Abstract
Pax5 controls the identity and development of B cells by repressing lineage-inappropriate genes and activating B-cell-specific genes. Here, we used genome-wide approaches to identify Pax5 target genes in pro-B and mature B cells. In these cell types, Pax5 bound to 40% of the cis-regulatory elements defined by mapping DNase I hypersensitive (DHS) sites, transcription start sites and histone modifications. Although Pax5 bound to 8000 target genes, it regulated only 4% of them in pro-B and mature B cells by inducing enhancers at activated genes and eliminating DHS sites at repressed genes. Pax5-regulated genes in pro-B cells account for 23% of all expression changes occurring between common lymphoid progenitors and committed pro-B cells, which identifies Pax5 as an important regulator of this developmental transition. Regulated Pax5 target genes minimally overlap in pro-B and mature B cells, which reflects massive expression changes between these cell types. Hence, Pax5 controls B-cell identity and function by regulating distinct target genes in early and late B lymphopoiesis.
Keywords: B-cell identity, cis-regulatory landscape, genome-wide Pax5 binding, Pax5-dependent target genes, pro-B and mature B cells
Introduction
Hematopoietic stem cells (HSCs) in the bone marrow give rise to all mature B-cell types in peripheral lymphoid organs by first differentiating to lymphoid-primed multipotent progenitors (LMPPs) and common lymphoid progenitors (CLPs), which consist of Ly6D− all-lymphoid progenitors (ALPs) and Ly6D+ B-cell-biased lymphoid progenitors (BLPs; Inlay et al, 2009). ALPs retain the full lymphoid potential as they are able to develop into B, T, NK and DC cells (Inlay et al, 2009). In contrast, BLPs initiate the B-cell gene expression programme and preferentially differentiate via the pre-pro-B cell stage to pro-B cells, which undergo B-lineage commitment and subsequent development to mature B cells (Hardy et al, 2007; Inlay et al, 2009).
B-cell commitment depends on the sequential activity of the instructive transcription factors E2A, EBF1 and Pax5 during the development of CLPs to pro-B cells (Nutt and Kee, 2007). The helix-loop-helix protein E2A and the early B-cell factor EBF1 specify the B-cell lineage by activating the expression of B-lymphoid genes in pre-pro-B cells (Lin et al, 2010; Treiber et al, 2010). Pax5 subsequently controls B-cell commitment at the transition to the pro-B cell stage by restricting the developmental potential of lymphoid progenitors to the B-cell lineage, as shown by the fact that Pax5-deficient pro-B cells are still able to differentiate into most hematopoietic cell types in vitro and in vivo (Nutt et al, 1999; Medvedovic et al, 2011). At the molecular level, Pax5 fulfills a dual role by repressing B-lineage-inappropriate genes to suppress alternative lineage options and by simultaneously activating B-cell-specific genes to promote B-cell development (Nutt et al, 1999; Medvedovic et al, 2011). Gene expression analyses of wild-type and Pax5-deficient pro-B cells identified 110 Pax5-repressed and 170 Pax5-activated genes, which code for key regulatory and structural proteins involved in transcriptional control, receptor signalling, adhesion, migration and immune function (Delogu et al, 2006; Schebesta et al, 2007; Pridans et al, 2008). Pax5 regulates these gene expression changes by inducing active chromatin at activated target genes and eliminating active chromatin at repressed genes in pro-B cells (McManus et al, 2011). Notably, Pax5 induces these chromatin and transcription changes by recruiting chromatin-remodelling, histone-modifying and basal transcription factor complexes to its target genes, which identifies Pax5 as an epigenetic regulator of B-cell commitment (McManus et al, 2011).
Pax5 is expressed throughout B-cell development from pro-B cells in the bone marrow to mature B cells in peripheral lymphoid organs (Fuxa and Busslinger, 2007), where it plays an important role in the generation and function of distinct mature B-cell types (Horcher et al, 2001; Medvedovic et al, 2011). Pax5 is essential for maintaining the B-cell gene expression programme in late B lymphopoiesis, as conditional inactivation of Pax5 leads to the down-regulation of B-cell-specific genes and reactivation of lineage-inappropriate genes in mature B cells (Horcher et al, 2001; Delogu et al, 2006; Schebesta et al, 2007). Importantly, the conditional loss of Pax5 results in the conversion of mature B cells into functional T cells by dedifferentiation to uncommitted progenitors in the bone marrow (Cobaleda et al, 2007). Loss of the B-cell phenotype upon conditional Pax5 inactivation highlights an important role of Pax5 in the maintenance of B-cell identity throughout B lymphopoiesis (Mikkola et al, 2002; Cobaleda et al, 2007). The question therefore arises whether Pax5 regulates a similar or different set of target genes to control the identity and function of B lymphocytes in early and late B-cell development. Our current knowledge about the molecular function of Pax5 in B lymphopoiesis is, however, too fragmentary to answer this question, as very few Pax5-regulated genes have so far been identified in mature B cells (Delogu et al, 2006; Schebesta et al, 2007) and only 1.6% of the mouse genome has recently been screened by chromatin immunoprecipitation (ChIP)-chip analysis for Pax5 target genes (defined as Pax5-bound genes) in pro-B cells (McManus et al, 2011).
Here, we have used genome-wide sequencing approaches to define the cis-regulatory landscape and to identify regulated Pax5 target genes in committed pro-B and mature B cells. Whereas Pax5 bound to a large part (40%) of the cis-regulatory genome, it regulated only a small subset of its target genes in early and late B-cell development. However, the regulated Pax5 target genes in pro-B cells accounted for a relatively large part (23%) of the gene expression changes occurring during B-cell commitment. Notably, only a minimal overlap was observed between regulated Pax5 target genes in pro-B and mature B cells, indicating that Pax5 controls the identity and function of B cells by regulating distinct target genes in early and late B-cell development.
Results
Genome-wide identification of regulatory elements in pro-B and mature B cells
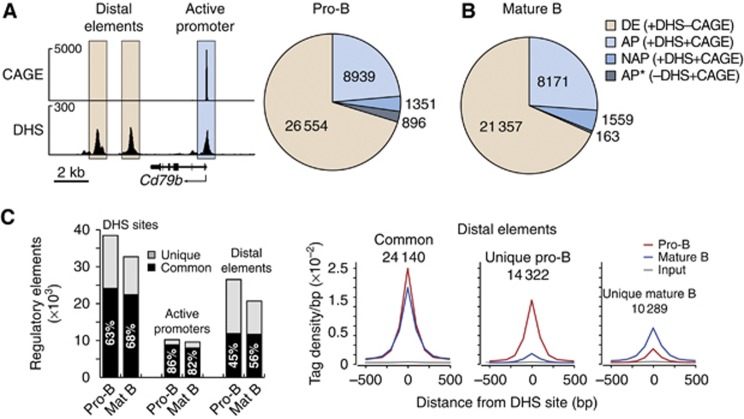
To define the cis-regulatory landscape, we characterized the genome-wide pattern of protein–DNA interactions by identifying DNase I hypersensitive (DHS) sites in early and late B-cell development. To this end, short-term cultured Rag2−/− pro-B cells were treated with DNase I to excise DHS sequences followed by deep sequencing of the released DNase I ‘double-hit’ fragments (Sabo et al, 2006; Hesselberth et al, 2009). This large-scale analysis resulted in the identification of 36 844 DHS sites, which were called with a stringent P-value of <10−10 as shown for the Cd79b gene (Figure 1A). To identify active promoters (AP) among these DHS sites, we performed cap analysis of gene expression (CAGE) coupled with deep sequencing. CAGE is a cap-trapping-based method that allows the systematic 5′ end profiling of capped mRNAs to generate comprehensive maps of transcription start sites (TSSs) at single-nucleotide resolution (Shiraki et al, 2003; Carninci et al, 2006). To identify active TSSs defining active promoters, we applied a cutoff of 2.6 RPM (reads per CAGE tag cluster per million mapped sequence tags), which distinguishes transcriptionally active (>2.6 RPM) from inactive (<2.6 RPM) genes (Supplementary Figure S1A and B). CAGE analysis of short-term cultured Rag2−/− pro-B cells thus identified 10 290 active promoters, which were defined by colocalization of CAGE tag clusters (>2.6 RPM) with DHS sites as exemplified for the Cd79b gene (Figure 1A). Among these active promoters, 1351 (13.1%) correspond to newly identified active promoters (NAP) that have not been annotated in the RefSeq database as demonstrated by Arntl, Spib and Bach2 (Supplementary Figure S1C). Moreover, 896 RefSeq-annotated promoters (AP*) contained active TSSs with CAGE tags above 2.6 RPM, although DHS sites were not called at the stringent P-value of <10−10 (Figure 1A). We refer to all other DHS sites (26 554) that do not overlap with active TSSs as distal elements (DE), which account for 72% of all accessible cis-regulatory elements in pro-B cells (Figure 1A). We next isolated quiescent mature B cells from lymph nodes (Supplementary Figure S1D) for CAGE and DHS site mapping, which identified a similar number and relative distribution of active promoters (9730; 31%) and distal elements (21 357; 69%) in mature B cells (Figure 1B; Supplementary Figure S1C) compared with pro-B cells (Figure 1A).
Figure 1.
Genome-wide identification of cis-regulatory elements in pro-B and mature B cells. (A) Identification of active promoters and distal elements in pro-B cells. Short-term cultured Rag2−/− pro-B cells (CD19+B220+) were used for CAGE and DHS site analysis combined with deep sequencing as described in Supplementary Materials and Methods and Supplementary Table S1. Active promoters were defined by the co-occurrence of DHS sites with active TSSs, which were determined by the presence of CAGE sequence tags above 2.6 RPM (reads per CAGE tag cluster per million mapped sequence tags; Supplementary Figure S1A and B), as shown for the Cd79b gene (left). Distal elements (DE) were defined by the presence of DHS site in the absence of transcription initiation (CAGE sequence tags below 2.6 RPM). A pie chart (right) indicates the total number of distal elements, active annotated promoters (AP, present in RefSeq) and non-annotated active promoters (NAP, absent in RefSeq), which were defined by the presence of a DHS site and active TSS. Due to the stringent P-value (<10−10) used, DHS sites were not called at 896 annotated promoters (AP*, present in RefSeq) that qualified, however, as active promoters due to the presence of active TSSs. The genomic coordinates of the identified promoters and distal elements are provided in Supplementary Table S2. (B) Identification of active promoters and distal elements in mature B cells as described in (A) and Supplementary Table S3. Quiescent mature B cells were MACS-sorted from lymph nodes (Supplementary Figures S1D [DHS sites] and S3A [CAGE]). (C) Overlap of regulatory elements in pro-B and mature B cells. Black bars indicate common DHS sites, active promoters and distal elements between pro-B and mature B cells (left), and grey bars denote unique regulatory elements in the respective cell type (left). Average sequence tag density profiles aligned at the central position of the DHS sites are shown for common and unique distal elements in the two cell types (right). Similar data were obtained by analysing a second DHS site mapping experiment of pro-B and mature B cells, whereas the CAGE analysis was performed only once for each cell type (Supplementary Table S1).
A large fraction (63–68%) of the identified DHS sites were present in both pro-B and mature B cells (Figure 1C). Almost all active promoters (82–86%) were common to both cell types, whereas only half of the distal elements (45–56%) were shared between pro-B and mature B cells (Figure 1C) consistent with previous findings that enhancers (among the distal elements) are highly cell type specific in contrast to the largely invariant promoter usage observed across different cell types (Heintzman et al, 2009). As shown by density plot analysis, the common distal elements exhibited a similar degree of DNase I hypersensitivity in both cell types, whereas the unique distal elements were characterized by a higher tag density in pro-B or mature B cells, respectively (Figure 1C, right). By assigning the distal elements to their closest gene, we identified six distinct gene classes with different assortments of common and unique distal elements (Supplementary Figure S2). In summary, the genome-wide analysis of DHS sites and TSSs has defined the cis-regulatory landscape by identifying distal elements and active promoters in pro-B and mature B cells.
Identification of active enhancers and inactive distal elements in pro-B and mature B cells
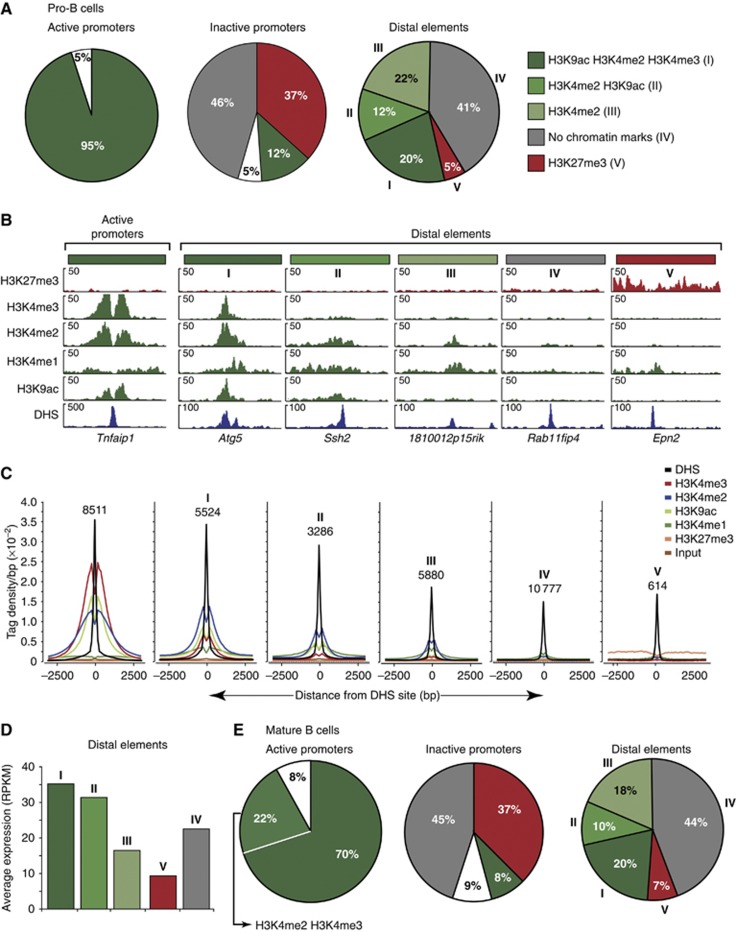
As active promoters and enhancers are characterized by specific chromatin signatures (Zhou et al, 2011), we further characterized the identified active promoters and distal elements by genome-wide profiling of active (H3K4me1, H3K4me2, H3K4me3, H3K9ac) and repressive (H3K27me3) histone modifications using ChIP coupled with deep sequencing (ChIP-seq). By calling these histone modifications with a stringent P-value of <10−10, we could demonstrate that almost all active promoters (95%) in Rag2−/− pro-B cells carried high levels of the three active histone marks H3K4me2, H3K4me3 and H3K9ac adjacent to their DHS site, as shown by global analysis (Figure 2A), a specific example (Figure 2B) and quantitative profiles of sequence tag densities (Figure 2C). These data confirmed that the co-occurrence of DHS sites and CAGE tag clusters correctly identified active promoters, which are known to contain the three active histone modifications H3K4me2, H3K4me3 and H3K9ac (Bernstein et al, 2005; Kim et al, 2005; Heintzman et al, 2007; Wang et al, 2008). A similar picture was observed for active promoters in mature B cells (Supplementary Figure S3A) except that we could not detect significant H3K9 acetylation at 22% of these promoters due to a generally low level of H3K9ac, which may reflect the quiescent state of mature B cells (Figure 2E; Supplementary Figure S3B and C). In contrast, silent promoters, which were defined as RefSeq-annotated promoters lacking active TSSs (CAGE tags <2.6 RPM), exhibited a radically different chromatin profile (Figure 2A and E).
Figure 2.
Identification of active enhancers and inactive distal elements in pro-B and mature B cells by chromatin profiling. (A) Characterization of promoters and distal elements by ChIP-seq mapping of histone modifications in short-term cultured Rag2−/− pro-B cells. Pie diagrams indicate the percentage of each combination of histone modifications (shown by different colours) at active promoters and distal elements, which were defined by the presence of DHS sites with or without CAGE sequence tags (>2.6 RPM; Figure 1A), respectively. White sectors refer to promoters lacking or containing different combinations of the three active histone modifications. Inactive promoters were defined as RefSeq-annotated promoters lacking active TSSs (CAGE tags <2.6 RPM). (B) Histone modifications at promoters and different distal elements in Rag2−/− pro-B cells. The H3K4me1 pattern and corresponding DHS site are additionally shown. Distal elements are labelled with the name of their closest gene. (C) Average sequence tag density profiles indicating the extent and intensity of the different histone modifications at regulatory elements in Rag2−/− pro-B cells. The profiles were aligned at the centre of the DHS sites, and the number of elements in each class is indicated. (D) Expression of genes associated with distinct classes of distal elements. The mRNA abundance of each gene was determined as reads per kilobase of exon per million mapped sequence reads (RPKM) by RNA sequencing of short-term cultured Rag2−/− pro-B cells. The average expression value (RPKM/gene) of all genes linked to a distinct class of distal elements is shown by the same colour code as in (A). (E) Chromatin signatures at promoters and distal elements in mature B cells isolated from lymph nodes (Supplementary Figure S3A). Representative chromatin profiles and density plots of each class of elements are shown in Supplementary Figure S3B and C. Results similar to (A–E) were obtained by analysing a second ChIP-seq experiment for each histone modification in pro-B and mature B cells (Supplementary Table S1). Supplementary Tables S4 and S5 contain the genomic coordinates of the different DE identified in pro-B and mature B cells, respectively.
The distal elements could be divided into five distinct classes according to the presence of different combinations of histone modifications at their DHS site in Rag2−/− pro-B cells. Three classes (I–III) contained significant amounts of H3K4 monomethylation (H3K4me1; Figure 2B and C), which has previously been associated with enhancers (Heintzman et al, 2007). These three classes differed, however, in their abundance of active histone modifications. Class I comprising 20% of all distal elements contained the three active marks H3K4me2, H3K4me3 and H3K9ac, class II (12%) was characterized by the presence of H3K4me2 and H3K9ac, and class III (22%) carried only significant levels of H3K4me2 (Figure 2A and B). A large part (41%) of the distal elements (class IV) contained none of the histone modifications analysed, whereas the presence of the repressive H3K27me3 mark defined the smallest (5%) class (V) of distal elements. Importantly, the five different classes of distal elements could be identified in similar relative proportions also in mature B cells (Figure 2E; Supplementary Figure S3B and C).
Recent studies used the occurrence of active histone modifications combined with the expression level of the nearby gene to predict active enhancers in the genome (Creyghton et al, 2010; Rada-Iglesias et al, 2011). We therefore analysed the transcriptome of Rag2−/− pro-B cells by RNA sequencing (Mortazavi et al, 2008) to determine the expression level of the genes associated with distinct classes of distal elements. The average transcript level of genes containing elements of class I and II was high, indicating that these distal elements qualify as active enhancers (Figure 2D). In contrast, the average expression of genes comprising class V elements was low, suggesting that these elements are inactive in pro-B cells (Figure 2D). Whereas the genes characterized by class III and IV DHS sites were expressed at intermediate levels (Figure 2D), only elements of class III significantly changed their chromatin signature during development to mature B cells, thus identifying them as poised distal elements (Supplementary Figure S3D). Interestingly, half of the class IV elements (corresponding to 20% of all distal elements) contained both CTCF- and cohesin-binding sites (Supplementary Figure S3E) and thus likely function in gene insulation, chromatin looping or chromosome cohesion (Peters et al, 2008; Phillips and Corces, 2009). In conclusion, our genome-wide analyses of DHS sites, TSSs, histone modifications and gene transcripts identified active and inactive promoters, active enhancers (l, II), poised (III) and inactive (V) distal elements as well as DHS sites (IV) enriched in CTCF/cohesin-binding sites in the absence of histone modifications, which together constitute the cis-regulatory landscape of committed pro-B and mature B cells.
Pax5 binds to a large part of the cis-regulatory elements in B cells
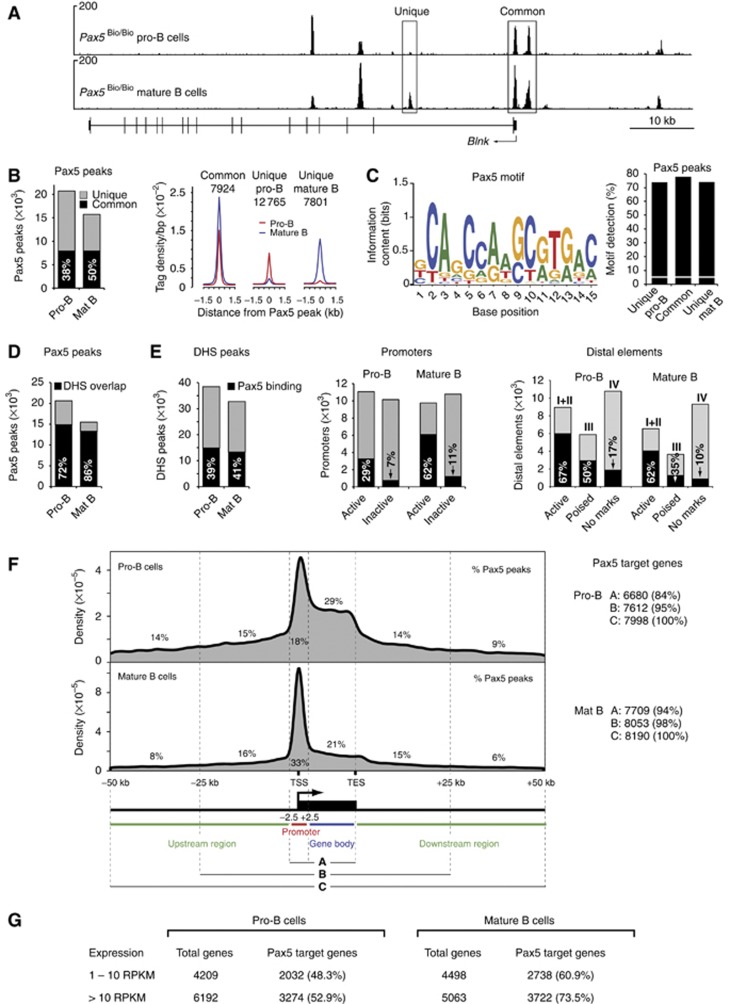
To map the genomic pattern of Pax5 binding, we took advantage of Pax5Bio/Bio mice, which exhibit normal B-cell development, although they carry a C-terminal biotin acceptor sequence together with an IRES-BirA gene insertion in the 3' untranslated region of Pax5 (McManus et al, 2011). These mice simultaneously express the biotin-tagged Pax5 protein and its modifying biotin ligase BirA (from Escherichia coli), which results in efficient biotinylation of Pax5 in all B lymphocytes (McManus et al, 2011). We thus used short-term cultured pro-B cells from Pax5Bio/Bio Rag2−/− mice and ex vivo sorted mature B cells from lymph nodes of Pax5Bio/Bio mice for streptavidin-mediated chromatin precipitation coupled with deep sequencing (referred to as Bio-ChIP-seq). As shown in Figure 3A, Bio-ChIP sequencing identified Pax5 peaks with strong enrichment over a low background at the Blnk locus in pro-B and mature B cells. By using a P-value of <10−10 for peak calling (Supplementary Figure S4A), we identified 20 613 and 15 468 Pax5-binding regions with an overlap of 7924 common peaks in pro-B and mature B cells, respectively (Figure 3B). By analysing the sequences of common Pax5 peaks with de-novo motif discovery programmes, we identified a complex and degenerate Pax5-binding motif of 15-bp length (Figure 3C) that closely resembles the previously described Pax5 consensus recognition sequence (Czerny et al, 1993; Czerny and Busslinger, 1995). Despite its degeneracy, the Pax5-binding motif could be found on average in 75% of all common and unique Pax5 peaks of pro-B and mature B cells in contrast to its detection in only 5.6% of random DNA sequences (Figure 3C). These data therefore confirmed the specificity of Pax5 binding at the peaks identified by Bio-ChIP sequencing.
Figure 3.
Pax5-binding pattern and identification of Pax5 target genes in pro-B and mature B cells. (A) Pax5 binding at the Blnk locus as defined by Bio-ChIP sequencing of Pax5Bio/Bio Rag2−/− pro-B cells and mature Pax5Bio/Bio B cells. Unique and common Pax5-binding sites are indicated together with the exon–intron structure of Blnk and a scale bar (in kb). (B) Overlap of Pax5 peaks in pro-B and mature B cells. Total numbers of 20 613 and 15 468 Pax5 peaks with an overlap of 7924 common peaks (black bar) were identified in pro-B and mature B cells, respectively, by peak calling relying on a P-value of <10−10 (Supplementary Figure S4A). Average sequence tag density profiles aligned at the centre of the Pax5 peaks are shown for common and unique Pax5-binding sites (right). The genomic coordinates of the Pax5 peaks are provided in Supplementary Table S6. (C) Consensus Pax5 recognition sequence identified by de-novo motif discovery, as described in Supplementary Materials and Methods. The Pax5-binding motif is shown with its information content indicating the size and complexity of the predicted binding. This motif was detected at the indicated frequency (%) in common and unique Pax5 peaks (right), whereas it was found only in 5.6% of random DNA sequences (white line). (D) Overlap of Pax5 peaks with DHS sites (black bar) in pro-B and mature B cells. (E) Pax5 binding at DHS sites, promoters and distal elements in pro-B and mature B cells. The percentage of Pax5 binding (black bar) to the different elements is indicated. (F) Density profiles indicating the distribution of Pax5 peaks relative to RefSeq-annotated genes in pro-B and mature B cells. DNA sequences from −2.5 kb to +2.5 kb relative to the TSS are referred to as promoter region. TES, transcription end site. The numbers of Pax5 target genes (right) were defined by the three criteria shown. (G) Expression of Pax5 target genes as determined by RNA sequencing of ex vivo sorted Rag2−/− pro-B cells and mature B cells. Similar results (A–F) were obtained by analysing a second Bio-ChIP-seq experiment for each cell type (Supplementary Table S1).
Analysis of Pax5 binding at cis-regulatory elements demonstrated that a majority (72 and 86%) of the Pax5 peaks coincides with DHS sites in pro-B and mature B cells, respectively (Figure 3D). Moreover, Pax5 binds to ∼40% of all DHS sites and thus to a large part of the cis-regulatory landscape in both cell types (Figure 3E). Although Pax5 preferentially interacts with active promoters at the two developmental stages, it binds to twice as many active promoters in mature B cells (62%) as compared with pro-B cells (29%). In contrast to distal elements lacking active histone modifications (IV), Pax5 peaks colocalize with a large part of active enhancers (I+II; 67 and 62%) and poised elements (III; 50 and 35%) in pro-B cells and mature B cells, respectively. Representative examples of Pax5 binding at active enhancers and promoters are shown for selected key transcription factor genes in Supplementary Figure S4B. In summary, these data demonstrate that Pax5 preferentially interacts with active and poised regulatory elements in early and late B lymphocytes.
Expression of a large number of Pax5 target genes in B lymphocytes
The distribution of Pax5 peaks along RefSeq-annotated gene loci is different in the two B-cell types, as the largest number of peaks is localized in the gene body (29%) in pro-B cells and in the promoter region (33%) in mature B cells (Figure 3F). Assigning the Pax5 peaks in the promoter and gene body (criterion A) to the respective gene is the least ambiguous way of determining Pax5 target genes, which already resulted in 6680 and 7709 target genes in pro-B and mature B cells, respectively (Figure 3F). Relaxing the criterion of peak assignment to a region extending from −50 kb to +50 kb beyond the gene (criterion C) identified a similar number of Pax5 target genes in pro-B cells (7998) and mature B cells (8190), which we used for all further analyses (Figure 3F). Given this large number, the identified Pax5 target genes correspond to ∼38% of all currently annotated genes (21 428) in the RefSeq database (for details see Supplementary Materials and methods).
Transcriptome analyses by RNA sequencing identified similar numbers of lowly (1–10 RPKM) and more highly (>10 RPKM) expressed genes in Rag2−/− pro-B and mature B cells (Figure 3G). Notably, Pax5 target genes account for 48 and 61% of all lowly and 53 and 73% of all highly expressed genes in pro-B and mature B cells, respectively (Figure 3G). In summary, the genome-wide binding of Pax5 identified ∼8000 Pax5 target genes, most of which are expressed in pro-B and mature B cells.
Identification of a relatively small number of regulated Pax5 target genes in pro-B cells
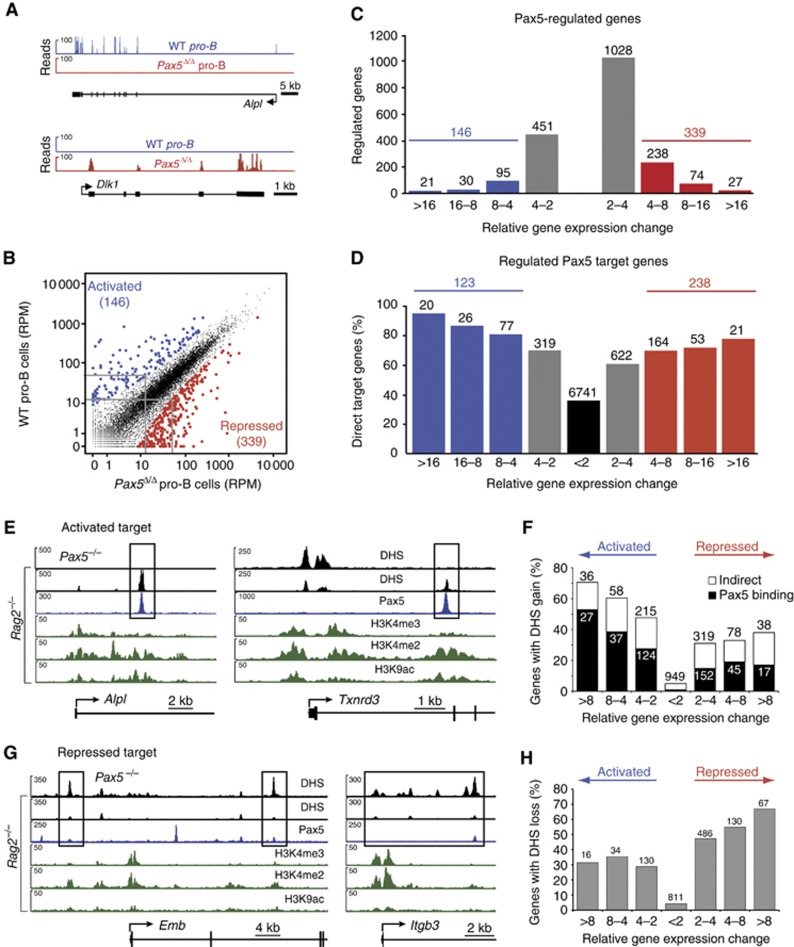
To determine which Pax5 target genes also require Pax5 for their expression, we used RNA sequencing to compare the expression level of each gene in short-term cultured Pax5fl/fl (WT) pro-B cells and Pax5Δ/Δ pro-B cells from Vav-Cre Pax5fl/fl mice (Supplementary Figure S5A), which delete Pax5 in the entire hematopoietic system. As shown in Figure 4A, the alkaline phosphatase gene Alpl was identified as a Pax5-activated gene by the presence of cDNA sequence reads at its exons in wild-type pro-B cells, whereas the Delta-like 1 (Dlk1) gene qualified as a Pax5-repressed gene by the specific accumulation of cDNA sequence reads in Pax5Δ/Δ pro-B cells. Scatter plot analysis of the normalized expression values (RPM) of all genes in the two pro-B-cell types revealed that the transcription of most genes (indicated in black) was not or only minimally affected by the presence or absence of Pax5 (Figure 4B). In contrast, 146 genes (indicated in blue) were more than four-fold activated, and 339 genes (indicated in red) were similarly repressed in wild-type pro-B cells compared with Pax5Δ/Δ pro-B cells (Figure 4B and C; Supplementary Figure S5B and C). The number of regulated genes strongly declined with increasing expression differences between the two pro-B-cell types (Figure 4C). However, when we additionally considered Pax5 binding at the regulated genes, we observed a positive correlation between the percentage of Pax5 target genes and the magnitude of differential gene expression (Figure 4D; Supplementary Figure S5B and C). This correlation was strongest for activated Pax5 target genes, demonstrating that most activated genes were also directly regulated by Pax5 (Figure 4D; Supplementary Figure S5B). Our analysis thus identified 123 activated and 238 repressed Pax5 target genes, which were more than four-fold regulated by Pax5. Interestingly, 74 and 95% of the activated and repressed Pax5 target genes were novel, as only 32 and 12 of these activated and repressed genes were recently identified as Pax5 target gene by ChIP-chip analysis (McManus et al, 2011). Importantly, the 361 regulated Pax5 target genes account for 4.5% of all 7998 target genes, which indicates that only a small subset of the Pax5-bound genes also require Pax5 for their expression in pro-B cells.
Figure 4.
Regulation of Pax5 target genes by gain and loss of enhancers in pro-B cells. (A) Expression of the Pax5-activated Alpl and Pax5-repressed Dlk1 genes in pro-B cells, as determined by RNA sequencing of short-term cultured Pax5fl/fl (WT) and Vav-Cre Pax5fl/fl (Pax5Δ/Δ) pro-B cells (Supplementary Figure S5A). (B) Scatter plot of gene expression differences observed between WT and Pax5Δ/Δ pro-B cells. The normalized expression value of each gene in the two pro-B-cell types was plotted as reads per gene per million mapped sequence reads (RPM) as described in Supplementary Materials and Methods. Genes with an expression difference corresponding to a significance value of >20 or <−20 are highlighted in blue and red, respectively. The cutoff value of 20 corresponds to a four-fold expression change for a gene that is expressed at 15 RPM in the lower expressing cell type, as indicated by grey lines. (C) Pax5-regulated gene expression. The number of differentially expressed genes in wild-type pro-B cells compared with Pax5Δ/Δ pro-B cells is shown for the indicated fold expression changes corresponding to a gene that is expressed at 15 RPM in the lower expressing cell type (see Supplementary Materials and methods). (D) Identification of activated and repressed Pax5 target genes in pro-B cells. The number and percentage of Pax5 target genes are shown for the indicated gene expression differences between wild-type and Pax5Δ/Δ pro-B cells. The expression of the most highly activated (20) and repressed (21) Pax5 target genes is shown in Supplementary Figure S5B and C. Results similar to (A–D) were obtained by analysing a second RNA-seq experiment for each pro-B-cell type (Supplementary Table S1). The RNA-seq data of the regulated Pax5 target genes are shown in Supplementary Table S7. (E) Induction of enhancers at the activated Pax5 target genes Alpl and Txnrd3 in pro-B cells. DHS sites (black) are shown for Rag2−/− and Pax5−/− pro-B cells together with the active chromatin profile (green) in Rag2−/− pro-B cells and the Pax5-binding pattern (blue) analysed in Pax5Bio/Bio Rag2−/− pro-B cells. (F) Gain of DHS sites at Pax5-activated genes. The number and percentage of Pax5-activated and Pax5-repressed genes with gain of a DHS site at distal elements in Rag2−/− pro-B cells are shown for the indicated expression changes observed between Rag2−/− and Pax5−/− pro-B cells (see C). Black bars indicate the presence of Pax5 binding at the induced DHS site in Rag2−/− pro-B cells, whereas white bars denote the absence of Pax5 peaks. (G) Loss of DHS sites at the repressed Pax5 target genes Emb and Itgb3 in Rag2−/− pro-B cells. Supplementary Figure S6C documents the transient binding of Pax5 at the DHS sites of Emb and Itgb3 in 4-hydroxytamoxifen-treated Pax5−/− pro-B cells expressing a Pax5–oestrogen receptor fusion protein, which was determined by ChIP–qPCR analysis with primers shown in Supplementary Table S9. (H) Loss of DHS sites at Pax5-repressed genes in Rag2−/− pro-B cells. Pax5 binding was not evaluated, as Pax5 peaks in Rag2−/− pro-B cells were often small or absent at the position, where a DHS site was present in Pax5−/− pro-B cells.
Target gene activation by Pax5-mediated induction of active enhancers
We have recently demonstrated that Pax5 is able to induce active chromatin at genomic Pax5-binding sites by recruiting chromatin-remodelling and histone-modifying enzymes (McManus et al, 2011). Here, we investigated whether Pax5 may activate its target genes by inducing DHS sites in committed pro-B cells. To this end, we additionally mapped DHS regions in Pax5−/− Rag2−/− pro-B cells, which resulted in a very similar number (38 576) of DHS sites compared with those (38 462) of committed Rag2−/− pro-B cells (Figure 1A). Interestingly, the emergence of DHS sites with characteristic enhancer chromatin in Rag2−/− pro-B cells coincided with Pax5 binding at the Alpl, Txnrd3, Bcat1 and Enpep loci (Figure 4E; Supplementary Figure S6A), suggesting that these newly formed enhancers (distal elements II) contribute to gene activity in committed pro-B cells. The appearance of active enhancers and promoters at Pax5-activated genes was a more general phenomenon, as the percentage of genes with a gain of DHS site increased with higher gene activation in wild-type pro-B cells compared with Pax5Δ/Δ pro-B cells (Figure 4F). Importantly, this proportional increase was exclusively due to activated Pax5 target genes containing newly formed enhancers coinciding with a Pax5 peak (black bar, Figure 4F), which strongly argues that Pax5 activates these genes by inducing the formation of active enhancers. In contrast, DHS sites that were present at enhancer positions of repressed Pax5 target genes in Pax5−/− pro-B cells were lost in Rag2−/− pro-B cells as exemplified by the Emb, Itgb3, Flt3 and Pdcd1lg2 genes (Figure 4G; Supplementary Figure S6B). The elimination of enhancers may be causally linked to gene silencing as repressed genes with loss of DHS sites proportionally increased with stronger transcriptional repression in wild-type pro-B cells (Figure 4H). However, Pax5 peaks were either very small or could not be detected at the eliminated DHS site in Rag2−/− pro-B cells, which may be explained in two ways. Pax5 may indirectly eliminate DHS regions by activating the expression of other transcriptional represssors. Alternatively, Pax5 binding initiates the shutdown of DHS regions at repressed target genes, but, after loss of the DHS site, Pax5 can no longer interact with the closed chromatin. Consistent with the latter possibility, we observed transient Pax5 binding with concomitant loss of the active H3K9ac mark at DHS sites of the repressed target genes Emb and Itgb3 upon oestrogen-mediated induction of Pax5 activity in Pax5−/− pro-B cells expressing a Pax5–oestrogen receptor fusion protein (Supplementary Figure S6C; Nutt et al, 1998). Together, these data revealed an important function of Pax5 in transcriptional activation and repression by inducing or eliminating active enhancers at B-cell commitment.
Role of Pax5 during the differentiation of CLPs to committed pro-B cells
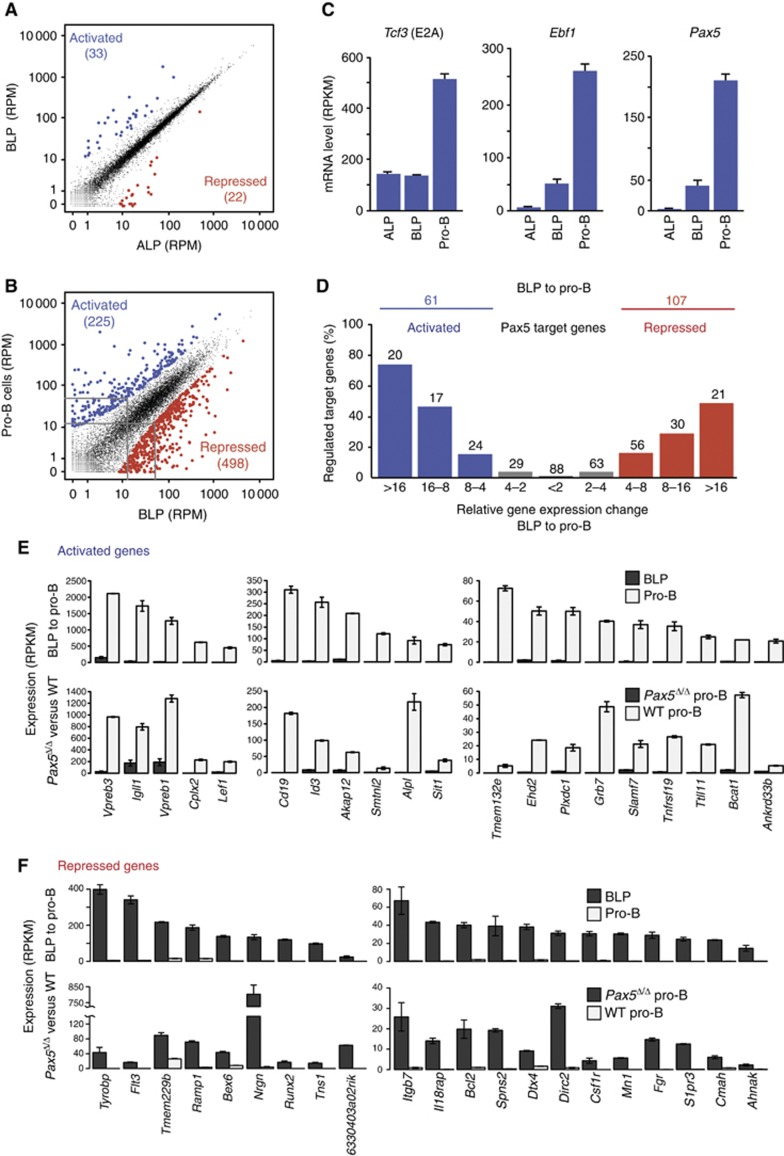
To investigate the in vivo contribution of the regulated Pax5 target genes to early B-cell development, we next determined how many genes are activated and repressed during the development of ALPs via BLPs to committed pro-B cells (Inlay et al, 2009). To this end, we isolated ALPs, BLPs and Rag2−/− pro-B cells with a purity of >93% by FACS sorting of bone marrow cells (Supplementary Figure S7A and B) and characterized their transcriptome by RNA sequencing. Scatter plot analysis of normalized expression values (RPM) revealed that 33 genes (indicated in blue) were more than four-fold activated and 22 genes (indicated in red) were similarly repressed in BLPs compared with ALPs (Figure 5A). Previous expression microarray analysis of ALPs and BLPs identified 21 activated genes (Inlay et al, 2009), 14 of which were also present among the 33 activated genes identified by RNA sequencing. Some of the activated genes code for lymphoid transcription factors (EBF1, Pax5, OBF1 [Pou2af1], SpiB, Foxo1), cell surface receptors (Ly6D, CD79a [Igα], VpreB2, VpreB3, λ5 [Igll1]) and signal transducers (BLK, PDE2a, PKCβ [Prkcb]). However, many more genes were activated and repressed during the next developmental transition from BLPs to committed pro-B cells (Figure 5B; Supplementary Figure S7C). Analysis of the instructive transcription factors E2A, EBF1 and Pax5 revealed that the Tcf3 (E2A) gene was equally expressed in ALPs and BLPs, whereas both Ebf1 and Pax5 were strongly activated in BLPs (Figure 5C) in agreement with published data (Inlay et al, 2009). The three genes were further activated ∼4-fold in pro-B cells (Figure 5C), consistent with the notion that all three transcription factors contribute to gene regulation during the BLP to pro-B-cell transition.
Figure 5.
Pax5 function during the development of BLPs to committed pro-B cells. (A, B) Gene expression changes during the developmental transitions from ALPs to pro-B cells. ALPs, BLPs and Rag2−/− pro-B cells were FACS-sorted with a purity of >93% (Supplementary Figure S7A) prior to RNA preparation and RNA sequencing. Normalized expression values (RPM) were calculated for each gene in the three cell types, and the gene expression differences between ALPs and BLPs (A) and between BLPs and Rag2−/− pro-B cells (B) were analysed as described in Figure 4B. The RNA-seq data of the genes regulated during these developmental transitions are shown in Supplementary Table S8. (C) Expression of Tcf3, Ebf1 and Pax5 at the ALP, BLP and pro-B cell stages. Average expression values (RPKM) with standard deviations were obtained from two RNA-seq experiments. (D) Genes with high expression differences between the BLP and pro-B cell stage (Supplementary Figure S7C) are enriched for activated (blue) and repressed (red) Pax5 target genes. Similar results (A–D) were obtained by analysing a second RNA-seq experiment for each cell type (Supplementary Table S1). (E) Expression of the most strongly (>16-fold) activated genes during the BLP to pro-B-cell transition that are also activated Pax5 target genes. Average expression values (RPKM) with standard deviations are shown for ex vivo sorted BLPs and Rag2−/− pro-B cells (top row) and short-term cultured Pax5Δ/Δ and wild-type (WT) pro-B cells (bottom row). (F) Expression of the most strongly (>16-fold) repressed genes during the BLP to pro-B-cell transition that are also repressed Pax5 target genes.
We next investigated to what extent Pax5 controls the 225 activated and 498 repressed genes that are regulated more than four-fold during the BLP to pro-B-cell transition (Figure 5B; Supplementary Figure S7C). As shown in Figure 5D, we found a strong correlation between the percentage of regulated Pax5 target genes and the magnitude of differential gene expression observed during the BLP to pro-B-cell transition. Hence, genes with high expression differences in this developmental transition were strongly enriched for regulated Pax5 target genes. Analysis of the normalized expression values (RPKM) for the most highly activated and repressed genes (>16-fold) unequivocally demonstrated that these genes are not only strongly regulated during the BLP to pro-B-cell transition, but also depend on Pax5 for their regulation in pro-B cells (Figure 5E and F). This conclusion was confirmed by analysing the 8- to 16-fold regulated gene class (Supplementary Figure S7D and E). These analyses furthermore revealed that about half of all regulated Pax5 target genes (61 (50%) of 123 activated and 107 (45%) of 238 repressed target genes) are also differentially expressed during the BLP to pro-B-cell transition (Figures 4D and 5D). Importantly, Pax5 directly activates 27% (61 Pax5 target genes) of 225 activated genes and represses 22% (107 Pax5 target genes) of 498 repressed genes during the BLP to pro-B-cell transition (Figure 5B and D). Hence, Pax5 controls a relatively large part of the gene expression changes occurring during the BLP to pro-B-cell transition.
Pax5 regulates distinct target genes in pro-B and mature B cells
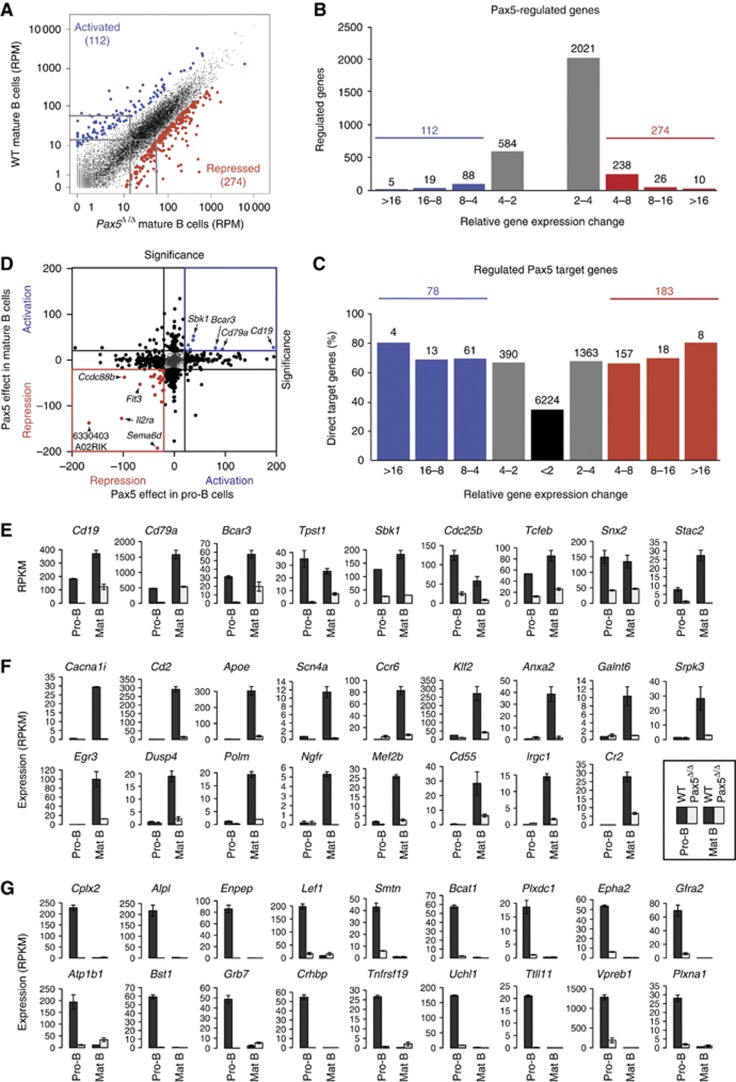
As Pax5 plays an essential role throughout B-cell development (Horcher et al, 2001; Cobaleda et al, 2007), we identified Pax5-regulated genes in late B lymphopoiesis by conditional mutagenesis using the Cd23-Cre line, which resulted in complete deletion of the floxed Pax5 allele in mature B cells (Kwon et al, 2008). We thus isolated quiescent mature B cells from the lymph nodes of wild-type and Cd23-Cre Pax5fl/fl (Pax5Δ/Δ) mice by FACS sorting (Supplementary Figures S3A and S8A) and identified Pax5-activated and Pax5-repressed genes in mature B cells by RNA sequencing, as shown for the activated Stac2 and repressed Car2 genes (Supplementary Figure S8B). Scatter plot analysis of the normalized expression values (RPM) of all genes revealed that 112 genes (indicated in blue) were more than four-fold activated, and 274 genes (indicated in red) were similarly repressed in wild-type compared with Pax5-deficient mature B cells (Figure 6A and B). Notably, fewer genes showed strong Pax5-dependent activation or repression in mature B cells (Figure 6A and B) compared with pro-B cells (Figure 4B and C), which likely reflects the reduced viability of Pax5-deficient mature B cells (Horcher et al, 2001). By additionally considering Pax5 binding, we identified 78 activated and 183 repressed Pax5 target genes in mature B cells (Figure 6C). Surprisingly however, we observed a minimal overlap between the Pax5-regulated genes identified in pro-B and mature B cells, as only very few genes were more than four-fold activated or repressed at both developmental stages (Figure 6D).
Figure 6.
Identification of regulated Pax5 target genes in mature B cells. (A, B) Gene expression differences between wild-type (WT) and Pax5Δ/Δ mature B cell that were FACS-sorted from lymph nodes of wild-type and Cd23-Cre Pax5fl/fl mice, respectively (Supplementary Figures S3A and S8A). The RNA-seq data of the two cell types are shown by scatter plot analysis (A) and gene expression (significance) differences (B) as described in Figure 4B and C. (C) Identification of activated and repressed Pax5 target genes in mature B cells. The number and percentage of Pax5 target genes are shown for the indicated gene expression differences between wild-type and Pax5Δ/Δ mature B cells. Results similar to (A–C) were obtained by analysing a second RNA-seq experiment for each cell type (Supplementary Table S1). The RNA-seq data of the regulated Pax5 target genes are shown in Supplementary Table S7. (D) Minimal overlap between Pax5-regulated genes in pro-B and mature B cells. Significance values describe the differential expression of each gene between wild-type and Pax5Δ/Δ pro-B cells on the x-axis and between wild-type and Pax5Δ/Δ mature B cells on the y-axis. (E–G) Different classes of activated Pax5 target genes. The expression of the different genes is shown as normalized expression value (RPKM) with standard deviation, as determined by two RNA-seq experiments. Black bars indicate expression in wild-type (WT) pro-B and mature B cells and grey bars in Pax5Δ/Δ pro-B and mature B cells. (E) Commonly activated target genes. Nine of 13 genes, which were activated at least three-fold in pro-B and mature B cells, are shown with Gsn, Edaradd, Lpcat2 and Rrm2 being the remaining four genes. (F) Activated Pax5 target genes in mature B cells with no or very low expression in pro-B cells. (G) Activated Pax5 target genes in pro-B cells with no or very low expression in mature B cells.
To investigate the reason for the low overlap among the regulated Pax5 target genes, we first analysed the 3569 common Pax5 target genes, which were similarly expressed in pro-B and mature B cells as shown by transcriptome comparison of the two cell types (Supplementary Figure S9A and B). As expected, the majority of these target genes were not regulated by Pax5 in either cell type (Supplementary Figure S9C) and largely coded for ‘house-keeping’ functions as shown by Gene Ontology analysis (Supplementary Figure S9D and E). The few Pax5-dependent genes among the common target genes (Supplementary Figure S9C) could be further distinguished as to whether they were similarly or differently regulated by Pax5 in pro-B and mature B cells. Surprisingly, we identified only 18 commonly repressed genes, which corresponded to 7.4 and 9.7% of all repressed Pax5 target genes in pro-B and mature B cells, respectively (Supplementary Figure S8G). Likewise, only 13 Pax5 target genes were activated in both cell types, thus constituting 10.6 and 16.7% of the activated Pax5 target genes in pro-B and mature B cells (Figure 6E). A second class of activated Pax5 target genes was regulated only in pro-B cells (30 genes) or mature B cells (23 genes), corresponding to 24.3 and 29.5% of the activated target genes in the respective cell type (Supplementary Figure S8C and D). The loss of Pax5-dependent activation in one of the two cell types could be caused by the absence of certain Pax5 peaks and their associated DHS sites, as exemplified for Ebf1 and Trp53i11 in mature B cells (Supplementary Figure S8E) and Fam43a and Sh3bp2 in pro-B cells (Supplementary Figure S8F).
The most striking finding of the transcriptome analysis of pro-B and mature B cells was the observed massive change of gene expression within the B-cell lineage, as 417 genes were more than four-fold activated and 1076 genes were similarly repressed during the development of pro-B cells to mature B cells (Supplementary Figure S9A and B). The large number of differentially expressed genes in pro-B and mature B cells therefore suggested the possibility that a majority of the activated Pax5 target genes might be significantly expressed only at one of the two developmental stages. Indeed, 31 (40%) of the activated Pax5 target genes in mature B cells showed no or only very low expression in pro-B cells, thus indicating that they could not qualify as activated Pax5 target genes in pro-B cells (Figure 6F). Consistent with this finding, most of these genes contained only significant levels of the H3K4me2 mark in pro-B cells, which is indicative of poised chromatin (Supplementary Figure S10A). Moreover, 62 (50.4%) of the activated Pax5 target genes in pro-B cells were either not at all or only lowly expressed in mature B cells (Figure 6G). Active chromatin and DHS sites were absent or strongly reduced at most genes of this category in mature B cells, demonstrating that they are epigenetically silenced in late B lymphopoiesis (Supplementary Figure S10B). In summary, we conclude that Pax5 largely regulates different target genes in early and late B-cell development.
The observed differential gene regulation could be explained by the selective binding of important transcription factors to Pax5 target genes in pro-B or mature B cells. To investigate this possibility, we searched for transcription factor-binding motifs, which are specifically enriched at Pax5 peaks in pro-B and mature B cells. The DNA-binding motifs for Foxo1, Lmo2 and EBF1 were moderately enriched at unique Pax5 peaks and Pax5-binding sites of activated target genes in pro-B cells, whereas stronger enrichment was observed for motifs of the five transcription factors Sp1, Myc, Cnot3, Zfx and Maz at unique Pax5 peaks and Pax5-binding sites of activated target genes in mature B cells (Supplementary Figure S10C; see Materials and methods). For one of these transcription factors, EBF1, we could directly determine the overlap of binding with Pax5 peaks by analysing EBF1 ChIP-seq data of pro-B and mature B cells (Vilagos et al, 2012). EBF binding was detected at 2913 (14%) of all Pax5 peaks in pro-B cells (Supplementary Figure S10D and E). Importantly, the frequency (23.9%) of Pax5 peaks with EBF1 binding significantly increased at activated Pax5 target genes in pro-B cells, which suggests a critical role for EBF1 in the regulation of these genes in early B-cell development (Supplementary Figure S10E). In contrast, we identified only 281 EBF1 target genes in mature B cells (Vilagos et al, 2012), which resulted in the colocalization of only 1.2% of all Pax5 peaks with EBF1-binding sites in mature B cells (Supplementary Figure S10D–F). Together, these data indicate that EBF1 is involved in the regulation of activated Pax5 target genes in early B lymphopoiesis, whereas other transcription factors, such as Sp1, Myc, Cnot3, Zfx and Maz, likely contribute to the activation of Pax5 target genes in late B-cell development.
Function of regulated Pax5 target genes
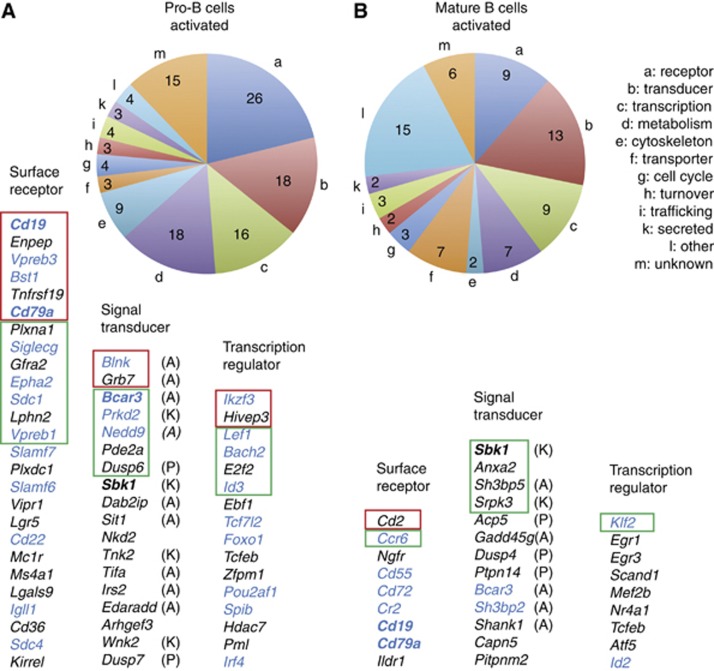
The 123 activated Pax5 target genes identified in pro-B cells (Figure 4D) code for proteins of distinct functions with three large classes consisting of 26 cell surface and adhesion receptors, 18 intracellular signal transducers and 16 transcriptional regulators (Figure 7A). The 78 activated Pax5 target genes in mature B cells (Figure 6C) revealed a similar distribution of protein function, coding for 9 cell surface receptors, 13 signal transducers and 9 transcriptional regulators (Figure 7A). These three classes contain only four genes (Cd19, Cd79a, Bcar3 and Sbk1; bold in Figure 7), which are commonly activated in pro-B and mature B cells, further demonstrating that Pax5 mainly regulates different genes in the two cell types. Several activated target genes in pro-B cells have known functions during B-cell development (blue; Figure 7A), as they code for structural or regulatory proteins involved in pre-BCR signalling (Cd19, Cd79a, Vpreb1, Vpreb3, Igll1, Blnk, Prkd2), inhibition of signalling (Siglecg, Cd22, Slamf6, Slamf7), B-cell adhesion and migration (Bst1, Epha2, Sdc1, Sdc4, Bcar3, Nedd9) or transcriptional regulation (Ikzf3, Lef1, Bach2, Id3, Tcf7l2, Foxo1, Pou2af1, Spib, Irf4). In mature B cells, we identified fewer activated target genes (blue; Figure 7B) with known functions in BCR signalling (Cr2 [Cd21], Cd19, Cd79a, Sh3bp2), inhibition of signalling (Cd72), B-cell trafficking and homing (Ccr6, Cd55, Bcar3) and transcriptional regulation (Klf2, Id2).
Figure 7.
Function of activated Pax5 target genes in pro-B and mature B cells. (A, B) Pie diagram indicating the different functional classes of activated Pax5 target genes in pro-B cells (A) and mature B cells (B). The activated Pax5 target genes shown below are ranked according to the significance of their differential expression as determined by RNA sequencing of Rag2−/− and Pax5-deficient pro-B cells (Figure 4D) or wild-type and Pax5-deficient mature B cells (Figure 6C). Genes with a significance value of >83 (>16-fold) or between 83 and 47 (16- to 8-fold) are indicated by red and green boxes, respectively, whereas the remaining genes are characterized by a significance value ranging from 47 to 20 (eight- to four-fold). Genes that are commonly activated in both cell types are indicated in bold, and genes with a known function in B lymphopoiesis are shown in blue. Signalling adaptors (A), kinases (K) and phosphatases (P) are indicated.
Protein functions were also assigned to the 238 and 183 repressed Pax5 target genes identified in pro-B cells (Figure 4D) and mature B cells (Figure 6C), respectively (Supplementary Figure S11). Cell surface receptors, signal transducers and transcriptional regulators are encoded by 39, 45 and 26 repressed target genes in pro-B cells and 23, 35 and 24 repressed target genes in mature B cells, respectively (Supplementary Figure S11). Hence, Pax5 down-regulates the expression of multiple receptors, signalling molecules and transcription factors, many of which play important roles in other hematopoietic cell types. Only seven genes of these three classes are commonly repressed in pro-B and mature B cells. One of these genes is Pax5, suggesting that Pax5 is involved in fine-tuning of its own expression (Supplementary Figures S8G and S11).
Discussion
The transcription factor Pax5 controls the development, identity and function of B cells by repressing B-lineage-inappropriate genes and activating B-cell-specific genes (Medvedovic et al, 2011). Here, we have investigated the direct transcriptional effects of Pax5 by genome-wide analyses, demonstrating that Pax5 binds to a large part of the cis-regulatory genome corresponding to ∼8000 Pax5 target genes in pro-B and mature B cells. However, only a small subset of these target genes also depend on Pax5 for their expression in pro-B cells (4.5%) and mature B cells (3.2%). Nevertheless, the regulated Pax5 target genes in pro-B cells account for a large part of the gene expression changes that occur during the developmental transition from BLPs to committed pro-B cells. Notably, we observed only a minimal overlap between regulated Pax5 target genes in pro-B and mature B cells, indicating that Pax5 controls the identity and function of B lymphocytes by regulating a relatively large but different spectrum of target genes in early and late B-cell development.
Genome-wide profiling of histone modifications has successfully been used for defining cis-regulatory elements and predicting gene activity in different cell types (Heintzman et al, 2007; Wang et al, 2008; Creyghton et al, 2010; Lin et al, 2010; Ernst et al, 2011; Northrup and Zhao, 2011; Rada-Iglesias et al, 2011). Here, we have employed a novel approach for analysing the cis-regulatory landscape by genome-wide mapping of all protein–DNA interaction sites (DHS sites) and active TSSs (CAGE) to define distal elements and active promoters in pro-B and mature B cells. The profiling of histone modifications was subsequently used to categorize the distal elements according to their chromatin signature as active enhancers or poised and inactive elements. The overall organization of the cis-regulatory landscape is comparable in pro-B and mature B cells containing 36 844 and 31 087 DHS sites, respectively. Among these DHS sites, active promoters (28 and 31%), active enhancers (23 and 21%), poised elements (16 and 13%) and inactive distal elements (3 and 5%) are present in similar proportions in pro-B and mature B cells, respectively. Surprisingly, 30% of all DHS sites (distal element class IV) in both cell types do not contain any significant amount of the active or repressive histone modifications analysed, but are enriched in CTCF- and cohesin-binding sites, indicating that they may be involved in gene insulation, chromatin looping or chromosome cohesion (Peters et al, 2008; Phillips and Corces, 2009).
Pax5 interacts with a large part (40%) of the cis-regulatory landscape in both B-cell types, as it binds to 20 613 and 15 468 genomic sites defining ∼8000 target genes in pro-B and mature B cells, respectively. Pax5 is not unique in this respect, as even higher numbers of binding sites have been reported for the transcription factor PU.1 in B cells (∼33 000; Heinz et al, 2010), CTCF in pro-B cells (∼49 000; Ebert et al, 2011) and MyoD in myoblasts (∼60 000; Cao et al, 2010). In contrast, fewer binding sites and thus less target genes were identified for EBF1 and E2A in pro-B cells (Lin et al, 2010; Treiber et al, 2010; Vilagos et al, 2012), GATA3 in T cells (Wei et al, 2011) and GATA1 in erythroblasts (Soler et al, 2010). The high number of genomic Pax5-binding sites could reflect the flexible sequence recognition by the DNA-binding paired domain of Pax5. While we previously identified a degenerate Pax5-binding sequence of 15-bp length (Czerny et al, 1993; Czerny and Busslinger, 1995), ChIP sequencing has now refined this degenerate recognition motif. The paired domain of Pax5 is known to recognize DNA through three different binding modules, as the N- and C-terminal helix-turn-helix domains recognize distinct DNA-binding motifs in adjacent major grooves of the DNA helix, whereas the linker between the two domains contacts the minor groove (Xu et al, 1995; Garvie et al, 2001). The crystal structure of the paired domain–DNA complex (Garvie et al, 2001) indicates that the residues 1–3 and 13–15 of the Pax5 recognition sequence (Figure 3C) mediate the major groove contacts and residues 5–10 the minor groove contacts of the paired domain. The degeneracy of sequence recognition is best explained by the observation that none of the three DNA-binding modules recognize their motif with highest affinity, which allows for considerable variability of the recognition sequence (Czerny et al, 1993).
Although Pax5 preferentially binds to active promoters, active enhancers and poised distal elements, it is not required for the expression of most (>95%) of its target genes. A similar discrepancy between transcription factor binding and gene regulation has recently been observed for EBF1 and GATA3 in pro-B and T cells, respectively (Treiber et al, 2010; Wei et al, 2011). This disparity could be explained in different ways. Transcription factors recognizing a simple or degenerate sequence motif like Pax5 may fortuitously interact with binding sites in accessible chromatin at active enhancers and promoters, although other transcription factors open up and control these regulatory elements in pro-B or mature B cells. Alternatively, Pax5 may be essential for the regulation of some of its target genes at another developmental stage or under specific stimulation conditions that we did not investigate. It has been suggested that lineage-determining factors such as PU.1, MyoD and Pax5 bind to a large part of the cis-regulatory landscape to maintain cell identity through global organization of the genome in the one- and three-dimensional space (Natoli, 2010). However, functional evidence for this hypothesis is currently missing particularly in view of the fact that Pax5 is dispensable for the expression of most of its target genes.
A remarkable aspect of our analysis is the identification of only a small number of activated (1.54 and 1.0%) and repressed (3.0 and 2.2%) Pax5 target genes in pro-B and mature B cells, respectively. Our genome-wide data also demonstrate that Pax5 represses twice as many genes as it activates in the two B-cell types. Importantly, we analysed the spectrum of regulated Pax5 target genes in pro-B cells in the context of the gene expression changes that are responsible for B-cell specification and commitment during the transition of ALPs to committed pro-B cells. This process is controlled by the transcription factors E2A, EBF1 and Pax5, whereby E2A activates Ebf1 possibly through Foxo1 induction (Welinder et al, 2011) and EBF1 in turn activates the Pax5 gene to accomplish B-cell commitment (Nutt and Kee, 2007; Decker et al, 2009). Surprisingly, the pattern of Ebf1 and Pax5 expression is quite similar in ALPs, BLPs and pro-B cells (this study; Inlay et al, 2009), suggesting that EBF1 rapidly induces Pax5 expression consistent with the fact that the intronic Pax5 enhancer is already activated in multipotent hematopoietic progenitors (Decker et al, 2009). Interestingly, Pax5 regulates a large part of the genes that are repressed (22%) or activated (27%) during the developmental transition from BLPs to committed pro-B cells. Hence, these genome-wide molecular data further document the importance of Pax5 for B-cell commitment.
Recently, Pax5 was shown to rapidly induce active chromatin marks at activated target genes and to remove active histone modifications at repressed target genes by recruiting chromatin-remodelling and histone-modifying protein complexes (McManus et al, 2011). At the global scale, we now demonstrate that Pax5 binding correlates with the induction of active enhancers and promoters at activated Pax5 target genes, whereas the loss of DHS sites is associated with repression of Pax5 target genes in committed pro-B cells. Together, these data indicate that Pax5 functions as an epigenetic regulator to control target gene expression by activating enhancers at activated target genes and by eliminating DHS sites at repressed target genes.
An unexpected finding of our study was the discovery that Pax5 predominantly regulates different genes in early and late B-cell development, as only 13 target genes are commonly activated and 18 target genes are commonly repressed in pro-B and mature B cells. However, Pax5 interacts with a similarly high number of genomic binding sites defining 8000 target genes in both cell types consistent with the fact that Pax5 is similarly expressed in pro-B and mature B cells (Fuxa and Busslinger, 2007). Surprisingly, the development of pro-B cells to mature B lymphocytes is accompanied by a two-fold larger change of gene expression within the B-cell lineage (417 up- and 1076 down-regulated genes) as compared with the developmental transition of BLPs to pro-B cell during B-cell commitment (225 up- and 498 down-regulated genes). This massive change of gene expression largely explains the different spectrum of activated Pax5 target genes in pro-B and mature B cells, as half of all activated target genes in one B-cell type do not qualify as regulated target genes in the other cell type, where they are not at all or only very lowly expressed. As a consequence, Pax5 regulates genes with different functions in pro-B and mature B cells.
The observed differential gene regulation by Pax5 in early and late B lymphopoiesis can be explained by epigenetic and transcriptional mechanisms. First, target genes, which are activated by Pax5 only in mature B cells, predominantly contain poised chromatin in pro-B cells, indicating that their chromatin structure does not support expression in early B-cell development. Moreover, target genes, which are activated by Pax5 exclusively in pro-B cells, are epigenetically silenced by the loss of active chromatin and DHS sites in mature B cells. Second, motif enrichment analyses identified a few transcription factors that may selectively bind to Pax5 target genes in pro-B or mature B cells. The binding of one of these transcription factors, EBF1, was shown by ChIP sequencing to be strongly enriched at the Pax5 peaks of a significant fraction of activated target genes in pro-B cells, which suggests a critical role for EBF1 in the regulation of these genes in early B-cell development.
Expression microarray analyses of pro-B cells previously demonstrated that Pax5 represses B-lineage-inappropriate genes involved in receptor signalling, cell adhesion, migration, transcriptional control and cellular metabolism in other hematopoietic lineages (Delogu et al, 2006), while simultaneously activating genes encoding regulatory and structural proteins important for B-cell signalling, trafficking, differentiation and immune function (Schebesta et al, 2007). By extending these expression analyses to the genome-wide scale and by defining the spectrum of regulated target genes in pro-B and mature B cells, we now demonstrate that Pax5 directly activates and represses different genes encoding signalling and adhesion receptors, intracellular signal transducers and transcriptional regulators in early and late B-cell development. We therefore conclude that Pax5 controls B-cell identity not by regulating the same target genes throughout B lymphopoiesis, but rather by controlling a relatively large but different set of genes involved in receptor signalling and transcriptional regulation in pro-B and mature B cells.
In summary, we have deciphered the role of Pax5 in gene transcription at the genome-wide level during normal B lymphopoiesis. Importantly, Pax5 is also implicated as a haploinsufficient tumour suppressor or oncogenic translocation fusion protein in the generation of human B-cell precursor acute lymphoblastic leukaemia (B-ALL; Mullighan et al, 2007; Nebral et al, 2009). Hence, our genome-wide characterization of Pax5 target genes has set the stage for future investigations to identify the spectrum of Pax5 target genes that contribute to B-ALL development upon deregulation by distinct PAX5 mutations.
Materials and methods
Mice
All mouse strains were maintained on the C57BL/6 background. Animal experiments were carried out according to valid project licences, which were approved and regularly controlled by the Austrian Veterinary Authorities.
Flow cytometry
Bone marrow and lymph node cells were stained for flow cytometry with the following antibodies: anti-B220/CD45R (RA3-6B2), CD3ε (145-2C11), CD4 (GK1.5), CD5 (53-7.3), CD8a (2.43), CD11b/Mac1 (M1/70), CD11c (HL3), CD19 (1D3), CD21 (7G6), CD23 (B3B4), CD25/IL-2Rα (PC61), CD43 (S7), CD49b (DX5), CD117/c-Kit (ACK4), CD127/IL-7Rα (A7R34), CD135/Flt3 (A2F10.1), Gr1 (RB6-8C5), IgD (1.19), IgMa (Igh-6a/DS-1), IgMb (AF6-78), Ly6C (6C3), Ly6D (49H4.3), NK1.1 (PK136), Sca1/Ly6A (D7), TCRβ (H57-597) and Ter119 (TER119) antibodies.
FACS and MACS sorting
For isolating ALPs and BLPs, lineage-positive cells (Lin+) cells were first depleted from the bone marrow by MACS sorting using the following lineage (Lin) marker antibodies: CD19, CD3ε, CD4, CD8a, DX5, Gr1, Ly6C, Mac1 and Ter119. Subsequently, ALPs (Lin−B220−IL-7Rα+Flt3+Sca1loc-KitloLy6D−) and BLPs (Lin−B220−IL-7Rα+ Flt3+Sca1loc-KitloLy6D+) were sorted with a FACS Aria machine (Becton Dickinson; Supplementary Figure S7A). Wild-type and Rag2−/− pro-B cells were sorted from the bone marrow as CD19+B220+c-Kit+ CD25−IgM− cells (Supplementary Figure S7B). Wild-type mature B cells were purified from lymph nodes by MACS depletion of non-B cells with anti-PE beads after staining with PE-labelled TCRβ, CD4, CD8a, Mac1, Gr1, DX5 and Ly6C antibodies (Supplementary Figure S3A). Pax5Δ/Δ mature B cells were FACS-sorted as B220+IgDlo/−CD25+TCRβ− cells (Horcher et al, 2001) from the lymph nodes of Cd23-Cre Pax5fl/fl mice (Supplementary Figure S8A).
In-vitro culture of lymphoid progenitors
Sorted pro-B cells were cultured for 4–5 days on OP9 cells in IL-7 containing IMDM as described (Nutt et al, 1997).
CAGE analysis
RNA was isolated from short-term cultured and sorted Rag2−/− pro-B cells as well as from sorted mature B cells of wild-type lymph nodes. Total RNA (∼50 μg) of Rag2−/− pro-B and mature B cells was sent to ImaGenes (Berlin) for preparation of CAGE libraries, which were analysed by Solexa deep sequencing.
DHS site mapping
Pro-B and mature B cells were enriched by density gradient purification using Lympholyte M (Cedarlane Laboratories) prior to DNase I treatment and isolation of DNA fragments between 150 and 250 bp by sucrose gradient centrifugation as described (Sabo et al, 2006). Fractions, which were shown by quantitative PCR to be enriched for DHS sequences, were pooled and analysed by Solexa deep sequencing.
ChIP-seq analysis of histone modifications
ChIP with histone modification-specific antibodies was performed as described in detail (Schebesta et al, 2007). Rabbit polyclonal antibodies recognizing the following histone tail modifications were used for ChIP analysis: H3K4me1 (ab8895) and H3K4me3 (ab8580) from Abcam, H3K4me2 (07-030) and H3K9ac (07-352) from Millipore and H3K27me3 from T Jenuwein (MPI Freiburg). The precipitated DNA was quantified by real-time PCR analysis (Decker et al, 2009) prior to Solexa deep sequencing.
Bio-ChIP-seq analysis of Pax5 binding
About 5 × 107 of in vitro cultured Pax5Bio/Bio Rag2−/− pro-B cells or 1 × 107 sorted mature Pax5Bio/Bio B cells were used for chromatin precipitation by streptavidin pulldown (Bio-ChIP) as described (Ebert et al, 2011). The precipitated DNA was quantified by real-time PCR prior to Solexa deep sequencing.
cDNA preparation for RNA sequencing
Total RNA was isolated using the RNeasy Plus Mini Kit (Qiagen) and was processed for deep sequencing as described (Mortazavi et al, 2008). mRNA was purified by one or two rounds of poly(A) selection and was then fragmented to a size of 200–500 nucleotides by heating at 94°C for 3 min. Fragmented mRNA was used for the first-strand cDNA synthesis with random hexamers, using the Superscript III First-Strand Synthesis System for RT–PCR (Invitrogen) followed by purification with a Mini Quick Spin Column for DNA (Roche). The second-strand synthesis was performed in the presence of E. coli DNA polymerase I, E. coli DNA ligase and RNase H, and the cDNA was purified using the MinElute Reaction Cleanup kit (Qiagen) prior to Solexa deep sequencing.
Solexa deep sequencing
About 5 ng of ChIP-precipitated DNA, DNA excised from DHS sites or cDNA prepared from mRNA was used as starting material for generating single-end or paired-end sequencing libraries according to Illumina’s ChIP Sequencing sample preparation protocol. Completed libraries were quantified using the Agilent QPCR NGS library quantification kit. Cluster generation and sequencing was carried out by using the Illumina/Solexa Genome Analyser (GA) II and IIx systems according to the manufacturer’s guidelines.
Sequence alignment
Sequence reads that passed the Illumina quality filtering were considered for alignment. In case of RNA-seq experiments, reads corresponding to mouse ribosomal RNA (BK000964.1) were removed. The remaining reads were aligned to the mouse genome assembly version of July 2007 (NCBI37/mm9) using the Bowtie program version 12.5.
Peak calling of ChIP-seq data
Peaks were called using the MACS program version 1.3.6.1 (Zhang et al, 2008) with default parameters, a read length of 36 or 76, a genome size of 2 654 911 517 bp (mm9) and the appropriate input control sample. For analysis of DHS and chromatin data, the read shifting model building was turned off. Peaks were further filtered for P-values of <10−10.
Peak-to-gene assignment
Peaks were assigned to genes in a stepwise manner by prioritizing genes containing peaks in their promoter and/or gene body. For this, peaks overlapping with the promoter (−2.5 kb to +2.5 kb relative to TSS) or gene body (+2.5 kb to TES) were first assigned to the corresponding gene. Other peaks within a specified region upstream of the TSS or downstream of the TES were assigned to the gene containing peaks in the promoter or gene body. All other peaks within the same specified region were assigned to the nearest gene, and all non-assigned peaks were classified as intergenic.
Analysis of RNA-seq data
The number of reads aligned to the gene models in each RNA sequencing experiment was first normalized by using the edgeR program (Robinson et al, 2010). Normalized expression values were calculated as RPKMs (Mortazavi et al, 2008). RPM values were used for analysing the gene expression changes between samples by calculating the differential expression significances [−log10 (P-value)] using the edgeR program.
De-novo motif discovery
For de-novo discovery of the Pax5-binding motif, peaks were called for pro-B and mature B cell tracks using the SISSRs program (Jothi et al, 2008) and only common SISSRs peaks between pro-B and mature B cells were used for de-novo motif discovery using the MEME program (Bailey et al, 2009).
Motif analysis of Pax5-binding regions
Motifs enriched in pro-B or mature B cells were identified by analysing common and unique Pax5 peaks of pro-B and mature B cells as well as Pax5 peaks associated with activated Pax5 target genes in pro-B cells or mature B cells by using the MEME Suite program (Bailey et al, 2009). Sequence motif databases were downloaded from the JASPAR and TRANSFAC (version 2010.2) databases or motifs were taken from Chen et al (2008) or our own ChIP-seq analyses (unpublished data). Motifs were searched in the different Pax5 peak sets using the MotifLocator program (Thijs et al, 2002), and the number and percentage of peak sequences containing at least one motif was calculated for each motif in the different Pax5 peak sets.
Accession numbers
All sequencing data discussed in this paper are available at the Gene Expression Omnibus (GEO) repository at NCBI under the accession numbers GSE 38046.
Supplementary Material
Acknowledgments
We thank G Stengl and G Schmauß for FACS sorting, M Sonntagsbauer and C Czepe for Solexa sequencing at the Campus Science Support Facilities (CSF Vienna). This research was supported by Boehringer Ingelheim, the Austrian GEN-AU initiative (financed by the Bundesminsterium für Bildung und Wissenschaft) and the European Union Sixth Framework Programme FP6 (funding the EuTRACC project and the Epigenome Network of Excellence). Roger Revilla-i-Domingo and Anja Ebert were recipients of Marie-Curie and EMBO fellowships.
Author contributions: IB, BV and RR performed the chromatin ChIP experiments; AE and RR carried out the Pax5 ChIP analysis; RR and BV sorted early progenitors; RR prepared all RNA sample for RNA sequencing; HT performed the DHS site mapping experiments; MJ together with IT and RR planned and established the bioinformatics pipelines; MJ and IT bioinformatically analysed the DHS site and ChIP-seq data together with IB and helped with the RNA-seq data analysis performed by RR; LS provided additional bioinformatic support; MJ and JT performed the motif analyses; AS was responsible for sample preparation and Solexa deep sequencing; RR, IB, BV, HT and AE together with MB planned and designed the experiments; MB wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS (2009) MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37: W202–W208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK, Kulbokas EJ 3rd, Gingeras TR, Schreiber SL, Lander ES (2005) Genomic maps and comparative analysis of histone modifications in human and mouse. Cell 120: 169–181 [DOI] [PubMed] [Google Scholar]
- Cao Y, Yao Z, Sarkar D, Lawrence M, Sanchez GJ, Parker MH, MacQuarrie KL, Davison J, Morgan MT, Ruzzo WL, Gentleman RC, Tapscott SJ (2010) Genome-wide MyoD binding in skeletal muscle cells: a potential for broad cellular reprogramming. Dev Cell 18: 662–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P, Sandelin A, Lenhard B, Katayama S, Shimokawa K, Ponjavic J, Semple CA, Taylor MS, Engstrom PG, Frith MC, Forrest AR, Alkema WB, Tan SL, Plessy C, Kodzius R, Ravasi T, Kasukawa T, Fukuda S, Kanamori-Katayama M, Kitazume Y et al. (2006) Genome-wide analysis of mammalian promoter architecture and evolution. Nat Genet 38: 626–635 [DOI] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, Loh YH, Yeo HC, Yeo ZX, Narang V, Govindarajan KR, Leong B, Shahab A, Ruan Y, Bourque G, Sung WK et al. (2008) Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133: 1106–1117 [DOI] [PubMed] [Google Scholar]
- Cobaleda C, Jochum W, Busslinger M (2007) Conversion of mature B cells into T cells by dedifferentation to uncommitted progenitors. Nature 449: 473–477 [DOI] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, Boyer LA, Young RA, Jaenisch R (2010) Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci USA 107: 21931–21936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czerny T, Busslinger M (1995) DNA-binding and transactivation properties of Pax-6: three amino acids in the paired domain are responsible for the different sequence recognition of Pax-6 and BSAP (Pax-5). Mol Cell Biol 15: 2858–2871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czerny T, Schaffner G, Busslinger M (1993) DNA sequence recognition by Pax proteins: bipartite structure of the paired domain and its binding site. Genes Dev 7: 2048–2061 [DOI] [PubMed] [Google Scholar]
- Decker T, Pasca di Magliano M, McManus S, Sun Q, Bonifer C, Tagoh H, Busslinger M (2009) Stepwise activation of enhancer and promoter regions of the B cell commitment gene Pax5 in early lymphopoiesis. Immunity 30: 508–520 [DOI] [PubMed] [Google Scholar]
- Delogu A, Schebesta A, Sun Q, Aschenbrenner K, Perlot T, Busslinger M (2006) Gene repression by Pax5 in B cells is essential for blood cell homeostasis and is reversed in plasma cells. Immunity 24: 269–281 [DOI] [PubMed] [Google Scholar]
- Ebert A, McManus S, Tagoh H, Medvedovic J, Salvagiotto G, Novatchkova M, Tamir I, Sommer A, Jaritz M, Busslinger M (2011) The distal VH gene cluster of the Igh locus contains distinct regulatory elements with Pax5 transcription factor-dependent activity in pro-B cells. Immunity 34: 175–187 [DOI] [PubMed] [Google Scholar]
- Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, Ku M, Durham T, Kellis M, Bernstein BE (2011) Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 473: 43–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuxa M, Busslinger M (2007) Reporter gene insertions reveal a strictly B lymphoid-specific expression pattern of Pax5 in support of its B cell identity function. J Immunol 178: 8222–8228 [DOI] [PubMed] [Google Scholar]
- Garvie CW, Hagman J, Wolberger C (2001) Structural studies of Ets-1/Pax5 complex formation on DNA. Mol Cell 8: 1267–1276 [DOI] [PubMed] [Google Scholar]
- Hardy RR, Kincade PW, Dorshkind K (2007) The protean nature of cells in the B lymphocyte lineage. Immunity 26: 703–714 [DOI] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, Ching KA, Antosiewicz-Bourget JE, Liu H, Zhang X, Green RD, Lobanenkov VV, Stewart R, Thomson JA, Crawford GE, Kellis M et al. (2009) Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459: 108–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, Wang W, Weng Z, Green RD, Crawford GE, Ren B (2007) Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet 39: 311–318 [DOI] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK (2010) Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38: 576–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesselberth JR, Chen X, Zhang Z, Sabo PJ, Sandstrom R, Reynolds AP, Thurman RE, Neph S, Kuehn MS, Noble WS, Fields S, Stamatoyannopoulos JA (2009) Global mapping of protein-DNA interactions in vivo by digital genomic footprinting. Nat Methods 6: 283–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horcher M, Souabni A, Busslinger M (2001) Pax5/BSAP maintains the identity of B cells in late B lymphopoiesis. Immunity 14: 779–790 [DOI] [PubMed] [Google Scholar]
- Jothi R, Cuddapah S, Barski A, Cui K, Zhao K (2008) Genome-wide identification of in vivo protein-DNA binding sites from ChIP-Seq data. Nucleic Acids Res 36: 5221–5231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inlay MA, Bhattacharya D, Sahoo D, Serwold T, Seita J, Karsunky H, Plevritis SK, Dill DL, Weissman IL (2009) Ly6d marks the earliest stage of B-cell specification and identifies the branchpoint between B-cell and T-cell development. Genes Dev 23: 2376–2381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Barrera LO, Zheng M, Qu C, Singer MA, Richmond TA, Wu Y, Green RD, Ren B (2005) A high-resolution map of active promoters in the human genome. Nature 436: 876–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon K, Hutter C, Sun Q, Bilic I, Cobaleda C, Malin S, Busslinger M (2008) Instructive role of the transcription factor E2A in early B lymphopoiesis and germinal center B cell development. Immunity 28: 751–762 [DOI] [PubMed] [Google Scholar]
- Lin YC, Jhunjhunwala S, Benner C, Heinz S, Welinder E, Mansson R, Sigvardsson M, Hagman J, Espinoza CA, Dutkowski J, Ideker T, Glass CK, Murre C (2010) A global network of transcription factors, involving E2A, EBF1 and Foxo1, that orchestrates B cell fate. Nat Immunol 11: 635–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McManus S, Ebert A, Salvagiotto G, Medvedovic J, Sun Q, Tamir I, Jaritz M, Tagoh H, Busslinger M (2011) The transcription factor Pax5 regulates its target genes by recruiting chromatin-modifying proteins in committed B cells. EMBO J 30: 2388–2404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medvedovic J, Ebert A, Tagoh H, Busslinger M (2011) Pax5: a master regulator of B cell development and leukemogenesis. Adv Immunol 111: 179–206 [DOI] [PubMed] [Google Scholar]
- Mikkola I, Heavey B, Horcher M, Busslinger M (2002) Reversion of B cell commitment upon loss of Pax5 expression. Science 297: 110–113 [DOI] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5: 621–628 [DOI] [PubMed] [Google Scholar]
- Mullighan CG, Goorha S, Radtke I, Miller CB, Coustan-Smith E, Dalton JD, Girtman K, Mathew S, Ma J, Pounds SB, Su X, Pui C-H, Relling MV, Evans WE, Shurtleff SA, Downing JR (2007) Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature 446: 758–764 [DOI] [PubMed] [Google Scholar]
- Natoli G (2010) Maintaining cell identity through global control of genomic organization. Immunity 33: 12–24 [DOI] [PubMed] [Google Scholar]
- Nebral K, Denk D, Attarbaschi A, König M, Mann G, Haas OA, Strehl S (2009) Incidence and diversity of PAX5 fusion genes in childhood acute lymphoblastic leukemia. Leukemia 23: 134–143 [DOI] [PubMed] [Google Scholar]
- Northrup DL, Zhao K (2011) Application of ChIP-Seq and related techniques to the study of immune function. Immunity 34: 830–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutt SL, Heavey B, Rolink AG, Busslinger M (1999) Commitment to the B-lymphoid lineage depends on the transcription factor Pax5. Nature 401: 556–562 [DOI] [PubMed] [Google Scholar]
- Nutt SL, Kee BL (2007) The transcriptional regulation of B cell lineage commitment. Immunity 26: 715–725 [DOI] [PubMed] [Google Scholar]
- Nutt SL, Morrison AM, Dörfler P, Rolink A, Busslinger M (1998) Identification of BSAP (Pax-5) target genes in early B-cell development by loss- and gain-of-function experiments. EMBO J 17: 2319–2333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutt SL, Urbánek P, Rolink A, Busslinger M (1997) Essential functions of Pax5 (BSAP) in pro-B cell development: difference between fetal and adult B lymphopoiesis and reduced V-to-DJ recombination at the IgH locus. Genes Dev 11: 476–491 [DOI] [PubMed] [Google Scholar]
- Peters JM, Tedeschi A, Schmitz J (2008) The cohesin complex and its roles in chromosome biology. Genes Dev 22: 3089–3114 [DOI] [PubMed] [Google Scholar]
- Phillips JE, Corces VG (2009) CTCF: master weaver of the genome. Cell 137: 1194–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pridans C, Holmes ML, Polli M, Wettenhall JM, Dakic A, Corcoran LM, Smyth GK, Nutt SL (2008) Identification of Pax5 target genes in early B cell differentiation. J Immunol 180: 1719–1728 [DOI] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J (2011) A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470: 279–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabo PJ, Kuehn MS, Thurman R, Johnson BE, Johnson EM, Cao H, Yu M, Rosenzweig E, Goldy J, Haydock A, Weaver M, Shafer A, Lee K, Neri F, Humbert R, Singer MA, Richmond TA, Dorschner MO, McArthur M, Hawrylycz M et al. (2006) Genome-scale mapping of DNase I sensitivity in vivo using tiling DNA microarrays. Nat Methods 3: 511–518 [DOI] [PubMed] [Google Scholar]
- Schebesta A, McManus S, Salvagiotto G, Delogu A, Busslinger GA, Busslinger M (2007) Transcription factor Pax5 activates the chromatin of key genes involved in B cell signaling, adhesion, migration and immune function. Immunity 27: 49–63 [DOI] [PubMed] [Google Scholar]
- Shiraki T, Kondo S, Katayama S, Waki K, Kasukawa T, Kawaji H, Kodzius R, Watahiki A, Nakamura M, Arakawa T, Fukuda S, Sasaki D, Podhajska A, Harbers M, Kawai J, Carninci P, Hayashizaki Y (2003) Cap analysis gene expression for high-throughput analysis of transcriptional starting point and identification of promoter usage. Proc Natl Acad Sci USA 100: 15776–15781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soler E, Andrieu-Soler C, de Boer E, Bryne JC, Thongjuea S, Stadhouders R, Palstra RJ, Stevens M, Kockx C, van Ijcken W, Hou J, Steinhoff C, Rijkers E, Lenhard B, Grosveld F (2010) The genome-wide dynamics of the binding of Ldb1 complexes during erythroid differentiation. Genes Dev 24: 277–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thijs G, Marchal K, Lescot M, Rombauts S, De Moor B, Rouze P, Moreau Y (2002) A Gibbs sampling method to detect overrepresented motifs in the upstream regions of coexpressed genes. J Computational Biol 9: 447–464 [DOI] [PubMed] [Google Scholar]
- Treiber T, Mandel EM, Pott S, Györy I, Firner S, Liu ET, Grosschedl R (2010) Early B cell factor 1 regulates B cell gene networks by activation, repression, and transcription-independent poising of chromatin. Immunity 32: 714–725 [DOI] [PubMed] [Google Scholar]
- Vilagos B, Hoffmann M, Souabni A, Sun Q, Werner B, Medvedovic J, Bilic I, Minnich M, Axelsson E, Jaritz M, Busslinger M (2012) Essential role of EBF1 in the generation and function of distinct mature B cell types. J Exp Med 209: 775–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh T-Y, Peng W, Zhang MQ, Zhao K (2008) Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet 40: 897–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei G, Abraham BJ, Yagi R, Jothi R, Cui K, Sharma S, Narlikar L, Northrup DL, Tang Q, Paul WE, Zhu J, Zhao K (2011) Genome-wide analyses of transcription factor GATA3-mediated gene regulation in distinct T cell types. Immunity 35: 299–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welinder E, Mansson R, Mercer EM, Bryder D, Sigvardsson M, Murre C (2011) The transcription factors E2A and HEB act in concert to induce the expression of FOXO1 in the common lymphoid progenitor. Proc Natl Acad Sci USA 108: 17402–17407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Rould MA, Jun S, Desplan C, Pabo CO (1995) Crystal structure of the paired domain-DNA complex at 2.5 Å resolution reveals structural basis for Pax developmental mutations. Cell 80: 639–650 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nussbaum C, Myers RM, Brown M, Li W, Liu XS (2008) Model-based analysis of ChIP-Seq (MACS). Genome Biol 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou VW, Goren A, Bernstein BE (2011) Charting histone modifications and the functional organization of mammalian genomes. Nat Rev Genet 12: 7–18 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.