHL deficient cells enhances tumor growth [68, 70]. Together these studies show support for HIF-1α as tumor suppressor gene in renal cancer development and HIF-2α as a key driver for renal cancer progression.
While there are a number of reasons to explain the contrasting properties of HIF-1α and HIF-2α in RCC pathogenesis, one intriguing observation is that HIF-1α and HIF-2α have opposing roles on the regulation of c-Myc activity. Specifically HIF-1α acts to suppress c-Myc activity while HIF-2α promotes the transactivation or transrepression of c-Myc specific target genes [59, 69, 71]. In keeping with this notion, RCC tumors that exclusively express HIF-2α have increased proliferation rates. Furthermore, intriguingly, a subset of clear cell RCC tumors appear to have copy number amplification of 8q24, where c-Myc resides [72].
VHL PROTEOSTASIS
Two pVHL isoforms exist in the cell, a 213 amino acid, 30 kilodalton form, and a 160 amino acid 19 kilodalton version[73]. In order to function, pVHL must fold to its native conformation. Proper folding and functionality of pVHL requires its tight association with elongin B and elongin C to give rise to a VHL-elongin BC complex (herein VBC). Failure of pVHL to fold and to interact with elongin BC results in misfolding and proteolytic degradation of pVHL [74]. This section summarizes pVHL protein homeostasis (also called proteostasis), and how disease causing mutations affect pVHL stability and functionality.
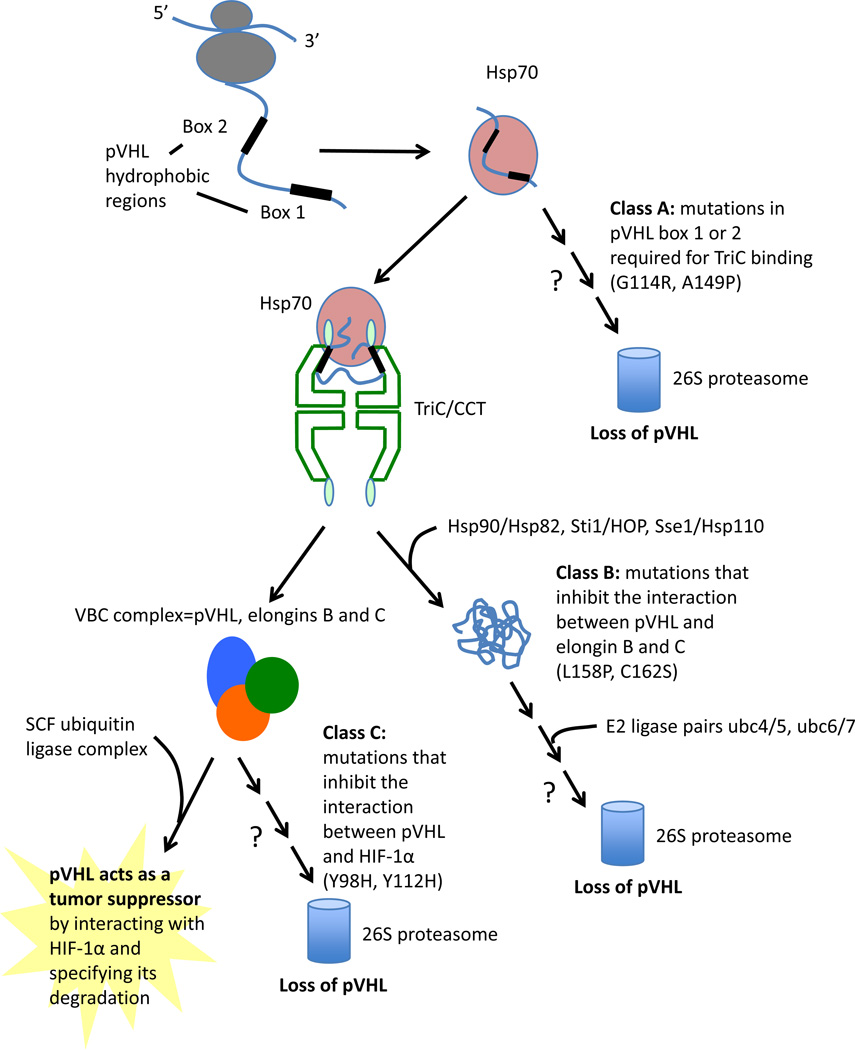
Molecular chaperones are essential mediators of protein folding and quality control of most proteins in the cell. Following synthesis on ribosomes, folding of functional pVHL protein is the result of a complex interplay between nascent pVHL and cellular chaperones. Nascent pVHL is shuttled from the ribosomal machinery with the assistance of heat shock protein 70 (HSP70)[75]. pVHL is then folded into its tertiary structure via association with the chaperonin complex TRiC (for TCP-1 Ring Complex), also called CCT (chaperonin containing TCP-1)[75–78]. This hetero-oligomeric complex consists of two stacked rings with a central chamber where unfolded polypeptides bind and fold. TRiC is responsible for folding a number of key proteins that, like pVHL, are also subunits of oligomeric complexes [76, 77, 79, 80]. Hsp70 likely functions to stabilize non-native forms of pVHL, while TRiC/CCT facilitates pVHL folding, which is coupled to its incorporation into assembly of VBC [81–83]. Upon VHL binding to elongin BC to form mature VBC, pVHL is released from TRiC [75] (Figure 2).
Figure 2. pVHL mutants are categorized as Class A, B and C depending on the affected step in pVHL protein quality control.
VHL proteostasis involves the chaperone mediated translocation of nascent VHL peptide from the ribosome to the TRiC/CCT chaperonin, where folding occurs in an ATP dependent process. The VBC complex is formed while VHL is bound to TRiC, and the mature complex is then released. Three different classes of mutation exist: Class A mutations prevent binding of VHL to TRiC, and abrogate folding into a mature complex. Class B mutations prevent association of Elongins C and B to VHL. Class C mutations inhibit interaction between VHL and HIF1 a.
Binding of VHL to TRiC occurs at amino acids 114–119 and 148–155, called Box 1 and Box 2, respectively[84]. Both motifs, located in adjacent strands of the beta-domain, harbor tumor-causing mutations that disrupt association with TRiC and lead to misfolding of newly translated pVHL. Mutations that block pVHL incorporation into a well folded VBC complex appear to result in destabilization and lower intracellular levels of pVHL, although residual functionality is maintained in some cases [85]. Further analysis of how specific mutations affect the interaction of pVHL with chaperones and chaperonins provides insight into targetable mechanisms of pVHL protein destabilization. Disease causing mutations in TRiC Box 1 and Box 2 binding sites [84, 86, 87], prevent association of pVHL to TRiC, resulting in a malfolded protein, and the absence of a mature VBC complex in the cell. Disease causing mutations also occur in the amino acid 155–181 elongin C binding region[86, 87]. This class of mutants can bind to TRiC, but cannot stably bind to elongin BC. Loss of elongin C binding capacity appears to prevent pVHL release from TRiC [84], resulting in a lack of mature VBC complex.
Failure to generate a properly folded pVHL or a mature VBC complex will result in pVHL degradation through the ubiquitin-proteasome system. Chaperones are also involved in this quality control process[84, 88]. pVHL degradation specifically requires another chaperone, Hsp90, which does not participate in pVHL folding[88]. The identification of two distinct pathways of chaperone interactions for pVHL, one leading to folding and one to degradation, suggests that the fate of pVHL may be controlled by a hierarchy of chaperone interactions. Understanding the mechanism of how destabilized pVHL mutants are targeted for proteasomal degradation may lead to strategies of refolding and stabilization of pVHL that is functional and competent to complex with elongins B and C. Bortezomib and MG132 are capable of increasing levels of VHL, and a cell-based Prestwick compound screen identified several compounds that up-regulate point mutated VHL[89]. Efforts to analyze functional consequences of pVHL upregulation using these compounds, as well as an expanded screening effort are underway.
In summary, our evolving understanding of proteostasis allows new therapeutic approaches to be developed for VHL disease. Recalibrating the interaction between point-mutated pVHL and the chaperones and chaperonins may alter disease phenotype and provide benefit for patients with lesions possessing either germline or sporadic VHL mutations.
RCC: ONE OF THE “CILIOPATHIES”
Together with Polycystic Kidney Disease (PKD), Tuberous Sclerosis Complex (TSC) and VHL Syndrome are considered ‘ciliopathies’ [90]. In PKD, TSC2 and VHL deficiency, renal cysts develop following loss of gene function, often as preneoplastic lesions. One of the hallmarks of cysts is dysfunctional primary cilia. All cells possess a single primary cilium, a non-motile organelle composed of a central microtubule axoneme anchored by the basal body, surrounded by the ciliary membrane (Figure 3). In the kidney, the primary cilium projects from the apical surface of renal epithelial cells into the kidney lumen, where it responds to fluid flow and acts as a chemo-, osmotic, mechano-sensor of the environment. Loss of primary cilia results in dysregulated cell signaling, cystogenesis in the kidney and several other organs, and is one of the hallmarks of many types of cancer, including RCC.
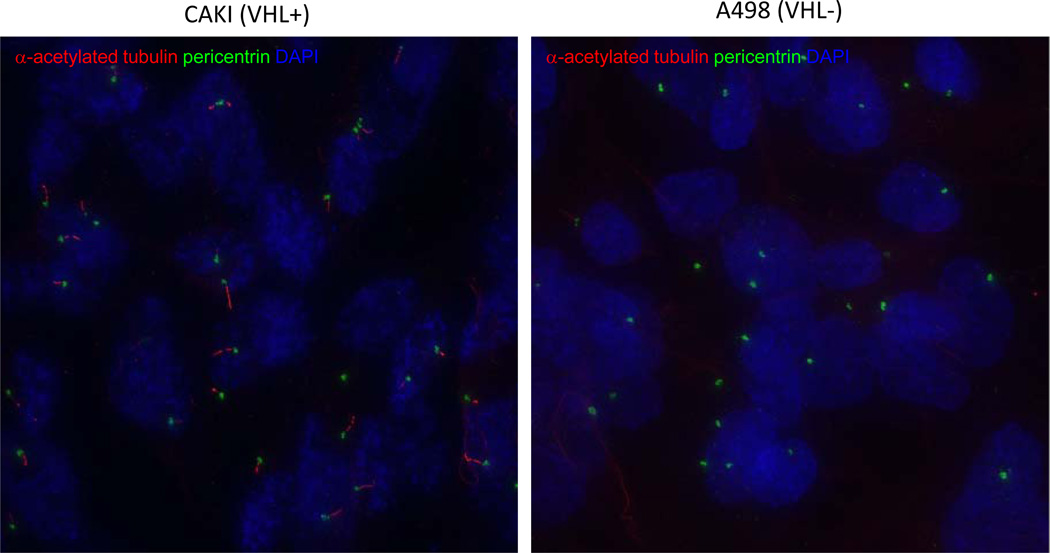
Figure 3. Immunofluorescent images of primary cilia in VHL+ and VHL− cells.
Immunofluorescent images of primary cilia in VHL+ and VHL− cells using ciliary marker alpha-acetylated-tubulin (red) and centrosomal marker antipericentrin (green), counterstained for DNA with DAPI (blue). Left panel shows three color merge VHL+ cells; right panel demonstrates the absence of cilia in VHL− cells. Individual cells are color coded by various shades of gray; cilium (red) and associated basal bodies (green) are also segmented by automated image analysis.
Several cell signaling pathways linked to tumorigenesis either localize specifically to the primary cilium and/or are spatially regulated by this organelle, including Wnt, Hedgehog and platelet-derived growth factor (PDGF) signaling [90, 91]. In addition to aberrant signaling, the microtubule organizing center (MTOC) that forms the foundation for the primary cilium, the basal body, also functions in the cell during mitosis as the centrosome [92]. The fact that this MTOC must shuttle between functioning as a basal body for the primary cilia and a centrosome for the mitotic spindle means that the cilia-centrosome cycle must be tightly coupled to cell division to maintain genomic stability. The cilia-centrosome cycle is important for maintaining genomic stability. Centrioles of the basal body, which serve as the microtubule organizing center (MTOC) for the ciliary axoneme, also serve as the MTOC for the mitotic spindle. There they function as the centrosome, which is comprised of a pair of centrioles responsible for spindle formation during mitosis. Thus, the centrioles serve two distinct and mutually exclusive functions in the cells, serving as the MTOC for either the mitotic spindle (during M phase) or the primary cilium (during G0/G1). The fact that this MTOC shuttles between these two functions means that the cilia-centrosome cycle must be tightly regulated to guarantee the fidelity of centrosome replication, spindle formation and genomic stability. Importantly, defects in cilia-centrosome cycle checkpoints have the potential to cause inappropriate centrosome replication, supernumerary centrosomes, and ultimately aneuploidy. Interestingly, it has recently been shown that ,VHL localizes to the mitotic spindle in mammalian cells and causes spindle misorientation and chromosomal instability when defective or absent [93]
pVHL, TSC and PKD-associated proteins also share a common function: regulation of the structure and function of the primary cilium. These “renal cystoproteins” have been localized at the primary cilium where they exert a variety of cellular responses [94, 95]. For instance, PKD-1 plays a critical role at the primary cilium where it is involved in ciliary mechanotransduction. Several studies have indicated that VHL is also involved in the biogenesis and function of the primary cilia [94, 96, 97] and biallelic inactivation of this gene is associated with loss of cilia [98]. Consistent with this observation, RCC of the clear cell type, associated with loss of pVHL, shows markedly reduced cilia formation when compared to papillary carcinoma [99]. In addition, pVHL binds to microtubules [93, 100], and co-localizes with the acetylated tubulin in the cilia where its mobility is dependent on its association with Kif3A [101]. Studies linking TSC2 deficiency to ciliary defects are more recent where loss of TSC2 is linked specifically to the development of aberrant primary cilia. This abnormal ciliary phenotype is also associated with loss of TSC1, which localizes to the basal body [102].
REGULATION OF THE EXTRACELLULAR MATRIX
The extracellular matrix (ECM) is a complex structural component that surrounds the cells and provides support. It is composed of proteoglycans, hyaluronic acid and glycoproteins such as fibronectin and many types of collagens [103, 104]. Disruption of its regular architecture has been associated with tumor growth, angiogenesis, and metastasis. pVHL plays an important role in the regulation of the ECM. It was shown to interact directly with fibronectin and collagen IV, resulting in their assembly into the ECM and suppression of tumorigenesis, angiogenesis and cell invasion [105–109]. Most pVHL mutants fail to bind and degrade HIF-α, however all pVHL mutants tested to date fail to bind fibronectin and collagen IV and lose the ability to assemble an ECM [105–111]. The interaction of pVHL with fibronectin is mediated by pVHL neddylation which acts as a molecular switch conferring selectivity to fibronectin binding over CUL2 [109, 112], while its interaction with collagen IV is dependent on endoplasmic reticulum (ER) hydroxylation [107]. The VHL-collagen IV interaction was shown to occur at the ER membrane, with pVHL binding to a 70 kDa fragment of the collagen IV amino terminus that protrudes out of the ER into the cytosol [107]. The mechanistic significance of these interactions is still not clear but it was shown that pVHL did not affect fibronectin and collagen IV production or secretion nor resulted in collagen IV proteosomal degradation [106, 107].
The role of pVHL in ECM regulation is independent of its role in HIF-α regulation. Indeed, it was shown that inactivation of the VHL-ECM assembly pathway results in tumors that are highly vascularized, have a remodeled fibronectin and collagen IV matrix and show increased invasive ability. Loss of the VHL-HIF-α regulation pathway resulted in tumors with high VEGF levels but with decreased angiogenesis, a tightly assembled fibronectin and collagen IV matrix and low invasive capacity. Therefore while both pathways cooperate in supporting tumorigenicity, ECM remodeling may promote angiogenesis by providing a path for blood vessels to infiltrate tumors [106].
Tumor cell invasion is dependent on adhesion and proteolytic remodeling of the ECM, both of which are influenced by pVHL activity. pVHL was shown to regulate adhesion molecules; its inactivation leads to downregulation of the adherens junction protein E-cadherin and stimulation of invasion in RCC [113–115]. Loss of pVHL function also leads to downregulation of tight junction proteins occludin and claudin, in an E-cadherin independent manner [116]. In these studies, disruption of both adherens and tight junctions were mediated by loss of the pVHL-HIF-α regulation pathway. In another study, pVHL was found to downregulate integrins in a HIF-α independent manner and this correlated with restoration of tight and adherens junctions [117]. Cells lacking pVHL also fail to form β1 fibrillar adhesions, possibly contributing to the increased cell motility and invasiveness seen in the absence of a functional pVHL [118].
VHL pathways also regulate matrix metalloproteinases (MMPs), a family of matrix-degrading enzymes involved in ECM turnover. RCC cell lines lacking pVHL showed increased invasiveness in growth factor-reduced Matrigel, overproduced MMPs 2 and 9 and displayed an extensive branching morphogenesis phenotype in response to hepatocyte growth factor/scatter factor as compared to those with wild-type pVHL[119]. Activation of MMPs upon loss of pVHL activity can be attributed to disruption of both VHL-ECM and VHL-HIF-α pathways. Loss of VHL-ECM pathway regulation in RCC cells resulted in increased cell invasiveness and activation of MMP-2 [106], and HIF-α was also shown to influence RCC cell invasiveness by regulating membrane type-1 MMP expression [120, 121]. Proteolytic remodeling of the ECM by MMPs was shown to expose cryptic sites in collagen IV, normally hidden within the triple helical structure, leading to loss of integrin α1β1 binding and a gain of binding to the αvβ3 integrin, resulting in stimulation of angiogenesis [122]. Antibodies directed towards collagen IV cryptic sites led to inhibition of angiogenesis, tumor growth and metastasis in vivo suggesting the importance of collagen IV matrix remodeling in these processes [122–125].
The role of pVHL in maintaining ECM integrity and suppression of tumorigenesis, angiogenesis and invasiveness is multi-faceted and complex. It may result from interplay between several mechanisms that still remain unresolved. It is possible that pVHL mediates fibronectin and collagen IV modification, allowing their proper assembly into the ECM. Loss of these interactions would lead to an aberrant ECM, activation of MMPs, ECM remodeling, release of ECM sequestered growth factors and stimulation of tumorigenesis, angiogenesis and invasion. Disruption of integrins and cell adhesion molecule regulation would further enhance the invasive RCC phenotype. Understanding the mechanisms of ECM regulation by pVHL could provide additional or alternate therapy for RCC patients distinct from tyrosine kinase inhibitors.
RENAL CELL CARCINOMA AND METABOLISM
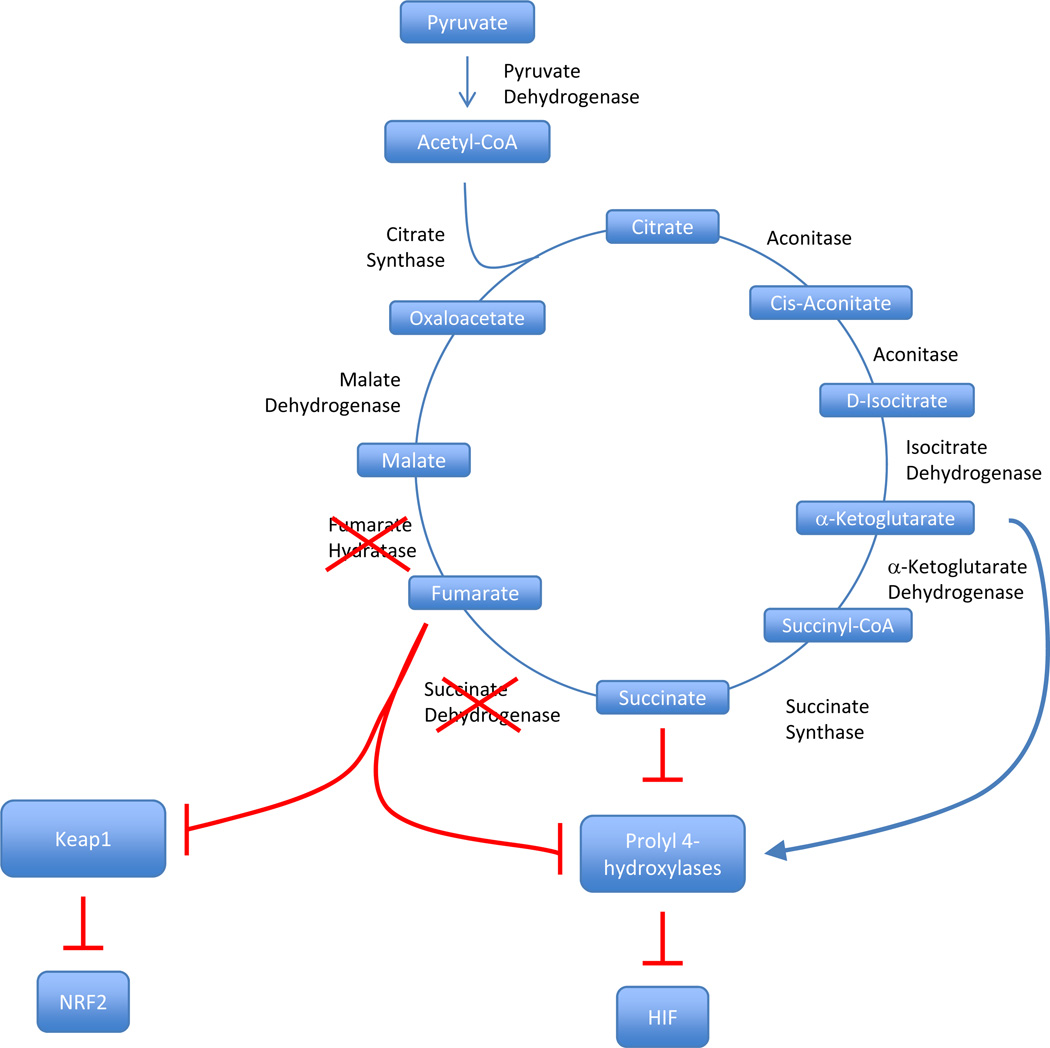
An intimate link between metabolism and renal cancer was established by the discovery that genes encoding enzymes of the Krebs cycle suppress tumor formation in kidney cells [126, 127]. The Krebs cycle refers to 9 sequential enzymatic reactions implicated in oxidizing acetyl-CoA generated from glucose, fatty acids and amino acids to CO2 (Figure 4). This cycle is essential to the process of mitochondrial ATP generation. Succinate dehydrogenase (SDH), a complex of 4 different polypeptides (SDHA-D) that is also involved in electron transfer, catalyzes the conversion of succinate to fumarate. Heterozygous germline mutations in SDH subunits predispose to pheochromocytoma/paraganglioma and mutations in SDHB and SDHD have also been associated with renal cell carcinoma (RCC) [6, 128].
Figure 4. Regulation of Prolyl Hydroxylases and Keap1 by Krebs cycle.
Regulation of Prolyl Hydroxylases by Tricarboxylic Acid (TCA) Cycle Intermediates. Prolyl hydroxylases use TCA cycle intermediates to help catalyze the oxygen, iron and ascorbate dependent- addition of a hydroxyl side chain to a Pro402 and Pro564 of HIF alpha subunits, leading to VHL binding and degradation. Defects in either fumarate hydratase or succinate dehydrogenase will drive up levels of fumarate and succinate, which competitively bind prolyl hydroxylases, and prevent HIF prolyl hydroxylation. This results in higher intracellular HIF levels.
Fumarate hydratase (FH) catalyzes the next reaction of the Krebs cycle, the conversion of fumarate to malate. Heterozygous germline FH mutations cause hereditary leiomyomatosis and renal cell cancer (HLRCC), a syndrome characterized by cutaneous and uterine leiomyomas as well as RCC [3, 129]. RCCs occur in 20–50% of HLRCC families, are typically papillary-type 2 (pRCC-2)[130], and tend to be very aggressive [131].
The FH and SDH genes function as two-hit tumor suppressor genes [55, 127]. Loss-of-function mutations in the germline are usually accompanied by loss of heterozygosity (LOH) in the tumor causing the truncation of the cycle and the accumulation of intermediates [132, 133]. The accumulation of succinate or fumarate causes the inhibition of a family of 2-oxoglutarate-dependent dioxygeneases normally implicated in HIF-α hydroxylation [134–136]. In the absence of this modification, HIF-α evades recognition by pVHL and accumulates, leading to increased HIF activity and tumor development [57]. In addition, the accumulation of succinate and fumarate results in the succination of proteins, including Keap1 [137, 138]. Keap1, is a component of an E3 ubiquitin ligase that targets NRF2 for degradation, and its succination blocks NRF2 degradation resulting in its accumulation and increased expression of stress-response and antioxidant genes [137–139].
Truncation of the Krebs cycle results in a compensatory increase in glucose uptake and glycolysis[132, 140–142]. Accordingly, HLRCC-associated pRCC-2 are intensely FDG-PET positive [142, 143]. Unlike other tumor cells, FH-deficient pRCC-2 cells are unable to grow in low glucose concentrations [142]. This dependency on glucose offers an opportunity for therapeutic intervention and we recently reported an attempt to treat an HLRCC patient with advanced pRCC-2 refractory to mammalian target of rapamycin complex 1 (mTORC1) inhibition with an inhibitor of glycolysis [143].
Metabolic derangements are also associated with mutations in VHL. Germline VHL mutations predispose to clear-cell RCC (ccRCC) [1], referred to in this manner because of the accumulation of lipid and glycogen which gives a clear appearance to the tumor cells in the tissue after processing. By contrast to VHL, SDH and FH genes are seldom mutated in the sporadic setting [12, 13]. Interestingly, while Vhl mutations do not cause RCC in the mouse, disruption in the liver phenocopies the accumulation of lipid and glycogen observed in ccRCC [61, 144–148]. Thus, hepatocytes may serve as a model to study the role of VHL in metabolism. Acute Vhl disruption in hepatocytes results in a Hif-dependent inhibition of mitochondrial respiration [148]. Deprived of Vhl, glucose and ketone production by hepatocytes drops and the mice die within days [148]. While the relative contribution of Hif-1 and Hif-2 remains to be fully determined, Hif-2 may play an important role [147–149]. Should a similar inhibition of mitochondrial respiration occur in ccRCC, these tumors may be exquisitely sensitive to glycolysis inhibitors.
Vulnerabilities arising from VHL loss in ccRCC are also being exploited through synthetic lethal screens [150, 151]. This was illustrated genetically by screening VHL-deficient ccRCC cell lines with shRNAs against kinase targets. This screen identified several kinases synthetic lethal with VHL including cyclin-dependent kinase 6 (CDK6), hepatocyte growth factor receptor (MET), and dual specificity mitogen-activated protein kinase kinase 1 (MEK1) [151]. Small molecule inhibitors of CDK6 also proved to reduce the viability of VHL-deficient ccRCC tumor cells [151]. In addition to the shRNA approach, small molecule screening has also been fruitful in identifying new targets exhibiting enhanced cytotoxicity against VHL-deficient ccRCC. The compound STF-62247 significantly reduced the survival of VHL-deficient ccRCC in cell culture as well as in transplanted tumors in immunodeficient mice. STF-62247 induces autophagy and disrupts Golgi trafficking, which in VHL-deficient cells leads to cell death [150]. From the same screen, a second compound, STF-31, was identified that also exhibits enhanced cytotoxicity against VHL-deficient ccRCC. STF-31 inhibits glucose uptake by the Glut-1 transporter and induces necrotic cell death in VHL-deficient ccRCC [152]. This small molecule provides evidence that targeting glucose metabolism directly in VHL-deficient ccRCC could provide a therapeutic gain clinically.
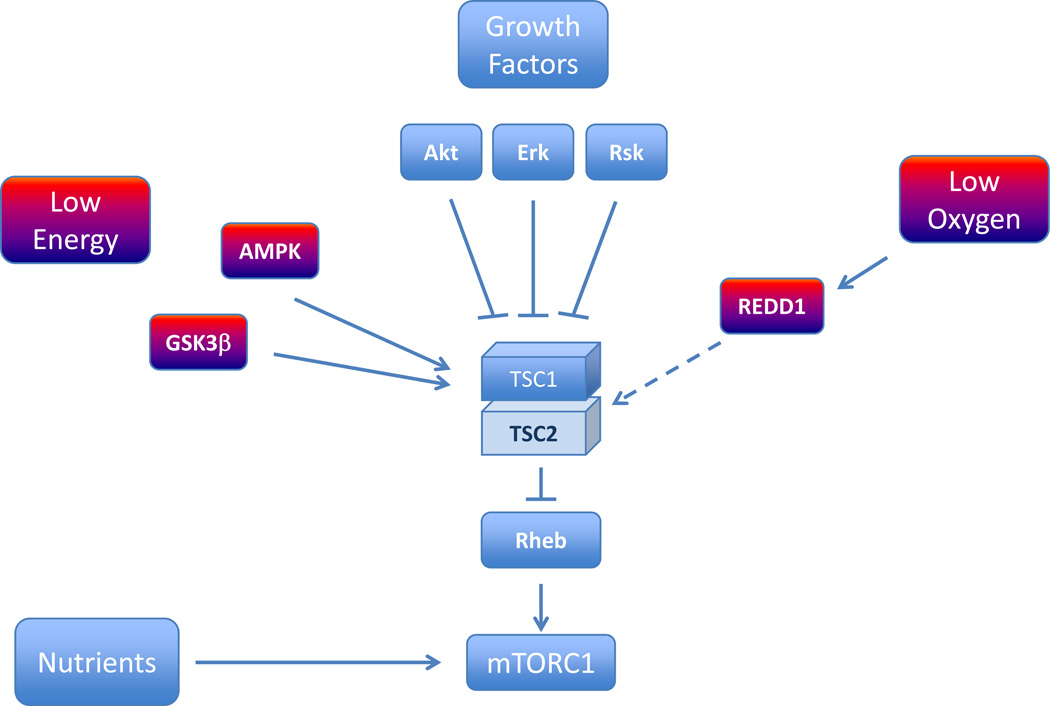
Another pathway implicated in RCC pathogenesis and which plays an important role in metabolism is the mTORC1 pathway (Figure 5). mTORC1 is the target of two FDA approved drugs, temsirolimus and everolimus, and is a master regulator of cell growth. mTORC1 integrates environmental and cellular cues with the cell growth machinery [153]. Signals from energy stores [154], oxygen [155] and growth factors [156] are largely transduced to mTORC1 through a protein complex formed by the proteins tuberous sclerosis complex 1 and 2 (TSC1/TSC2). By contrast, nutrients regulate the subcellular localization of mTORC1 [157]. Only in the presence of nutrients is mTORC1 receptive to signals funneled through TSC1/TSC2 [160, 161]. The best characterized function of mTORC1 is in promoting protein translation, a process mediated, at least in part, by the phosphorylation of S6K and the eukaryotic initiation factor 4E-binding protein 1 (4E-BP1) [158, 162, 163]. However, mTORC1 plays also an important role in suppressing autophagy [159, 163] and regulating mitochondria [164]. In addition, several transcription factors are regulated by mTORC1. mTORC1 regulates HIF-1 [165–170] coupling thereby trophic functions to angiogenesis. mTORC1 also regulates sterol regulatory element binding protein1 (SREBP1), a master regulator of lipogenesis [171, 172]. Finally, we recently reported that mTORC1 regulates the transcription factor EB (TFEB)[173], a controller of lysosome biogenesis [174]. Interestingly, the TFEB gene is translocated in a subset of RCCs [175, 176] and the regulation of TFEB by mTORC1 may provide opportunities for therapeutic intervention.
Figure 5. Regulation of mTORC1.
HIF regulation and mTOR pathway connections. Hypoxia blocks HIF expression in a TSC1/2 and REDD dependent pathway [155]. HIF1α appears to be both TORC1 and TORC2 dependent, whereas HIF2α is only TORC2 dependent [275]. Signaling via TORC2 appears to upregulate HIF2α in an AKT dependent manner [69].
DEFINING NEW MOLECULAR SUBTYPES OF RCC
It has become increasingly clear that ccRCC is an incredibly heterogeneous disease. We have learned only in recent years how thoroughly distinct are the differences between ccRCC and non-clear cell histologies, that they in fact may be best served to be considered as different diseases, with distinct biology, prognosis, and response to treatment [177–180, 181, 182]. Indeed, even within the less common papillary type RCC, two distinct subtypes emerge, papillary types 1 and 2 [183–188]. These two papillary subtypes are associated with distinct familial syndromes: hereditary papillary RCC (associated with type 1 papillary RCC), caused by germline mutations in the Met proto-oncogene [2], and hereditary leiomyosarcoma and RCC (HLRCC, associated with type 2 papillary RCC) [129], in this case fumarate hydratase, are discussed above. The underlying genetic events in sporadic versions of these two histologically defined subtypes are undergoing investigation.
Within the category of clear cell RCC, heterogeneity has also been widely appreciated, despite studies revealing an increasingly tight connection with mutation of VHL. As discussed above, VHL mutation provides a permissive setting for the deregulation of HIF family members, notably HIF-1α and HIF-2α [189]. Tumors can be classified, therefore, into tumors expressing both factors (H1H2) or those expressing only HIF-2α (H2), or a smaller number that produce a functional pVHL [69]. These definitions demonstrate distinct patterns of gene expression and signal transduction, and suggest that HIF profile may be important for selecting therapy. This method of subclassifying ccRCC tumors has been hindered by assay inconsistency for the highly labile HIF proteins. Future studies may be best served to consider a transcriptionally based instrument for assigning H1H2 vs H2 status.
Indeed, ccRCC provides an outstanding tumor model for expression-based analyses, and numerous groups laid the groundwork for defining the heterogeneity of this tumor classification based on transcriptional measurements. Studies based on supervised gene expression profiling of primary tumors vs metastases, early vs late recurrences, or short vs long survival have consistently shown differentially expressed genes [179, 190, 191]. Recently, Rini, et al reported on a transcriptional profile indicative of poor risk for recurrence developed from paraffin embedded specimens[192], which indicates that expression-based biomarkers are ready for translation to the clinic for prospective evaluation.
In parallel, several groups have performed unsupervised analyses to determine if inherent subtypes exist within ccRCC that can be defined based on purely molecular means [193, 194]. Two primary subgroups are found in relatively equal abundance in unselected tumors, suggesting that ccRCC may be represented by two major subclassifications, termed ccA and ccB in recent analyses [195]. The ccA and ccB subclassifications share many similarities to the gene sets identified in good risk or poor risk tumors described above, respectively, in particular gene sets involved in local invasion, and epithelial to mesenchymal transition. Moreover, when the clinical outcomes are examined, ccA cases display a long median survival of 8.6 years, whereas their ccB counterparts display median survival of only 2 years (p=0.002). The advantages these emerging strategies of subclassification include the potential to assign the profile of an individual tumor; capture of molecular information which is tied to genetic events which may be critical for selection of targeted therapy; and prognostic models which also consider clinically intermediate disease categories. A recent validation by meta-analysis confirmed the presence of these ccA and ccB subtypes, but further identified a subset defined by gene expression indicative of a wild type VHL and variant histology consistent with the newly described clear cell papillary subtype [196, 197].
In spite of the hurdles ahead, it seems likely that molecular strategies to classify individual tumors are on the horizon (Figure 6). In fact, the emerging data from clinically supervised strategies to find risk-associated biomarkers, and molecularly driven strategies to identify patterns within unselected tumors suggest that these two differing top-down, and bottom-up approaches are converging on the same conclusion: That ccRCC is composed of two dominant subgroups, which are closely aligned with clinical outcome. How this information will enable physicians and patients to make wise decisions in the management of ccRCC, or eventually select optimal pharmaceutical therapy remains to be seen, but in the light of many emerging targeted therapies, such information is likely to be highly valuable.
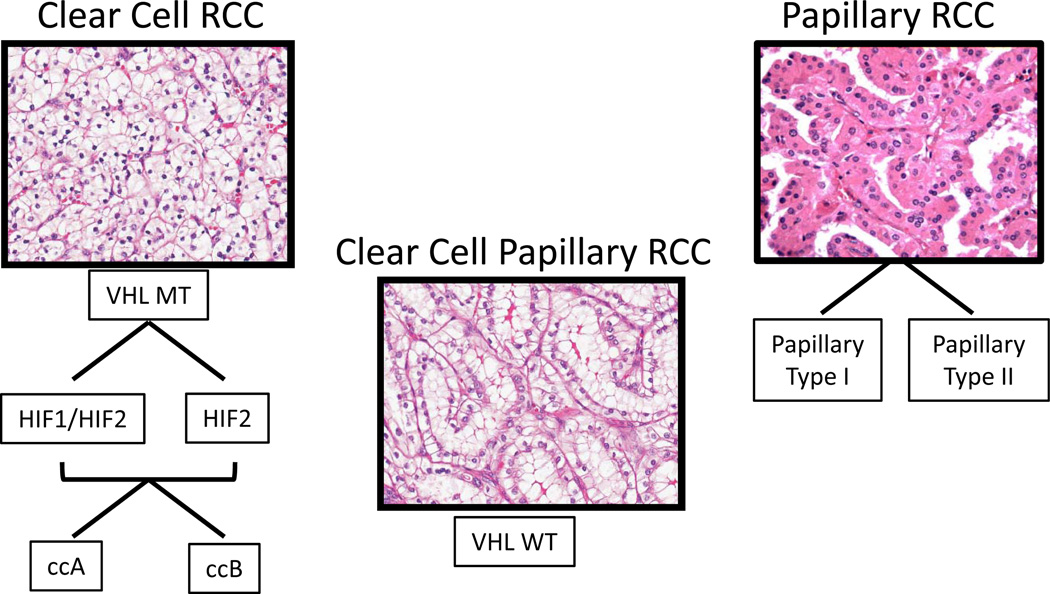
Figure 6. Subtypes of clear cell and papillary RCC, and a new subtype, clear cell papillary.
Different subtypes of clear cell RCC can be defined by HIF patterns as well as by transcriptomic expression as defined by ccA and ccB subtypes. Papillary RCC also demonstrates distinct histological subtypes. A recently described variant denoted as clear cell papillary RCC is VHL wildtype (VHL WT), while other clear cell tumors are characterized by VHL mutation, loss, or inactivation (VHL MT).
BIOMARKERS
The modern emergence of therapeutic options based on the increased understanding of the genetics and molecular biology of the RCC group of diseases has intensified the need for biomarkers to accurately assess prognosis, identify patients likely to benefit from therapy and specific drugs or classes of drugs, and understand mechanisms of resistance. Here we present a succinct overview of the most recent advances in the development of biomarkers for RCC, in particular for the clear cell subtype. Although some are promising, it is important to note that none of these biomarkers are available for clinical testing.
In clinically localized ccRCC, the emphasis has been placed on biomarkers of prognosis expressed in tumor tissue. Some have been found independently prognostic, such as the HIF-1α-regulated hypoxia marker carbonic anhydrase IX [198], the antiapoptotic protein survivin [199–202], the cell proliferation protein KI-67 [203–206], and the immune inhibitory family of ligands B7-H [207–209], but their clinical value is still in question due to lack of independent and prospective validation. IMP3 (one of the insulin-like growth factor II mRNA binding proteins), whose immunohistochemical expression in tumor cells was found associated with short metastasis-free and OS, is a rare exception since findings were subsequently validated in an independent patient cohort [210, 211].
Cytogenetic and gene expression profiling studies have also shown some potential to deliver prognostic information in non-metastatic ccRCC. In mostly small cohorts of patients, specific chromosomal abnormalities have been linked to good (5q gain) or poor prognosis (9p, 14q loss) [212–214]. However, the relation of 9p loss with poor outcome including prognostic value for small renal masses has been repeatedly observed [215, 216], making this a logical candidate to incorporate to available prognostic algorithms. A number of potential biomarkers related to tumor development and progression have additionally emerged from gene expression analyses, several of which have identified gene signatures associated with significant survival differences in patients [217–219]. However, as with the immunohistochemical and cytogenetic markers, these signatures have not yet been validated. Work to validate these gene signatures to predict risk of recurrence is ongoing.
In patients with advanced ccRCC, the availability of effective treatments targeting the VEGF and mTOR pathways has shifted the focus towards the search of biomarkers, predominantly in tumor tissue but also blood, capable of predicting therapy response and resistance. While the analysis of VHL gene status has not resulted in consistent data to support either a prognostic or predictive value [220–224], the activation state of the HIF subunits [69] and multiple HIF-responsive genes are being examined. One HIF target, VEGF, and other angiogenesis-related and tumorigenic factors in serum or plasma have been evaluated across multiple clinical trials of targeted agents in RCC. It has been established that higher baseline VEGF levels are associated with worse tumor stage and grade, performance status, and overall prognosis [225–230]. Moreover, in a phase III trial of sorafenib v placebo, patients with VEGF in the highest concentration quartile obtained greater relative benefit from sorafenib than those with lower concentrations [230]. However, studies addressing whether VEGF is a predictive marker for identifying RCC patients likely to benefit from VEGF-targeted therapies have yielded inconsistent results [225, 230, 231]. Preliminary evidence supports the premise that proteomic plasma profiling of cytokines and angiogenic factors (CAFs) in plasma or serum may be used to develop prognostic and predictive biomarkers, and potentially also contribute to molecularly improve RCC classification [232]. Using this approach, two broad groups of metastatic ccRCC patients were identified, one predominantly expressing angiogenesis/hypoxia-related markers and a second one showing an alternative expression of inflammatory markers. Regarding clinical benefit by VEGF inhibitors, a recent study in plasma samples collected in subsequent phase II and III studies of pazopanib identified low concentrations of IL-8, HGF, OPN and TIMP-1 with improved PFS on pazopanib [233]. IL-8 had previously been implicated in resistance to sunitinib [234]. Unfortunately, no biomarkers predictive of differential benefit between available and active drugs in RCC have been validated. In a randomized phase II study of sorafenib v sorafenib in combination with interferon that yielded no differences in PFS, a candidate 6-CAF signature consistent of markers in the angiogenic/hypoxia group (OPN, VEGF, collagen-IV, soluble CAIX, TRAIL, soluble VEGF receptor-2) predicted for distinct PFS in the 2 arms [232]. Results of similar analyses in larger patient sets are eagerly awaited.
IMMUNOTHERAPY
The ability of some renal tumors to evoke an immune response and the possibility that this may lead to spontaneous regression of metastatic renal cell carcinoma (RCC) in some patients have spurred the idea of developing immunotherapy as an effective treatment for patients with RCC [235–237]. Various immunotherapeutic strategies have been tested with many showing some evidence of activity [238, 239]. Established therapies consist of cytokines such as interferon alpha (IFN) and interleukin 2 (IL-2). IFN was reported to provide a survival benefit in a meta-analysis [240]. HD IL-2 has produced tumor responses in ~10–20% of patients with some patients achieving long-term response off treatment [241–245] The FDA approved HD IL-2 as treatment for metastatic RCC in 1992 based on Phase II data [241]. However, both IFN and HD IL-2 are associated with substantial toxicities, which have limited their use [246, 247]. In addition, due to the emergence of novel VEGF and mTOR targeted therapies, which are easier to administer, better tolerated and proven to provide clinical benefit in Phase III clinical trials [248, 249], the use of IFN and HD IL-2 as treatment for metastatic RCC has diminished. However, a subset of patients clearly exists who develop significant clinical benefit from immunotherapy. Efforts are ongoing to understand the mechanisms of action and identify predictors of response to cytokine therapies such as IFN and HD IL-2 in an attempt to better select patients for treatment. In addition, novel immunotherapeutic strategies are being developed due to advances in the field of basic immunology, which have provided strong scientific and pre-clinical data to enable successful immunotherapy trials.
Improved understanding of various mechanisms by which T-cell activation can be positively or negatively regulated (Figure 7) has led to the development of agents which can enhance anti-tumor T cell responses. The first such agent, and prototype, is anti-CTLA-4 antibody, which laid the foundation for the development of other immune checkpoint agents such as anti-PD-1 antibody.
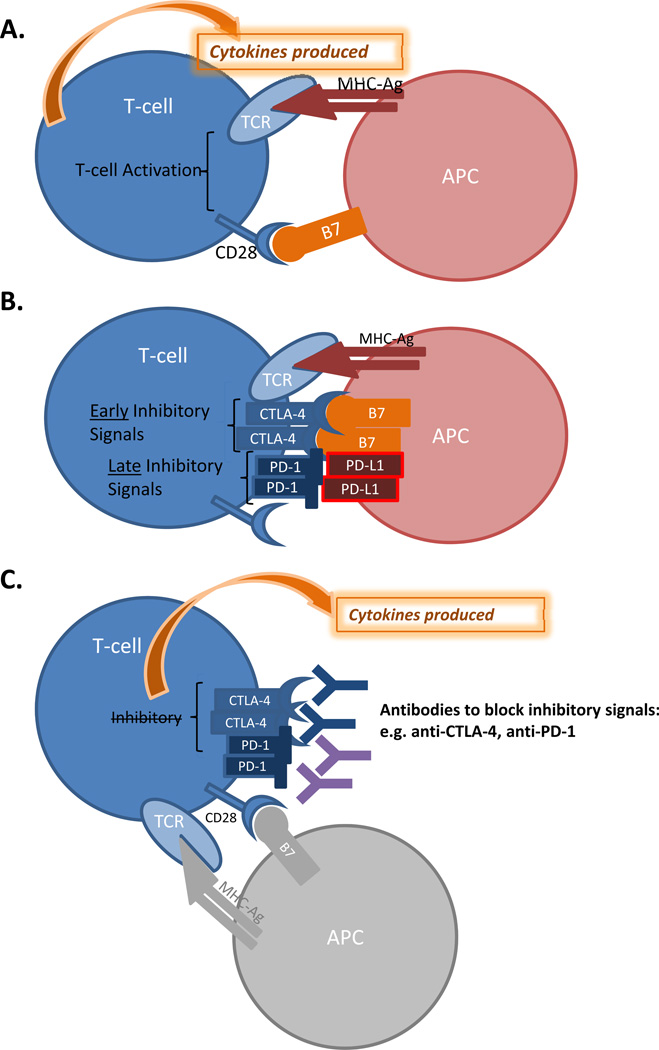
Figure 7. T cell regulation in RCC.
Immune regulation of renal tumor cells. A: When an antigen presenting cell (APC) engages a T-cell via a cognate T-cell receptor (TCR) and CD28, T-cell cell activation occurs. B: Early and late T-cell inhibitory signals are mediated via CTLA-4 and PD-1 receptors, and this occurs via engagement of the APC via B7 and PD-L1, respectively. C: Inhibitory antibodies against CTLA-4 and PD-1 can overcome T-cell downregulation and once again allow cytokine production.
Upon engagement of T cell receptor with antigen bound by MHC (signal 1) and co-stimulation provided by CD28 interacting with B7-1 and B7-2 (signal 2), T cells become activated to produce cytokines and proliferate[250, 251]. However, T cell activity must be regulated in order to prevent damage to normal cells and tissues. Therefore, when T cells are turned ‘on”, a series of signals within the cells also generate an “off” mechanism. This “off” switch is known as CTLA-4. CTLA-4 acts to limit T cell responses [252, 253]. The understanding of how CTLA-4 functions led to the idea that an antibody that blocks CTLA-4, thereby temporarily disengaging the “off” switch, would allow for enhanced T cell responses against tumors. This idea was validated in pre-clinical models [254, 255] and then tested in clinical trials [256–258]. Two Phase III randomized clinical trials were completed and documented a survival benefit for patients with metastatic melanoma who were treated with the anti-CTLA-4 antibody known as ipilimumab (Bristol-Myers Squibb)[259, 260]. Based on these data, ipilimumab was recently FDA-approved in March 2011 as treatment for patients with metastatic melanoma. Since anti-CTLA-4 targets a molecule expressed on T cells, as opposed to a molecule on tumor cells, this therapy is potentially applicable to multiple tumor types.
Anti-CTLA-4 has been evaluated in patients with metastatic RCC. In a Phase II trial, two cohorts of patients with advanced RCC received two different dosing schedules of ipilimumab: a 3mg/kg loading dose followed by either 1mg/kg or 3mg/kg maintenance doses every 3 weeks [261]. Of the 21 patient receiving the 1mg/kg maintenance dose, one patient (4.7%) experienced a partial response. Of 40 patients treated with the 3mg/kg maintenance dose, 5 (12.5%) experienced partial responses. Importantly, responses were observed in patients who had failed prior HD IL-2 suggesting that there is no clear cross resistance. Given its recent FDA approval for melanoma, ipilimumab will likely be investigated further in patients with RCC.
PD-1 is another receptor that is expressed on activated T cells [262]. Interactions with PD-1 and its ligands (PD-L1 and PD-L2) can act to inhibit T cell responses. PD-L1 was shown to be over-expressed in many RCC and greater expression was associated with worse prognosis [263]. MDX-1106 is a monoclonal antibody directed against PD-1 which was recently assessed in Phase I trials including many patients with advanced RCC[264, 265]. Anti-tumor activity was seen in a patient with RCC in the initial trial involving a single dose of the PD1antibody[264]. In a subsequent study [265], MDX-1106 was administered in doses of 1, 3, and 10mg/kg given every 2 weeks. Of 16 patients with RCC treated at various doses, 5 patients (31%) achieved objective responses, including one complete response. This promising activity coupled with a mild toxicity profile has prompted the initiation of a phase II trial MDX-1106 in patients with advanced RCC. Over the next several years, agents such as ipilimumab and MDX-1106 will likely be assessed, possibly with cytokines and other therapies, in various sequences and combinations with the goal of achieving higher rates of durable responses than is possible with currently available therapies.
MOLECULARLY TARGETED THERAPY
The biology of RCC as elucidated above has led to the development of multiple agents targeting elements of the relevant VEGF and mTOR pathways [266]. Table 1 outlines that major phase III trials of targeted therapy in RCC that have led to regulatory approval of several agents. There are several points to be made about key discriminating features of these agents. VEGF-targeted therapy produces more robust RECIST-defined objective response rates than cytokine therapy, on the order of 30% to nearly 50% for the most active agents. Within the class of VEGF receptor inhibitors, the response rate can vary from 10% to nearly 50%, with the higher rates observed with the drugs that more potently inhibit the VEGF-Receptor. It is also recognized that anti-tumor activity, especially of VEGF-targeting agents, is not entirely captured by size changes alone, as tumor necrosis (reduced perfusion on a contrast-enhanced CT scan) is felt to be indicative of drug effect and may or may not be accompanied by tumor size reduction. mTOR-targeted therapy in general produces more modest response rates of 2–10%, although to date studied in different populations than the VEGF-targeted therapies [267, 268]. When considering the percentage of patients who have at least some tumor burden reduction on therapy (i.e. including patients with 1%-29% reduction not meeting the arbitrary 30% reduction required for a RECIST-defined response), VEGF-targeted therapy shrinks tumors in about 75% of patients while mTOR-targeted therapy shrinks tumors in about 50–60% of patients.
TABLE 1.
Phase III Trials of Targeted Therapy in Metastatic Renal Cell Carcinoma
| Trial | Number of patients |
Clinical setting | RR (%) | PFS (months) | OS (months) |
|---|---|---|---|---|---|
| VEGF-Targeted Therapy | |||||
| *AVOREN Bevacizumab + IFNa vs.IFNa[270] |
649 | First-line | 31 vs. 12 | 10.2 vs. 5.5 (p<0.001) |
23.3 vs. 21.3 (p=0.129) |
| *CALBG 90206 Bevacizumab + IFNa vs.IFNa[271] |
732 | First-line | 25.5 vs. 13 | 8.4 vs. 4.9 (p<0.001) |
18.3 vs. 17.4 (p=0.069) |
| Sunitinib vs. IFNa[248] |
750 | First-line | 47 vs. 12 | 11 vs. 5 (p=0.0001) |
26.4 vs. 21.8 (p=0.051) |
| *TARGET Sorafenib vs. Placebo[272] |
903 | Second-line (post-cytokine) |
10 vs. 2 | 5.5 vs. 2.8 (p<0.01) |
17.8vs.15.2 (p=0.88) |
| Pazopanib vs. placebo[273] |
435 | First line/second line (post-cytokine) |
30 vs. 3 | 9.2 vs. 4.2 (p<0.0001) |
22.9 vs. 20.5 (p=0.224) |
| *AXIS Axitinib vs. sorafenib [269] |
723 | Second line (post-sunitinib, cytokine, bevacizumab or temsirolimus) |
19 vs. 9 (p=0.0001) |
6.7 vs. 4.7 (p<0.0001) |
Not reported |
| mTOR-Targeted Therapy | |||||
| *ARCC Temsirolimus vs. Tem + IFNa vs. IFNa[249] |
624 | First line, ≥ 3 poor risk featuresa |
9 vs. 5 | 3.8 vs. 1.9 for IFNa monotherapy (p=0.0001) |
10.9 vs. 7.3 for IFNa (p=0.008) |
| *RECORD-1 Everolimus vs. placebo [274] |
410 | Second line (post sunitinib and/or sorafenib) |
2 vs. 0 | 4.9 vs. 1.9 (p<0.0001) |
14.8 vs. 14.5 |
RCC renal cell carcinoma, RR response rate, OS overall survival, PFS progression free survival, VEGF vascular endothelial growth factor, IFNa interferon alpha, mTOR mammalian target of rapamycin. AVOREN AVastin fOr RENal cell cancer, CALBG Cancer and Leukemia Group B. TARGET Treatment Approaches in Renal Cancer Global Evaluation Trial. AXIS Axitinib in Second Line. ARCC Advanced Renal-Cell Carcinoma. RECORD-1 REnal Cell cancer treatment withOral RAD001 given Daily.
Including serum lactate dehydrogenase level of more than 1.5 times the upper limit of the normal range, a hemoglobin level below the lower limit of the normal range; a corrected serum calcium level of more than 10 mg per deciliter (2.5 mmol per liter), a time from initial diagnosis of renal-cell carcinoma to randomization of less than 1 year, a Karnofsky performance score of 60 or 70, or metastases in multiple organs.
Progression-free survival is generally doubled with targeted therapy compared with placebo/cytokines. Again, here we see important differences among the VEGF-R inhibitors, with the biochemically more potent agents producing a PFS of approximately 11 months in untreated patients, compared to 5 months for the biochemically weaker agent, sorafenib. Axitinib is the most biochemically potent, but to date results are only available in previously-treated patients [269]. Of note, in the subset of patients not exposed to prior VEGF-targeted therapy in the AXIS trial (the cytokine-refractory subgroup), the median PFS was over 12 months. In regards to PFS, the mTOR-targeting agents have been studied in unique patient circumstances, poor-risk for temsirolimus and VEGF-R TKI-refractory RCC for everolimus [267, 268]. The PFS in each was modest (approximately 5 months), but the effect of these agents front-line in good/intermediate risk patients awaits further study. In addition, the clinical activity of drugs is more robust in untreated patients than in cytokine-refractory, and even less in patients who have already failed targeted therapy. Last, overall survival in these trials is of note for several reasons. The front-line trials of VEGF-targeted agents have produced an OS of approximately 2 years, roughly double that of historical cytokine-treated controls. Nonetheless, no single trial (with the exception of the temsirolimus trial in poor risk RCC) has shown a statistically significant overall survival benefit (despite a numerical advantage in the median OS). This is largely felt to be due to the high percentage of patients who cross over from initial therapy on trial (placebo or cytokine) and receive one or more active targeted therapies at progression. The efficacy of such a sequential ‘salvage’ strategy has confounded interpretation of OS from these trials, although there is general consensus that targeted therapy has meaningfully extended the lives of metastatic RCC patients.
There is no consensus on the ‘best’ drug for initial therapy or the optimal sequence of agents. Ongoing trials are beginning to approach these questions, but the multitude of agents and the relative rarity of RCC make definitive trials not possible. Future research in targeted therapy in RCC is focused on such questions of relative toxicity/efficacy among agents, the importance of switching mechanism at progression, and biomarkers of response and resistance that may allow for improvement upon the current standard of an empiric sequence of monotherapies.
CONCLUSIONS
There has been a clear and important evolution in our understanding of RCC biology. We are now challenged with converting this newly acquired information into actionable items that will alter our approach to prevention, diagnosis and management of RCC. Several new rare types of cancers are now recognized to occur in the kidney, which will both challenge the urologic oncology community to maintain up-to-date guidelines for the management of these tumors and provide new opportunities to develop effective personalized therapies.
By comparing genomic, transcriptomic and epigenetic data from precursor lesions and early ccRCC, we will be able to establish a roadmap of tumor ontogeny for this more common subtype. Use of material from patients with hereditary VHL disease will be essential to achieve this goal. These data, in conjunction with epidemiological and laboratory based studies, will allow the investigator to identify driver mutations and epigenetic changes, allowing development of markers that permit early identification of ccRCC. In the same manner, study of the cilia centrosome cycle and HIF regulation will build a mechanistic, molecular biological understanding of early cancer development, with resultant opportunities for therapeutic intervention.
In more advanced disease, the study of genomics, transcriptomics and molecular biology will provide investigators with insight into mechanisms of tumor progression, especially if performed in parallel with in vitro and in vivo models employing potential driver pathways identified in ccRCC or other rare variant cancers. The hope is that by understanding both the cause and the consequence of the complex interactions between genomic and epigenetic changes, and assigning significance to the output of these alterations, we will be able to replicate RCC tumor diversity, identify subgroups, and develop more specific therapeutic interventions. To achieve this goal will require coordinated interaction between high-throughput platform experts, molecular biologists and computational scientists capable of controlling and codifying the complex systems that arise from these collaborations. The recognition that the output of these changes is profoundly influence by host genomic and phenotypic characteristics, and that the tumor microenvironment varies as a function of these characteristics requires the development of precise tools to measure the tumor microenvironment.
Finally, the renaissance of tumor immunology was fueled by the recognition that tumors can take advantage of the innate regulatory pathways built into T-cells and other immune effectors. As we begin to understand the impact of tumor biology on T-cell regulation as well as on the recruitment of bone marrow derived immunological precursors, significantly better treatments will become available for patients with ccRCC in the next few years. Understanding the interface between evolving tumor biology and host genomic determinants of stromal end endothelial phenotype will further advance this field.
We are poised to make very significant advances in RCC research in the next few years. With the right team and the right tools, the potential for a truly personalized approach to treatment is within reach.
Acknowledgement
We would like to acknowledge Ruhee Dere for assistance in figure production.
REFERENCES
- 1.Latif F, et al. Identification of the von Hippel-Lindau disease tumor suppressor gene. Science. 1993;260(5112):1317–1320. doi: 10.1126/science.8493574. [DOI] [PubMed] [Google Scholar]
- 2.Schmidt L, et al. Germline and somatic mutations in the tyrosine kinase domain of the MET proto-oncogene in papillary renal carcinomas. Nat Genet. 1997;16(1):68–73. doi: 10.1038/ng0597-68. [DOI] [PubMed] [Google Scholar]
- 3.Tomlinson IP, et al. Germline mutations in FH predispose to dominantly inherited uterine fibroids, skin leiomyomata and papillary renal cell cancer. Nat Genet. 2002;30(4):406–410. doi: 10.1038/ng849. [DOI] [PubMed] [Google Scholar]
- 4.Nickerson ML, et al. Mutations in a novel gene lead to kidney tumors, lung wall defects, and benign tumors of the hair follicle in patients with the Birt-Hogg-Dubé syndrome. Cancer Cell. 2002;2(2):157–164. doi: 10.1016/s1535-6108(02)00104-6. [DOI] [PubMed] [Google Scholar]
- 5.Bjornsson J, et al. Tuberous sclerosis-associated renal cell carcinoma. Clinical, pathological, and genetic features. Am J Pathol. 1996;149(4):1201–1208. [PMC free article] [PubMed] [Google Scholar]
- 6.Ricketts CJ, et al. Tumor risks and genotype–phenotype–proteotype analysis in 358 patients with germline mutations in SDHB and SDHD. Human Mutation. 2010;31(1):41–51. doi: 10.1002/humu.21136. [DOI] [PubMed] [Google Scholar]
- 7.Purdue MP, et al. Genome-wide association study of renal cell carcinoma identifies two susceptibility loci on 2p21 and 11q13.3. Nat Genet. 2011;43(1):60–65. doi: 10.1038/ng.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gnarra JR, et al. Mutations of the VHL tumour suppressor gene in renal carcinoma. Nat Genet. 1994;7(1):85–90. doi: 10.1038/ng0594-85. [DOI] [PubMed] [Google Scholar]
- 9.Nickerson ML, et al. Improved Identification of von Hippel-Lindau Gene Alterations in Clear Cell Renal Tumors. Clin Cancer Res. 2008;14(15):4726–4734. doi: 10.1158/1078-0432.CCR-07-4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Herman JG, et al. Silencing of the VHL tumor-suppressor gene by DNA methylation in renal carcinoma. Proc Natl Acad Sci U S A. 1994;91(21):9700–9704. doi: 10.1073/pnas.91.21.9700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herman JG, et al. Silencing of the VHL tumor-suppressor gene by DNA methylation in renal carcinoma. Proceedings of the National Academy of Sciences. 1994;91(21):9700–9704. doi: 10.1073/pnas.91.21.9700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kiuru M, et al. Few FH Mutations in Sporadic Counterparts of Tumor Types Observed in Hereditary Leiomyomatosis and Renal Cell Cancer Families. Cancer Research. 2002;62(16):4554–4557. [PubMed] [Google Scholar]
- 13.Morris MR, et al. Molecular genetic analysis of FIH-1, FH, and SDHB candidate tumour suppressor genes in renal cell carcinoma. Journal of Clinical Pathology. 2004;57(7):706–711. doi: 10.1136/jcp.2003.011767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parry L, et al. Analysis of the TSC1and TSC2genes in sporadic renal cell carcinomas. Br J Cancer. 2001;85(8):1226–1230. doi: 10.1054/bjoc.2001.2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kucejova B, et al. Interplay between pVHL and mTORC1 pathways in clear-cell renal cell carcinoma. Mol Cancer Res. 2011;9(9):1255–1265. doi: 10.1158/1541-7786.MCR-11-0302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Furge KA, et al. Detection of DNA Copy Number Changes and Oncogenic Signaling Abnormalities from Gene Expression Data Reveals MYC Activation in High-Grade Papillary Renal Cell Carcinoma. Cancer Research. 2007;67(7):3171–3176. doi: 10.1158/0008-5472.CAN-06-4571. [DOI] [PubMed] [Google Scholar]
- 17.El-Hariry I, et al. Amplification of epidermal growth factor receptor gene in renal cell carcinoma. European Journal of Cancer. 2010;46(5):859–862. doi: 10.1016/j.ejca.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 18.Dalgliesh GL, et al. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature. 2010;463(7279):360–363. doi: 10.1038/nature08672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Haaften G, et al. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nat Genet. 2009;41(5):521–523. doi: 10.1038/ng.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Varela I, et al. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature. 2011;469(7331):539–542. doi: 10.1038/nature09639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thompson M. Polybromo-1: The chromatin targeting subunit of the PBAF complex. Biochimie. 2009;91(3):309–319. doi: 10.1016/j.biochi.2008.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reisman D, Glaros S, Thompson EA. The SWI/SNF complex and cancer. Oncogene. 2009;28(14):1653–1668. doi: 10.1038/onc.2009.4. [DOI] [PubMed] [Google Scholar]
- 23.Gerlinger M, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366(10):883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Niu X, et al. The von Hippel-Lindau tumor suppressor protein regulates gene expression and tumor growth through histone demethylase JARID1C. Oncogene. 2011 doi: 10.1038/onc.2011.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lemon B, et al. Selectivity of chromatin-remodelling cofactors for ligand-activated transcription. Nature. 2001;414(6866):924–928. doi: 10.1038/414924a. [DOI] [PubMed] [Google Scholar]
- 26.Kenneth NS, et al. SWI/SNF regulates the cellular response to hypoxia. J Biol Chem. 2009;284(7):4123–4131. doi: 10.1074/jbc.M808491200. [DOI] [PubMed] [Google Scholar]
- 27.Xia X, Kung AL. Preferential binding of HIF-1 to transcriptionally active loci determines cell-type specific response to hypoxia. Genome Biol. 2009;10(10):R113. doi: 10.1186/gb-2009-10-10-r113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duns G, et al. Histone methyltransferase gene SETD2 is a novel tumor suppressor gene in clear cell renal cell carcinoma. Cancer Res. 2010;70(11):4287–4291. doi: 10.1158/0008-5472.CAN-10-0120. [DOI] [PubMed] [Google Scholar]
- 29.Sun XJ, et al. Identification and characterization of a novel human histone H3 lysine 36-specific methyltransferase. J Biol Chem. 2005;280(42):35261–35271. doi: 10.1074/jbc.M504012200. [DOI] [PubMed] [Google Scholar]
- 30.Carrozza MJ, et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123(4):581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 31.Keogh MC, et al. Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell. 2005;123(4):593–605. doi: 10.1016/j.cell.2005.10.025. [DOI] [PubMed] [Google Scholar]
- 32.Lee MG, et al. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science. 2007;318(5849):447–450. doi: 10.1126/science.1149042. [DOI] [PubMed] [Google Scholar]
- 33.Hong S, et al. Identification of JmjC domain-containing UTX and JMJD3 as histone H3 lysine 27 demethylases. Proc Natl Acad Sci U S A. 2007;104(47):18439–18444. doi: 10.1073/pnas.0707292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Agger K, et al. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449(7163):731–734. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- 35.Issaeva I, et al. Knockdown of ALR (MLL2) reveals ALR target genes and leads to alterations in cell adhesion and growth. Mol Cell Biol. 2007;27(5):1889–1903. doi: 10.1128/MCB.01506-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tahiliani M, et al. The histone H3K4 demethylase SMCX links REST target genes to X-linked mental retardation. Nature. 2007;447(7144):601–605. doi: 10.1038/nature05823. [DOI] [PubMed] [Google Scholar]
- 37.Seligson DB, et al. Global levels of histone modifications predict prognosis in different cancers. Am J Pathol. 2009;174(5):1619–1628. doi: 10.2353/ajpath.2009.080874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wellmann S, et al. Hypoxia upregulates the histone demethylase JMJD1A via HIF-1. Biochem Biophys Res Commun. 2008;372(4):892–877. doi: 10.1016/j.bbrc.2008.05.150. [DOI] [PubMed] [Google Scholar]
- 39.Beyer S, et al. The histone demethylases JMJD1A and JMJD2B are transcriptional targets of hypoxia-inducible factor HIF. J Biol Chem. 2008;283(52):36542–36552. doi: 10.1074/jbc.M804578200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krieg AJ, et al. Regulation of the histone demethylase JMJD1A by hypoxia-inducible factor 1 alpha enhances hypoxic gene expression and tumor growth. Mol Cell Biol. 2010;30(1):344–353. doi: 10.1128/MCB.00444-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xia X, et al. Integrative analysis of HIF binding and transactivation reveals its role in maintaining histone methylation homeostasis. Proc Natl Acad Sci U S A. 2009;106(11):4260–4265. doi: 10.1073/pnas.0810067106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ivan M, et al. HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science. 2001;292(5516):464–468. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- 43.Jaakkola P, et al. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292(5516):468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- 44.Zhou X, et al. Hypoxia induces trimethylated H3 lysine 4 by inhibition of JARID1A demethylase. Cancer Res. 2010;70(10):4214–4221. doi: 10.1158/0008-5472.CAN-09-2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morrissey C, et al. Epigenetic inactivation of the RASSF1A 3p21.3 tumor suppressor gene in both clear cell and papillary renal cell carcinoma. Cancer Res. 2001;61(19):7277–7281. [PubMed] [Google Scholar]
- 46.Dreijerink K, et al. The candidate tumor suppressor gene, RASSF1A, from human chromosome 3p21.3 is involved in kidney tumorigenesis. Proc Natl Acad Sci U S A. 2001;98(13):7504–7509. doi: 10.1073/pnas.131216298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Herman JG, et al. Inactivation of the CDKN2/p16/MTS1 gene is frequently associated with aberrant DNA methylation in all common human cancers. Cancer Res. 1995;55(20):4525–4530. [PubMed] [Google Scholar]
- 48.Clifford SC, et al. Inactivation of the von Hippel-Lindau (VHL) tumour suppressor gene and allelic losses at chromosome arm 3p in primary renal cell carcinoma: evidence for a VHL-independent pathway in clear cell renal tumourigenesis. Genes Chromosomes Cancer. 1998;22(3):200–209. doi: 10.1002/(sici)1098-2264(199807)22:3<200::aid-gcc5>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 49.Kawamoto K, et al. DNA methylation and histone modifications cause silencing of Wnt antagonist gene in human renal cell carcinoma cell lines. Int J Cancer. 2008;123(3):535–542. doi: 10.1002/ijc.23514. [DOI] [PubMed] [Google Scholar]
- 50.Esteller M, et al. A gene hypermethylation profile of human cancer. Cancer Res. 2001;61(8):3225–3229. [PubMed] [Google Scholar]
- 51.Baldewijns MM, et al. Genetics and epigenetics of renal cell cancer. Biochim Biophys Acta. 2008;1785(2):133–155. doi: 10.1016/j.bbcan.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 52.Ibanez de Caceres I, et al. Identification of novel target genes by an epigenetic reactivation screen of renal cancer. Cancer Res. 2006;66(10):5021–5028. doi: 10.1158/0008-5472.CAN-05-3365. [DOI] [PubMed] [Google Scholar]
- 53.Morris MR, et al. Genome-wide methylation analysis identifies epigenetically inactivated candidate tumour suppressor genes in renal cell carcinoma. Oncogene. 2011;30(12):1390–1401. doi: 10.1038/onc.2010.525. [DOI] [PubMed] [Google Scholar]
- 54.Ellinger J, et al. Prognostic relevance of global histone H3 lysine 4 (H3K4) methylation in renal cell carcinoma. Int J Cancer. 2010;127(10):2360–2366. doi: 10.1002/ijc.25250. [DOI] [PubMed] [Google Scholar]
- 55.Mahalingam D, et al. Vorinostat enhances the activity of temsirolimus in renal cell carcinoma through suppression of survivin levels. Clin Cancer Res. 2010;16(1):141–153. doi: 10.1158/1078-0432.CCR-09-1385. [DOI] [PubMed] [Google Scholar]
- 56.Hainsworth JD, et al. A Phase II Trial of Panobinostat, a Histone Deacetylase Inhibitor, in the Treatment of Patients with Refractory Metastatic Renal Cell Carcinoma. Cancer Invest. 2011;29(7):451–455. doi: 10.3109/07357907.2011.590568. [DOI] [PubMed] [Google Scholar]
- 57.Kaelin WG, Jr, Ratcliffe PJ. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol Cell. 2008;30(4):393–402. doi: 10.1016/j.molcel.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 58.Semenza GL. Targeting HIF-1 for cancer therapy. Nat Rev Cancer. 2003;3(10):721–732. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- 59.Gordan JD, Simon MC. Hypoxia-inducible factors: central regulators of the tumor phenotype. Curr Opin Genet Dev. 2007;17(1):71–77. doi: 10.1016/j.gde.2006.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim WY, Kaelin WG. Role of VHL Gene Mutation in Human Cancer. Journal of Clinical Oncology. 2004;22(24):4991–5004. doi: 10.1200/JCO.2004.05.061. [DOI] [PubMed] [Google Scholar]
- 61.Kim WY, et al. Failure to prolyl hydroxylate hypoxia-inducible factor alpha phenocopies VHL inactivation in vivo. Embo J. 2006;25(19):4650–4662. doi: 10.1038/sj.emboj.7601300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rankin EB, et al. Inactivation of the arylhydrocarbon receptor nuclear translocator (Arnt) suppresses von Hippel-Lindau disease-associated vascular tumors in mice. Mol Cell Biol. 2005;25(8):3163–3172. doi: 10.1128/MCB.25.8.3163-3172.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rankin EB, Tomaszewski JE, Haase VH. Renal Cyst Development in Mice with Conditional Inactivation of the von Hippel-Lindau Tumor Suppressor. Cancer Research. 2006;66(5):2576–2583. doi: 10.1158/0008-5472.CAN-05-3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kondo K, et al. Inhibition of HIF2alpha is sufficient to suppress pVHL-defective tumor growth. PLoS Biol. 2003;1(3):E83. doi: 10.1371/journal.pbio.0000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kondo K, et al. Inhibition of HIF is necessary for tumor suppression by the von Hippel-Lindau protein. Cancer Cell. 2002;1(3):237–246. doi: 10.1016/s1535-6108(02)00043-0. [DOI] [PubMed] [Google Scholar]
- 66.Zimmer M, et al. Inhibition of hypoxia-inducible factor is sufficient for growth suppression of VHL−/− tumors. Mol Cancer Res. 2004;2(2):89–95. [PubMed] [Google Scholar]
- 67.Maranchie JK, et al. The contribution of VHL substrate binding and HIF1-alpha to the phenotype of VHL loss in renal cell carcinoma. Cancer Cell. 2002;1(3):247–255. doi: 10.1016/s1535-6108(02)00044-2. [DOI] [PubMed] [Google Scholar]
- 68.Shen C, et al. Genetic and Functional Studies Implicate HIF1alpha as a 14q Kidney Cancer Suppressor Gene. Cancer Discov. 2011;1(3):222–235. doi: 10.1158/2159-8290.CD-11-0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gordan JD, et al. HIF-alpha effects on c-Myc distinguish two subtypes of sporadic VHL-deficient clear cell renal carcinoma. Cancer Cell. 2008;14(6):435–446. doi: 10.1016/j.ccr.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Biswas S, et al. Effects of HIF-1alpha and HIF2alpha on Growth and Metabolism of Clear-Cell Renal Cell Carcinoma 786-0 Xenografts. J Oncol. 2010;2010:757908. doi: 10.1155/2010/757908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gordan JD, et al. HIF-2alpha promotes hypoxic cell proliferation by enhancing c-myc transcriptional activity. Cancer Cell. 2007;11(4):335–347. doi: 10.1016/j.ccr.2007.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Beroukhim R, et al. Patterns of gene expression and copy-number alterations in von-hippel lindau disease-associated and sporadic clear cell carcinoma of the kidney. Cancer Res. 2009;69(11):4674–4681. doi: 10.1158/0008-5472.CAN-09-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schoenfeld A, Davidowitz EJ, Burk RD. A second major native von Hippel-Lindau gene product, initiated from an internal translation start site, functions as a tumor suppressor. Proc Natl Acad Sci U S A. 1998;95(15):8817–8822. doi: 10.1073/pnas.95.15.8817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schoenfeld AR, Davidowitz EJ, Burk RD. Elongin BC complex prevents degradation of von Hippel-Lindau tumor suppressor gene products. Proc Natl Acad Sci U S A. 2000;97(15):8507–8512. doi: 10.1073/pnas.97.15.8507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Feldman DE, et al. Formation of the VHL-elongin BC tumor suppressor complex is mediated by the chaperonin TRiC. Mol Cell. 1999;4(6):1051–1061. doi: 10.1016/s1097-2765(00)80233-6. [DOI] [PubMed] [Google Scholar]
- 76.Rommelaere H, et al. Eukaryotic cytosolic chaperonin contains t-complex polypeptide 1 and seven related subunits. Proc Natl Acad Sci U S A. 1993;90(24):11975–11979. doi: 10.1073/pnas.90.24.11975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Frydman J, et al. Function in protein folding of TRiC, a cytosolic ring complex containing TCP-1 and structurally related subunits. Embo J. 1992;11(13):4767–4778. doi: 10.1002/j.1460-2075.1992.tb05582.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Melki R, Cowan NJ. Facilitated folding of actins and tubulins occurs via a nucleotide-dependent interaction between cytoplasmic chaperonin and distinctive folding intermediates. Mol Cell Biol. 1994;14(5):2895–2904. doi: 10.1128/mcb.14.5.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Llorca O, et al. Eukaryotic type II chaperonin CCT interacts with actin through specific subunits. Nature. 1999;402(6762):693–696. doi: 10.1038/45294. [DOI] [PubMed] [Google Scholar]
- 80.Llorca O, et al. Eukaryotic chaperonin CCT stabilizes actin and tubulin folding intermediates in open quasi-native conformations. Embo J. 2000;19(22):5971–5979. doi: 10.1093/emboj/19.22.5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Feldman DE, et al. Formation of the VHL-elongin BC tumor suppressor complex is mediated by the chaperonin TRiC. Molecular Cell. 1999;4(6):1051–1061. doi: 10.1016/s1097-2765(00)80233-6. [DOI] [PubMed] [Google Scholar]
- 82.Hansen WJ, et al. Diverse effects of mutations in exon II of the von Hippel-Lindau (VHL) tumor suppressor gene on the interaction of pVHL with the cytosolic chaperonin and pVHL-dependent ubiquitin ligase activity. Mol Cell Biol. 2002;22(6):1947–1960. doi: 10.1128/MCB.22.6.1947-1960.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Melville MW, et al. The Hsp70 and TRiC/CCT chaperone systems cooperate in vivo to assemble the von Hippel-Lindau tumor suppressor complex. Molecular And Cellular Biology. 2003;23:3141–3151. doi: 10.1128/MCB.23.9.3141-3151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Feldman DE, et al. Tumorigenic mutations in VHL disrupt folding in vivo by interfering with chaperonin binding. Mol Cell. 2003;12(5):1213–1224. doi: 10.1016/s1097-2765(03)00423-4. [DOI] [PubMed] [Google Scholar]
- 85.Hacker KE, Lee CM, Rathmell WK. VHL type 2B mutations retain VBC complex form and function. PLoS One. 2008;3(11):e3801. doi: 10.1371/journal.pone.0003801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Beroud C, et al. Software and database for the analysis of mutations in the VHL gene. Nucleic Acids Res. 1998;26(1):256–258. doi: 10.1093/nar/26.1.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nordstrom-O'Brien M, et al. Genetic analysis of von Hippel-Lindau disease. Hum Mutat. 31(5):521–537. doi: 10.1002/humu.21219. [DOI] [PubMed] [Google Scholar]
- 88.McClellan AJ, Scott MD, Frydman J. Folding and quality control of the VHL tumor suppressor proceed through distinct chaperone pathways. Cell. 2005;121(5):739–748. doi: 10.1016/j.cell.2005.03.024. [DOI] [PubMed] [Google Scholar]
- 89.Ding Z, et al. High throughput screening to identify molecules that alter VHL stability in AACR Annual Meeting. Orlando, Fla: 2011. [Google Scholar]
- 90.Hildebrandt F, Benzing FT, Katsanis N. Ciliopathies. The New England journal of medicine. 2011;364(16):1533–1543. doi: 10.1056/NEJMra1010172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ishikawa H, Marshall WF. Ciliogenesis: building the cell's antenna. Nature reviews. Molecular cell biology. 2011;12(4):222–234. doi: 10.1038/nrm3085. [DOI] [PubMed] [Google Scholar]
- 92.Kobayashi T, Dynlacht BD. Regulating the transition from centriole to basal body. The Journal of cell biology. 2011;193(3):435–444. doi: 10.1083/jcb.201101005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Thoma CR, et al. VHL loss causes spindle misorientation and chromosome instability. Nature cell biology. 2009;11(8):994–1001. doi: 10.1038/ncb1912. [DOI] [PubMed] [Google Scholar]
- 94.Esteban MA, et al. Formation of primary cilia in the renal epithelium is regulated by the von Hippel-Lindau tumor suppressor protein. Journal of the American Society of Nephrology : JASN. 2006;17(7):1801–1806. doi: 10.1681/ASN.2006020181. [DOI] [PubMed] [Google Scholar]
- 95.Thoma CR, et al. pVHL and GSK3beta are components of a primary cilium-maintenance signalling network. Nature cell biology. 2007;9(5):588–595. doi: 10.1038/ncb1579. [DOI] [PubMed] [Google Scholar]
- 96.Lutz MS, Burk RD. Primary cilium formation requires von hippel-lindau gene function in renal-derived cells. Cancer research. 2006;66(14):6903–6907. doi: 10.1158/0008-5472.CAN-06-0501. [DOI] [PubMed] [Google Scholar]
- 97.Schermer B, et al. The von Hippel-Lindau tumor suppressor protein controls ciliogenesis by orienting microtubule growth. The Journal of cell biology. 2006;175(4):547–554. doi: 10.1083/jcb.200605092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wiesener MS, Maxwell PH, Eckardt KU. Novel insights into the role of the tumor suppressor von Hippel Lindau in cellular differentiation, ciliary biology, and cyst repression. Journal of molecular medicine. 2009;87(9):871–877. doi: 10.1007/s00109-009-0504-x. [DOI] [PubMed] [Google Scholar]
- 99.Schraml P, et al. Sporadic clear cell renal cell carcinoma but not the papillary type is characterized by severely reduced frequency of primary cilia. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc. 2009;22(1):31–36. doi: 10.1038/modpathol.2008.132. [DOI] [PubMed] [Google Scholar]
- 100.Hergovich A, et al. Regulation of microtubule stability by the von Hippel-Lindau tumour suppressor protein pVHL. Nature cell biology. 2003;5(1):64–70. doi: 10.1038/ncb899. [DOI] [PubMed] [Google Scholar]
- 101.Mans DA, et al. Mobility of the von Hippel-Lindau tumour suppressor protein is regulated by kinesin-2. Experimental cell research. 2008;314(6):1229–1236. doi: 10.1016/j.yexcr.2007.12.020. [DOI] [PubMed] [Google Scholar]
- 102.Hartman TR, et al. The tuberous sclerosis proteins regulate formation of the primary cilium via a rapamycin-insensitive and polycystin 1-independent pathway. Hum Mol Genet. 2009;18(1):151–163. doi: 10.1093/hmg/ddn325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Singh P, Carraher C, Schwarzbauer JE. Assembly of fibronectin extracellular matrix. Annu Rev Cell Dev Biol. 26:397–419. doi: 10.1146/annurev-cellbio-100109-104020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kalluri R. Basement membranes: structure, assembly and role in tumour angiogenesis. Nat Rev Cancer. 2003;3(6):422–433. doi: 10.1038/nrc1094. [DOI] [PubMed] [Google Scholar]
- 105.Ohh M, et al. The von Hippel-Lindau tumor suppressor protein is required for proper assembly of an extracellular fibronectin matrix. Mol Cell. 1998;1(7):959–968. doi: 10.1016/s1097-2765(00)80096-9. [DOI] [PubMed] [Google Scholar]
- 106.Kurban G, et al. Characterization of a von Hippel Lindau pathway involved in extracellular matrix remodeling, cell invasion, and angiogenesis. Cancer Res. 2006;66(3):1313–1319. doi: 10.1158/0008-5472.CAN-05-2560. [DOI] [PubMed] [Google Scholar]
- 107.Kurban G, et al. Collagen matrix assembly is driven by the interaction of von Hippel-Lindau tumor suppressor protein with hydroxylated collagen IV alpha 2. Oncogene. 2008;27(7):1004–1012. doi: 10.1038/sj.onc.1210709. [DOI] [PubMed] [Google Scholar]
- 108.Grosfeld A, et al. Interaction of hydroxylated collagen IV with the von hippel-lindau tumor suppressor. J Biol Chem. 2007;282(18):13264–13269. doi: 10.1074/jbc.M611648200. [DOI] [PubMed] [Google Scholar]
- 109.Stickle NH, et al. pVHL modification by NEDD8 is required for fibronectin matrix assembly and suppression of tumor development. Mol Cell Biol. 2004;24(8):3251–3261. doi: 10.1128/MCB.24.8.3251-3261.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hoffman MA, et al. von Hippel-Lindau protein mutants linked to type 2C VHL disease preserve the ability to downregulate HIF. Hum Mol Genet. 2001;10(10):1019–1027. doi: 10.1093/hmg/10.10.1019. [DOI] [PubMed] [Google Scholar]
- 111.Clifford SC, et al. Contrasting effects on HIF-1alpha regulation by disease-causing pVHL mutations correlate with patterns of tumourigenesis in von Hippel-Lindau disease. Hum Mol Genet. 2001;10(10):1029–1038. doi: 10.1093/hmg/10.10.1029. [DOI] [PubMed] [Google Scholar]
- 112.Russell RC, Ohh M. NEDD8 acts as a 'molecular switch' defining the functional selectivity of VHL. EMBO Rep. 2008;9(5):486–491. doi: 10.1038/embor.2008.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Evans AJ, et al. VHL promotes E2 box-dependent E-cadherin transcription by HIF-mediated regulation of SIP1 and snail. Mol Cell Biol. 2007;27(1):157–169. doi: 10.1128/MCB.00892-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Esteban MA, et al. Regulation of E-cadherin expression by VHL and hypoxia-inducible factor. Cancer Res. 2006;66(7):3567–3575. doi: 10.1158/0008-5472.CAN-05-2670. [DOI] [PubMed] [Google Scholar]
- 115.Krishnamachary B, et al. Hypoxia-inducible factor-1-dependent repression of E-cadherin in von Hippel-Lindau tumor suppressor-null renal cell carcinoma mediated by TCF3, ZFHX1A, and ZFHX1B. Cancer Res. 2006;66(5):2725–2731. doi: 10.1158/0008-5472.CAN-05-3719. [DOI] [PubMed] [Google Scholar]
- 116.Harten SK, et al. Regulation of renal epithelial tight junctions by the von Hippel-Lindau tumor suppressor gene involves occludin and claudin 1 and is independent of E-cadherin. Mol Biol Cell. 2009;20(3):1089–1101. doi: 10.1091/mbc.E08-06-0566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ji Q, Burk RD. Downregulation of integrins by von Hippel-Lindau (VHL) tumor suppressor protein is independent of VHL-directed hypoxia-inducible factor alpha degradation. Biochem Cell Biol. 2008;86(3):227–234. doi: 10.1139/o08-035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Esteban-Barragan MA, et al. Role of the von Hippel-Lindau tumor suppressor gene in the formation of beta1-integrin fibrillar adhesions. Cancer Res. 2002;62(10):2929–2936. [PubMed] [Google Scholar]
- 119.Koochekpour S, et al. The von Hippel-Lindau tumor suppressor gene inhibits hepatocyte growth factor/scatter factor-induced invasion and branching morphogenesis in renal carcinoma cells. Mol Cell Biol. 1999;19(9):5902–5912. doi: 10.1128/mcb.19.9.5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Petrella BL, Lohi J, Brinckerhoff CE. Identification of membrane type-1 matrix metalloproteinase as a target of hypoxia-inducible factor-2 alpha in von Hippel-Lindau renal cell carcinoma. Oncogene. 2005;24(6):1043–1052. doi: 10.1038/sj.onc.1208305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Petrella BL, Brinckerhoff CE. Tumor cell invasion of von Hippel Lindau renal cell carcinoma cells is mediated by membrane type-1 matrix metalloproteinase. Mol Cancer. 2006;5:66. doi: 10.1186/1476-4598-5-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Xu J, et al. Proteolytic exposure of a cryptic site within collagen type IV is required for angiogenesis and tumor growth in vivo. J Cell Biol. 2001;154(5):1069–1079. doi: 10.1083/jcb.200103111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Roth JM, et al. Inhibition of experimental metastasis by targeting the HUIV26 cryptic epitope in collagen. Am J Pathol. 2006;168(5):1576–1586. doi: 10.2353/ajpath.2006.050176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Pernasetti F, et al. Novel anti-denatured collagen humanized antibody D93 inhibits angiogenesis and tumor growth: An extracellular matrix-based therapeutic approach. Int J Oncol. 2006;29(6):1371–1379. [PubMed] [Google Scholar]
- 125.Cretu A, et al. Disruption of endothelial cell interactions with the novel HU177 cryptic collagen epitope inhibits angiogenesis. Clin Cancer Res. 2007;13(10):3068–3078. doi: 10.1158/1078-0432.CCR-06-2342. [DOI] [PubMed] [Google Scholar]
- 126.Eng C, et al. A role for mitochondrial enzymes in inherited neoplasia and beyond. Nat Rev Cancer. 2003;3(3):193–202. doi: 10.1038/nrc1013. [DOI] [PubMed] [Google Scholar]
- 127.Gottlieb E, Tomlinson IP. Mitochondrial tumour suppressors: a genetic and biochemical update. Nat Rev Cancer. 2005;5(11):857–866. doi: 10.1038/nrc1737. [DOI] [PubMed] [Google Scholar]
- 128.Vanharanta S, et al. Early-onset renal cell carcinoma as a novel extraparaganglial component of SDHB-associated heritable paraganglioma. Am J Hum Genet. 2004;74(1):153–159. doi: 10.1086/381054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Toro JR, et al. Mutations in the fumarate hydratase gene cause hereditary leiomyomatosis and renal cell cancer in families in North America. Am J Hum Genet. 2003;73(1):95–106. doi: 10.1086/376435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lehtonen HJ. Hereditary leiomyomatosis and renal cell cancer: update on clinical and molecular characteristics. Fam Cancer. 10(2):397–411. doi: 10.1007/s10689-011-9428-z. [DOI] [PubMed] [Google Scholar]
- 131.Grubb RL, 3rd, et al. Hereditary leiomyomatosis and renal cell cancer: a syndrome associated with an aggressive form of inherited renal cancer. J Urol. 2007;177(6):2074–2079. doi: 10.1016/j.juro.2007.01.155. discussion 2079-80. [DOI] [PubMed] [Google Scholar]
- 132.Isaacs JS, et al. HIF overexpression correlates with biallelic loss of fumarate hydratase in renal cancer: novel role of fumarate in regulation of HIF stability. Cancer Cell. 2005;8(2):143–153. doi: 10.1016/j.ccr.2005.06.017. [DOI] [PubMed] [Google Scholar]
- 133.Pollard PJ, et al. Accumulation of Krebs cycle intermediates and over-expression of HIF1α in tumours which result from germline FH and SDH mutations. Human Molecular Genetics. 2005;14(15):2231–2239. doi: 10.1093/hmg/ddi227. [DOI] [PubMed] [Google Scholar]
- 134.Pollard P, et al. Evidence of increased microvessel density and activation of the hypoxia pathway in tumours from the hereditary leiomyomatosis and renal cell cancer syndrome. J Pathol. 2005;205(1):41–49. doi: 10.1002/path.1686. [DOI] [PubMed] [Google Scholar]
- 135.Selak MA, et al. Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIF-alpha prolyl hydroxylase. Cancer Cell. 2005;7(1):77–85. doi: 10.1016/j.ccr.2004.11.022. [DOI] [PubMed] [Google Scholar]
- 136.MacKenzie ED, et al. Cell-permeating alpha-ketoglutarate derivatives alleviate pseudohypoxia in succinate dehydrogenase-deficient cells. Mol Cell Biol. 2007;27(9):3282–3289. doi: 10.1128/MCB.01927-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ooi A, et al. An antioxidant response phenotype shared between hereditary and sporadic type 2 papillary renal cell carcinoma. Cancer Cell. 2011;20(4):511–523. doi: 10.1016/j.ccr.2011.08.024. [DOI] [PubMed] [Google Scholar]
- 138.Adam J, et al. Renal cyst formation in Fh1-deficient mice is independent of the Hif/Phd pathway: roles for fumarate in KEAP1 succination and Nrf2 signaling. Cancer Cell. 2011;20(4):524–537. doi: 10.1016/j.ccr.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kinch L, Grishin NV, Brugarolas J. Succination of Keap1 and activation of Nrf2-dependent antioxidant pathways in FH-deficient papillary renal cell carcinoma type 2. Cancer Cell. 2011;20(4):418–420. doi: 10.1016/j.ccr.2011.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Pollard PJ, et al. Targeted inactivation of fh1 causes proliferative renal cyst development and activation of the hypoxia pathway. Cancer Cell. 2007;11(4):311–319. doi: 10.1016/j.ccr.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 141.Sudarshan S, et al. Fumarate hydratase deficiency in renal cancer induces glycolytic addiction and hypoxia-inducible transcription factor 1alpha stabilization by glucose-dependent generation of reactive oxygen species. Mol Cell Biol. 2009;29(15):4080–4090. doi: 10.1128/MCB.00483-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Yang Y, et al. UOK 262 cell line, fumarate hydratase deficient (FH-/FH-) hereditary leiomyomatosis renal cell carcinoma: in vitro and in vivo model of an aberrant energy metabolic pathway in human cancer. Cancer Genet Cytogenet. 196(1):45–55. doi: 10.1016/j.cancergencyto.2009.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Yamasaki T, et al. Exploring a glycolytic inhibitor for the treatment of an FH-deficient type-2 papillary RCC. Nat Rev Urol. 8(3):165–171. doi: 10.1038/nrurol.2010.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Haase VH, et al. Vascular tumors in livers with targeted inactivation of the von Hippel-Lindau tumor suppressor. Proc Natl Acad Sci U S A. 2001;98(4):1583–1588. doi: 10.1073/pnas.98.4.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Park SK, Haase VH, Johnson RS. von Hippel Lindau tumor suppressor regulates hepatic glucose metabolism by controlling expression of glucose transporter 2 and glucose 6-phosphatase. Int J Oncol. 2007;30(2):341–348. [PubMed] [Google Scholar]
- 146.Peyssonnaux C, et al. Regulation of iron homeostasis by the hypoxia-inducible transcription factors (HIFs) J Clin Invest. 2007;117(7):1926–1932. doi: 10.1172/JCI31370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Rankin EB, et al. Hypoxia-inducible factor 2 regulates hepatic lipid metabolism. Mol Cell Biol. 2009;29(16):4527–4538. doi: 10.1128/MCB.00200-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Kucejova B, et al. Uncoupling hypoxia signaling from oxygen sensing in the liver results in hypoketotic hypoglycemic death. Oncogene. 30(18):2147–2160. doi: 10.1038/onc.2010.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Qu A, et al. Hypoxia-inducible transcription factor 2α promotes steatohepatitis through augmenting lipid accumulation, inflammation, and fibrosis. Hepatology. 54(2):472–483. doi: 10.1002/hep.24400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Turcotte S, et al. A molecule targeting VHL-deficient renal cell carcinoma that induces autophagy. Cancer Cell. 2008;14(1):90–102. doi: 10.1016/j.ccr.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Bommi-Reddy A, et al. Kinase requirements in human cells: III. Altered kinase requirements in VHL-/- cancer cells detected in a pilot synthetic lethal screen. Proc Natl Acad Sci U S A. 2008;105(43):16484–16489. doi: 10.1073/pnas.0806574105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Chan DA, et al. Targeting GLUT1 and the Warburg effect in renal cell carcinoma by chemical synthetic lethality. Sci Transl Med. 2011;3(94):94ra70. doi: 10.1126/scitranslmed.3002394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 12(1):21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Inoki K, Zhu T, Guan KL. TSC2 mediates cellular energy response to control cell growth and survival. Cell. 2003;115(5):577–590. doi: 10.1016/s0092-8674(03)00929-2. [DOI] [PubMed] [Google Scholar]
- 155.Brugarolas J, et al. Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/TSC2 tumor suppressor complex. Genes Dev. 2004;18(23):2893–2904. doi: 10.1101/gad.1256804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Huang J, Manning BD. The TSC1-TSC2 complex: a molecular switchboard controlling cell growth. Biochem J. 2008;412(2):179–190. doi: 10.1042/BJ20080281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Sancak Y, et al. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320(5882):1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol. 2009;10(5):307–318. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- 159.He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Sancak Y, et al. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141(2):290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Interferon-alpha and survival in metastatic renal carcinoma: early results of a randomised controlled trial. Medical Research Council Renal Cancer Collaborators. Lancet. 1999;353(9146):14–17. [PubMed] [Google Scholar]
- 162.Sancak Y, et al. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 141(2):290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Sancak Y, et al. The Rag GTPases Bind Raptor and Mediate Amino Acid Signaling to mTORC1. Science. 2008;320(5882):1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Blagosklonny MV, Hall MN. Growth and aging: a common molecular mechanism. Aging (Albany NY) 2009;1(4):357–362. doi: 10.18632/aging.100040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Zhong H, et al. Modulation of hypoxia-inducible factor 1alpha expression by the epidermal growth factor/phosphatidylinositol 3-kinase/PTEN/AKT/FRAP pathway in human prostate cancer cells: implications for tumor angiogenesis and therapeutics. Cancer Res. 2000;60(6):1541–1545. [PubMed] [Google Scholar]
- 166.Zundel W, et al. Loss of PTEN facilitates HIF-1-mediated gene expression. Genes Dev. 2000;14(4):391–396. [PMC free article] [PubMed] [Google Scholar]
- 167.Jiang BH, et al. Phosphatidylinositol 3-kinase signaling controls levels of hypoxia-inducible factor 1. Cell Growth Differ. 2001;12(7):363–369. [PubMed] [Google Scholar]
- 168.Hudson CC, et al. Regulation of hypoxia-inducible factor 1alpha expression and function by the mammalian target of rapamycin. Mol Cell Biol. 2002;22(20):7004–7014. doi: 10.1128/MCB.22.20.7004-7014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Brugarolas JB, et al. TSC2 regulates VEGF through mTOR-dependent and - independent pathways. Cancer Cell. 2003;4(2):147–158. doi: 10.1016/s1535-6108(03)00187-9. [DOI] [PubMed] [Google Scholar]
- 170.Thomas GV, et al. Hypoxia-inducible factor determines sensitivity to inhibitors of mTOR in kidney cancer. Nat Med. 2006;12(1):122–127. doi: 10.1038/nm1337. [DOI] [PubMed] [Google Scholar]
- 171.Porstmann T, et al. SREBP activity is regulated by mTORC1 and contributes to Akt-dependent cell growth. Cell Metab. 2008;8(3):224–236. doi: 10.1016/j.cmet.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Duvel K, et al. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol Cell. 39(2):171–183. doi: 10.1016/j.molcel.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Pena-Llopis S, et al. Regulation of TFEB and V-ATPases by mTORC1. EMBO J. 30(16):3242–3258. doi: 10.1038/emboj.2011.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Sardiello M, et al. A gene network regulating lysosomal biogenesis and function. Science. 2009;325(5939):473–477. doi: 10.1126/science.1174447. [DOI] [PubMed] [Google Scholar]
- 175.Davis IJ, et al. Cloning of an Alpha-TFEB fusion in renal tumors harboring the t(6;11)(p21;q13) chromosome translocation. Proc Natl Acad Sci U S A. 2003;100(10):6051–6056. doi: 10.1073/pnas.0931430100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Kuiper RP, et al. Upregulation of the transcription factor TFEB in t(6;11(p21;q13)-positive renal cell carcinomas due to promoter substitution. Hum Mol Genet. 2003;12(14):1661–1669. doi: 10.1093/hmg/ddg178. [DOI] [PubMed] [Google Scholar]
- 177.Furge KA, et al. Combining differential expression, chromosomal and pathway analyses for the molecular characterization of renal cell carcinoma. Can Urol Assoc J. 2007;1(2 Suppl):S21–S27. doi: 10.5489/cuaj.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Furge KA, et al. Robust classification of renal cell carcinoma based on gene expression data and predicted cytogenetic profiles. Cancer Res. 2004;64(12):4117–4121. doi: 10.1158/0008-5472.CAN-04-0534. [DOI] [PubMed] [Google Scholar]
- 179.Takahashi M, et al. Gene expression profiling of clear cell renal cell carcinoma: gene identification and prognostic classification. Proc Natl Acad Sci U S A. 2001;98(17):9754–9759. doi: 10.1073/pnas.171209998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Tan MH, et al. Genomic expression and single-nucleotide polymorphism profiling discriminates chromophobe renal cell carcinoma and oncocytoma. BMC Cancer. 2010;10:196. doi: 10.1186/1471-2407-10-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Yang XJ, et al. Classification of renal neoplasms based on molecular signatures. J Urol. 2006;175(6):2302–2306. doi: 10.1016/S0022-5347(06)00255-2. [DOI] [PubMed] [Google Scholar]
- 182.Junker K, et al. Genetic subtyping of renal cell carcinoma by comparative genomic hybridization. Recent Results Cancer Res. 2003;162:169–175. doi: 10.1007/978-3-642-59349-9_15. [DOI] [PubMed] [Google Scholar]
- 183.Allory Y, et al. Papillary renal cell carcinoma. Prognostic value of morphological subtypes in a clinicopathologic study of 43 cases. Virchows Arch. 2003;442(4):336–342. doi: 10.1007/s00428-003-0787-1. [DOI] [PubMed] [Google Scholar]
- 184.Antonelli A, et al. Cytogenetic features, clinical significance and prognostic impact of type 1 and type 2 papillary renal cell carcinoma. Cancer Genet Cytogenet. 2010;199(2):128–133. doi: 10.1016/j.cancergencyto.2010.02.013. [DOI] [PubMed] [Google Scholar]
- 185.Gontero P, et al. Prognostic factors in a prospective series of papillary renal cell carcinoma. BJU Int. 2008;102(6):697–702. doi: 10.1111/j.1464-410X.2008.07756.x. [DOI] [PubMed] [Google Scholar]
- 186.Gunawan B, et al. Cytogenetic and morphologic typing of 58 papillary renal cell carcinomas: evidence for a cytogenetic evolution of type 2 from type 1 tumors. Cancer Res. 2003;63(19):6200–6205. [PubMed] [Google Scholar]
- 187.Okon K, Sinczak-Kuta A, Stachura J. Renal papillary carcinoma classification into subtypes may be reproduced by nuclear morphometry. Anal Quant Cytol Histol. 2009;31(2):109–117. [PubMed] [Google Scholar]
- 188.Klatte T, et al. Cytogenetic and molecular tumor profiling for type 1 and type 2 papillary renal cell carcinoma. Clin Cancer Res. 2009;15(4):1162–1169. doi: 10.1158/1078-0432.CCR-08-1229. [DOI] [PubMed] [Google Scholar]
- 189.Kaelin WG., Jr The von Hippel-Lindau tumor suppressor protein and clear cell renal carcinoma. Clin Cancer Res. 2007;13(2 Pt 2):680s–684s. doi: 10.1158/1078-0432.CCR-06-1865. [DOI] [PubMed] [Google Scholar]
- 190.Wuttig D, et al. Gene signatures of pulmonary metastases of renal cell carcinoma reflect the disease-free interval and the number of metastases per patient. Int J Cancer. 2009;125(2):474–482. doi: 10.1002/ijc.24353. [DOI] [PubMed] [Google Scholar]
- 191.Sanjmyatav J, et al. A specific gene expression signature characterizes metastatic potential in clear cell renal cell carcinoma. J Urol. 2011;186(1):289–294. doi: 10.1016/j.juro.2011.03.033. [DOI] [PubMed] [Google Scholar]
- 192.Rini BI, et al. Identification of prognostic genomic markers in patients with localized clear cell renal cell carcinoma (ccRCC) J Clin Oncol. 2010;28(15s) p. Auppl; abstr 4501. [Google Scholar]
- 193.Zhao H, et al. Gene expression profiling predicts survival in conventional renal cell carcinoma. PLoS Med. 2006;3(1):e13. doi: 10.1371/journal.pmed.0030013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Zhao H, et al. Alteration of gene expression signatures of cortical differentiation and wound response in lethal clear cell renal cell carcinomas. PLoS One. 2009;4(6):e6039. doi: 10.1371/journal.pone.0006039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Brannon AR, et al. Molecular Stratification of Clear Cell Renal Cell Carcinoma by Consensus Clustering Reveals Distinct Subtypes and Survival Patterns. Genes Cancer. 2010;1(2):152–163. doi: 10.1177/1947601909359929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Brannon AR, et al. Meta-analysis of clear cell renal cell carcinoma gene expression defines a variant subgroup and identifies gender influences on tumor biology. Eur Urol. 2012;61(2):258–268. doi: 10.1016/j.eururo.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Rohan SM, et al. Clear-cell papillary renal cell carcinoma: molecular and immunohistochemical analysis with emphasis on the von Hippel-Lindau gene and hypoxia-inducible factor pathway-related proteins. Mod Pathol. 2011;24(9):1207–1220. doi: 10.1038/modpathol.2011.80. [DOI] [PubMed] [Google Scholar]
- 198.Sandlund J, et al. Prognostic impact of carbonic anhydrase IX expression in human renal cell carcinoma. BJU international. 2007;100(3):556–560. doi: 10.1111/j.1464-410X.2007.07006.x. [DOI] [PubMed] [Google Scholar]
- 199.Zamparese R, et al. Survivin expression in renal cell carcinoma. Cancer investigation. 2008;26(9):929–935. doi: 10.1080/07357900802017553. [DOI] [PubMed] [Google Scholar]
- 200.Baytekin F, et al. Significance of P-glycoprotein, P53, and survivin expression in renal cell carcinoma. Urologic oncology. 2009 doi: 10.1016/j.urolonc.2009.09.001. [DOI] [PubMed] [Google Scholar]
- 201.Lei Y, et al. Prognostic significance of survivin expression in renal cell cancer and its correlation with radioresistance. Molecular and cellular biochemistry. 2010;344(1–2):23–31. doi: 10.1007/s11010-010-0525-3. [DOI] [PubMed] [Google Scholar]
- 202.Parker AS, et al. High expression levels of survivin protein independently predict a poor outcome for patients who undergo surgery for clear cell renal cell carcinoma. Cancer. 2006;107(1):37–45. doi: 10.1002/cncr.21952. [DOI] [PubMed] [Google Scholar]
- 203.Dudderidge TJ, et al. Mcm2, Geminin, and KI67 define proliferative state and are prognostic markers in renal cell carcinoma. Clinical cancer research : an official journal of the American Association for Cancer Research. 2005;11(7):2510–2517. doi: 10.1158/1078-0432.CCR-04-1776. [DOI] [PubMed] [Google Scholar]
- 204.Lam JS, et al. Clinicopathologic and molecular correlations of necrosis in the primary tumor of patients with renal cell carcinoma. Cancer. 2005;103(12):2517–2525. doi: 10.1002/cncr.21127. [DOI] [PubMed] [Google Scholar]
- 205.Bui MH, et al. Prognostic value of carbonic anhydrase IX and KI67 as predictors of survival for renal clear cell carcinoma. The Journal of urology. 2004;171(6 Pt 1):2461–2466. doi: 10.1097/01.ju.0000116444.08690.e2. [DOI] [PubMed] [Google Scholar]
- 206.Tollefson MK, et al. Ki-67 and coagulative tumor necrosis are independent predictors of poor outcome for patients with clear cell renal cell carcinoma and not surrogates for each other. Cancer. 2007;110(4):783–790. doi: 10.1002/cncr.22840. [DOI] [PubMed] [Google Scholar]
- 207.Thompson RH, et al. Tumor B7-H1 is associated with poor prognosis in renal cell carcinoma patients with long-term follow-up. Cancer research. 2006;66(7):3381–3385. doi: 10.1158/0008-5472.CAN-05-4303. [DOI] [PubMed] [Google Scholar]
- 208.Crispen PL, et al. Tumor cell and tumor vasculature expression of B7-H3 predict survival in clear cell renal cell carcinoma. Clinical cancer research : an official journal of the American Association for Cancer Research. 2008;14(16):5150–5157. doi: 10.1158/1078-0432.CCR-08-0536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Frigola X, et al. Identification of a soluble form of B7-H1 that retains immunosuppressive activity and is associated with aggressive renal cell carcinoma. Clinical cancer research : an official journal of the American Association for Cancer Research. 2011;17(7):1915–1923. doi: 10.1158/1078-0432.CCR-10-0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Jiang Z, et al. Analysis of RNA-binding protein IMP3 to predict metastasis and prognosis of renal-cell carcinoma: a retrospective study. The lancet oncology. 2006;7(7):556–564. doi: 10.1016/S1470-2045(06)70732-X. [DOI] [PubMed] [Google Scholar]
- 211.Hoffmann NE, et al. External validation of IMP3 expression as an independent prognostic marker for metastatic progression and death for patients with clear cell renal cell carcinoma. Cancer. 2008;112(7):1471–1479. doi: 10.1002/cncr.23296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Gunawan B, et al. Prognostic impacts of cytogenetic findings in clear cell renal cell carcinoma: gain of 5q31-qter predicts a distinct clinical phenotype with favorable prognosis. Cancer research. 2001;61(21):7731–7738. [PubMed] [Google Scholar]
- 213.Klatte T, et al. Cytogenetic profile predicts prognosis of patients with clear cell renal cell carcinoma. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2009;27(5):746–753. doi: 10.1200/JCO.2007.15.8345. [DOI] [PubMed] [Google Scholar]
- 214.Monzon FA, et al. Chromosome 14q loss defines a molecular subtype of clear-cell renal cell carcinoma associated with poor prognosis. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc. 2011 doi: 10.1038/modpathol.2011.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Brunelli M, et al. Loss of chromosome 9p is an independent prognostic factor in patients with clear cell renal cell carcinoma. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc. 2008;21(1):1–6. doi: 10.1038/modpathol.3800967. [DOI] [PubMed] [Google Scholar]
- 216.La Rochelle J, et al. Chromosome 9p deletions identify an aggressive phenotype of clear cell renal cell carcinoma. Cancer. 2010;116(20):4696–4702. doi: 10.1002/cncr.25279. [DOI] [PubMed] [Google Scholar]
- 217.Zhao H, et al. Gene expression profiling predicts survival in conventional renal cell carcinoma. PLoS medicine. 2006;3(1):e13. doi: 10.1371/journal.pmed.0030013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Brannon AR, et al. Molecular Stratification of Clear Cell Renal Cell Carcinoma by Consensus Clustering Reveals Distinct Subtypes and Survival Patterns. Genes & cancer. 2010;1(2):152–163. doi: 10.1177/1947601909359929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Rini BI, et al. Identification of prognostic genomic biomarkers in patients with localized clear cell renal cell carcinoma. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2010;28 (suppl; abstr 4501) [Google Scholar]
- 220.Banks RE, et al. Genetic and epigenetic analysis of von Hippel-Lindau (VHL) gene alterations and relationship with clinical variables in sporadic renal cancer. Cancer research. 2006;66(4):2000–2011. doi: 10.1158/0008-5472.CAN-05-3074. [DOI] [PubMed] [Google Scholar]
- 221.Patard JJ, et al. Low CAIX expression and absence of VHL gene mutation are associated with tumor aggressiveness and poor survival of clear cell renal cell carcinoma. Int J Cancer. 2008;123(2):395–400. doi: 10.1002/ijc.23496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Smits KM, et al. Genetic and epigenetic alterations in the von hippel-lindau gene: the influence on renal cancer prognosis. Clinical cancer research : an official journal of the American Association for Cancer Research. 2008;14(3):782–787. doi: 10.1158/1078-0432.CCR-07-1753. [DOI] [PubMed] [Google Scholar]
- 223.Choueiri TK, et al. von Hippel-Lindau gene status and response to vascular endothelial growth factor targeted therapy for metastatic clear cell renal cell carcinoma. The Journal of urology. 2008;180(3):860–865. doi: 10.1016/j.juro.2008.05.015. discussion 865-6. [DOI] [PubMed] [Google Scholar]
- 224.Hutson TE, et al. Biomarker analysis and final efficacy and safety results of a phase II renal cell carcinoma trial with pazopanib (GW786034), a multi-kinase angiogenesis inhibitor. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2008;26 p. abstr 5046. [Google Scholar]
- 225.Escudier B, et al. Sorafenib for treatment of renal cell carcinoma: Final efficacy and safety results of the phase III treatment approaches in renal cancer global evaluation trial. J Clin Oncol. 2009;27(20):3312–3318. doi: 10.1200/JCO.2008.19.5511. [DOI] [PubMed] [Google Scholar]
- 226.Dosquet C, et al. Are angiogenic factors, cytokines, and soluble adhesion molecules prognostic factors in patients with renal cell carcinoma? Clin Cancer Res. 1997;3(12 Pt 1):2451–2458. [PubMed] [Google Scholar]
- 227.Jacobsen J, et al. Vascular endothelial growth factor as prognostic factor in renal cell carcinoma. J Urol. 2000;163(1):343–347. [PubMed] [Google Scholar]
- 228.Negrier S, et al. Interleukin-6, interleukin-10, and vascular endothelial growth factor in metastatic renal cell carcinoma: prognostic value of interleukin-6--from the Groupe Francais d'Immunotherapie. J Clin Oncol. 2004;22(12):2371–2378. doi: 10.1200/JCO.2004.06.121. [DOI] [PubMed] [Google Scholar]
- 229.Negrier S, et al. Serum level of vascular endothelial growth factor (VEGF) as an independent prognostic factor in metastatic renal cell carcinoma (MRCC) J Clin Oncol. 2007;25(18S):5044. [Google Scholar]
- 230.Pena C, et al. Biomarkers predicting outcome in patients with advanced renal cell carcinoma: Results from sorafenib phase III Treatment Approaches in Renal Cancer Global Evaluation Trial. Clinical cancer research : an official journal of the American Association for Cancer Research. 2010;16(19):4853–4863. doi: 10.1158/1078-0432.CCR-09-3343. [DOI] [PubMed] [Google Scholar]
- 231.Escudier BJ, et al. Update on AVOREN trial in metastatic renal cell carcinoma (mRCC): efficacy and safety in subgroups of patients (pts) and pharmacokinetic (PK) analysis. J Clin Oncol. 2008;26:15S. (suppl; abstr 5025) [Google Scholar]
- 232.Zurita AJ, et al. A cytokine and angiogenic factor (CAF) analysis in plasma for selection of sorafenib therapy in patients with metastatic renal cell carcinoma. Annals of oncology : official journal of the European Society for Medical Oncology / ESMO. 2011 doi: 10.1093/annonc/mdr047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Liu Y, et al. Circulating baseline plasma cytokines and angiogenic factors (CAF) as markers of tumor burden and therapeutic response in a phase III study of pazopanib for metastatic renal cell carcinoma (mRCC) Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2011;29 (suppl; abstr 4553) [Google Scholar]
- 234.Huang D, et al. Interleukin-8 mediates resistance to antiangiogenic agent sunitinib in renal cell carcinoma. Cancer research. 2010;70(3):1063–1071. doi: 10.1158/0008-5472.CAN-09-3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Gleave ME, et al. Interferon Gamma-1b Compared with Placebo in Metastatic Renal-Cell Carcinoma. New England Journal of Medicine. 1998;338(18):1265–1271. doi: 10.1056/NEJM199804303381804. [DOI] [PubMed] [Google Scholar]
- 236.Vogelzang NJ, Priest ER, Borden L. Spontaneous regression of histologically proved pulmonary metastases from renal cell carcinoma: a case with 5-year followup. The Journal of urology. 1992;148(4):1247–1248. doi: 10.1016/s0022-5347(17)36874-x. [DOI] [PubMed] [Google Scholar]
- 237.Oliver RTD, Nethersell ABW, Bottomley JM. Unexplained Spontaneous Regression and Alpha-interferon as Treatment for Metastatic Renal Carcinoma. British Journal of Urology. 1989;63(2):128–131. doi: 10.1111/j.1464-410x.1989.tb05147.x. [DOI] [PubMed] [Google Scholar]
- 238.Marten A, et al. Therapeutic vaccination against metastatic renal cell carcinoma by autologous dendritic cells: preclinical results and outcome of a first clinical phase I/II trial. Cancer Immunol Immunother. 2002;51(11–12):637–644. doi: 10.1007/s00262-002-0324-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.McDermott DF, Rini BI. Immunotherapy for metastatic renal cell carcinoma. BJU Int. 2007;99(5 Pt B):1282–1288. doi: 10.1111/j.1464-410X.2007.06818.x. [DOI] [PubMed] [Google Scholar]
- 240.Coppin C, et al. Immunotherapy for advanced renal cell cancer. Cochrane Database Syst Rev. 2005;(1):CD001425. doi: 10.1002/14651858.CD001425.pub2. [DOI] [PubMed] [Google Scholar]
- 241.Fyfe G, et al. Results of treatment of 255 patients with metastatic renal cell carcinoma who received high-dose recombinant interleukin-2 therapy. J Clin Oncol. 1995;13(3):688–696. doi: 10.1200/JCO.1995.13.3.688. [DOI] [PubMed] [Google Scholar]
- 242.Fisher RI, Rosenberg SA, Fyfe G. Long-term survival update for high-dose recombinant interleukin-2 in patients with renal cell carcinoma. Cancer J Sci Am. 2000;6(Suppl 1):S55–S57. [PubMed] [Google Scholar]
- 243.Yang JC, et al. Randomized Study of High-Dose and Low-Dose Interleukin-2 in Patients With Metastatic Renal Cancer. Journal of Clinical Oncology. 2003;21(16):3127–3132. doi: 10.1200/JCO.2003.02.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.McDermott DF, et al. Randomized phase III trial of high-dose interleukin-2 versus subcutaneous interleukin-2 and interferon in patients with metastatic renal cell carcinoma. J Clin Oncol. 2005;23(1):133–141. doi: 10.1200/JCO.2005.03.206. [DOI] [PubMed] [Google Scholar]
- 245.McDermott DF, et al. The high-dose Aldesleukin (HD IL-2) “SELECT” trial in patients with metastatic renal cell carcinoma (mRCC) J Clin Oncol. 2010;28:15s. doi: 10.3816/CGC.2009.n.014. abstr 4514. [DOI] [PubMed] [Google Scholar]
- 246.Oudard S, et al. Phase II study of vinorelbine in patients with androgen-independent prostate cancer. Annals of oncology : official journal of the European Society for Medical Oncology / ESMO. 2001;12(6):847–852. doi: 10.1023/a:1011141611560. [DOI] [PubMed] [Google Scholar]
- 247.Escudier B, et al. Cytokines in Metastatic Renal Cell Carcinoma: Is It Useful to Switch to Interleukin-2 or Interferon After Failure of a First Treatment? Journal of Clinical Oncology. 1999;17(7):2039. doi: 10.1200/JCO.1999.17.7.2039. [DOI] [PubMed] [Google Scholar]
- 248.Motzer RJ, et al. Sunitinib versus interferon alfa in metastatic renal-cell carcinoma. N Engl J Med. 2007;356(2):115–124. doi: 10.1056/NEJMoa065044. [DOI] [PubMed] [Google Scholar]
- 249.Hudes G, et al. Temsirolimus, interferon alfa, or both for advanced renal-cell carcinoma. N Engl J Med. 2007;356(22):2271–2281. doi: 10.1056/NEJMoa066838. [DOI] [PubMed] [Google Scholar]
- 250.Harding FA, et al. CD28-mediated signalling co-stimulates murine T cells and prevents induction of anergy in T-cell clones. Nature. 1992;356(6370):607–609. doi: 10.1038/356607a0. [DOI] [PubMed] [Google Scholar]
- 251.Peggs KS, et al. Cancer Immunotherapy, in Cancer Medicine. 2010 [Google Scholar]
- 252.Krummel MF, Allison JP. CD28 and CTLA-4 have opposing effects on the response of T cells to stimulation. The Journal of Experimental Medicine. 1995;182(2):459–465. doi: 10.1084/jem.182.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Mittendorf EA, Sharma P. Mechanisms of T-cell inhibition: implications for cancer immunotherapy. Expert Rev Vaccines. 9(1):89–105. doi: 10.1586/erv.09.144. [DOI] [PubMed] [Google Scholar]
- 254.Leach DR, Krummel MF, Allison JP. Enhancement of Antitumor Immunity by CTLA-4 Blockade. Science. 1996;271(5256):1734–1736. doi: 10.1126/science.271.5256.1734. [DOI] [PubMed] [Google Scholar]
- 255.van Elsas A, Hurwitz AA, Allison JP. Combination immunotherapy of B16 melanoma using anti-cytotoxic T lymphocyte-associated antigen 4 (CTLA-4) and granulocyte/macrophage colony-stimulating factor (GM-CSF)-producing vaccines induces rejection of subcutaneous and metastatic tumors accompanied by autoimmune depigmentation. J Exp Med. 1999;190(3):355–366. doi: 10.1084/jem.190.3.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Hodi FS, et al. Biologic activity of cytotoxic T lymphocyte-associated antigen 4 antibody blockade in previously vaccinated metastatic melanoma and ovarian carcinoma patients. Proc Natl Acad Sci U S A. 2003;100(8):4712–4717. doi: 10.1073/pnas.0830997100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Small EJ, et al. A pilot trial of CTLA-4 blockade with human anti-CTLA-4 in patients with hormone-refractory prostate cancer. Clin Cancer Res. 2007;13(6):1810–1815. doi: 10.1158/1078-0432.CCR-06-2318. [DOI] [PubMed] [Google Scholar]
- 258.Carthon BC, et al. Preoperative CTLA-4 blockade: tolerability and immune monitoring in the setting of a presurgical clinical trial. Clin Cancer Res. 16(10):2861–2871. doi: 10.1158/1078-0432.CCR-10-0569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Hodi FS, et al. Improved Survival with Ipilimumab in Patients with Metastatic Melanoma. New England Journal of Medicine. 363(8):711–723. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Robert C, et al. Ipilimumab plus Dacarbazine for Previously Untreated Metastatic Melanoma. New England Journal of Medicine. 364(26):2517–2526. doi: 10.1056/NEJMoa1104621. [DOI] [PubMed] [Google Scholar]
- 261.Yang JC, et al. Ipilimumab (anti-CTLA4 antibody) causes regression of metastatic renal cell cancer associated with enteritis and hypophysitis. J Immunother. 2007;30(8):825–830. doi: 10.1097/CJI.0b013e318156e47e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Keir ME, et al. PD-1 and its ligands in tolerance and immunity. Annu Rev Immunol. 2008;26:677–704. doi: 10.1146/annurev.immunol.26.021607.090331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Thompson RH, et al. Tumor B7-H1 Is Associated with Poor Prognosis in Renal Cell Carcinoma Patients with Long-term Follow-up. Cancer Research. 2006;66(7):3381–3385. doi: 10.1158/0008-5472.CAN-05-4303. [DOI] [PubMed] [Google Scholar]
- 264.Brahmer J, et al. Safety and activity of MDX-1106 (ONO-4538), an anti-PD-1 monoclonal antibody, in patients with selected refractory or relapsed malignancies. J Clin Oncol. 2008;26 (May 20 suppl):abstr 3006. [Google Scholar]
- 265.Sznol M, et al. Safety and antitumor activity of biweekly MDX-1106 (Anti-PD-1, BMS-936558/ONO-4538) in patients with advanced refractory malignancies. J Clin Oncol. 2010;28:15s. (suppl; abstr 2506) [Google Scholar]
- 266.Rini BI. New strategies in kidney cancer: therapeutic advances through understanding the molecular basis of response and resistance. Clin Cancer Res. 2010;16(5):1348–1354. doi: 10.1158/1078-0432.CCR-09-2273. [DOI] [PubMed] [Google Scholar]
- 267.Motzer RJ, et al. Efficacy of everolimus in advanced renal cell carcinoma: a double-blind, randomised, placebo-controlled phase III trial. Lancet. 2008;372(9637):449–456. doi: 10.1016/S0140-6736(08)61039-9. [DOI] [PubMed] [Google Scholar]
- 268.Hudes G, et al. A phase 3, randomized, 3-arm study of temsirolimus (TEMSR) or interferon-alpha (IFN) or the combination of TEMSR + IFN in the treatment of first-line, poor-risk patients with advanced renal cell carcinoma (adv RCC) Journal of Clinical Oncology, 2006 ASCO Annual Meeting Proceedings Part I. 2006;24(18S):LBA4. [Google Scholar]
- 269.Rini BI, et al. Comparative effectiveness of axitinib versus sorafenib in advanced renal cell carcinoma (AXIS): a randomised phase 3 trial. Lancet. 2011;378(9807):1931–1939. doi: 10.1016/S0140-6736(11)61613-9. [DOI] [PubMed] [Google Scholar]
- 270.Escudier B, et al. Bevacizumab plus interferon alfa-2a for treatment of metastatic renal cell carcinoma: a randomised, double-blind phase III trial. Lancet. 2007;370(9605):2103–2111. doi: 10.1016/S0140-6736(07)61904-7. [DOI] [PubMed] [Google Scholar]
- 271.Rini BI, et al. Bevacizumab plus interferon alfa compared with interferon alfa monotherapy in patients with metastatic renal cell carcinoma: CALGB 90206. J Clin Oncol. 2008;26(33):5422–5428. doi: 10.1200/JCO.2008.16.9847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Escudier B, et al. Sorafenib in advanced clear-cell renal-cell carcinoma. N Engl J Med. 2007;356(2):125–134. doi: 10.1056/NEJMoa060655. [DOI] [PubMed] [Google Scholar]
- 273.Sternberg CN, et al. Pazopanib in locally advanced or metastatic renal cell carcinoma: results of a randomized phase III trial. J Clin Oncol. 2010;28(6):1061–1068. doi: 10.1200/JCO.2009.23.9764. [DOI] [PubMed] [Google Scholar]
- 274.Motzer RJ, et al. Efficacy of everolimus in advanced renal cell carcinoma: a double-blind, randomised, placebo-controlled phase III trial. Lancet. 2008 doi: 10.1016/S0140-6736(08)61039-9. [DOI] [PubMed] [Google Scholar]
- 275.Toschi A, et al. Differential dependence of hypoxia-inducible factors 1 alpha and 2 alpha on mTORC1 and mTORC2. J Biol Chem. 2008;283(50):34495–34499. doi: 10.1074/jbc.C800170200. [DOI] [PMC free article] [PubMed] [Google Scholar]