Highlights
► miR-191 expression is upregulated in senescencent human epidermal keratinocytes. ► miR-191 overexpression is sufficient per se to induce senescence in keratinocytes. ► SATB1 and CDK6 are downregulated in senescence and are direct miR-191 targets. ► SATB1 and CDK6 silencing by siRNA triggers senescence in HEKn cells.
Abbreviations: microRNA, miRNA; HEKn, human epidermal keratinocytes neonatal; SA-β-gal, senescence-associated beta-galactosidase; HAT, Histone Acetyl Transferase
Keywords: Keratinocytes, Senescence, Proliferation, MicroRNA-191, CDK6, SATB1
Abstract
Keratinocyte replicative senescence has an important role in time-dependent changes of the epidermis, a tissue with high turnover. Senescence encompasses growth arrest during which cells remain metabolically active but acquire a typical enlarged, vacuolar and flattened morphology. It is also accompanied by the expression of endogenous senescence-associated-β-galactosidase and specific gene expression profiles. MicroRNAs levels have been shown to be modulated during keratinocytes senescence, playing key roles in inhibiting proliferation and in the acquisition of senescent markers. Here, we identify miR-191 as an anti-proliferative and replicative senescence-associated miRNA in primary human keratinocytes. Its overexpression is sufficient per se to induce senescence, as evaluated by induction of several senescence-associated markers. We show that SATB1 and CDK6 3′UTRs are two miR-191 direct targets involved in this pathway. Cdk6 and Satb1 protein levels decrease during keratinocytes replicative senescence and their silencing by siRNA is able to induce a G1 block in cell cycle, accompanied by an increase in senescence-associated markers.
1. Introduction
The epidermis is a tissue with a high turnover in which the correct regulation of keratinocyte proliferation and differentiation passages, occurring in its upper layers, is crucial [1,2]. For continuously dividing cells, like those of the epithelia, replicative senescence is considered as a consequence of the accumulation of cellular damage, such as telomere shortening and DNA mutations, that inevitably happens during the process of cell division [3–7]. These events are particularly relevant in adult stem cells that, dividing throughout the life, undergo both chronological and replicative aging [8]. As the incidence of mutations and damage increases with age, the probability that a cell will start senescence, apoptosis, or malignant transformation pathways also increases [9].
Therefore, cellular senescence is also a process aimed to prevent cellular transformation and to arrest the progression of malignant phenomena resulting from age-related genomic instability, thereby protecting the organism from cancer [10]. Senescent cells are blocked in their growth, but remain metabolically active and acquire a typical morphology: they become enlarged, vacuolar, flattened and express endogenously the senescence-associated-β-galactosidase (SA-β-galactosidase) enzyme [11,12]. Normal cells enter in a state of replicative senescence stopping the progression from the G1 to the S phase of the cell cycle and remaining quiescent [13]. All the changes described are the consequence of new gene expression profiles [14–16]. Recent studies have shown a diffuse chromatin remodeling in senescent versus proliferating cells and in general during adult stem cells aging [17,18]. These epigenetic alterations implicate both histones modifications (acetylation/deacetylation, methylation/demethylation) [19,20] and DNA methylation senescence-associated changes [21,22].
MicroRNAs (miRNAs) are conserved small non-coding RNAs (19–22nt) that recognize the 3′-untranslated (3′UTR) region of target messenger RNAs (mRNAs) inhibiting their translation and/or inducing their degradation [23]. miRNAs have been described to act both as classical oncogenes and as tumor suppressors in different malignancies, because of their tight modulation of key players of cell cycle progression and apoptotic pathways [24]. miRNAs have been shown also to be involved in skin development and pathologies regulating keratinocytes stemness, proliferation, differentiation and death [25,26], as demonstrated by the generation of mice strains with conditional ablations of Dicer or DGCR8. The absence of these key components of miRNAs biosynthesis complexes is sufficient to induce severe developmental and structural defects in mouse skin [25,27]. miRNA expression regulation during senescence and aging represents an emerging field in miRNA studies and the list of miRNAs and corresponding target mRNAs involved in these pathways is rapidly increasing [28–30]. Recently, in a model of in vitro replicative senescence of normal human epidermal keratinocytes neonatal (HEKn), we identified also a ΔNp63a-miRNAs regulatory loop that represents a “stemness master gene”-mediated strategy to promote proliferation and to counteract senescence [30]. A new role in senescence-associated transcriptional gene repression was also proposed for endogenous AGO-2/miRNAs complexes that, interacting with RB1/E2F target promoters and recruiting co-repressor factors, trigger heterochromatin formation [31].
Here, we investigated the role of miR-191 in HEKn. By microarray profiling of miRNAs levels modulated during HEKn senescence, we selected miR-191 as one of the most upregulated miRNAs [30]. We provide evidence that miR-191 overexpression is sufficient to induce senescence in HEKn cells and that the direct targets, involved in this process, are the Special AT-rich Binding protein 1 (SATB1) and the Cyclin Dependent Kinase 6 (CDK6) mRNAs.
2. Materials and methods
2.1. Cell culture and transfection
Primary Human Epidermal Keratinocytes neonatal (HEKn, Cascade, Invitrogen, Carlsbad, California, USA) were cultured in Epilife medium with HKGS growth supplements (Cascade). Cells were constantly kept sub-confluent in order to avoid triggering of differentiation. At each passage cells were collected to extract RNA and protein and an aliquot was submitted to senescence associated-β-galactosidase staining. Cells were induced to differentiate by adding 1.2 mM CaCl2 to the culture medium. For microRNA overexpression, human primary keratinocytes were transfected with human pre-miRNAs or a scrambled sequence as a negative control (Ambion, Texas, USA). For CDK6 and SATB1 silencing, HEKn were transfected with Hs-CDK6-siRNA GeneSolution and Hs-SATB1-siRNA GeneSolution (Qiagen, Hilden, Germany). AllStars negative control siRNA (Qiagen) was used as negative control. All transfections were performed using the Lipofectamine RNAimax transfection reagent (Invitrogen) according to manufacturer protocols. HEK 293E cells were grown in D-MEM with 10% FBS, 100 U penicillin, 100 μg/ml streptomycin (GIBCO, Invitrogen) and transfected using Lipofectamine 2000 according to manufacturer protocols (Invitrogen).
2.2. RNA extraction and Real Time PCR analysis
Total RNA from cells was isolated using the mirVana mirRNA Isolation Kit (Ambion, Applied Biosystems) following the manufacturer’s protocol. TaqMan MicroRNA assays were used for miRNA quantification by Real Time RT-qPCR; RNA was reverse transcribed using the TaqMan MicroRNA Reverse Transcription kit; Real Time q-PCR was performed with the TaqMan universal master mix and U18 was used as housekeeping gene for normalization (Applied Biosystems).
For determination of mRNA expression levels, total RNA was reverse transcribed with GoScript™ Reverse Transcription System (Promega, Madison, WI, USA) according to manufacturer’s protocols. Real Time PCR was then performed with CDK6 or SATB1 specific primers by using the Platinum SYBR Green qPCR SuperMix UDG (Invitrogen). The sequences of the primers used in this study were as follow: hCDK6F 5′-CGTGGTCAGGTTGTTTGATGTG-3′, hCDK6R 5′-ACTCGGTGTGAATGAAGAAAGTCC-3′, hSATB1F 5′-GCCTTGGGAATCCTCCAGAGTT TC-3′, hSATB1R 5′-AGTTTGCCGTGG TGCTTGAGATAG-3′. β-Actin was used as a housekeeping gene for normalization. The expression of each gene and miRNA was defined from the threshold cycle (Ct), and relative expression levels were calculated by using the 2−ΔΔCt method after normalization with reference to expression of housekeeping genes.
2.3. Senescence-associated β-galactosidase staining
1.5 × 105 cells were seeded into six-well culture plates the day before transfection. Fourty eight hours after transfection, HEKn cells were washed with PBS and fixed with 0.2% glutaraldehyde/2% formaldehyde/2 mM MgCl2 in PBS and then incubated with β-galactosidase staining solution (2 mM MgCl2, 5 mM potassium ferricyanide, 5 mM potassium ferrocyanide, 1 mg/ml X-gal, pH 6.0) for 24 h at 37 °C. The reaction was stopped by replacing the staining solution with 70% glycerol.
2.4. Cell proliferation and cell cycle analysis
Incorporation of bromodeoxyuridine (BrdU) during DNA synthesis was evaluated with the Click-iT™ EdU flow cytometry assay kit, following the manufacturer’s protocol (Molecular Probes, Eugene, OR, USA). Cell cycle was analysed using a FACS Calibur flow cytometer (BD Biosciences, San Jose, CA, USA). Fifteen thousand events were evaluated using the Cell Quest (BD) software.
2.5. Luciferase assay and constructs
The 3′-UTRs of miR-191 target mRNAs were amplified by PCR from human genomic DNA using the following primer pairs: hSATB1–3′UTR-F 5′-GGCCTCTAGAGATAAAAGTA TTTGTTTCGTTCAAC-3′; hSATB1–3′UTR-R 5′-GGCCTCTAGAGTGTAGTTACAGTCAA TAACCACTC-3′; hCDK6–3′UTR-F 5′-GGCCTCTAGATGCTCATGGCACCCATTAGA-3′; hCDK6–3′UTR-R 5′-GGCCTCTAGACTATACCATACCTGAGGCCA-3′. PCR fragments were restricted and ligated to a compatible XbaI-linearized pGL3Control vector (Promega). Cells were transfected with 100 ng of pGL3 vectors, 12 pmol of pre-miR or a scrambled sequence (Ambion), and 10 ng of Renilla luciferase pRL-CMV vector (Promega). Luciferase assays were then performed as described before [32].
2.6. Western Blotting
Total cell extracts were resolved on SDS polyacrylamide gels and blotted onto a Hybond P PVDF membrane (G&E Healthcare, UK). The following antibodies were used: anti-p63 (Ab4, Neomarkers, Fremont, California, USA; dilution 1:500), anti-β actin (Sigma, St Louis, Minnesota, USA; dilution 1:5000), anti-p16 (Santa Cruz Biotechnology, California, USA; dilution 1:1000), anti-PML (Santa Cruz Biotechnology; dilution 1:400) anti-K10 (Covance, Princeton, NJ, USA; dilution 1:1000), anti-SATB1 (Sigma, dilution 1:400), anti-CDK6 (Santa Cruz Biotechnology, dilution 1:1000).
2.7. Bioinformatics
Analysis of microRNA target sites were performed using the TargetScan 5.1 software available at <http://www.targetscan.org.>.
3. Results
3.1. MiR-191 expression is upregulated in senescencent human epidermal keratinocytes
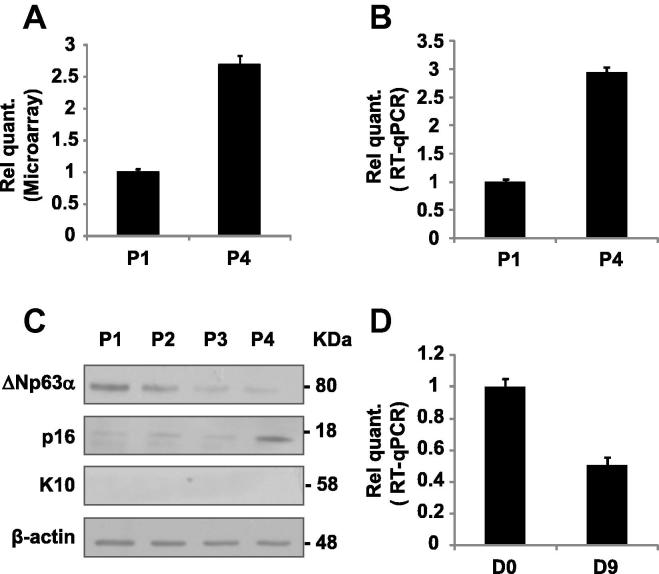
To investigate miRNAs levels modulated during replicative senescence, we used the microarray approach described in a previous study [30], comparing miRNA expression profiles of proliferating HEKn at passage 1 (P1) to senescent HEKn at passage 4 (P4). Among the most consistently upregulated miRNAs at P4 we selected miR-191 (fold change 2.7 ± 0.05, Fig. 1A) and validated the array result by Real Time RT-qPCR (Fig. 1B). As a control, a Western Blot analysis was also perfomed on protein extracts from HEKn cells collected at the same passages (P1–P4). As expected, ΔNp63α, important in maintaining the proliferative potential of the epithelial cells, decreases upon serial passages, while p16 expression increases (Fig 1C). We also excluded the possibility that the HEKn cells after serial passages in culture underwent differentiation by verifying that the cytokeratin 10 (K10), an early marker of keratinocyte differentiation, was and remained undetectable from P1 to P4 (Fig. 1C). To confirm that miR-191 was an anti-proliferative miRNA directly involved in the senescence process and not a pro-differentiation miRNA, we checked also its expression in HEKn cells differentiated in vitro by CaCl2 addition showing that its expression levels did not increase in these specific conditions (Fig. 1D).
Fig. 1.
MiR-191 expression is upregulated during HEKn cells replicative senescence. (A) MiR-191 was identified by microarray screening in proliferating (P1) versus senescent (P4) HEKn cells [30]. (B) Real Time RT-qPCR was employed to analyze the expression levels of miR-191 in HEKn cells at a growing number of passages in culture (P1–P4). Values reported are the average ± SD of three independent experiments. (C) Western Blot of HEKn cells protein extracts collected as described in (B). Proliferation (ΔNp63α), senescence (p16) and differentiation (K10) markers are shown. β-actin was used as a loading control. (D) Real Time RT-qPCR was employed to analyze the expression levels of miR-191 in HEKn cells at day 0 (D0) and 9 (D9) after induction of differentiation by calcium addiction to culture medium. Values reported are the average ± SD of three independent experiments.
3.2. MiR-191 overexpression is sufficient per se to induce senescence in proliferating HEKn cells
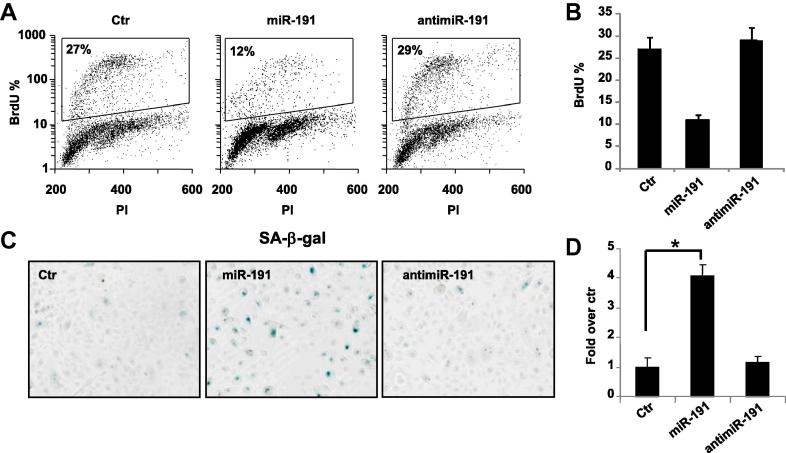
To understand miR-191 role’s in keratinocyte senescence we set up its overexpression and depletion in proliferating HEKn cells performing transfections with pre-miR-191, anti-miR-191 or scrambled control sequences. Forty-eight hours after transfection, we observed a decrease in the percent of BrdU incorporating cells in presence of miR-191 overexpression (12% versus 27% of scrambled control) indicating a cell cycle G1-arrest, while anti-miR-191 transfection had no effects on proliferation (29%; Fig. 2A, B). Under identical conditions, we also found an increase of more than 3-folds in the number of cells positive for SA-β-galactosidase staining as shown by the images and blue-cell quantification (Fig. 2C, D). These results demonstrated that miR-191 overexpression is sufficient per se to induce keratinocytes senescence.
Fig. 2.
MiR-191 overexpression is sufficient to decrease HEKn proliferation and induce senescence. (A) 48 h after transfection of HEKn cells with either a scrambled control (Ctr), miR-191 or antimiR-191, cells were subjected to a 4 h BrdU-pulse, collected, PI stained and analyzed by flow cytometry. BrdU positive cells are indicated as S phase fluorescent populations and are assessed by PI staining of DNA content of 2n or 4n (fixed to values of 200 and 400 in the plots). (B) BrdU positive cells percentage (BrdU%) of each sample in A) is reported in the histogram. Values represent the average ± SD of three independent transfections. (C) SA-β-galactosidase staining of HEKn cells 48 h after transfection with either a scrambled control (Ctr), miR-191 or antimiR-191. (D) Relative SA-β-galactosidase positive cells quantified by blue cell counting/field (as fold over control). Values reported are average ± SD of three independent stains. ∗p-Value < 0.01 by Student’s t test.
3.3. SATB1 and CDK6 expression is downregulated in senescent HEKn cells and their 3′UTRs are direct miR-191 targets
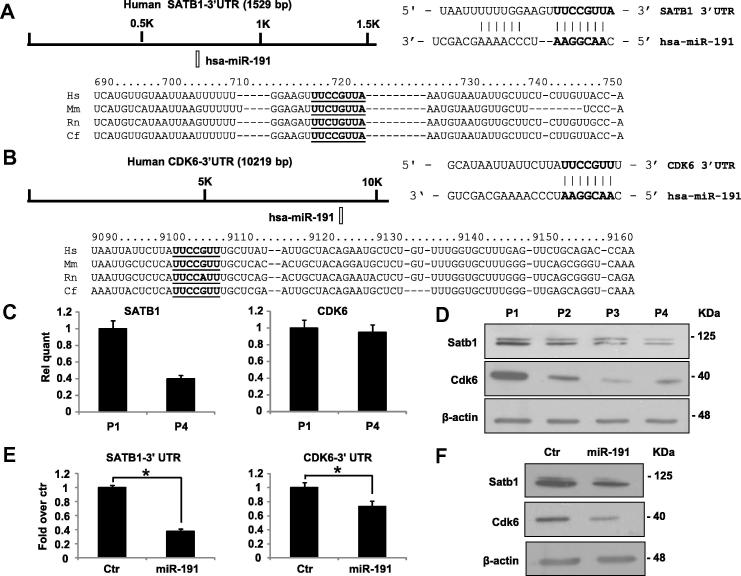
To elucidate miR-191 role’s in senescence pathways, we performed an analysis of its putative molecular targets. From an in silico prediction, by TargetScan 5.1 software, we selected SATB1–3′UTR and CDK6–3′UTR as putative miR-191 targets. These 3′UTRs harbor at least one miR-191 target site that is highly conserved among vertebrates as shown by the sequence alignments in Fig. 2A, B. At the mRNA level, we found a significant decrease (about 60%) of SATB1 mRNA by real-time RT-qPCR, while CDK6 expression did not change, between P1 and P4 keratinocytes. However, at the protein level, it was possible to confirm a downregulation of both miR-191 putative targets (Fig. 3C). The observed inverse correlation between miR-191 expression and its putative targets strengthened the hypothesis of their direct regulation.
Fig. 3.
SATB1 and CDK6 expression is downregulated in senescent HEKn cells and their 3′UTRs are direct miR-191 targets. (A,B) Predicted miR-191 target sites on human SATB1 and CDK6 3′UTRs were identified by TargetScan 5.1 software. The putative miRNA target sequences are conserved between the 3’-UTRs of human (Hs), mouse (Mm), rat (Rn) and dog (Cf) mRNAs. (C) Real time RT-qPCR was employed to analyze the expression levels of SATB1 and CDK6 in HEKn cells at a growing number of passages in culture (P1–P4). Values reported are the average ± SD of three independent experiments. (D) Western Blot of HEKn cells protein extracts collected at increasing passage number in culture (P1–P4). Satb1 and Cdk6 protein levels are shown and β-actin was used as loading control. (E) Insertion of SATB-1 or CDK6 3′UTR target sequences in a luciferase reporter vector leads to diminished luciferase activity in presence of miR-191 in HEK293 cells 24 h after co-transfection. Histograms show the values resulting as the average ± SD from three independent co-transfections. (F) Western Blot analysis of protein extracts of HEKn transfected with miR-191 versus a scrambled control sequence (Ctr). miR-191 overexpression decreases Satb1 and Cdk6 protein levels; β-actin was used as a loading control.
CDK6 is a member of the cyclin-dependent protein kinase family and a main regulator of cell cycle progression. In particular, this kinase is the catalytic subunit of the complex that regulates G1 phase progression and G1/S transition during cell cycle. CDK6, as well as CDK4, has been shown to phosphorylate, and thus regulate the activity of Rb. This cell cycle check point is frequently deregulated in different malignancies [33–36].
The Specialized Adenine and Thymine-rich Binding protein 1 (SATB1) functions as a genome organizer that regulates higher-order chromatin remodeling in order to establish particular spatial organization of genes within the nuclei [37]. SATB1 recruits transcription factors and chromatin remodeling complexes to specific genomic regions establishing an epigenetic modification status that sometimes has a key role in tissue specific gene expression [38–40]. In this way, SATB1 is able to coordinate the regulation of a high percentage of genes (more than 10% of the total) and it is presumable that it may be crucial for the changes in gene expression patterns that happen in human cancers [41].
To confirm that the selected mRNAs are direct miR-191 targets, we cloned part of SATB1 and CDK6–3′UTRs sequences upstream of luciferase cDNA, to use them in luc-reporter assays. Transfecting SATB1–3′UTR and CDK6–3′UTR reporter constructs in presence of pre-miR-191 or a scrambled control sequence, we obtained a significant downregulation of more than 60% and about 25%, respectively, in relative luciferase activity (Fig. 3D). Overexpression of miR-191 by transfection in proliferating HEKn led to a significant decrease in SATB1 and CDK6 protein levels (Fig. 3F), thus demonstrating that miR-191 is able to repress the selected targets endogenously expressed in keratinocytes.
3.4. SATB1 and CDK6 silencing by siRNA triggers senescence in HEKn cells
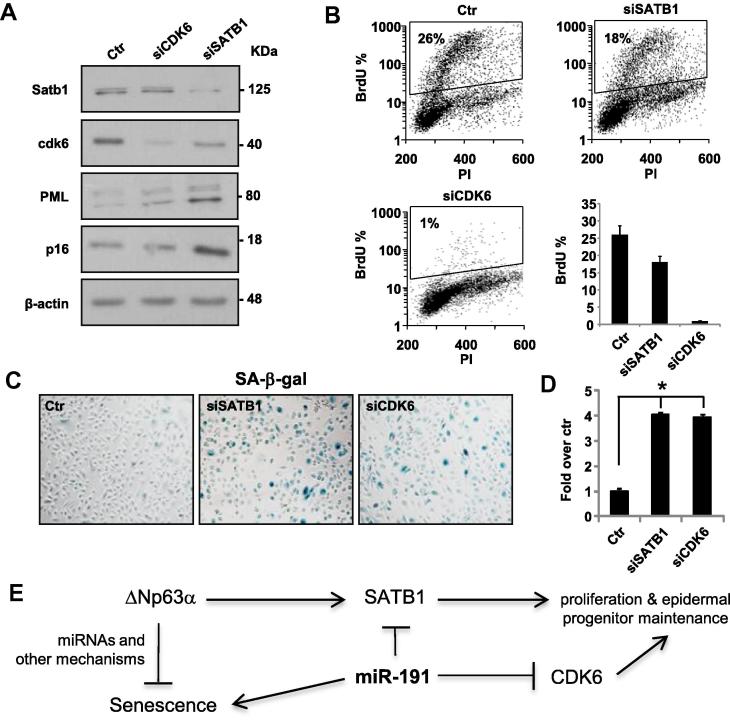
To establish that miR-191’s ability to trigger senescence is mainly mediated by the down regulation of these two targets, we performed their silencing by transfection using specific siRNAs or scrambled control sequences. Forty-eight hours after transfection we managed to reduce both SATB1 and CDK6 expression at the protein level as shown in the Western Blots (Fig. 4A). In the same conditions, we observed a significant reduction in the percentage of BrdU incorporation of SATB1-silenced HEKn cells (18%) compared to scramble transfected cells (26%) and a total block in the G1 cell cycle phase of CDK6-silenced cells (1%) as clearly visible in Fig. 4B. The decreased proliferation was followed by a marked staining for SA-β-galactosidase (Fig. 4C) as indicated by a 3-fold increase in blue cells counts (Fig. 4D). These data, together with the induction of the senescence markers p16 and PML (Fig. 4A), indicated that SATB1 and CDK6-silenced HEKn cells undergo senescence. The effects of silencing miR-191’s targets in HEKn were coherent with the consequences of miR-191 overexpression in the same model (Fig. 2), but the results of the specific CDK6 silencing in proliferating HEKn seemed to be more effective both in cell cycle block and in increase SA-β-galactosidase staining (Fig. 4B–D). This difference was probably due to an incomplete silencing of CDK6 upon miR-191 transfection in comparison to CDK6 silencing by siRNA (compare Figs. 3F and 4A).
Fig. 4.
SATB1 and CDK6 silencing by siRNA induces senescence in HEKn cells. (A) Western Blot analysis of protein extracts of HEKn transfected with a scrambled control sequence (Ctr), SATB1-siRNA or CDK6-siRNA. Satb1, Cdk6 and senescence (PML and p16) markers protein levels are shown. β-actin was used as a loading control. (B) 48 h after transfection of HEKn cells with either a scrambled control (Ctr) or siRNA for SATB1 or CDK6, cells were subjected to a 4 h BrdU-pulse, collected, PI stained and analyzed by flow cytometry as described in Fig. 2. Values represent the average ± SD of three independent transfections. (C) SA-β-galactosidase staining of HEKn cells 48 h after transfection with a scrambled control sequence (Ctr), SATB1-siRNA or CDK6-siRNA. (D) Relative SA-β-galactosidase positive cells quantification by blue cell counting/field (Fold over control). Values reported are average ± SD of three independent stains. ∗p-value < 0.01 by Student’s t test. (E) Schematic representation of miR-191 role’s in primary human keratinocyte replicative senescence.
4. Discussion
Our data indicate that miR-191 levels are upregulated during keratinocyte replicative senescence and that miR-191 overexpression in proliferating HEKn is sufficient per se to trigger growth inhibition and to induce SA-β-galactosidase activity. We demonstrated that SATB1 and CDK6 mRNAs are directly regulated by this miRNA and that their silencing, by siRNA, leads HEKn to senescence. These findings suggest that miR-191 acts at least in two different ways: inhibiting proliferation and altering gene expression by organizing chromatin remodeling. In particular, by targeting CDK6, miR-191 blocks G1-S phase transition leading to a cell cycle arrest and to a quiescent state that results in starting the senescence processes. Additionally, the downregulation of the “genome organizer” SATB1, suggests that miR-191 upregulation establishes senescence-associated epigenetic modifications followed by new gene expression profile also leading to senescence. On the other hand, the observation that SATB1 is linked with increased lifespan and contrasted age-related pathologies in mice, through the recruitment of HAT activity complexes [42], demonstrated the general involvement of SATB1 in counteracting the senescence and/or aging pathways. Recently, SATB1 has been characterized as a new p63 target gene demonstrating an important role of SATB1 as a part of the p63-dependent epidermal developmental program through a skin-specific chromatin organization and a proper epidermal stem cell gene expression pattern [40]. Beside its role in epidermal morphogenesis, p63-deficiency causes cellular senescence in proliferating keratinocytes [43,44,30] and leads to accelerated aging in p63 heterozygous mice in vivo [45,46]. Further studies have shown that the ΔNp63α isoform counteracts keratinocyte replicative senescence by inhibiting at least four senescence-associated miRNAs [30]. In this context, our data indicate that the senescence-associated miR-191, through SATB1 silencing opposed to p63 function adding a new regulatory step to p63-mediated function in keratinocytes (Fig. 4E). In summary, we have studied replicative senescence of human keratinocytes and our results indicate that CDK6 and SATB1 are two novel miR-191 targets strongly linked to keratinocytes proliferation thus opening up new horizons for future studies regarding their involvement in skin aging and pathologies.
Acknowledgments
The work reported in this manuscript has been supported by Grants from Telethon (GGP09133), AIRC (2743), MIUR, MinSan, ACC12 to GM and partially supported by RF73, RF57 and ACC12 to EC.
Contributor Information
G. Melino, Email: gerry.melino@uniroma2.it.
E. Candi, Email: candi@uniroma2.it.
References
- 1.Fuchs E., Horsley V. More than one way to skin. Genes Dev. 2008;22:976–985. doi: 10.1101/gad.1645908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barton C.E., Johnson K.N., Mays D.M. Novel p63 genes involved in paracrine signaling and keratinocyte differentiation. Cell Death Dis. 2010;1:e74. doi: 10.1038/cddis.2010.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kuilman T., Michaloglou C., Mooi W.J. The essence of senescence. Genes Dev. 2010;24:2463–2479. doi: 10.1101/gad.1971610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buckingham E.M., Klingelhutz A.J. The role of telomeres in the ageing of human skin. Exp. Dermatol. 2011;20:297–302. doi: 10.1111/j.1600-0625.2010.01242.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Artandi S.E., DePinho R.A. Telomeres and telomerase in cancer. Carcinogenesis. 2010;31:9–18. doi: 10.1093/carcin/bgp268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campisi J., Vijg J. Does damage to DNA and other macromolecules play a role in aging? If so, how? J. Gerontol. A Biol. Sci. Med. Sci. 2009;64A:175–178. doi: 10.1093/gerona/gln065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garinis G.A., van der Horst G.T.J., Vijg J. DNA damage and ageing: new-age ideas for an age-old problem. Nat. Cell Biol. 2008;10:1241–1247. doi: 10.1038/ncb1108-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu L., Rando T.A. Manifestations and mechanisms of stem cell aging. J. Cell Biol. 2011;193:257–266. doi: 10.1083/jcb.201010131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seviour E.G., Lin S.Y. The DNA damage response: balancing the scale between cancer and ageing. Aging. 2010;2:900–907. doi: 10.18632/aging.100248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Campisi J. Senescent cells, tumor suppression, and organismal aging: good citizens, bad neighbors. Cell. 2005;120:513–522. doi: 10.1016/j.cell.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 11.Adams P.D. Healing and hurting: molecular mechanisms, functions, and pathologies of cellular senescence. Mol. Cell. 2009;36:2–14. doi: 10.1016/j.molcel.2009.09.021. [DOI] [PubMed] [Google Scholar]
- 12.Dimri G.P., Lee X., Basile G. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl. Acad. Sci. USA. 1995;92:9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blagonsklonny M. Cell senescence: hypertrophic arrest beyond the restriction point. J. Cell. Physiol. 2006;209:592–597. doi: 10.1002/jcp.20750. [DOI] [PubMed] [Google Scholar]
- 14.de Magalhães J.P., Curado J., Church G.M. Meta-analysis of age-related gene expression profiles identifies common signatures of aging. Bioinformatics. 2009;25:875–881. doi: 10.1093/bioinformatics/btp073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zahn J.M., Poosala S., Owen A.B. AGEMAP: a gene expression database for aging in mice. PLoS Genet. 2007;3:e201. doi: 10.1371/journal.pgen.0030201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lener T., Moll P.R., Rinnerthaler M. Expression profiling of aging in the human skin. Exp. Gerontol. 2006;41:387–397. doi: 10.1016/j.exger.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 17.Rando T.A., Chang H.Y. Aging, rejuvenation, and epigenetic reprogramming: resetting the aging clock. Cell. 2012;148:46–57. doi: 10.1016/j.cell.2012.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pollina E.A., Brunet A. Epigenetic regulation of aging stem cells. Oncogene. 2011;30:3105–3126. doi: 10.1038/onc.2011.45. [DOI] [PubMed] [Google Scholar]
- 19.Dang W., Steffen K.K., Perry R. Histone H4 lysine 16 acetylation regulates cellular lifespan. Nature. 2009;459:802–807. doi: 10.1038/nature08085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sarg B., Koutzamani E., Helliger W. Postsynthetic trimethylation of histone H4 at lysine 20 in mammalian tissues is associated with aging. J. Biol. Chem. 2002;277:39195–39201. doi: 10.1074/jbc.M205166200. [DOI] [PubMed] [Google Scholar]
- 21.Maegawa S., Hinkal G., Kim H.S. Widespread and tissue specific age-related DNA methylation changes in mice. Genome Res. 2010;3:332–340. doi: 10.1101/gr.096826.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murgatroyd C., Wu Y., Bockmühl Y. The Janus face of DNA methylation in aging. Aging. 2010;2:107–110. doi: 10.18632/aging.100124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krol J., Loedige I., Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 24.Di Leva G., Croce C.M. Roles of small RNAs in tumor formation. Trends Mol. Med. 2010;16:257–267. doi: 10.1016/j.molmed.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yi R., O’Carroll D., Pasolli H.A. Morphogenesis in skin is governed by discrete sets of differentially expressed microRNAs. Nat. Genet. 2006;38:356–362. doi: 10.1038/ng1744. [DOI] [PubMed] [Google Scholar]
- 26.Yi R., Fuchs E. MicroRNA-mediated control in the skin. Cell Death Differ. 2010;17:229–235. doi: 10.1038/cdd.2009.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yi R., Pasolli H.A., Landthaler M. DGCR8-dependent microRNA biogenesis is essential for skin development. Proc. Natl. Acad. Sci. USA. 2009;106:498–502. doi: 10.1073/pnas.0810766105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gorospe M., Abdelmohsen K. MicroRegulators come of age in senescence. Trends Genet. 2011;27:233–241. doi: 10.1016/j.tig.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith-Vikos T., Slack F.J. MicroRNAs and their roles in aging. J. Cell Sci. 2012;125:7–17. doi: 10.1242/jcs.099200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rivetti di Val Cervo P., Lena A.M., Nicoloso M. P63-microRNA feedback in keratinocyte senescence. Proc. Natl. Acad. Sci. USA. 2012;109:1133–1138. doi: 10.1073/pnas.1112257109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Benhamed M., Herbig U., Ye T. Senescence is an endogenous trigger for microRNA-directed transcriptional gene silencing in human cells. Nat. Cell Biol. 2012;14:266–275. doi: 10.1038/ncb2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lena A.M., Shalom-Feuerstein R., Rivetti di Val Cervo P. MiR-203 represses “stemness” by repressing DeltaNp63. Cell Death Differ. 2008;15:1187–1195. doi: 10.1038/cdd.2008.69. [DOI] [PubMed] [Google Scholar]
- 33.Ruas M., Gregory F., Jones R. CDK4 and CDK6 delay senescence by kinase-dependent and p16INK4a-independent mechanisms. Mol. Cell Biol. 2007;27:4273–4282. doi: 10.1128/MCB.02286-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ohtani N., Yamakoshi K., Takahashi A. The p16INK4a-RB pathway: molecular link between cellular senescence and tumor suppression. J. Med. Invest. 2004;51:146–153. doi: 10.2152/jmi.51.146. [DOI] [PubMed] [Google Scholar]
- 35.Paternot S., Bockstaele L., Bisteau X. Rb inactivation in cell cycle and cancer: the puzzle of highly regulated activating phosphorylation of CDK4 versus constitutively active CDK-activating kinase. Cell cycle. 2010;9:689–699. doi: 10.4161/cc.9.4.10611. [DOI] [PubMed] [Google Scholar]
- 36.Massagué J. G1 cell-cycle control and cancer. Nature. 2004;432:298–306. doi: 10.1038/nature03094. [DOI] [PubMed] [Google Scholar]
- 37.Galande S., Purbey P.K., Notani D. The third dimension of gene regulation: organization of dynamic chromatin loopscape by SATB1. Curr. Opin. Genet. Dev. 2007;17:408–414. doi: 10.1016/j.gde.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 38.Cai S., Han H.J., Kohwi-Shigematsu T. Tissue-specific nuclear architecture and gene expression regulated by SATB1. Nat. Genet. 2003;34:42–51. doi: 10.1038/ng1146. [DOI] [PubMed] [Google Scholar]
- 39.Yasui D., Miyano M., Cai S. SATB1 targets chromatin remodelling to regulate genes over long distances. Nature. 2002;419:641–645. doi: 10.1038/nature01084. [DOI] [PubMed] [Google Scholar]
- 40.Fessing M.Y., Mardaryev A.N., Gdula M.R. P63 regulates SATB1 to control tissue-specific chromatin remodeling during development of the epidermis. J. Cell Biol. 2011;194:825–839. doi: 10.1083/jcb.201101148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Han H.J., Russo J., Kohwi Y. SATB1 reprogrammes gene expression to promote breast tumour growth and metastasis. Nature. 2008;452:187–193. doi: 10.1038/nature06781. [DOI] [PubMed] [Google Scholar]
- 42.Zhang M., Poplawski M., Yen K. Role of CBP and SATB-1 in aging, dietary restriction, and insulin-like signaling. PLoS Biol. 2009;7:e1000245. doi: 10.1371/journal.pbio.1000245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Senoo M., Pinto F., Crum C.P. P63 is essential for the proliferative potential of stem cells in stratified epithelia. Cell. 2007;129:523–536. doi: 10.1016/j.cell.2007.02.045. [DOI] [PubMed] [Google Scholar]
- 44.Shalom-Feuerstein R., Lena A.M., Zhou H. ΔNp63 is an ectodermal gatekeeper morphogenesis. Cell Death Differ. 2011;18:887–896. doi: 10.1038/cdd.2010.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Keyes W.M., Wu Y., Vogel H. P63 deficiency activates a program of cellular senescence and leads to accelerated aging. Genes Dev. 2005;19:1986–1999. doi: 10.1101/gad.342305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Paris M., Rouleau M., Pucéat M., Aberdam D. Regulation of skin aging and heart development by TAp63. Cell Death Differ. 2012;19:186–193. doi: 10.1038/cdd.2011.181. [DOI] [PMC free article] [PubMed] [Google Scholar]