Summary
Previous work has shown that primary skin-derived fibroblasts from long-lived pituitary dwarf mutants resist the lethal effects of many forms of oxidative and non-oxidative stress. We hypothesized that increased autophagy may protect fibroblasts of Pit-1dw/dw (Snell dwarf) mice from multiple forms of stress. We found dwarf-derived fibroblasts had higher levels of autophagy, using LC3 and p62 as markers, in responses to amino acid deprivation, hydrogen peroxide, and paraquat. Fibroblasts from dwarf mice also showed diminished phosphorylation of mTOR, S6K and 4EBP1, consistent with the higher levels of autophagy in these cells after stress. Similar results were also observed in fibroblasts from mutant mice lacking growth hormone receptor (GHRKO mice) after amino acid withdrawal. Our results suggested that increased autophagy, regulated by TOR-dependent processes, may contribute to stress resistance in fibroblasts from long-lived mutant mice.
Keywords: Autophagy, Snell dwarf, GHRKO, Aging, Oxidative Stress, Amino Acid Deprivation
Introduction
Single gene mutations that impair the strength of insulin and IGF-1 signals slow aging and extend lifespan in mice, worms, and flies (McCulloch & Gems, 2003; Partridge et al., 2011; Bartke, 2011), and there are at least six mutations that extend mouse lifespan through modulation of levels or effects of growth hormone (GH) and/or its mediator IGF-1 (Brown-Borg et al., 1996; Zhou et al., 1997; Bartke et al., 2001; Holzenberger et al., 2003; Conover, 2010). Working out the cellular mechanisms that connect altered hormonal signals to delay of aging and multiple late-life diseases is a principal challenge for biological gerontology.
Snell dwarf mice (dw/dw) carry a recessive mutation in the gene encoding a transcription factor, Pit-1, critical to embryonic development of the anterior pituitary. Snell dwarf mice have an abnormal hormone profile characterized by primary deficiencies in growth hormone (GH), thyroid-stimulating hormone (TSH), and prolactin, and consequently secondary deficiencies in insulin-like growth factor I (IGF-1) and thyroxine (Snell, 1929; Bartke, 2011; Dozmorov et al., 2002). Snell dwarf mice, like the closely related Prop1 mutant Ames dwarf and mice lacking the GH receptor (GHRKO or Laron dwarf mice), exhibit extended lifespan and show deceleration of multiple aspects of aging (Bartke, 2011).
Prior investigations from our laboratory involved analysis of dermal fibroblast cell lines derived from young adult Snell dwarf mice (Salmon et al., 2005; Sun et al., 2009; Leiser and Miller 2010). These cells were found to be resistant, in vitro, to many types of cytotoxic stress, including agents that kill cells at least in part via reactive oxygen species (ROS), such as paraquat (PQ), hydrogen peroxide (H2O2), cadmium (Cd), and others that cause cell death through other pathways, including UV light, the DNA alkylating agent methyl methanesulfonate (MMS), and heat. Similar resistance profiles were also seen in fibroblast cells derived from two other related long-lived mutants, the Ames dwarf mouse, and the GHRKO mice (Salmon et al., 2005). The mechanism of resistance might involve the NF-E2-related factor 2 (Nrf-2) sensitive anti-oxidant genes and mitogen-activated protein kinases (MAPKs) / extracellular signal-regulated protein kinases (Erk) pathway (Sun et al., 2009; Leiser et al., 2010). A good deal of evidence indicates that autophagy may help to maintain cellular homeostasis and cell survival (Reggiori & Klionsky, 2002; Scott et al., 2011). The three types of long-lived dwarf mice all abrogate the strength of GH signals, and diminish plasma levels of IGF-1 and insulin. These alterations in hormones and metabolites may regulate autophagy via the mTOR pathway (Sobolewska et al., 2008). Thus we considered the idea that dwarf-derived cells might show altered control of autophagy with or without stress exposure in vitro.
Autophagy is a catabolic lysosome-dependent process for the degradation of misfolded proteins and damaged organelles such as mitochondria (Reggiori & Klionsky, 2002). Autophagy is maintained at a basal level in most tissues benefiting the balance of cellular homeostasis (Scott et al., 2011). During aging, production of reactive oxygen species (ROS) increases, resulting in elevated levels of oxidative stress, with subsequent cellular stress responses (Finkel & Holbrook, 2000). Increased ROS or oxidative stress response has been shown to induce autophagy (Scherz-Shouval et al., 2007). The primary functions of autophagy are housekeeping and quality control of proteins and organelles. Autophagy can promote cell survival by recycling amino acids and fatty acids for energy utilization during nutrient withdrawal and by removal of oxidazed proteins or damaged organelles during stress. Alternatively, autophagy can lead to cell death if cells are severely damaged. Multiple lines of evidence have demonstrated that the efficiency of autophagic degradation declines with aging and contributes to accumulation of intracellular waste (Brunk & Terman, 2002; Cuervo et al., 2005). The accumulation of impaired macromolecules and organelles, a consequence of impaired autophagy, is a frequent characteristic of age-related change in many cell types. Some findings have suggested that modulation of autophagy might contribute to preserved cellular function in several models of delayed aging and extended longevity. In mice, damaged proteins and organelles accumulate in cytoplasm as a consequence of manipulations that knock down function of several autophagy related genes (Komatsu et al., 2006). In C. elegans, abrogation of autophagy abolished lifespan extension of daf-2 mutants (Melendez et al., 2003). In Drosophila, adult mutants lacking autophagy related gene 7 (Atg7−/−) are short-lived and hypersensitive to nutrient and oxidative stress (Juhász et al., 2007). SIRT1, mTOR, Foxo3, NF-kB and P53, each of which is thought to have a role in longevity determination, all modulate autophagy, though the connections between autophagy and aging are still not well-defined in mammals (Salminen & Kaarniranta, 2009). Rapamycin, which activates autophagy by down-regulating the mTOR pathway, enhances mouse lifespan (Harrison et al., 2009).
These findings in C. elegans, Drosophila and mammals suggest an important role for autophagy as a regulator of the aging process and cellular stress responses, but little is known about autophagic function in mouse mutants in which longevity is increased by reduction of pathways controlled by GH and/or IGF-1. In our study, we tested the hypothesis that fibroblasts grown from Snell dwarf or GHRKO mice might have enhanced autophagy when subjected to oxidative stress or deprivation of amino acids. We also evaluated mTOR/S6k pathways, which regulate autophagy, in fibroblast cells from mutant and control mice.
Results
1, Increased autophagy in fibroblasts derived from Snell dwarf mice after amino acid deprivation
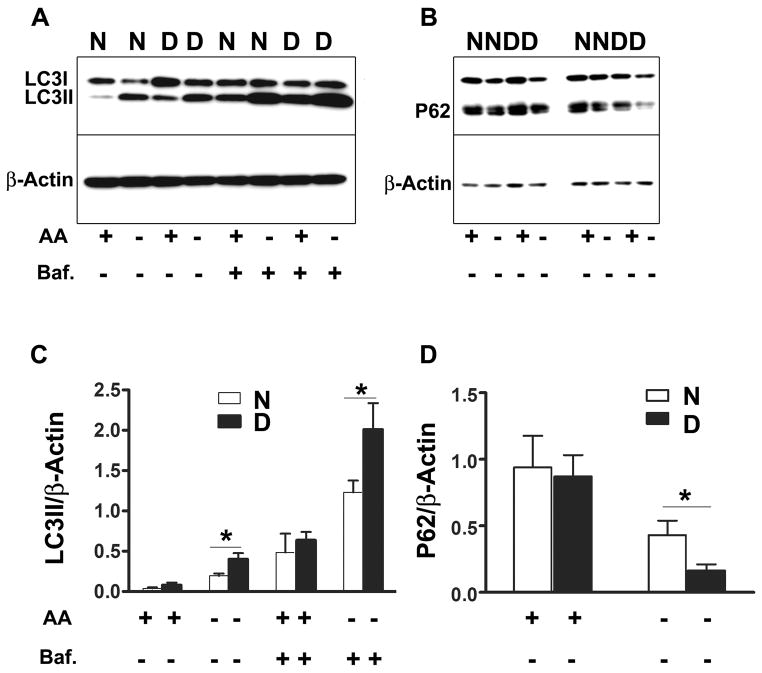
We evaluated autophagy in fibroblasts grown from Snell dwarf and control mice by measuring isoforms of LC3II, with and without Bafilomycin A1 as an inhibitor of lysosomal degradation of LC3II. One hour of incubation in medium without amino acids induced autophagy in fibroblasts of both Snell dwarf and littermate control mice, with higher levels of LC3II accumulation in the cells from the Snell dwarf mice (Figure 1A and C). Bafilomycin A1 increased levels of LC3II in both cell types, again with higher levels in dwarf-derived compared to control cells (Figure 1A and C). An evaluation using immunofluorescence to quantitate LC3 punctae showed a similar pattern of results (Supplemental Figure 1). To confirm these findings, we used an assay based on levels of the ubiquitin binding protein p62, which binds to autophagosomal membrane protein LC3/Atg8. Lysosomal degradation of autophagosomes leads to a decrease in p62 levels during autophagy (Bjørkøy et al., 2005). Consistent with the LC3 results, cells from Snell dwarf mice showed increased degradation of p62 after amino acid withdrawal (Figure 1B and D).
Figure 1. LC3II and p62 protein levels after amino acid deprivation in fibroblasts from Snell dwarf and control mice.
Cells were incubated with Minimum Essential Medium (MEM) supplemented with 10% dialyzed fetal bovine serum (FBS). Amino acid deprivation was initiated by replacing the medium with Earle’s Balance Salt Solution (EBSS) including 10% dialyzed FBS and vitamin mix for 1 hour. Autophagic flux was studied by measuring LC3II with and without the presence of bafilomycin A at 10 nM. Abbreviations: N, Normal cells; D, Snell dwarf cells; AA+, amino acids present; AA−, amino acids absent; Baf., bafilomycin. N=7, Mean ± SEM, *P<0.05.
2, Increased autophagy in fibroblasts derived from Snell dwarf mice after exposure to oxidative stress inducers H2O2 and paraquat
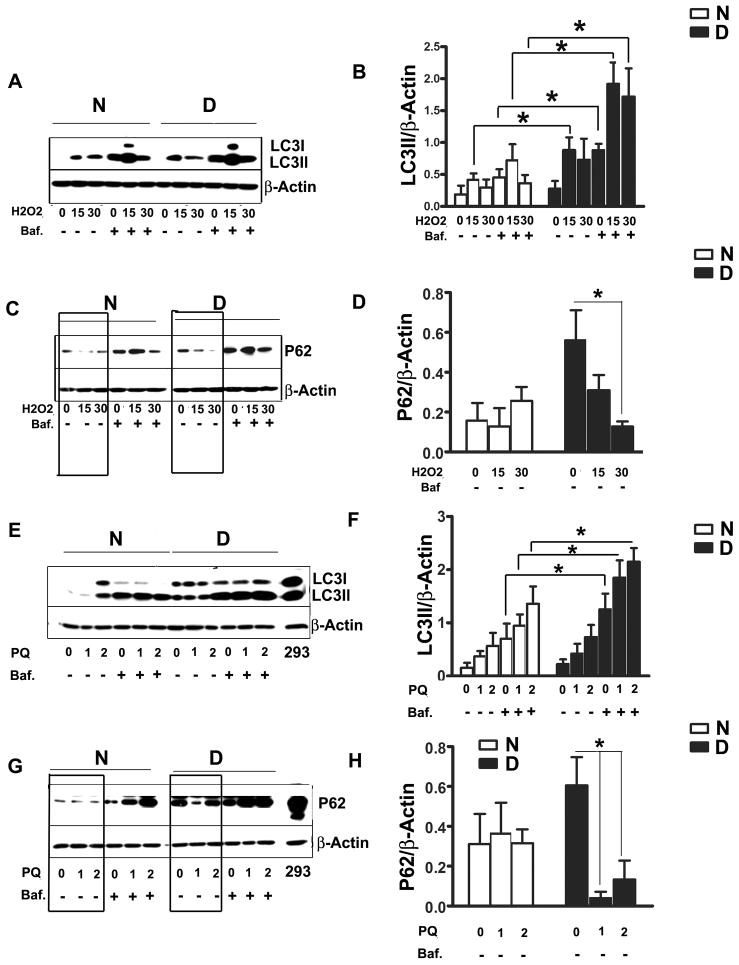
Cells were washed once with PBS and cultured in serum-free medium (DMEM with 2% BSA) for 14–16 hr prior to initiating oxidative stress treatments. H2O2, PQ, or Cd was then added, with or without Bafilomycin A1, in fresh serum free DMEM for 2 hours prior to measurement of LC3II or p62 levels (Figure 2). Peroxide induced higher levels of LC3II in dwarf cells, compared to control cells, at each tested dose and regardless of bafilomycin treatment; the results were statistically significant except at the higher peroxide dose without bafilomycin. The level of LC3II was higher in bafilomycin-exposed dwarf cells than in control cells even prior to peroxide treatment (Figure 2, A and B). Similarly results were observed using PQ as a source of oxidative stress (Figure 2, E and F), with significant differences noted at either PQ dose in the presence of bafilomycin. A similar pattern was also seen using LC3 immunofluorescence (Supplemental Figure 2). P62 was also evaluated in parallel for further confirmation. Prior to stress exposure, dwarf fibroblasts had higher p62 levels, but exposure to H2O2 or PQ, in the absence of bafilomycin led to a decline in p62 levels, an index of autophagy, mainly in the dwarf cells (Figure 2, C, D, G, and H). Neither cells from Snell dwarf mice nor cells from control mice showed any accumulation of LC3II after exposure to cadmium over a range of 1 μM to 20 μM (data not shown).
Figure 2. LC3II and p62 protein levels under oxidative stress conditions in fibroblasts from Snell dwarf and control mice.
Cells were grown in medium without serum (DMEM with 2% BSA) for 14–16 hours prior to oxidative stress. Then cells were treated by H2O2 (15μM or 30μM) or paraquat (PQ, at 1 mM or 2 mM) for 2 hours. LC3II and p62 were measured by immunoblotting. Bafilomycin A at10 nM was added to some cultures as indicated. Human Embryonic Kidney 293 cell lysate was used as a positive control in panels G and H. N=8, Mean ± SEM, *P<0.05.
3, Diminished phosphorylation of mTOR, S6k, and 4EBP1 in Snell dwarf fibroblasts after amino acid deprivation
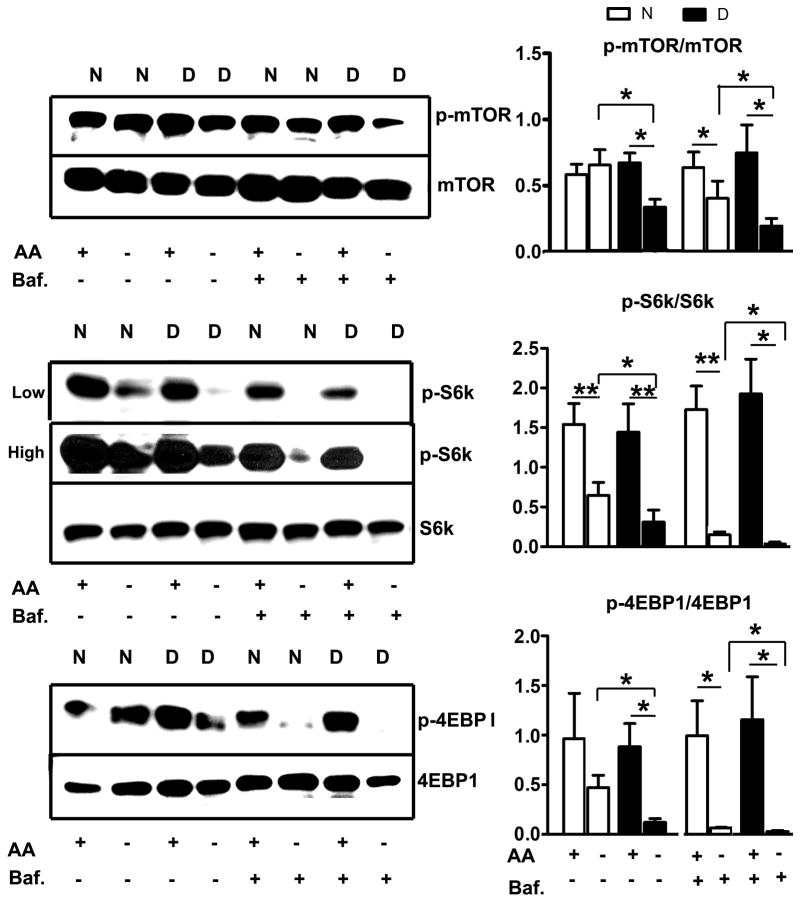
The mTOR signaling pathway is known to regulate autophagy, and inhibition of mTOR results in activation of autophagy (Ravikumar et al., 2004). We therefore evaluated the phosphorylation of mTOR and two of its substrates, S6K and 4EBP1, to see if these might contribute to the increased tendency of dwarf cells to initiate autophagy after amino acid reduction. In control cells, removal of amino acids led to little if any change in mTOR phosphorylation, and a modest decline in phosphorylation of S6K and 4EBP-1 (Figure 3). Cells from dwarf mice, in contrast, showed a significant decline in mTOR phosphorylation, and a more dramatic decline in phosphorylation of S6K and 4EBP-1 as compared to control cells. Dwarf and control cells showed no difference in the total amount of mTOR protein. Interestingly, addition of bafilomycin during amino acid deprivation further decreased the levels of phosphorylation of mTOR, S6K and 4EBP1 in both control and dwarf fibroblasts.
Figure 3. Phosphorylation of mTOR, S6K and 4EBP1 after amino acid deprivation in fibroblasts from Snell dwarf and control mice.
Cells were deprived of amino acids for 1 hour. Phosphorylation and total protein levels of mTOR, S6K and 4EBP1 were evaluated by western blot. N=6, Mean ± SEM, *P<0.05, **P<0.01.
4, Diminished phosphorylation of mTOR/S6K/4EBP1 in fibroblasts derived from Snell dwarf mice exposed to H2O2
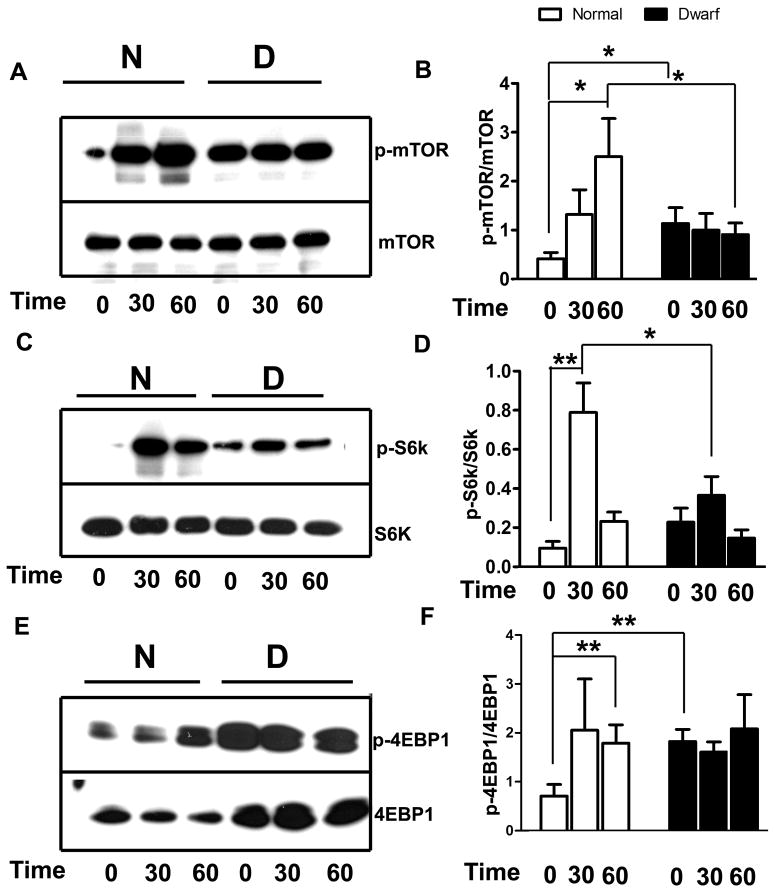
The phosphorylation of mTOR and its two substrates S6K and 4EBP1 was evaluated after exposure to 30μM H2O2 for 30 or 60 min (Figure 4). Prior to H2O2 exposure, levels of phosphorylated mTOR were higher in dwarf fibroblasts than in controls, presumably due to overnight incubation in serum-free medium prior to peroxide exposure. Consistent with this observation, levels of p-4EBP1 were higher in dwarf than in control cells prior to H2O2 exposure, and p-S6K showed a similar, though non-significant, trend. Peroxide exposure for 60 min increased mTOR phosphorylation in the control cells, but did not change mTOR phosphorylation in the dwarf cells, so that levels of phospho-mTOR were higher in control than in dwarf cells after 60 min exposure. We saw no difference in total levels of mTOR between cells derived from dwarf as compared to control cells. Exposure to 30μM H2O2 for 30 min enhanced phosphorylation of S6K in cells from both Snell dwarf and control mice, but the response in control cells was more robust (Figure 4C and D). Control cells, but not dwarf cells, also showed higher levels of 4EBP1 phosphorylation after 60 min of exposure to 30μM H2O2 (Figure 4E and F).
Figure 4. Phosphorylation of mTOR, S6K and 4EBP1 after exposure to H2O2 in fibroblasts from Snell dwarf and control mice.
Cells were cultured overnight in serum-free medium, and then challenged by 30 μM H2O2 for 0, 30 or 60 minutes. Phosphorylation and total protein levels of mTOR, S6K and 4EBP1 were examined. N=6, Mean ± SEM, *P<0.05, **P<0.01.
5, Increased autophagy in fibroblasts derived from GHRKO mice after amino acid deprivation
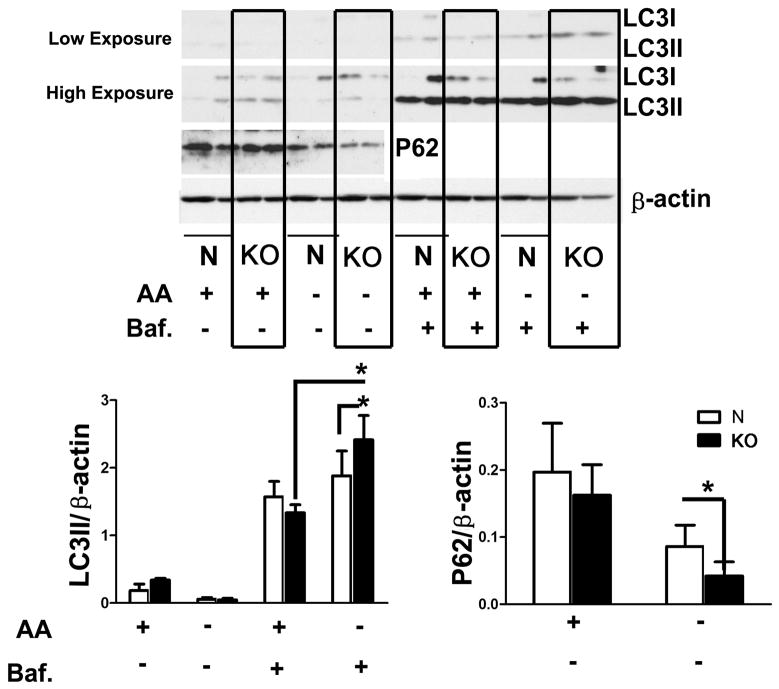
To see if similar effects were produced in cells from a different long-lived, stress resistant mouse stock, we evaluated autophagy in cells from GHRKO mice after withdrawal of amino acids. In contrast to cells from Snell dwarf mice (Figure 1), one hour amino acid withdrawal without bafilomycin did not lead to a pronounced increase in LC3II accumulation either in control or and GHRKO cells, perhaps as a result of more robust lysosomal degradation of LC3II in this system. When autophagic flux was evaluated after addition of bafilomycin, LC3II accumlation was enhanced both in control and GHRKO cells, was higher levels in the GHRKO cells (Figure 5), consistent with the results seen in Snell dwarf cells (Figure 1). A similar pattern of results was seen in the immunofluorescence system (Supplemental Figure 3). Analysis of P62 levels (Figure 5) supported the conclusion that amino acid withdrawal leads to stronger induction of autophagy in fibroblasts from GHRKO mice than in control cells.
Figure 5. LC3II and p62 protein levels after amino acid deprivation in fibroblasts from GHRKO and control mice.
Amino acid deprivation was performed by incubating the cells with EBSS including 10% dialyzed FBS and vitamin mix for 1 hour. Autophagic flux was studied by measuring LC3II with and without the presence of bafilomycin A at 10 nM. Abbreviations: N, Normal cells; KO, Growth Hormone Receptor Knockout cells; AA+, amino acids present; AA−, amino acids absent; Baf., bafilomycin. N=4, Mean ± SEM, *P<0.05.
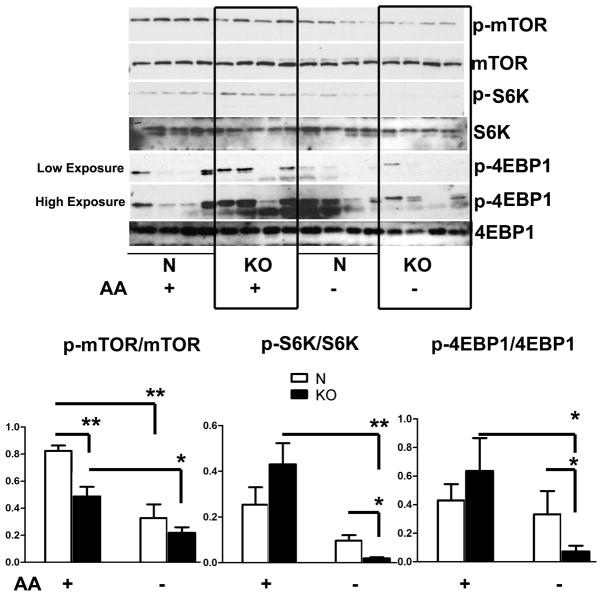
6, Diminished phosphorylation of S6K and 4EBP1 in GHRKO fibroblasts after amino acid deprivation
In parallel, we also evaluated mTOR signaling pathways in cells from GHRKO mice (Figure 6). mTOR phosphorylation was significantly lower in GHRKO cells even prior to amino acid withdrawal. Amino acid starvation for one hour decreased mTOR phosphorylation in control and KO cells, but no significant difference was seen between control and KO cells after amino acid withdrawal. As expected, incubation without amino acids led to lower levels of phosphorylation of both S6K and 4EBP1, and this decline was significantly more dramatic in GHRKO KO, consistent with the observations on cells from Snell dwarf mice (Figure 3).
Figure 6. Phosphorylation of mTOR, S6K and 4EBP1 after amino acid deprivation in fibroblasts from GHRKO and control mice.
Cells were deprived of amino acids for 1 hour. Phosphorylation and total protein levels of mTOR, S6K and 4EBP1 were evaluated by western blot. N=4, Mean ± SEM, *P<0.05, **P<0.01.
Discussion
We show here that fibroblasts from Snell dwarf mice differ from controls in several ways: (a) dwarf cells show more vigorous induction of autophagy in responses to amino acid deprivation and to oxidant stress; (b) amino acid withdrawal induces a more dramatic decline in phosphorylation of mTOR and its substrates in dwarf-derived cells; and (c) dwarf cells are more resistant than controls to mTOR activation induced by exposure to hydrogen peroxide. Consistent results were seen using three methods: LC3II accumulation, p62 reduction, and immunofluorescent assessment of LC3 punctae. Similar results were seen using fibroblasts from another long-lived mouse with altered GH and IGF-1 signals, the GHRKO mutant. These observations are consistent with a model in which in vivo exposure to the altered hormonal milieu of the Snell dwarf and GHRKO mice leads to long-lasting epigenetic changes, retained in fibroblast cells in vitro, that increase their resistance to multiple forms of lethal stress (Salmon et al., 2005), augment plasma membrane redox pumps and other Nrf-2 sensitive defences (Leiser et al., 2010), and enhance expression of some ERK-dependent immediate early genes despite the lower induction of ERK1/2 phosphorylation (Sun et al., 2009).
Autophagy serves housekeeping functions by self-digestion of long-lived proteins and organelles, and is essential for maintaining healthy cells. Once autophagy is initiated, cytoplasmic materials become enclosed in a double membrane structure, which subsequently fuses with a lysosome leading to the degradation of damaged or unwanted components. The formation of autophagosomes requires a process which is called Light chain 3 (LC3)-modification. During this process, LC3II is formed by lipidation of LC3I, an orthologue of yeast Atg8, and becomes incorporated into the autophagosomal membrane. Thus conversion from LC3I to LC3II has been used as an index of autophagy (Kabeya et al., 2004). However, LC3II itself is degraded by autophagy, and therefore, it is often helpful to measure autophagic flux by comparing the amount of LC3II in the presence and absence of lysosome protease inhibitors such as Bafilomycin A1, a specific inhibitor of vacuolar-type (H+)-ATPase (Mizushima & Yoshimori, 2007). P62, also named sequestosome 1 (SQSTM1), binds to LC3 and is involved in the navigation of polyubiquitinated protein aggregates to the autophagic machinery (Bjørkøy et al., 2005). The level of p62 can also be used as an index of autophagy, because autophagic degradation of p62 in autophagosomes leads to decline in p62 levels, and conversely, autophagy inhibitors stabilize p62 levels. Accumulation of LC3 into lysosomes can be evaluated by immunofluorescence microscopy, providing a third index of induced autophagy (Klionsky et al., 2008). Although each of these autophagy indicators is potentially subject to different forms of technical artifacts, the agreement among all of these assay systems increases our confidence that fibroblasts from dwarf mice are indeed more prone to autophagy than control cells in responses to both amino acid withdrawal and oxidative stress.
Insufficient autophagy is thought to be at least partly responsible for a range of age-related diseases (Brunk & Terman, 2002; Cuervo et al., 2005). Levels of expression of the autophagy-related gene Atg7 and of LC3 protein exhibit a significant decline, as levels of p62 and polyubiquitin accumulate concomitant with decreased autophagy in aging rat kidney (Cui et al., 2011). Similar phenomena have been documented in liver and thymus of aging mice (Uddin et al., 2011). Abolishment of several autophagy related genes results in increased aggregates of damaged proteins and organelles in mice (Komatsu et al., 2006), abrogates lifespan extension in daf-2 mutants of C. elegans (Melendez et al., 2003), and shortens the lifespan in Drosophila adults of stocks hypersensitive to nutrient and oxidative stress (Juhász et al., 2007). Many of the pathways implicated in control of lifespan in invertebrates, mice, and humans, including SIRT1, mTOR, Foxo3, NF-kB and P53, are known to modulate autophagy (Salminen & Kaarniranta, 2009). Similarly rapamycin, an activator of autophagy, extends life span from C. elegans to mammals (Harrison et al., 2009; Bjedov et al., 2010), suggesting that manipulation of autophagy may be able to provide insights into the molecular mechanism of aging processes.
Previous studies have also demonstrated a link between autophagy and oxidative stress. Cellular oxidative stress and increased generation of reactive oxygen species (ROS) have been identified as positive regulators of autophagy, which may lead the cell either to survival or death (Scherz-Shouval et al., 2007). Induced autophagy by oxidative stress contributes to the removal of damaged oxidized proteins, and supports cell survival (Xiong et al., 2007). Conversely, autophagic cell death occurs when oxidative stress is so severe that repair cannot compensate for cellular damage (Kirkland et al., 2002). Our results with peroxide and paraquat suggest that higher levels of autophagy may act as a rescue mechanism which cells from Snell dwarf mice can use to escape from cell death induced by oxidative stressors.
Amino acid withdrawal is a well-known inducer of autophagy. Through autophagy, amino acids are recycled or re-utilized, promoting cell survival (Mortimore & Schworer, 1977). In our experiment, as expected, amino acid withdrawal stimulated autophagy in cells from both Snell dwarf and control mice. Strikingly more robust autophagy activities were observed in Snell dwarf derived cells when amino acids were removed, suggesting that these cells may be prone to autophagy whether induced by nutrient or oxidative stress. Previous work from this laboratory has shown that fibroblasts from Snell dwarf mice are resistant to metabolic changes induced by incubation in medium with very low glucose levels (Leiser et al., 2006), probably reflecting differences in Nrf2-dependent regulation of the plasma membrane redox system (Leiser & Miller, 2010). Studies from other groups also demonstrated the involvement of autophagy in modulation of cell death after oxidative stress, including studies of Atg7 mutation in Drosophila exposed to starvation or oxidative stress. The mutant flies are shorter lived, and show progressive decline of neuronal function, accumulated ubiquitin positive particles in the brain, and have increased autophagic cell death (Juhász et al., 2007).
Target of rapamycin (TOR), an evolutionarily conserved serine/threonine kinase, is a negative regulator of autophagy (Ravikumar et al., 2004). Reduction of TOR signaling has been implicated in life span extension in the context of lower protein biosynthesis (Hansen et al., 2007). mTORC1 affects cell growth and translation via at least two important substrates: the ribosomal p70S6 protein kinase (S6K) and the eukaryotic initiation factor 4E-binding protein 1 (4EBP1) (Sonenberg and Hinnebusch, 2009). As an index of TOR pathway activation, we evaluated the phosphorylation of mTOR on Ser2448 after amino acid deprivation, and found dephosphorylation of mTOR only in Snell dwarf cells. Bafilomycin-mediated inhibition of lysosomal activity increased the sensitivity of TOR Ser2448 to amino acid withdrawal, but did not abrogate the difference between dwarf and control cells. Dwarf fibroblasts also showed a more pronounced decline in phosphorylation of both S6K and 4EBP1 after withdrawal of amino acids, consistent with the data on TOR phosphorylation. Similar results were noted after amino acid withdrawal using cells from GHRKO mice. Diminution of mTOR signaling pathways in response to nutrient deficits is considered to be an adjustment to physiological states that require cell maintenance and repair (Foster and Fingar, 2010), and so it seems likely that the more pronounced decline of mTOR signals seen in dwarf-derived cells may contribute to the enhanced autophagy seen in these cells after amino acid withdrawal. Inhibition of autophagy by bafilomycin rendered both dwarf and control cells more sensitive to the effects of amino acid withdrawal on phosphorylation of mTOR, S6K and 4EBP1. This observation implies that interplay between mTOR signaling and autophagy includes a positive feedback loop that amplifies autophagy activity.
Phosphorylation of mTOR, S6K and 4EBP1 were also evaluated in cells treated with H2O2. mTOR/S6K/4EBP1 signaling activities were intensified in cells from control mice by H2O2, but these responses to oxidant stress were blunted in Snell dwarf cells. Lower levels of TOR function after peroxide stress may also contribute to higher autophagy (Fig 2) and diminished cell death (Salmon et al., 2005) in Snell dwarf fibroblasts confronted with oxidant stress.
Our data thus suggest that the stimulation of autophagy-mediated clearance of damaged cellular components may protect Snell cells from various lethal and non-lethal sources of stress, perhaps through changes in baseline and/or stimulated levels of TOR signals. The stress resistance of primary fibroblast cultures grown from long-lived Snell dwarf mice presumably reflects long-lasting epigenetic change induced, in vivo, by maturation of fibroblast precursor cells in a post-natal environment lacking GH and IGF-1. Differences in stress resistance between Snell dwarf and control-derived cells are not apparent when cultures are established from mice that are only 7 days old (Salmon et al., 2005), and can be prevented or reversed by injecting Ames dwarf mice with GH initiated at 2 weeks of age (Panici et al., 2010). It remains to be seen to what extent the differences seen in cultured fibroblasts also affect cells of other lineages in living mice, although there is evidence (Sun et al., 2011) that some of the differences in stress-kinase function seen in cultured cells also affect liver cells in mice before and after exposure to diquat. Further studies of autophagy and its links to TOR function in intact mice may shed light on the cellular basis of disease resistance and longevity in pituitary dwarf mice.
Experimental Procedures
Animals
Snell dwarf (dw/dw) mice, and heterozygote (dw/+) controls were bred as the progeny of (DW/J × C3H/HeJ)F1 dw/+ males and (DW/J × C3H/HeJ)F1 dw/+ females. GHRKO mice and normal littermate controls were produced by mating heterozygous (+/−) carriers of the disrupted GHR/GHBP gene or by mating homozygous knockout (−/−) males with (+/−) females. Both male and female tail skin biopsies were taken from 4–6 months old mice as in our previous work. Mutant and control mice were gender-matched within each experiment.
Primary and secondary cell cultures
Tail skin biopsies (1.5–2 cm) were obtained and primary fibroblast cells were cultured as previously described (Salmon et al., 2005). Briefly, skin samples were rinsed, minced, and digested overnight in collagenase type II (400 U/ml; Gibco-Invitrogen, Carlsbad, CA) dissolved in Dulbecco’s modified Eagle’s medium (DMEM) at 37° with 5% CO2 in air. Then cells were dislodged, centrifuged, resuspended in DMEM with serum, and seeded at approximately 2.5 ×105 cells in 5 ml of medium into tissue culture flasks of 25cm2 surface area. These initial cultures were considered “passage 0.” Cells were fed at day 3 with replacement of 2/3rds of the medium, and then at day 7 were removed by trypsinization and transferred to 75cm2 flasks at 7.5 ×105 cells/flask in 12 ml complete medium (“passage 1”). This process was repeated, and cells at the third passage were plated at a confluent density of 105 cells/cm2 for assessment of responses to oxidative and nutritional stresses.
Antibodies
The following antibodies were used: LC3B Antibody with catalog #2775, p62/SQSTM1 antibody with catalog #p0067, p-mTOR (S2448) with catalog #2971, mTOR with catalog #2972, p-p70 S6 kinase (T389) with catalog #9205, and p70 S6 kinase with catalog #9202 are from Cell Signaling Technology, Inc. (Danvers, MA 01923); p-4ebp1 (Ser 65/Thr 70) with catalog #Sc-12884-R, 4ebp1 with catalog #Sc-6936, goat anti-rabbit IgG-HRP with catalog #Sc-2301, β-Actin conjugated with horseradish peroxidase (HRP) with catalog # Sc-47778 HRP are from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA 95060).
Cell treatments
For the amino acid deprivation assay, the growth medium was replaced by fresh minimum essential medium (MEM), with 10% dialyzed FBS 24 hours before assay. Amino acid deprivation was achieved by replacing the MEM with Earle’s Balanced Salt Solution (EBSS) supplemented with vitamin mix and dialyzed FBS for the indicated times (Kaushik et al., 2008). For H2O2, paraquat (PQ) and cadmium (Cd) treatment, cells were washed once with PBS and cultured in medium without serum (DMEM with 2% BSA) for 14–16 hr. The cells were then exposed to fresh serum free DMEM containing H2O2, PQ or Cd for the indicated times and doses (Hariharan et al., 2011). Bafilomycin A1 (10 nM), a lysosomal inhibitor, was added to the medium where indicated to compare steady state autophagic activity with measures of cumulative LC3-II concentrations (Mizushima & Yoshimori, 2007).
Immunoblots
Lysates for Western blots were prepared in radioimmuno-precipitation assay (RIPA) buffer (Leiser et al., 2010). Lysates were collected and stored at −80° prior to analysis. LC3 levels were evaluated using SDS-PAGE gels containing 12% acrylamide, and p-mTOR, mTOR, p-S6K and S6K blots were run on SDS-PAGE gels containing 6% acrylamide (Bio-Rad). After electrophoresis, proteins were transferred to an Immobilon-P Transfer Membrane (Millipore). Membranes were blocked in Tris buffered saline containing 0.05% Tween20 (TBS-T) and 5% (w/ v) non-fat dry milk (BioRad) for 1 h. After blocking, membranes were probed overnight with primary antibodies with shaking at 4°C, followed by incubation with secondary antibody for 1 hr, with shaking, at room temperature. Antibody detection was accomplished with SuperSignal West Pico Chemiluminescent Substrate (Pierce). Quantification was performed using ImageQuant software.
Statistical analysis
Statistical analyses were performed using paired t-tests (two-tailed) for one factor comparisons such as comparisons of treated to control cells. Analysis of variance (ANOVA) was used to compare effects and interactions of two factors, for example genotype and treatment. Graphs show the mean of combined experiments, and error bars represent the standard error of the mean.
Supplementary Material
The number of LC3 dots was counted by Image J software. Results shown represent the mean ± SEM for three images from fibroblasts per mice obtained from three mice per genotype. Blue: Nuclei DAPI, Green: LC3. Scale bars represent 50 μm. *P<0.05.
H2O2 (15μM) or PQ (2 mM) was used to induce oxidative stress. Bafilomycin A 10 nM was added to some cultures as indicated. Scale bars represent 50 μm. The number of LC3 dots was counted by Image j software. Results shown represent the mean ± SEM for three images from fibroblasts per mice obtained from three mice per genotype. Blue: Nuclei DAPI, Green: LC3. *P<0.05, **P<0.01.
Scale bars represent 50 μm. Blue: Nuclei DAPI, Green: LC3. The number of LC3 dots was counted by Image j software. Results shown represent the mean ± SEM for three images from fibroblasts per mice obtained from three mice per genotype. *P<0.05, , **P<0.01.
Acknowledgments
This work was supported by NIH grants AG031736 and AG019899, and by an award from the Glenn Foundation for Medical Research. The authors thank Sabrina Friedline and Lisa Burmeister for technical assistance, and thank Dr. John Kopchick for breeding pairs of the GHRKO mice.
References
- Bartke A, Wright C, Mattison JA, Ingram DK, Miller RA, George S, Roth GS. Longevity: Extending the lifespan of long-lived mice. Nature. 2001:414. doi: 10.1038/35106646. [DOI] [PubMed] [Google Scholar]
- Bartke A. Growth hormone, insulin and aging: the benefits of endocrine defects. Exp Gerontol. 2011;46:108–11. doi: 10.1016/j.exger.2010.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjørkøy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–14. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjedov I, Toivonen JM, Kerr F, Slack C, Jacobson J, Foley A, Partridge L. Mechanisms of life span extension by rapamycin in the fruit fly Drosophila melanogaster. Cell metabolism. 2010;11:35–46. doi: 10.1016/j.cmet.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown-Borg, Borg KE, Meliska CJ, Bartke A. Dwarf mice and the ageing process. Nature. 1996;384:33. doi: 10.1038/384033a0. [DOI] [PubMed] [Google Scholar]
- Brunk UT, Terman A. The mitochondrial-lysosomal axis theory of aging-Accumulation of damaged mitochondria as a result of imperfect autophagocytosis. Eur J Biochem. 2002;269:1996–2002. doi: 10.1046/j.1432-1033.2002.02869.x. [DOI] [PubMed] [Google Scholar]
- Conover CA. PAPP-A: a new anti-aging target? Aging Cell. 2010;9:942–946. doi: 10.1111/j.1474-9726.2010.00630.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuervo AM, Bergamini E, Brunk UT, Dröge W, Ffrench M, Terman A. Autophagy and aging: the importance of maintaining “clean” cells. Autophagy. 2005;3:131–40. doi: 10.4161/auto.1.3.2017. [DOI] [PubMed] [Google Scholar]
- Cui J, Bai XY, Shi SZ, Cui SY, Hong Q, Cai GY, Chen XM. Age-related changes in the function of autophagy in rat kidneys. Age (Dordr) 2011;1:21455601. doi: 10.1007/s11357-011-9237-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dozmorov I, Galecki A, Chang Y, Krzesicki R, Vergara M, Miller RA. Gene expression profile of long-lived snell dwarf mice. J Gerontol A Biol Sci Med Sci. 2002;57:B99–108. doi: 10.1093/gerona/57.3.b99. [DOI] [PubMed] [Google Scholar]
- Finkel T, Holbrook NJ. Oxidants, oxidative stress and the biology of ageing. Nature. 2000;9:239–47. doi: 10.1038/35041687. [DOI] [PubMed] [Google Scholar]
- Foster KG, Fingar DC. Mammalian Target of Rapamycin (mTOR): Conducting the Cellular Signaling Symphony. The Journal of Biological Chemistry. 2010;285:14071–14077. doi: 10.1074/jbc.R109.094003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M, Taubert S, Crawford D, Libina N, Lee SJ, Kenyon C. Lifespan extension by conditions that inhibit translation In Caenorhabditis elegans. Aging Cell. 2007;6:95–110. doi: 10.1111/j.1474-9726.2006.00267.x. [DOI] [PubMed] [Google Scholar]
- Harrison DE, Strong R, Sharp ZD, Nelson JF, Astle CM, Flurkey K, Nadon NL, Wilkinson JE, Frenkel K, Carter CS, Pahor M, Javors MA, Fernandez E, Miller RA. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460:392–395. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hariharan N, Zhou PY, Sadoshima J. Oxidative stress stimulates autophagic flux during ischemia/reperfusion. 2011;14(11) doi: 10.1089/ars.2010.3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzenberger M, Dupont J, Ducos B, Leneuve P, Géloën A, Even PC, Cervera P, Le Bouc Y. IGF-1 receptor regulates lifespan and resistance to oxidative stress in mice. Nature. 2003;421(6919):182–7. doi: 10.1038/nature01298. [DOI] [PubMed] [Google Scholar]
- Juhász G, Érdi B, Sass M, Neufeld TP. Atg7-dependent autophagy promotes neuronal health, stress tolerance, and longevity but is dispensable for metamorphosis in Drosophila. Genes Dev. 2007;21:3061–3066. doi: 10.1101/gad.1600707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y, Mizushima N, Yamamoto A, Oshitani-Okamoto S, Ohsumi Y, Yoshimori T. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J Cell Sci. 2004;117:2805–12. doi: 10.1242/jcs.01131. [DOI] [PubMed] [Google Scholar]
- Kaushik S, Massey AC, Mizushima N, Cuervo AM. Constitutive activation of chaperone-mediated autophagy in cells with impaired macroautophagy. Mol Biol Cell. 2008;19:2179–2192. doi: 10.1091/mbc.E07-11-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkland RA, Windelborn JA, Kasprzak JM, Franklin JL. A Bax-Induced Pro-Oxidant State Is Critical for Cytochrome c Release during Programmed Neuronal Death. The Journal of Neuroscience. 2002;22:6480–6490. doi: 10.1523/JNEUROSCI.22-15-06480.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4(2):151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Chiba T, Murata S, Iwata JI, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, Tanaka K. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- Leiser SF, Salmon AB, Miller RA. Correlated resistance to glucose deprivation and cytotoxic agents in fibroblast cell lines from long-lived pituitary dwarf mice. Mechanisms Of Ageing And Development. 2006;127:821–829. doi: 10.1016/j.mad.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Leiser SF, Miller RA. Nrf2 Signaling, a Mechanism for Cellular Stress Resistance in Long-Lived Mice. Molecular and Cellular Biology. 2010;30:871–884. doi: 10.1128/MCB.01145-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCulloch D, Gems D. Body size, insulin/IGF signalling and aging in the nematode Caenorhabditis elegans. Experimental Gerontology. 2003;38:129–136. doi: 10.1016/s0531-5565(02)00147-x. [DOI] [PubMed] [Google Scholar]
- Meléndez A, Tallóczy Z, Seaman M, Eskelinen EL, Hall DH, Levine B. Autophagy Genes Are Essential for Dauer Development and Life-Span Extension in C. elegans. Science. 2003;301:1387–1391. doi: 10.1126/science.1087782. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T. How to Interpret LC3 Immunoblotting. Autophagy. 2007;3:542–545. doi: 10.4161/auto.4600. [DOI] [PubMed] [Google Scholar]
- Mortimore GE, Schworer C. Induction of autophagy by amino-acid deprivation in perfused rat liver. Nature. 1977;270:174–176. doi: 10.1038/270174a0. [DOI] [PubMed] [Google Scholar]
- Panici JA, Harper JM, Miller RA, Bartke A, Spong A, Masternak MM. Early life growth hormone treatment shortens longevity and decreases cellular stress resistance in long-lived mutant mice. The FASEB Journal. 2010;24(12):5073–5079. doi: 10.1096/fj.10-163253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge L, Alic N, Bjedov I, Piper MDW. Ageing in Drosophila: The role of the insulin/Igf and TOR signalling network. Exp Gerontol. 2011;46:376–381. doi: 10.1016/j.exger.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravikumar B, Vacher C, Berger Z, Davies JE, Luo S, Oroz LG, Scaravilli F, Easton DF, Duden R, O’Kane CJ, Rubinsztein DC. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet. 2004;36:585–95. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- Reggiori F, Klionsky DJ. Autophagy in the eukaryotic cell. Eukaryot Cell. 2002;1:11–21. doi: 10.1128/EC.01.1.11-21.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon AB, Murakami S, Bartke A, Kopchick J, Yasumura K, Miller RA. Fibroblast cell lines from young adult mice of long-lived mutant strains are resistant to multiple forms of stress. Am J Physiol Endocrinol Metab. 2005;289:E23–E29. doi: 10.1152/ajpendo.00575.2004. [DOI] [PubMed] [Google Scholar]
- Salminen A, Kaarniranta K. Regulation of the aging process by autophagy. Trends Mol Med. 2009;15:217–24. doi: 10.1016/j.molmed.2009.03.004. [DOI] [PubMed] [Google Scholar]
- Scherz-Shouval R, Shvets E, Fass E, Shorer H, Gil L, Elazar Z. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J. 2007;26:1749–60. doi: 10.1038/sj.emboj.7601623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott PJ, Hanna O, Jeffrey R. Atg7 Induces Basal Autophagy and Rescues Autophagic Deficiency in CryABR120G Cardiomyocytes. Circulation Research. 2011;109:151. doi: 10.1161/CIRCRESAHA.110.237339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N, Hinnebusch AG. Regulation of Translation Initiation in Eukaryotes: Mechanisms and Biological Targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobolewska A, Gajewska M, Zarzynska J, Gajkowska B, Motyl T. IGF-1, EGF, and sex steroids regulate autophagy in bovine mammary epithelial cells via the mTOR pathway. European Journal of Cell Biology. 2008;88:117–130. doi: 10.1016/j.ejcb.2008.09.004. [DOI] [PubMed] [Google Scholar]
- Snell GD. Dwarf, a new Mendelian recessive character of the house mouse. Proc Natl Acad Sci. 1929;15:733–734. doi: 10.1073/pnas.15.9.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun LY, Bokov AF, Richardson A, Miller RA. Hepatic response to oxidative injury in long-lived Ames dwarf mice. The FASEB Journal. 2011;25:11/0025-0398. doi: 10.1096/fj.10-164376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun LY, Steinbaugh MJ, Masternak MM, Bartke A, Miller RA. Fibroblasts from long-lived mutant mice show diminished ERK1/2 phosphorylation but exaggerated induction of immediate early genes. Free Radical Biology and Medicine. 2009;47:1753–1761. doi: 10.1016/j.freeradbiomed.2009.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uddin MN, Nishio N, Ito S, Suzuki H, Isobe KI. Autophagic activity in thymus and liver during aging. Age (Dordr) 2011:9. doi: 10.1007/s11357-011-9221-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, Contento AL, Nguyen PQ, Bassham DC. Degradation of Oxidized Proteins by Autophagy during Oxidative Stress in Arabidopsis. Plant Physiology. 2007;143:291–299. doi: 10.1104/pp.106.092106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Xu BC, Maheshwari HG, He L, Reed M, Lozykowski M, Okada S, Cataldo L, Coschigamo K, Wagner TE, Baumann G, Kopchick JJ. A mammalian model for Laron syndrome produced by targeted disruption of the mouse growth hormone receptor/binding protein gene (the Laron mouse) Proc Natl Acad Sci. 1997;94:13215–13220. doi: 10.1073/pnas.94.24.13215. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The number of LC3 dots was counted by Image J software. Results shown represent the mean ± SEM for three images from fibroblasts per mice obtained from three mice per genotype. Blue: Nuclei DAPI, Green: LC3. Scale bars represent 50 μm. *P<0.05.
H2O2 (15μM) or PQ (2 mM) was used to induce oxidative stress. Bafilomycin A 10 nM was added to some cultures as indicated. Scale bars represent 50 μm. The number of LC3 dots was counted by Image j software. Results shown represent the mean ± SEM for three images from fibroblasts per mice obtained from three mice per genotype. Blue: Nuclei DAPI, Green: LC3. *P<0.05, **P<0.01.
Scale bars represent 50 μm. Blue: Nuclei DAPI, Green: LC3. The number of LC3 dots was counted by Image j software. Results shown represent the mean ± SEM for three images from fibroblasts per mice obtained from three mice per genotype. *P<0.05, , **P<0.01.