Abstract
Investigations into the genetic basis underlying antigenic variation in malaria parasites have primarily described transcriptional regulation of the large, multi-copy gene families that encode red cell surface antigens. In particular, extensive alterations to chromatin structure and subnuclear localization have been shown to play key roles in mutually exclusive expression, gene silencing and activation, and epigenetic memory. However the mechanisms responsible for the generation of sequence diversity within these gene families, a characteristic that is equally important for a parasite’s ability to avoid the host’s immune response, remains poorly understood in malaria. Recent work in model organisms suggests that the mechanisms controlling gene activation and silencing might also contribute to preferential recombination between antigen encoding genes, thus linking these two key processes.
Introduction
Antigenic variation refers to the ability of infectious organisms to systematically alter the immunogenic epitopes exposed to the immune system of their host, thus avoiding the antibody response and establishing a persistent infection. Many pathogens, including bacteria, fungi and protozoan parasites utilize antigenic variation to sustain an infection and thus increase the likelihood of successful transmission [1]. For the human malaria parasite Plasmodium falciparum, the primary antigen expressed on the surface of infected red blood cells (RBCs) is a highly variable protein called Plasmodium falciparum erythrocyte membrane protein one (PfEMP1). Different forms of PfEMP1 are encoded by different members of the var gene family [2–4]. The genome of P. falciparum contains approximately 60 var genes, thus parasites can vary their antigenic signature by changing which var gene is expressed, thereby cycling through their repertoire of surface proteins. Antigenic variation in P. falciparum is manifest clinically as a continuous series of waves of parasitemia, with each wave representing antigenically distinct populations that are not recognized by antibodies produced against previously expressed antigens [5]. PfEMP1 is key to parasite virulence; the protein is composed of a variable number of functional domains that interact with the surface of the host endothelium and thus acts as adhesins allowing the parasite to avoid systemic circulation and clearance by the spleen. Expression of different forms of PfEMP1 results in adhesion within different organs and consequently different disease manifestations [6].
Successful antigenic variation depends on the parasite’s ability to efficiently utilize its catalog of variant antigens over the course of an infection. For this purpose parasites have evolved a system of strict mutually exclusive expression, whereby one, but only one, var gene is expressed at any given time [7]. By limiting exposure to a single form of PfEMP1 at a time, and by switching to a new variant when the host has mounted an effective antibody response, parasites can maximize the utility of their repertoire of variant antigens. A persistent infection therefore requires both a highly diverse repertoire of antigen encoding genes and a sophisticated system of coordinated gene activation and silencing to ensure mutually exclusive expression. Recent work in higher eukaryotic systems suggests that these two key aspects of antigenic variation may be closely linked and share several mechanistic attributes.
Diversity of var genes
Within the genome of any given parasite, the var gene family displays extreme sequence diversity. Moreover, unlike the rest of the genome which is largely conserved, when var gene repertoires are compared from independent parasite isolates, the overall degree of diversity is virtually limitless [8•;9]. This extreme diversity is required so that each form of PfEMP1 is sufficiently unique to avoid cross-reactive antibodies generated earlier in an infection or during previous infections. The extraordinary diversity of var gene sequences within the background of a conserved genome suggests that a specific, systematic process is driving the rapid diversification of this gene family without risking disruption of open reading frames or the domain structure of the encoded proteins so as to maintain the proteins adhesive and virulence properties. Studies of large databases of var gene sequences have detected evidence for frequent gene conversion events [10;11], suggesting that this is the primary mechanism underlying the generation of the extensive variability found within this gene family.
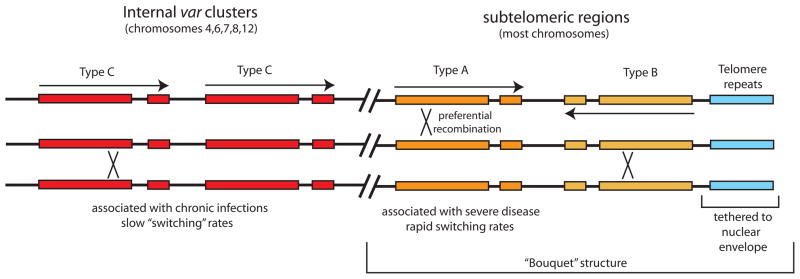
Much work has been invested in defining and categorizing var genes and their diversity. The classification of var genes into three major types, A, B and C, has been proposed and is based on domain architecture, chromosomal location and upstream non-coding sequence characteristics (Figure 1) [12;13]. These classifications have been borne out as relevant clinically; severe disease has been associated with the subtelomeric A type vars and chronic infection associated with the internally located C type vars [14–17]. Different rates of on and off switching of gene expression appear to correlate with var type as well [18–21•]. These var gene types are also relevant regarding the development of genetic diversity. There appears to be preferential recombination between members of each var gene type, resulting in three different co-evolving groups [10;12;13]. In addition, each parasite has a similar complement of var genes with roughly equal numbers of each type [8]. However, there are notable exceptions, termed strain transcendent var genes that are significantly less recombinogenic and appear to only be expressed in unique situations. For example, the gene var2csa appears to seldom recombine with other var genes and to be virtually exclusively expressed by parasites infecting pregnant women [22].
Figure 1.
Arrangement of var genes within the P. falciparum genome. There are ~60 var genes found in the parasite’s genome, divided into three basic types, A (yellow), B (orange) and C (red). Type B genes are typically found immediately adjacent to the telomeric repeats (blue) and transcribed away from the telomere. Type A genes are usually also found within the subtelomeric domains, but transcribed toward the telomeres. Type C genes are located in tandem arrays in the central regions of chromosomes 4,6,7,8 and 12. The telomeres are tethered to the nuclear envelope and are gathered into clusters of 6–8 telomere within a “bouquet” structure that results in physical alignment of the Type A and B genes. This alignment is proposed to aid in preferential recombination between genes of the same type. A similar structure may also contribute to recombination between Type C genes.
The chromatin environment surrounding var genes
Early investigations into var gene regulation indicated that it is an epigenetic process that depends on specific histone modifications. In particular, modifications at histone 3 lysine 9 (H3K9) have been shown to be directly associated with transcriptional activity. Specifically, the promoter of the single active var gene is associated with acetylated H3K9, while in silent genes this region is occupied by tri-methylated H3K9 [23;24]. While these are common histone marks for silent and active chromatin in most higher eukaryotes, in P. falciparum the trimethylated H3K9 mark appears to be devoted solely to genes involved in antigenic variation [25;26]. In addition to the H3K9 marks, the active var locus is enriched in hyperacetylated H4 as well as di- and tri-methylated H3K4 while the histone binding protein HP1 is found specifically at silent loci [25–27]. Active var genes also incorporate the variant histone H2A.Z at the promoter [28•;29•]. The various histone marks are perpetuated through the multiple rounds of DNA replication and chromatin assembly that are a part of schizogeny, thereby contributing to what is referred to as “epigenetic memory”. This term describes the tendency of a gene to remain transcriptionally active for numerous cycles of schizogeny once it has switched to the active state, a property that enables the parasites to expand to a large wave of parasitemia in which all parasites express the same form of PfEMP1.
The observation that there is a particular histone modification (H3K9me3) associated specifically with antigenically variant gene families suggests that the overall chromatin structure within these regions of the genome is unique. It is interesting to speculate that the unique chromatin structure found at the large hyper-variable gene families involved in antigenic variation might also contribute to accelerated recombination, thus acting as a driver of diversification as well as an important component of transcriptional regulation. No studies have investigated this possibility in P. falciparum, however studies in yeast and higher eukaryotes have demonstrated that chromatin structure is a key aspect of the regulation of double strand break (DSB) repair, including homology-directed repair (HR), the molecular mechanism underlying gene conversions like those frequently observed in var genes. For example, in C. elegans H3K9me3 is associated with chromosomal regions that undergo elevated levels of HR during meiosis [30;31]. Similarly, alterations in the normal chromatin structure of immunoglobulin genes significantly changes how these genes undergo regulated sequence diversification via gene conversion during chicken B cell development [32;33]. It has been proposed that parallels can be drawn between the role of the “histone code” in regulating transcription and what is being learned about the relationship between chromatin modifications and DNA repair. Chromatin state directly influences which repair factors are recruited to a site of DNA damage and thus chromatin markers direct the ensuing type of repair [34]. Given how conserved many aspects of DNA repair and chromatin modifications are between even distantly related eukaryotic organisms, it is tempting to think that in addition to regulating expression of variant antigen encoding genes, the unique histone modifications found within these regions of the parasite’s genome might also be contributing to their accelerated diversification.
The unique environment of the nuclear periphery
Changes in subnuclear localization have also been implicated in contributing to transcriptional activation and silencing of individual genes. Multiple var genes are found located within the subtelomeric domains of most chromosomes in addition to tandem arrays of var genes found within the internal regions of the chromosomes (Figure 1). Analyses using fluorescent in situ hybridization (FISH) have shown that these chromosomal domains are tethered in some way to the nuclear periphery [35], thus forming subnuclear regions containing the silent var loci. Upon activation, the single expressed var gene moves away from the clusters of silent var loci, yet remains associated with the nuclear periphery [36]. These observations have led to the hypothesis that there exists a specific subnuclear “expression site” where var genes become positioned when activated, and there is some evidence that members of other hyper-variable gene families, including rifins and stevors, might also localize to this site when actively transcribed [37]. There is recent evidence that an element within var introns, a sequence previously implicated in var gene transcriptional regulation, contributes to the subnuclear positioning of var genes [38•]. The exact nature of the putative “expression site” or the role that the nuclear periphery plays in var gene silencing or activation remain unclear. In higher eukaryotes, transcriptionally silent heterochromatin tends to be found in the nuclear periphery [39], and telomeres have been shown to cluster into “bouquets” located at the nuclear membrane [40], providing possible parallels. In addition, actively transcribed genes can be found juxtaposed to nuclear pores, in theory to provide ready access for mRNA export, however the pores themselves have also been reported to influence chromatin structure [41]. All of these observations strongly suggest that subnuclear structure and organization play a significant role in var gene regulation, although the details are yet to be resolved.
Subnuclear structure and gene localization can also strongly influence DNA recombination and repair. For example, repair of DNA DSBs in the telomeric regions in S. cerevisiae is dependent both on components of nuclear pores and the act of anchoring telomeres to the nuclear membrane [42]. Broken ends are “shunted” to the pore complex [43;44] where homologous DNA sequences that serve as donors for HR are tethered, thus placing them in close proximity to aid in efficient repair. This model draws obvious parallels to the proposed clustering of parasite telomeres at the nuclear periphery within bouquets in which the var genes are aligned according to type (Figure 1). The propensity of var genes to undergo gene conversions [10;11], a process that requires the physical alignment of DNA sequences, suggests that subnuclear localization could play a similarly important role for the generation of diversity as has been proposed for regulating expression.
Pathways for DNA DSB repair in P. falciparum: HR versus NHEJ
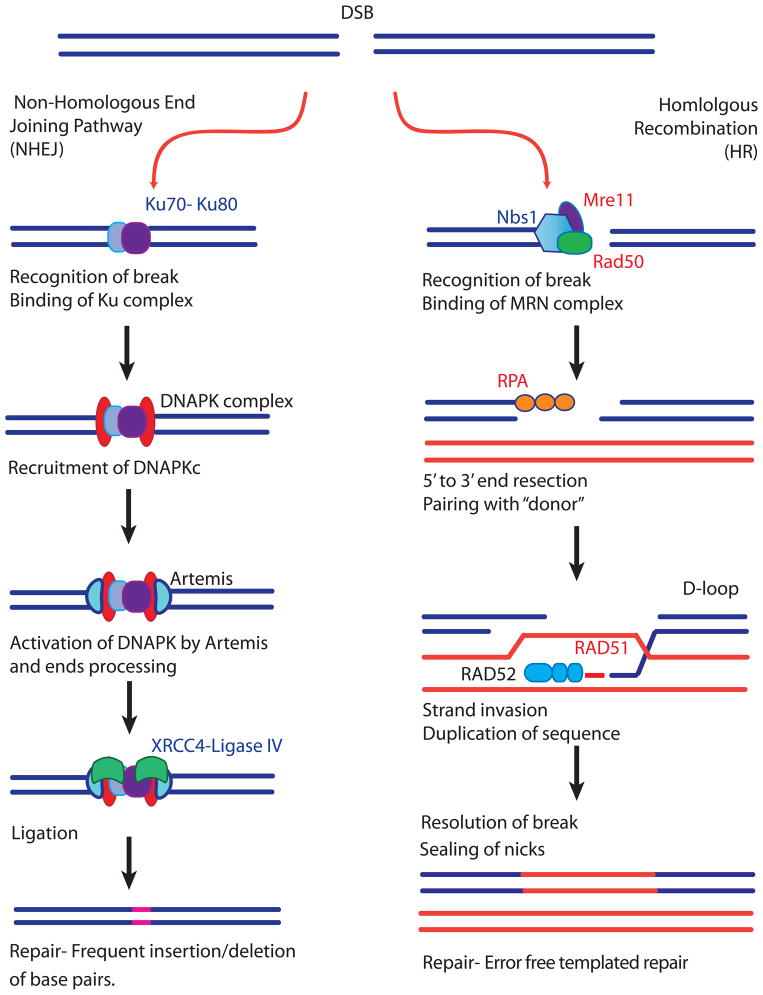
For pathogens, the ability to repair DNA damage can be considered an integral part of immune avoidance. Malaria parasites face all of the same sources of DSBs as other eukaryotic organisms, with additional sources of oxidative damage resulting from heme metabolism and from products of the host’s immune system [45]. Therefore, efficient DSB repair is expected to be an important aspect of parasite biology. Plasmodium falciparum’s response to DNA damage has not been characterized in detail, with the exception of work describing particular aspects of mismatch repair and base excision repair [45–47]. In particular, the parasite’s response to DSBs has not been extensively studied; however it is notable that many of the key components required for classical non homologous end joining (NHEJ)-the predominant DSB repair pathway utilized by higher eukaryotes, are not present in the parasite genome [48;49]. Several of these proteins, for example Ku 70 and 80, are present and active within closely related parasites like Toxoplasma, suggesting they have been recently lost by malaria parasites, a somewhat surprising finding considering that this repair pathway is conserved evolutionarily. In the absence of a classical NHEJ pathway, parasites must rely on alternative means of DSB repair and by assessment of repair proteins known to be present in the genome (Table 1), it is likely that HR plays a dominant role. This could result in a distinct skew toward the products of HR, specifically gene conversions, when DSBs are encountered (Figure 2).
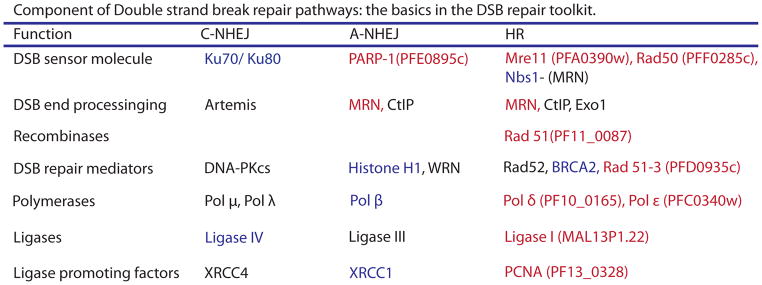
Table 1.
Summary of DSB repair proteins. Listed are the basic DNA DSB repair proteins as defined in model organisms. Those proteins listed in blue are notably absent from the P. falciparum genome and those in red have identified orthologues. Those in black have not been identified as present or missing form the parasite’s genome. Alternative Non Homologous End Joining (A- NHEJ), otherwise known as Microhomology Mediated End Joining, is defined as a Ku independent pathway for end joining and is characterized by end resection and ligation at areas of homology. Repair via this pathway occurs as a backup pathway in most model organisms and is associated with deletions of intervening sequence. The HR pathway appears intact in malaria parasites however there have been no investigations into other DSB repair pathways to date. HR is notable for its accuracy and maintenance of open reading frames compared to the deletions and insertions commonly associated with end joining pathways. Modified from Mladenov and colleagues [57].
|
Figure 2.
Mechanisms of DNA Double Strand Break Repair. Double Strand Breaks (DSBs) are repaired by two major pathways. Homologous Recombination (HR, right) or Nonhomologous End Joining (NHEJ, left). HR can result in different end products, but is defined by the use of a template to guide repair. Pictured is HR leading to a gene conversion. A DNA lesion is recognized by the MRN (MRE11/RAD50/NBS1) complex which generates single stranded DNA by resection. The newly generated single strand is bound by replication protein A (RPA), followed by RAD 52 which acts as a mediator between RPA and RAD51. RAD51 catalyzes the invasion of the single strand to form a displacement loop (D- loop) which is resolved through alternative pathways to result in accurate repair. HR frequently results in a gene conversion event. In contrast, NHEJ does not require a template and instead simply ligates the two free DNA ends together to repair the break. In classical NHEJ, Ku 70/80 sense the DSB and recruit DNA dependent protein kinase complex (DNAPKc) which likely regulates the processing of DNA ends in addition to recruiting the other components of repair including XRCC4, DNA ligase IV, XLF and Artemis. This type of repair frequently results in deletions or insertions at the site of repair (shown as a red insertion in the final product). Components of the repair pathways shown in blue have not been found in the P. falciparum genome, those in red have orthologues identified in the parasite genome. This is further detailed in Table 1.
Evidence for frequent gene conversion events have been reported for several gene families in addition to the var family, including those encoding the merozoite surface proteins 1 and 2 [50;51], the RBL family of reticulocyte binding proteins [52], proteins involved in red blood cell invasion [53], and the falcipain gene family [54], providing further support for the hypothesis that HR is a common pathway for repair. While gene conversions have been shown to occur during meiosis [35], HR could also function when the parasites are haploid if regions of significant sequence homology are present within the genome, for example within multi-copy gene families. Since the choice of template for HR depends both on the degree of sequence identity as well as genome location [55;56], reliance on this mechanism for DSB repair should result in the generation over time of var gene types that more frequently recombine with one another, consistent with the previously described A, B and C types of var genes (Figure 1). In addition, the use of HR rather than NHEJ ensures that repaired genes maintain open reading frames and encode functional proteins. These observations highlight the potential selective advantages for the use of HR as the primary method of DSB repair within large antigen encoding gene families, and could provide one possible explanation for the apparent loss of classical NHEJ.
Conclusions
To survive the antibody response of its human host, P. falciparum has evolved a complex system of antigenic variation that relies on two key aspects: 1) tight transcriptional regulation of large multi-copy families of antigen encoding genes and 2) the continuous generation of sequence variation within these families. Recent work in both P. falciparum and model organisms suggest that these two aspects of antigenic variation are tightly linked and may in fact share many mechanistic components.
Highlights.
Antigenic variation enables parasites to avoid the antibody response of their host.
The multi-copy var gene family of P. falciparum encodes variant surface antigens.
Control of var gene expression depends on modifications to chromatin structure.
Recombination between var genes leads to extensive sequence diversity.
Chromatin structure and DSB repair likely contribute to var gene diversification.
Acknowledgments
Work in the laboratory of KWD is supported by a grant from the National Institutes of Health (AI 52390) and the United States-Israel Binational Science Foundation. The Department of Microbiology and Immunology at Weill Medical College of Cornell University acknowledges the support of the William Randolph Hearst Foundation. KWD is a Stavros S. Niarchos Scholar. LAK is supported by a grant from the National Institutes of Health (AI76635).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Deitsch KW, Lukehart SA, Stringer JR. Common strategies for antigenic variation by bacterial, fungal and protozoan pathogens. Nat Rev Microbiol. 2009;7:493–503. doi: 10.1038/nrmicro2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith JD, Chitnis CE, Craig AG, Roberts DJ, Hudson-Taylor DE, Peterson DS, Pinches R, Newbold CI, Miller LH. Switches in expression of Plasmodium falciparum var genes correlate with changes in antigenic and cytoadherent phenotypes of infected erythrocytes. Cell. 1995;82:101–110. doi: 10.1016/0092-8674(95)90056-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baruch DI, Pasloske BL, Singh HB, Bi X, Ma XC, Feldman M, Taraschi TF, Howard RJ. Cloning the P. falciparum gene encoding PfEMP1, a malarial variant antigen and adherence receptor on the surface of parasitized human erythrocytes. Cell. 1995;82:77–87. doi: 10.1016/0092-8674(95)90054-3. [DOI] [PubMed] [Google Scholar]
- 4.Su X, Heatwole VM, Wertheimer SP, Guinet F, Herrfeldt JV, Peterson DS, Ravetch JV, Wellems TE. A large and diverse gene family (var) encodes 200–350 kD proteins implicated in the antigenic variation and cytoadherence of Plasmodium falciparum-infected erythrocytes. Cell. 1995;82:89–100. doi: 10.1016/0092-8674(95)90055-1. [DOI] [PubMed] [Google Scholar]
- 5.Miller LH, Baruch DI, Marsh K, Doumbo OK. The pathogenic basis of malaria. Nature. 2002;415:673–679. doi: 10.1038/415673a. [DOI] [PubMed] [Google Scholar]
- 6.Montgomery J, Mphande FA, Berriman M, Pain A, Rogerson SJ, Taylor TE, Molyneux ME, Craig A. Differential var gene expression in the organs of patients dying of falciparum malaria. Mol Microbiol. 2007;65:959–967. doi: 10.1111/j.1365-2958.2007.05837.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scherf A, Lopez-Rubio JJ, Riviere L. Antigenic variation in Plasmodium falciparum. Annu Rev Microbiol. 2008;62:445–470. doi: 10.1146/annurev.micro.61.080706.093134. [DOI] [PubMed] [Google Scholar]
- 8•.Rask TS, Hansen DA, Theander TG, Gorm PA, Lavstsen T. Plasmodium falciparum erythrocyte membrane protein 1 diversity in seven genomes--divide and conquer. PLoS Comput Biol. 2010:6. doi: 10.1371/journal.pcbi.1000933. This paper took advantage of complete genome sequence information from seven parasite isolates to construct a dataset of 399 different PfEMP1 amino acid sequences. Computational analysis of this dataset enabled the authors to produce the most complete analysis yet of PfEMP1 domain classes as well as a potential hotspot for recombination. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barry AE, Leliwa-Sytek A, Tavul L, Imrie H, Migot-Nabias F, Brown SM, McVean GA, Day KP. Population Genomics of the Immune Evasion (var) Genes of Plasmodium falciparum. PLoS Pathog. 2007;3:e34. doi: 10.1371/journal.ppat.0030034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kraemer SM, Kyes SA, Aggarwal G, Springer AL, Nelson SO, Christodoulou Z, Smith LM, Wang W, Levin E, Newbold CI, Myler PJ, Smith JD. Patterns of gene recombination shape var gene repertoires in Plasmodium falciparum: comparisons of geographically diverse isolates. BMC Genomics. 2007;8:45. doi: 10.1186/1471-2164-8-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ward CP, Clottey GT, Dorris M, Ji DD, Arnot DE. Analysis of Plasmodium falciparum PfEMP-1/var genes suggests that recombination rearranges constrained sequences. Mol Biochem Parasitol. 1999;102:167–177. doi: 10.1016/s0166-6851(99)00106-1. [DOI] [PubMed] [Google Scholar]
- 12.Lavstsen T, Salanti A, Jensen ATR, Arnot DE, Theander TG. Sub-grouping of Plasmodium falciparum 3D7 var genes based on sequence analysis of coding and non-coding regions. Malaria Journal. 2003;2:27. doi: 10.1186/1475-2875-2-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kraemer SM, Smith JD. Evidence for the importance of genetic structuring to the structural and functional specialization of the Plasmodium falciparum var gene family. Molecular Microbiology. 2003;50:1527–1538. doi: 10.1046/j.1365-2958.2003.03814.x. [DOI] [PubMed] [Google Scholar]
- 14.Rottmann M, Lavstsen T, Mugasa JP, Kaestli M, Jensen AT, Muller D, Theander T, Beck HP. Differential expression of var gene groups is associated with morbidity caused by Plasmodium falciparum infection in Tanzanian children. Infect Immun. 2006;74:3904–3911. doi: 10.1128/IAI.02073-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaestli M, Cockburn IA, Cortes A, Baea K, Rowe JA, Beck HP. Virulence of malaria is associated with differential expression of Plasmodium falciparum var gene subgroups in a case-control study. J Infect Dis. 2006;193:1567–1574. doi: 10.1086/503776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Falk N, Kaestli M, Qi W, Ott M, Baea K, Cortes A, Beck HP. Analysis of Plasmodium falciparum var genes expressed in children from Papua New Guinea. J Infect Dis. 2009;200:347–356. doi: 10.1086/600071. [DOI] [PubMed] [Google Scholar]
- 17.Kyriacou HM, Stone GN, Challis RJ, Raza A, Lyke KE, Thera MA, Kone AK, Doumbo OK, Plowe CV, Rowe JA. Differential var gene transcription in Plasmodium falciparum isolates from patients with cerebral malaria compared to hyperparasitaemia. Mol Biochem Parasitol. 2006;150:211–218. doi: 10.1016/j.molbiopara.2006.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18•.Frank M, Dzikowski R, Amulic B, Deitsch K. Variable switching rates of malaria virulence genes are associated with chromosomal position. Mol Microbiol. 2007;64:1486–1498. doi: 10.1111/j.1365-2958.2007.05736.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19•.Enderes C, Kombila D, Dal Bianco M, Dzikowski R, Kremsner P, Frank M. Var Gene promoter activation in clonal Plasmodium falciparum isolates follows a hierarchy and suggests a conserved switching program that is independent of genetic background. J Infect Dis. 2011;204:1620–1631. doi: 10.1093/infdis/jir594. [DOI] [PubMed] [Google Scholar]
- 20•.Bachmann A, Predehl S, May J, Harder S, Burchard GD, Gilberger TW, Tannich E, Bruchhaus I. Highly co-ordinated var gene expression and switching in clinical Plasmodium falciparum isolates from non-immune malaria patients. Cell Microbiol. 2011;13:1397–1409. doi: 10.1111/j.1462-5822.2011.01629.x. [DOI] [PubMed] [Google Scholar]
- 21•.Recker M, Buckee CO, Serazin A, Kyes S, Pinches R, Christodoulou Z, Springer AL, Gupta S, Newbold CI. Antigenic variation in Plasmodium falciparum malaria involves a highly structured switching pattern. PLoS Pathog. 2011;7:e1001306. doi: 10.1371/journal.ppat.1001306. References 18–21 utilize extensive datasets of var gene expression patterns from both field samples and laboratory lines to identify switching patterns. Combined with mathematical modeling, these analyses describe a model for differential switching rates for different var gene types. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Salanti A, Dahlback M, Turner L, Nielsen MA, Barfod L, Magistrado P, Jensen AT, Lavstsen T, Ofori MF, Marsh K, Hviid L, Theander TG. Evidence for the involvement of VAR2CSA in pregnancy-associated malaria. J Exp Med. 2004;200:1197–1203. doi: 10.1084/jem.20041579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chookajorn T, Dzikowski R, Frank M, Li F, Jiwani AZ, Hartl DL, Deitsch KW. Epigenetic memory at malaria virulence genes. Proc Natl Acad Sci USA. 2007;104:899–902. doi: 10.1073/pnas.0609084103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lopez-Rubio JJ, Gontijo AM, Nunes MC, Issar N, Hernandez RR, Scherf A. 5' flanking region of var genes nucleate histone modification patterns linked to phenotypic inheritance of virulence traits in malaria parasites. Mol Microbiol. 2007;66:1296–1305. doi: 10.1111/j.1365-2958.2007.06009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lopez-Rubio JJ, Mancio-Silva L, Scherf A. Genome-wide analysis of heterochromatin associates clonally variant gene regulation with perinuclear repressive centers in malaria parasites. Cell Host Microbe. 2009;5:179–190. doi: 10.1016/j.chom.2008.12.012. [DOI] [PubMed] [Google Scholar]
- 26.Flueck C, Bartfai R, Volz J, Niederwieser I, Salcedo-Amaya AM, Alako BT, Ehlgen F, Ralph SA, Cowman AF, Bozdech Z, Stunnenberg HG, Voss TS. Plasmodium falciparum heterochromatin protein 1 marks genomic loci linked to phenotypic variation of exported virulence factors. PLoS Pathog. 2009;5:e1000569. doi: 10.1371/journal.ppat.1000569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perez-Toledo K, Rojas-Meza AP, Mancio-Silva L, Hernandez-Cuevas NA, Delgadillo DM, Vargas M, Martinez-Calvillo S, Scherf A, Hernandez-Rivas R. Plasmodium falciparum heterochromatin protein 1 binds to tri-methylated histone 3 lysine 9 and is linked to mutually exclusive expression of var genes. Nucleic Acids Res. 2009;37:2596–2606. doi: 10.1093/nar/gkp115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28•.Petter M, Lee CC, Byrne TJ, Boysen KE, Volz J, Ralph SA, Cowman AF, Brown GV, Duffy MF. Expression of P. falciparum var genes involves exchange of the histone variant H2A.Z at the promoter. PLoS Pathog. 2011;7:e1001292. doi: 10.1371/journal.ppat.1001292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29•.Bartfai R, Hoeijmakers WA, Salcedo-Amaya AM, Smits AH, Janssen-Megens E, Kaan A, Treeck M, Gilberger TW, Francoijs KJ, Stunnenberg HG. H2A.Z demarcates intergenic regions of the plasmodium falciparum epigenome that are dynamically marked by H3K9ac and H3K4me3. PLoS Pathog. 2010;6:e1001223. doi: 10.1371/journal.ppat.1001223. References 28 and 29 describe the role of variant histone H2A.Z in segregating different regions of the parasite genome and in regulating gene expression. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu T, Rechtsteiner A, Egelhofer TA, Vielle A, Latorre I, Cheung MS, Ercan S, Ikegami K, Jensen M, Kolasinska-Zwierz P, Rosenbaum H, Shin H, Taing S, Takasaki T, Iniguez AL, Desai A, Dernburg AF, Kimura H, Lieb JD, Ahringer J, Strome S, Liu XS. Broad chromosomal domains of histone modification patterns in C. elegans. Genome Res. 2011;21:227–236. doi: 10.1101/gr.115519.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gu SG, Fire A. Partitioning the C. elegans genome by nucleosome modification, occupancy, and positioning. Chromosoma. 2010;119:73–87. doi: 10.1007/s00412-009-0235-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cummings WJ, Bednarski DW, Maizels N. Genetic variation stimulated by epigenetic modification. PLoS ONE. 2008;3:e4075. doi: 10.1371/journal.pone.0004075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cummings WJ, Yabuki M, Ordinario EC, Bednarski DW, Quay S, Maizels N. Chromatin structure regulates gene conversion. PLoS Biol. 2007;5:e246. doi: 10.1371/journal.pbio.0050246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shi L, Oberdoerffer P. Chromatin dynamics in DNA double-strand break repair. Biochim Biophys Acta. 2012 doi: 10.1016/j.bbagrm.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Freitas-Junior LH, Bottius E, Pirrit LA, Deitsch KW, Scheidig C, Guinet F, Nehrbass U, Wellems TE, Scherf A. Frequent ectopic recombination of virulence factor genes in telomeric chromosome clusters of P. falciparum. Nature. 2000;407:1018–1022. doi: 10.1038/35039531. [DOI] [PubMed] [Google Scholar]
- 36.Ralph SA, Scheidig-Benatar C, Scherf A. Antigenic variation in Plasmodium falciparum is associated with movement of var loci between subnuclear locations. Proc Natl Acad Sci USA. 2005;102:5414–5419. doi: 10.1073/pnas.0408883102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Howitt CA, Wilinski D, Llinas M, Templeton TJ, Dzikowski R, Deitsch KW. Clonally variant gene families in Plasmodium falciparum share a common activation factor. Mol Microbiol. 2009;73:1171–1185. doi: 10.1111/j.1365-2958.2009.06846.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38•.Zhang Q, Huang Y, Zhang Y, Fang X, Claes A, Duchateau M, Namane A, Lopez-Rubio JJ, Pan W, Scherf A. A critical role of perinuclear filamentous actin in spatial repositioning and mutually exclusive expression of virulence genes in malaria parasites. Cell Host Microbe. 2011;10:451–463. doi: 10.1016/j.chom.2011.09.013. This paper describes a putative role for nuclear actin in positioning var genes at the nuclear periphery. It provides that latest step toward understanding how the different genes and chromosome fragments get arranged into different subnuclear domains. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sutherland H, Bickmore WA. Transcription factories: gene expression in unions? Nat Rev Genet. 2009;10:457–466. doi: 10.1038/nrg2592. [DOI] [PubMed] [Google Scholar]
- 40.Funabiki H, Hagan I, Uzawa S, Yanagida M. Cell cycle-dependent specific positioning and clustering of centromeres and telomeres in fission yeast. J Cell Biol. 1993;121:961–976. doi: 10.1083/jcb.121.5.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arib G, Akhtar A. Multiple facets of nuclear periphery in gene expression control. Curr Opin Cell Biol. 2011;23:346–353. doi: 10.1016/j.ceb.2010.12.005. [DOI] [PubMed] [Google Scholar]
- 42.Therizols P, Fairhead C, Cabal GG, Genovesio A, Olivo-Marin JC, Dujon B, Fabre E. Telomere tethering at the nuclear periphery is essential for efficient DNA double strand break repair in subtelomeric region. J Cell Biol. 2006;172:189–199. doi: 10.1083/jcb.200505159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nagai S, Dubrana K, Tsai-Pflugfelder M, Davidson MB, Roberts TM, Brown GW, Varela E, Hediger F, Gasser SM, Krogan NJ. Functional targeting of DNA damage to a nuclear pore-associated SUMO-dependent ubiquitin ligase. Science. 2008;322:597–602. doi: 10.1126/science.1162790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nagai S, Davoodi N, Gasser SM. Nuclear organization in genome stability: SUMO connections. Cell Res. 2011;21:474–485. doi: 10.1038/cr.2011.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nathan C, Shiloh MU. Reactive oxygen and nitrogen intermediates in the relationship between mammalian hosts and microbial pathogens. Proc Natl Acad Sci USA. 2000;97:8841–8848. doi: 10.1073/pnas.97.16.8841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bethke L, Thomas S, Walker K, Lakhia R, Rangarajan R, Wirth D. The role of DNA mismatch repair in generating genetic diversity and drug resistance in malaria parasites. Mol Biochem Parasitol. 2007;155:18–25. doi: 10.1016/j.molbiopara.2007.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haltiwanger BM, Matsumoto Y, Nicolas E, Dianov GL, Bohr VA, Taraschi TF. DNA base excision repair in human malaria parasites is predominantly by a long-patch pathway. Biochemistry. 2000;39:763–772. doi: 10.1021/bi9923151. [DOI] [PubMed] [Google Scholar]
- 48.Aravind L, Iyer LM, Wellems TE, Miller LH. Plasmodium biology: Genomic gleanings. Cell. 2003;115:771–785. doi: 10.1016/s0092-8674(03)01023-7. [DOI] [PubMed] [Google Scholar]
- 49.Gardner MJ, Hall N, Fung E, White O, Berriman M, Hyman RW, Carlton JM, Pain A, Nelson KE, Bowman S, Paulsen IT, James K, Eisen JA, Rutherford K, Salzberg SL, Craig A, Kyes S, Chan MS, Nene V, Shallom SJ, Suh B, Peterson J, Angiuoli S, Pertea M, Allen J, Selengut J, Haft D, Mather MW, Vaidya AB, Martin DM, Fairlamb AH, Fraunholz MJ, Roos DS, Ralph SA, McFadden GI, Cummings LM, Subramanian GM, Mungall C, Venter JC, Carucci DJ, Hoffman SL, Newbold C, Davis RW, Fraser CM, Barrell B. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hoffmann EH, Malafronte RS, Moraes-Avila SL, Osakabe AL, Wunderlich G, Durham AM, Ribolla PE, del Portillo HA, Ferreira MU. Origins of sequence diversity in the malaria vaccine candidate merozoite surface protein-2 (MSP-2) in Amazonian isolates of Plasmodium falciparum. Gene. 2006;376:224–230. doi: 10.1016/j.gene.2006.03.011. [DOI] [PubMed] [Google Scholar]
- 51.Ferreira MU, Ribeiro WL, Tonon AP, Kawamoto F, Rich SM. Sequence diversity and evolution of the malaria vaccine candidate merozoite surface protein-1 (MSP-1) of Plasmodium falciparum. Gene. 2003;304:65–75. doi: 10.1016/s0378-1119(02)01180-0. [DOI] [PubMed] [Google Scholar]
- 52.Rayner JC, Huber CS, Galinski MR, Barnwell JW. Rapid evolution of an erythrocyte invasion gene family: the Plasmodium reichenowi Reticulocyte Binding Like (RBL) genes. Mol Biochem Parasitol. 2004;133:287–296. doi: 10.1016/j.molbiopara.2003.10.017. [DOI] [PubMed] [Google Scholar]
- 53.Cortes A. A chimeric Plasmodium falciparum Pfnbp2b/Pfnbp2a gene originated during asexual growth. Int J Parasitol. 2005;35:125–130. doi: 10.1016/j.ijpara.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 54.Nielsen KM, Kasper J, Choi M, Bedford T, Kristiansen K, Wirth DF, Volkman SK, Lozovsky ER, Hartl DL. Gene conversion as a source of nucleotide diversity in Plasmodium falciparum. Mol Biol Evol. 2003;20:726–734. doi: 10.1093/molbev/msg076. [DOI] [PubMed] [Google Scholar]
- 55.LaRocque JR, Jasin M. Mechanisms of recombination between diverged sequences in wild-type and BLM-deficient mouse and human cells. Mol Cell Biol. 2010;30:1887–1897. doi: 10.1128/MCB.01553-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Miller D, Reynolds GE, Mejia R, Stark JM, Murnane JP. Subtelomeric regions in mammalian cells are deficient in DNA double-strand break repair. DNA Repair (Amst) 2011;10:536–544. doi: 10.1016/j.dnarep.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mladenov E, Iliakis G. Induction and repair of DNA double strand breaks: the increasing spectrum of non-homologous end joining pathways. Mutat Res. 2011;711:61–72. doi: 10.1016/j.mrfmmm.2011.02.005. [DOI] [PubMed] [Google Scholar]