Abstract
Photoactivated localization microscopy (PALM) is a powerful approach for investigating protein organization, yet tools for quantitative, spatial analysis of PALM datasets are largely missing. Combining pair-correlation analysis with PALM (PC-PALM), we provide a method to analyze complex patterns of protein organization across the plasma membrane without determination of absolute protein numbers. The approach uses an algorithm to distinguish a single protein with multiple appearances from clusters of proteins. This enables quantification of different parameters of spatial organization, including the presence of protein clusters, their size, density and abundance in the plasma membrane. Using this method, we demonstrate distinct nanoscale organization of plasma-membrane proteins with different membrane anchoring and lipid partitioning characteristics in COS-7 cells, and show dramatic changes in glycosylphosphatidylinositol (GPI)-anchored protein arrangement under varying perturbations. PC-PALM is thus an effective tool with broad applicability for analysis of protein heterogeneity and function, adaptable to other single-molecule strategies.
The plasma membrane is a semipermeable, protein-rich, membrane bilayer that mediates key cell functions. Many proteins in the plasma membrane have highly organized yet distinct patterns of distribution. These distribution patterns arise from the specific affinities of proteins for other proteins or lipids in the plasma membrane, or from physical barriers imposed by cytoskeletal elements in contact with the plasma membrane, which impede protein lateral diffusion, sometimes trapping proteins in particular plasma-membrane domains1,2. The overall result is that most plasma-membrane proteins distribute heterogeneously in domains of diverse size and composition. This complex arrangement of proteins and lipids in the plasma membrane is believed to be critical to various physiological processes3,4.
Understanding the activities of the plasma membrane requires an accurate description of how its associated proteins distribute, but methods for precisely characterizing this spatial organization have been less than satisfactory. Many plasma-membrane proteins show no enriched signal or specific spatial organization when visualized optically because the scale of their heterogeneity is too small to be resolved. Techniques such as electron microscopy5,6, near-field scanning optical microscopy7 and fluorescence resonance energy transfer8,9, all of which can be used for nanoscale interrogation, do not offer a detailed nanoscopic description of overall plasma-membrane protein organization at high density owing to several technical obstacles. In particular, high-density labeling of proteins in electron microscopy is difficult, antibodies used in electron microscopy and near-field scanning optical microscopy may cause artifactual cross-linking (or result in multiple labeling of single proteins)10, mechanical rip-off of the top portion of the cell to visualize plasma-membrane proteins in electron microscopy could well disrupt plasma-membrane nanoscale organization, and fluorescence resonance energy transfer cannot be used to interrogate protein organization over distances greater than ~10 nm.
A promising approach to overcome many of these limitations is single-molecule super-resolution imaging, involving single marker switching to enable light emission from only one fluorophore in a diffraction-limited spot. Depending on the fluorophore, the technique has been called photoactivated localization microscopy (PALM)11, fluorescence PALM12, stochastic optical reconstruction microscopy (STORM)13, ground state depletion microscopy followed by individual molecule return14 and direct STORM15. In the field of plasma-membrane organization, PALM has been particularly useful because proteins in this approach are precisely modified with a single fluorescent label by genetically tagging them with a photoactivatable fluorescent protein (PA-FP), and a high density of proteins can be achieved by controlling their expression. Consequently, PALM has been used to characterize various plasma-membrane proteins, including T-cell receptor and Lat distribution on plasma-membrane sheets from T cells16, and the distribution of hemagglutinin17, paxillin18, Gag19 and Src proteins20.
Despite these successes, use of PALM or other single-molecule imaging techniques to precisely characterize the spatial organization of proteins in the plasma membrane has been limited because of several factors. One factor is imprecise localization and blinking of the fluorophores. This can result in molecules being inappropriately detected and localized, making it difficult to draw conclusions about the number and density of molecules in particular areas of the plasma membrane. A second factor is the limitation in statistical approaches used to characterize PALM datasets. Here we present a pair-correlation method for quantitatively describing PALM datasets of proteins tagged with PA-FP (PC-PALM) that overcomes these limitations, enabling a more comprehensive picture of overall plasma-membrane heterogeneity. We used PC-PALM to describe how different plasma-membrane proteins are distributed by evaluating the extent of clustering, overall density patterns and changes in these characteristics upon cholesterol depletion, sphingomyelinase treatment, actin depolymerization and antibody-induced cross-linking. The findings demonstrate that PC-PALM is an effective method for obtaining detailed spatial analysis of protein heterogeneity and function in the plasma membrane. The technique also has broad applicability for clarifying protein organization in different cellular compartments.
RESULTS
PC-PALM
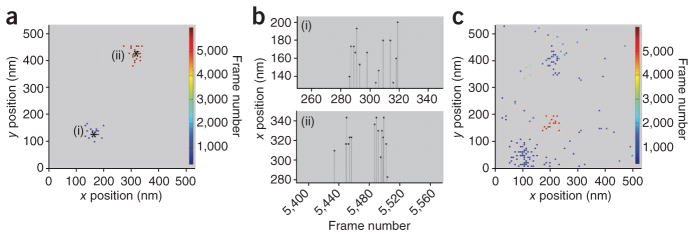
A single molecule in a PALM image typically appears in multiple frames with variable intervals of blinking (in which the PA-FP temporarily shifts to a dark state) before it is irreversibly photobleached. As the localization precision of a single molecule depends on the total number of photons it emits and this number varies stochastically from frame to frame, the centroid positions of the molecule’s diffraction limited spot (that is, peaks) in successive frames do not exactly coincide. This results in a single protein in a PALM image appearing as a cluster of peaks (Fig. 1a). Owing to blinking, the peaks appear in a time window that depends on the intervals of dark state and photobleaching rate (Fig. 1b). Because the cluster has a well-defined spatial dispersion (determined by the average localization precision, σs, of the peaks), it can be grouped and assigned to an individual protein (Fig. 1a). Grouping, however, becomes difficult when clusters of peaks belonging to different molecules are not well separated spatially and temporally. The variable time period of dark state creates additional difficulties, especially when proteins are expressed at high density and imaged at a high frame rate. Under these conditions, it is very challenging to appropriately group peaks to a single protein (Fig. 1c).
Figure 1.
Single molecules appear multiple times with variable blinking intervals. (a–c) TfR-PAGFP was expressed transiently at low or high density on the plasma membrane of COS-7 cells and imaged using PALM. Best fit coordinates of peak centers from individual frames were calculated and displayed as single dots in a and c. Shown are clusters of peaks corresponding to two spatially and temporally well-separated molecules in low expressing cell (a) and peaks corresponding to molecules that are not well separated in highly expressing cell (c). Note that the peaks arising from a single molecule are spatially confined in a region determined by average localization precision of the peaks. Center positions of molecules (asterisks) were determined by grouping peaks in the clusters. Color bars indicate frame number at which the molecules appeared. Graphs in b show multiple frame appearances of the single molecules (i) and (ii) from a. Frames in the sequence with no signal represent times when the molecule switched to a dark state.
We used pair-correlation–function analysis to distinguish individual proteins and clusters of proteins in a PALM image. Pair-correlation analysis provides a robust and objective method to characterize spatial scales of density fluctuations21. For this analysis, we considered peaks appearing in consecutive frames and within a radius covering 99% probability density of the two-dimensional (2D) Gaussian function for localization uncertainty to be a single peak, as they represent a single molecule appearance interrupted only because of the acquisition frame rate. Owing to blinking, the same protein will produce multiple peaks that are temporally isolated. To circumvent the problem of group assignment of these temporally isolated peaks to individual proteins, we subjected peaks in a PALM image assembled from all frames to PC-PALM analysis. Using the defined spatial signature of peaks belonging to a single molecule, we calculated correlation functions of peaks belonging to a single protein and those belonging to a group of proteins.
In the PC-PALM approach, the total pair-wise correlation function of all peaks in a PALM image can be represented as g(r)peaks = (g(r)centroid + g(r)protein) * g(r)PSF, in which g(r)PSF is the correlation function of the effective point spread function (PSF) of uncertainty in localization, and * represents convolution. The convolution of g(r)centroid (protein correlation function at r = 0) with g(r)PSF quantifies the correlation arising from multiple appearances of the same molecule and is defined as g(r)stoch. The convolution of g(r)protein (protein correlation function at r > 0) with g(r)PSF represents the correlation function of the relative spatial distribution of the proteins.
The pair-correlation function quantifies the increased probability of finding another protein at a distance r away from a protein compared to that expected from random distribution of proteins. Therefore, when a protein population is randomly distributed, the correlation function describing its organization, g(r)protein, is ~1. The total pair-wise correlation function under these conditions reduces to:
| (1) |
When proteins are nonrandomly distributed, showing clustering or aggregation, g(r)protein will be greater than 1. For randomly shaped clusters in a 2D system, the decay of probability density of proteins from the center of the cluster can be approximated with an exponential function (Supplementary Fig. 1). Furthermore, g(r)protein should decay to 1 at longer distances, where organization of proteins approaches a random distribution. Using the above criteria, we defined protein correlation as g(r)protein = A × exp(–r/ξ) + 1 (with exp representing exponential, the correlation length, ξ, being a measure of the domain size of the protein cluster and A being roughly the amplitude of the protein correlations extrapolated to r = 0). Thus, the total correlation will be given by:
| (2) |
If the computed pair-wise correlation of all peaks fits to equation (1), then the proteins are randomly organized with no spatial correlation. Otherwise, the computed pair-wise correlation function is fit to equation (2) to evaluate g(r)protein and the fit parameters ξ and A. This allows quantification of three different parameters to provide a physical description of the spatial organization of the proteins: (i) the increased local density of proteins appearing in a cluster or domain, ψcluster, (ii) correlation length of clusters, ξ, which gives a rough estimate of the radius of a cluster and (iii) the average number of proteins in a cluster, Ncluster. Ncluster is not an absolute number given the uncertainties in the efficiency of photoactivatable GFP (PAGFP) photoactivation, which can lead to an underestimation of the actual number of proteins expressed.
Validation of PC-PALM
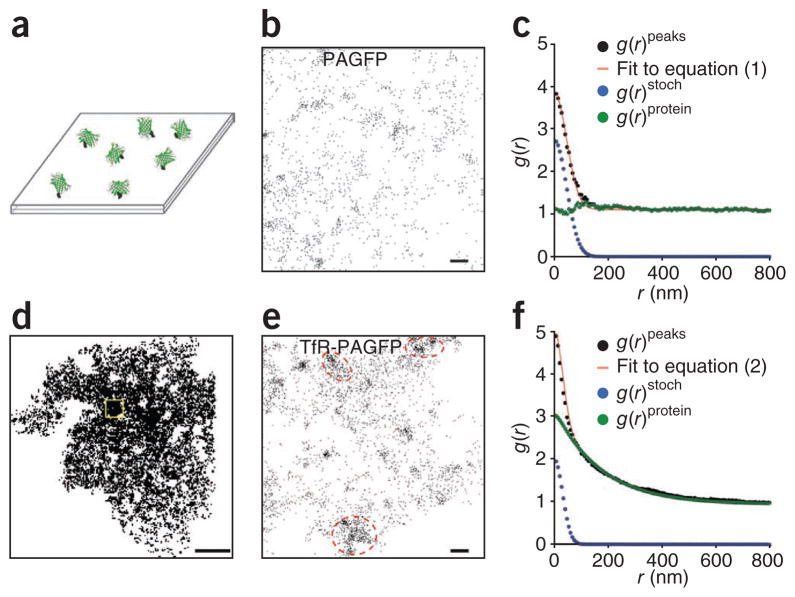
To validate the PC-PALM approach, we computed the pair-correlation functions for both random and clustered protein distribution patterns. In the random distribution condition, we analyzed the spatial distribution of PAGFP molecules covalently immobilized randomly on a glass coverslip (Fig. 2a). A PALM image of a region of the coverslip surface displaying all peaks (Fig. 2b) revealed clusters of peaks associated with individual molecules distributed widely across the surface. Computed g(r)peaks of this image could be well fit with equation (1) (Fig. 2c). Supporting a random distribution of the PAGFP molecules, g(r)protein did not deviate appreciably from 1 (Fig. 2c), indicating that the proteins were not clustered within the error bounds of our experiments.
Figure 2.
PC-PALM distinguishes random versus clustered distributions. (a) Diagram showing purified PAGFP molecules randomly immobilized on glass coverslip. (b) Actual spatial distribution of peak centers of PAGFP molecules in a section of this coverslip. (c) Plot of calculated autocorrelation function (g(r)peaks) of PAGFP molecules in b fit to equation (1). The correlation owing to multiple appearances of a single protein (g(r)stoch) and the protein correlation (g(r)protein) were evaluated from the fit. (d) Distribution of peak centers of TfR-PAGFP across the plasma membrane of a COS-7 cell. A representative section of the cell used for correlation analysis is marked by a yellow box. (e) Spatial distribution of peak centers of TfR-PAGFP in the section indicated in d. Beside peak clusters associated with individual molecules, there was noticeable grouping of these clusters (red dashed ovals). (f) Measured correlation function of all peaks (g(r)peaks) in e was well fit to equation (2). Corrected protein correlation function (g(r)protein) was evaluated by subtracting the contribution from multiple appearances of single protein (g(r)stoch) from the measured correlation function. Scale bars, 200 nm (b,e) and 5 μm (d).
In the clustered distribution condition, we examined the spatial distribution of transferrin receptor labeled with PAGFP (TfR-PAGFP) expressed on the plasma membrane of COS-7 cells. TfR-PAGFP is known to partially distribute on the plasma membrane in clathrin-coated pits (radius < 200 nm). A PALM image of a region of the plasma membrane displaying all peaks (Fig. 2d,e) supported this organization. Because of this secondary organization, we could not approximate pair-wise autocorrelation functions of TfR-PAGFP peaks with a random protein distribution model (equation (1)). However, the data could be well fit with a nonrandom protein distribution model (equation (2); Fig. 2f). Evaluation of g(r)protein and the parameters ξ and A from this fit revealed that a fraction of TfR-PAGFP resides in clusters with average radius of ~150 nm, which is within the appropriate size range of clathrin coated pits in the plasma membrane. We also performed cross-correlation analysis between TfR-PAGFP and TfR tagged with photoactivable monomeric Cherry 1 (TfR-PAmCh) transiently expressed in COS-7 cells, and imaged with PALM. We detected substantial cross-correlation between TfR-PAGFP and TfR-PAmCh (Supplementary Fig. 2), supporting the idea that TfR is present in clusters on the plasma membrane. These results demonstrate PC-PALM can correctly describe the 2D spatial organization of proteins, either randomly distributed or highly clustered.
Steady-state organization of plasma-membrane proteins
We next used PC-PALM to quantitatively interrogate the distribution of proteins with distinct membrane anchors. We studied the following proteins: an outer leaflet glycosylphosphatidylinositol (GPI)-anchored protein, a transmembrane protein (Lat), an inner leaflet lipid-anchored protein (Lyn) and the transmembrane protein vesicular stomatitis viral glycoprotein (VSVG). GPI proteins have been widely used as model, liquid order–preferring proteins and are thought to cluster in the plasma membrane into small, dense clusters of less than four proteins7,8. Lat and Lyn have a central role in signal transduction pathways, with their steady-state lateral organization important for signaling in these pathways22,23. VSVG is vital for viral entry and forms trimers at the plasma membrane24. We chose these proteins because each has little or no endogenous expression in the COS-7 cells in which we expressed them.
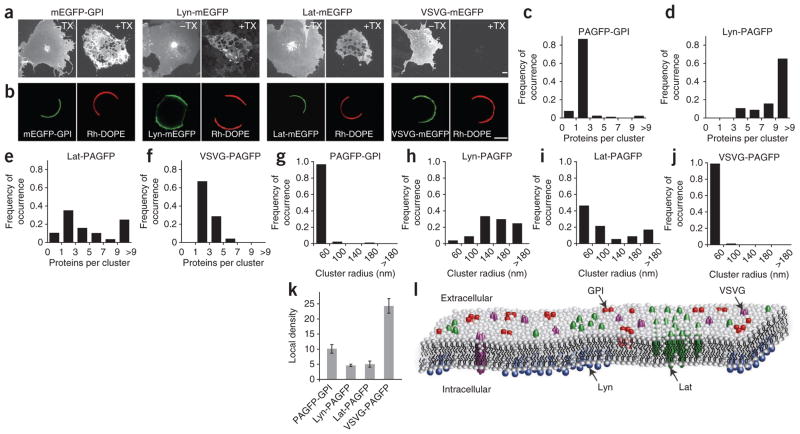
Diffraction-limited confocal images of monomeric enhanced GFP (mEGFP)-tagged versions of the proteins expressed in COS-7 cells revealed that they individually distribute uniformly on the plasma membrane (Fig. 3a). Nevertheless, the proteins showed substantial differences in their response to detergent extraction in cold Triton X-100, with mEGFP-GPI, Lat-mEGFP and Lyn-mEGFP resisting extraction, and VSVG-mEGFP being extracted (Fig. 3a). The proteins also had different lipid partitioning characteristics in giant plasma-membrane vesicles (GPMVs)25 induced to bleb off the plasma membrane in expressing cells (Fig. 3b). mEGFP-GPI and Lat-mEGFP partitioned into regions of the GPMVs lacking 1,2-dioleyl phosphatidylethanolamine-N- (lissamine rhodamine B) (Rh-DOPE), a liquid disorder–preferring lipid. This suggested they prefer residing in a membrane environment enriched in ordered lipids. In contrast, Lyn-mEGFP and VSVG-mEGFP localized together with Rh-DOPE in the liquid-disorder phase.
Figure 3.
Steady-state spatial distribution of plasma-membrane proteins assessed by PC-PALM. (a) Maximum-intensity projection confocal images of mEGFP-labeled proteins expressed in COS-7 cells. mEGFP-GPI, Lyn-mEGFP, Lat-mEGFP and VSVG-mEGFP were uniformly distributed across the plasma membrane (–TX). On extraction with 1% Triton X-100 at 4 °C (+TX), mEGFP-GPI, Lyn-mEGFP and Lat-mEGFP were present in detergent resistant membrane, whereas VSVG-mEGFP was solubilized. Scale bars, 5 μm. (b) Partitioning of mEGFP-labeled proteins and Rh-DOPE between coexisting liquid phases in GPMVs. (c–j) Cluster parameters of steady-state distribution of PAGFP-labeled proteins graphed as proteins per cluster (c–f) and as distribution of cluster radius (g–j) assessed by PC-PALM (n = 91 for PAGFP-GPI, n = 57 for Lyn-PAGFP, n = 88 for Lat-PAGFP and n = 73 for VSVG-PAGFP). (k) Density of proteins in clusters for PAGFP-GPI, Lyn-PAGFP, Lat-PAGFP and VSVG-PAGFP. Error bars, s.e.m. (n = 91 for PAGFP-GPI, n = 57 for Lyn-PAGFP, n = 88 for Lat-PAGFP and n = 73 for VSVG-PAGFP). (l) Model of steady-state spatial distribution of the four proteins based on PC-PALM analysis.
We next used PC-PALM to quantitatively interrogate nanoscale organization of these proteins in the intact plasma membrane. We expressed PAGFP-tagged proteins transiently in COS-7 cells and imaged the plasma membrane using PALM. We evaluated protein pair-wise autocorrelation functions, that is, g(r)protein, from correlation analysis of the peak centers, and obtained quantitative description of protein clustering patterns using the parameters ξ (correlation length of clusters), Ncluster (detected proteins per cluster) and ψcluster (density of proteins in cluster). Representative curve fits are shown in Supplementary Figure 3.
We observed clear differences in the clustering characteristics of the four proteins (Fig. 3c–j). PAGFP-GPI molecules were organized into clusters of radius < 60 nm containing on average ~2–3 detected proteins (Ncluster = 2.3 ± 0.3; s.e.m., n = 91) (Fig. 3c,g). Lyn-PAGFP molecules, in contrast, resided in clusters with three or more proteins, with a wide range of cluster sizes mostly above 60 nm (Fig. 3d,h). Lat-PAGFP clusters were also organized into domains with widely varying ξ and Ncluster values. Approximately 30% of the radii of Lat-PAGFP clusters were greater than 100 nm, and Ncluster value was 2–40 (Fig. 3e,i and Supplementary Fig. 4). VSVG-PAGFP was distributed in clusters with an average of 2.8 ± 0.4 (s.e.m.; n = 73) detected proteins with radius < 60 nm (Fig. 3f,j).
We obtained additional insight into the organization of these proteins by examining the ψcluster values obtained from the autocorrelation functions (Fig. 3k). The ψcluster value indicates how densely packed proteins are in a cluster compared to the rest of the plasma membrane. VSVG-PAGFP had the largest ψcluster value (24.3 ± 2.4; s.e.m., n = 73), suggesting the clusters consisted of tightly associated proteins. Given the protein number per cluster of VSVG-PAGFP was ~3.0 (Fig. 3f), this supports VSVG-PAGFP occurring primarily as a trimer in the plasma membrane with no higher-order organization, consistent with previous biochemical studies24. PAGFP-GPI had the next highest ψcluster value (10.1 ± 1.4; s.e.m., n = 91). Together with its ξ and Ncluster values, this suggested that PAGFP-GPI distributes in relatively dense clusters of uniform size. Both Lat-PAGFP and Lyn-PAGFP had low ψcluster values (≤5). Given these two proteins show a wide range of proteins per cluster and variable cluster radii, the data suggest they distribute in nonuniform clusters with low enrichment of proteins.
Based on the above quantitative descriptions, we can propose the following model for steady-state distributions of GPI, Lyn, Lat, and VSVG in the plasma membrane (Fig. 3l). VSVG exists as about three tightly associated detectable proteins, GPI shows clustering in domains less than 60 nm containing 2–3 detectable proteins, and Lyn and Lat cluster in less dense domains of variable sizes with variable numbers of detected proteins.
PAGFP-GPI reorganization by modulation of lipids
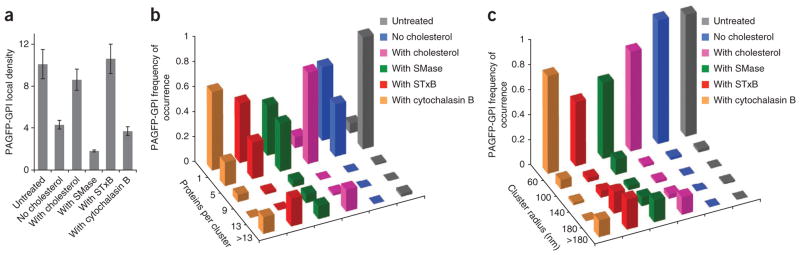
GPI-anchored proteins are attached to the plasma membrane solely by a lipid anchor, and their steady-state organization is sensitive to the membrane lipid environment8. Given this, we used PC-PALM to examine how nanoscale clusters of PAGFP-GPI are affected by perturbations in the lipid composition of the plasma membrane. Depletion of cholesterol with methyl-beta cyclodextrin (MβCD) decreased both ψcluster and Ncluster of PAGFP-GPI (Fig. 4a,b). Around 60% of the images had Ncluster of no more than 1 (Fig. 4b), suggesting that a large fraction of the proteins were randomly distributed. Cholesterol addition, in contrast, induced formation of additional clusters with larger radii, and Ncluster increased relative to that found for untreated conditions (Fig. 4b,c). These observations demonstrate that small nanoclusters of PAGFP-GPI are sensitive to the cholesterol content of the plasma membrane, with cholesterol depletion dispersing nanoclusters and cholesterol addition causing them to be larger.
Figure 4.
Reorganization of PAGFP-GPI induced by perturbations of plasma membrane evaluated by PC-PALM. (a) Local density of PAGFP-GPI in clusters under steady state and after perturbations. Error bars, s.e.m. (n = 91 for untreated, n = 60 for no cholesterol, n = 58 with cholesterol treatment, n = 58 with sphingomyelinase (SMase), n = 59 with STxB and n = 53 with cytochalasin B). (b) Distribution of PAGFP-GPI molecules per cluster. (c) Distribution of correlation lengths of PAGFP-GPI under steady state and upon perturbations.
We next examined the effect of sphingomyelinase treatment on the distribution of PAGFP-GPI by PC-PALM. Sphingomyelinase generates ceramide in the plasma membrane26, which, in turn, can alter cholesterol organization27. Sphingomyelinase treatment dramatically decreased ψcluster value of PAGFP-GPI by a factor of ~5 (Fig. 4a). PAGFP-GPI was either randomly distributed (~40% of the samples) or organized into larger clusters of radius > 120 nm with decreased Ncluster value (Fig. 4b,c). This indicated that sphingomyelinase disrupts the small, dense nanoclusters of PAGFP-GPI and leads to their partial inclusion into larger domains induced by ceramides.
PAGFP-GPI remodeling by toxin and actin depolymerization
Binding of pentameric B-subunit of Shiga toxin (STxB) to its glycosphingolipid receptor globotriaosyl ceramide (Gb3) triggers the formation of ordered membrane domains and tubular plasma-membrane invaginations28. We examined how this affects the lateral organization of PAGFP-GPI by PC-PALM. Values of Ncluster and ξ of PAGFP-GPI both increased substantially (Fig. 4b,c). ψcluster, however, remained largely the same (Fig. 4a). This suggests that long-range correlations of PAGFP-GPI may be due to inclusion of GPI-anchored nanoclusters into STxB-induced ordered domains and invaginations.
Previous work has shown that perturbation of the actin cytoskeleton affects the organization of GPI-anchored proteins8. To investigate this in more detail, we used PC-PALM to examine PAGFP-GPI distribution during treatment of cells with cytochalasin B, which depolymerizes the actin cytoskeleton. GPI nanoclusters were dispersed and proteins were randomly distributed in ~60% of the images examined. In addition, a fraction of the GPI proteins were present in larger, less dense domains with the domains having a wide range of spatial scales (Fig. 4). Thus, maintenance of the small nanoclusters of PAGFP-GPI is affected by actin depolymerization.
Cross-correlation analysis of PAGFP-GPI and actin
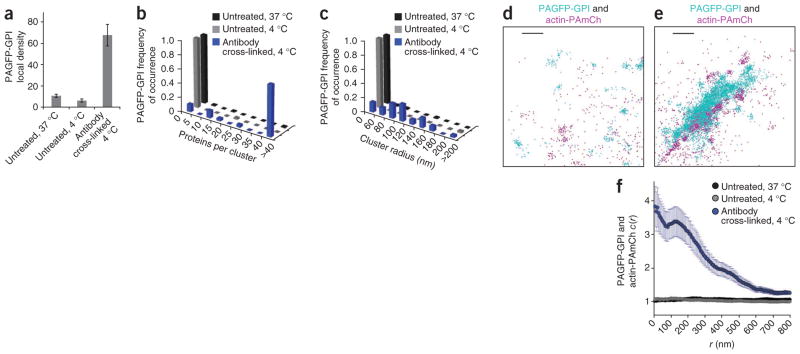
During receptor signaling, transport vesicle biogenesis, cell polarization and viral budding, critical clustering events occur that induce or stabilize plasma-membrane structures, leading to recruitment of downstream machinery29. Such clustering often induces reorganization of cortical actin cytoskeleton30. To investigate potential co-recruitment of actin to sites of protein clustering in the plasma membrane, we used PC-PALM to analyze cells expressing PAGFP-GPI exposed to a cross-linking antibody to GFP at 4 °C. We found this treatment induced PAGFP-GPI to become tightly packed in large structures, with increased ψcluster (about sixfold), Ncluster and long-range correlations (ξ) (Fig. 5a–c). There was no substantial change in ξ of PAGFP-GPI on lowering the temperature from 37 °C to 4 °C in absence of cross-linking, though there was some reduction in the extent of clustering (Fig. 5a–c and Supplementary Fig. 5).
Figure 5.
Actin-PAmCh localizes with clusters of antibody–cross-linked PAGFP-GPI. (a) Local density of PAGFP-GPI evaluated by PC-PALM in clusters at steady state at 37 °C (s.e.m.; n = 91), at 4 °C (s.e.m.; n = 29) and upon antibody-induced cross-linking of PAGFP-GPI at 4 °C (s.e.m.; n = 35). (b) Distribution of PAGFP-GPI molecules per cluster at 37 °C, 4 °C and after antibody cross-linking at 4 °C. (c) Distribution of PAGFP-GPI cluster sizes at 37 °C, 4 °C and upon antibody cross-linking at 4 °C. (d) Section of a COS-7 cell coexpressing PAGFP-GPI and actin-PAmCh in absence of cross-linking antibodies at 37 °C. Centers of peaks are shown. (e) Section of a cell expressing PAGFP-GPI and actin-PAmCh following antibody cross-linking of PAGFP-GPI at 4 °C. (f) Cross-correlation between PAGFP-GPI and actin-PAmCh with (at 4 °C; s.e.m., n = 61) and without (at 4 °C and 37 °C; s.e.m., n = 39 for 4 °C, n = 34 for 37 °C) antibody cross-linking. Scale bars, 200 nm.
We next applied cross-correlation analysis to examine the response of actin to cross-linking of PAGFP-GPI by antibody to GFP at 4 °C. We observed no appreciable spatial colocalization of PAGFP-GPI and actin-PAmCh in absence of antibody cross-linking (Fig. 5d). But actin-PAmCh dramatically reorganized in response to the cross-linking of PAGFP-GPI and localized with the clusters of PAGFP-GPI, as evident from the PALM image and from the increased value of c(r) (Fig. 5e,f). PAGFP-GPI and actin-PAmCh were spatially uncorrelated in the absence of antibody cross-linking at both 37 °C and 4 °C (c(r) ~1) (Fig. 5f). As GPI anchors are localized only on the outer plasma-membrane bilayer, whereas actin is cytoplasmically localized, clustering of GPI and actin therefore must involve some type of signaling across the bilayer.
DISCUSSION
Using an algorithm to distinguish single proteins from clusters of proteins, PC-PALM enables quantification of different parameters of protein spatial organization. This includes descriptions of cluster features of proteins across different size scales, such as detected number of proteins within a cluster, density of proteins in a cluster and overall size of a cluster. The approach avoids the problems associated with precise identification of single molecules, such as overcounting of labeled molecules in peak-grouping strategies. In doing so, it provides a quantitative approach for dissecting the spatial features of nanoscale protein organization.
A fundamental strength of PC-PALM is its relative simplicity and ability to add new details to spatial organization in a biological system. Using PC-PALM, we could clearly distinguish a random distribution of PAGFP molecules immobilized on coverslips from large-scale clustering of TfR-PAGFP proteins (~100–200 nm) on the plasma membrane. Moreover, we could explore current concepts of membrane organization and membrane remodeling in response to specific perturbations. The observed details of nanoscale clustering of GPI, Lat, Lyn and VSVG proteins (including cluster size, density and variability) provide a more complete picture of protein organization in the plasma membrane on a wide range of spatial scales (50 nm to 1 μm). In addition to characterization of the distribution patterns of distinct proteins, PC-PALM can be used to examine how two different proteins are spatially organized with respect to each other by performing cross-correlation analysis.
Because the key aspects of PC-PALM are straightforward and well-suited for characterization of both small oligomers and larger protein domains, the method is an ideal tool to study protein reorganization during various physiological processes. For example, PC-PALM could help clarify how signaling receptors on the plasma membrane cluster in response to ligand binding, uncover steps in the pathway for viral budding and dissect clustering of endocytic receptors. PC-PALM is not limited to examining protein organization in the plasma membrane but can be used to interrogate quantitative aspects of protein organization anywhere in the cell, including in organelles such as endoplasmic reticulum, mitochondria and Golgi apparatus. Finally, although in this study we used PA-FPs, the PC-PALM method is applicable to any single-molecule application, such as those using fluorescent dyes in STORM or gold-labeled antibodies in electron microscopy (S.L. Veatch, B. Machta, S. Shelby, E. Chiang, D. Holowka and B. Baird, arXiv:1106.6068).
ONLINE METHODS
Mammalian expression vectors
Plasmid encoding PAGFP-GPI was generated by exchanging YFP in YFP-GPI with PAGFP using AgeI and BsrGI restriction enzyme sites. To produce Lyn-PAGFP vector, EGFP in Lyn-EGFP was replaced with PAGFP using AgeI and NotI restriction sites. Lat-PAGFP vector was obtained by inserting EcoRI and KpnI–digested fragment of Lat from Lat-EGFP into PAGFP-N1 vector. Plasmid constructs for TfR-PAGFP, TfR-PAmCh, VSVG-PAGFP, and actin-PAmCh were provided by G. Patterson (National Institute of Biomedical Imaging and Bioengineering).
Sample preparation
We cleaned 18-mm #1.5 coverslips (Warner Instruments) with either 1% hydrofluoric acid (HF) for 5 min or 1% Hellmanex II (Fisher) for 3 h, followed by distilled water and 100% ethanol. Cleaned coverslips were then flamed and placed in sterile 35-mm tissue culture dishes. For PALM, cells were grown on fibronectin coated (2 μg ml−1 in PBS (pH 7.4); Sigma) cover-slips and transiently transfected 48 h after plating with Fugene 6 (Roche). Approximately 24 h after transfection, cells were washed twice with PBS, fixed with 4% paraformaldehyde, 0.2% glutaraldehyde (Electron Microscopy Sciences) in PBS for 35 min at room temperature (25 °C) (or for 15 min at 4 °C followed by 30 min at room temperature for cells incubated at 4 °C). Quenching was done with filter-sterilized 10 mg ml−1 BSA in PBS for 5 min, and cells were finally washed four times with PBS. In all cases, fixation was preformed just before the imaging. Cells expressing VSVG were incubated at 32 °C at least 8 h before fixation. To ensure high density of TfR-PAGFP on the plasma membrane, cells were incubated with 100 μM deferoxamine mesylate salt (DFO, Sigma) for 18 h after transfection. Coverslips were incubated with 1:4,000 diluted Tetraspec beads (Invitrogen) in PBS for 10 min that served as fiducial markers.
For confocal microscopy, cells were grown on coverslips (18-mm, #1.5) and transfected 24 h before imaging.
To generate GPMVs, COS-7 cells transiently expressed with mEGFP-tagged proteins were labeled with 200 μg ml−1 Rh-DOPE (Avanti Polar Lipids) dissolved in ethanol for 5 min, washed twice with GPMV buffer (2 mM CaCl2, 10 mM Hepes and 150 mM NaCl, pH 7.4), and incubated with freshly prepared GPMV active reagent consisting of 2 mM N-ethyl maleimide (Sigma) in GPMV buffer for 1 h at 37 °C with shaking (60 cycles min−1). GPMVs were then gently decanted into a tube and allowed to sit undisturbed on ice for 30 min to allow the larger GPMVs to sediment. Finally, GPMVs were collected from the bottom 20% of the total volume of the tube and imaged at 25 °C.
To covalently immobilize on coverslips, clean coverslips were first treated with 5% (wt/vol) 3-aminopropyl trimethoxysilane (APTMS) in acetone for 15 min at room temperature, washed with acetone and PBS in succession, and then incubated with 0.25% (wt/vol) glutaraldehyde in PBS for 30 min. PAGFP in PBS was centrifuged (100,000g in TLA 45 rotor for 2 h) to minimize any potential aggregation before the experiment. Next, functionalized coverslips were incubated with a combination of 10 nM PAGFP and 5 μM BSA in PBS for 30 min at room temperature in a humid chamber. Coverslips were extensively washed after the incubation and then imaged in PBS.
Cells and membrane perturbations
COS-7 cells were cultured in Phenol Red–free Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% FBS, 1 mM sodium pyruvate, 100 units ml−1 penicillin, 100 units ml−1 streptomycin and 2 mM glutamine (Invitrogen). Approximately 24 h after transfection, cells were washed quickly in PBS pre-warmed to 37 °C and immediately fixed in 4% (w/v) paraformaldehyde and 0.2% (w/v) glutaraldehyde (Electron Microscopy Sciences) for 35 min at room temperature in PBS (or for 15 min at 4 °C followed by 30 min at room temperature for cells incubated at 4 °C). These fixation conditions have been reported to immobilize most of the plasma-membrane proteins10. Plasma-membrane cholesterol levels were elevated or depleted by incubating cells with either 2 mM soluble cholesterol (Sigma) or 10 mM methyl-β-cyclodextrin (Sigma), respectively, in DMEM supplemented with 10 mM HEPES and 1 mg ml−1 BSA for 30 min at 37 °C. Ceramide was generated in the plasma membrane by incubating cells with sphingomyelinase from Streptococcus aureus (Sigma) at a final concentration of 10 units ml−1 for 30 min at 37 °C. To bind STxB to its lipid receptor globotriaosyl ceramide (Gb3), cells were incubated with 60 nM STxB for 1.5 h at 4 °C in medium supplemented with 10 mM HEPES and subsequently warmed up to 37 °C for 10 min. Cortical cytoskeleton was disrupted by treating cells with 1 μM cytochalasin B (Sigma) for 15 min at 37 °C. To cross-link PAGFP-GPI, cells were incubated with 5 μg ml−1 polyclonal antibody to GFP (Invitrogen) for 1 h at 10 °C, followed by 5 μg ml−1 horseradish peroxidase–conjugated secondary goat antibody to rabbit (Jackson Labs) for 1 h at 10 °C. Cells were then washed with medium, and incubated at 4 °C for an additional 30 min. Untreated cells were similarly treated in absence of antibodies. Triton X-100 (Sigma) extraction experiments, were performed as described previously31. Briefly, live cells were cooled to 4 °C, and subsequently incubated with either ice cold 1% Triton X-100 (Tx-100) in PBS or PBS (for control experiments) for 20 min at 4 °C.
Microscopy
PALM imaging was performed on an Olympus IX81 microscope using a 60 × 1.45 numerical aperture (NA) objective (Olympus, PlanApoN). Cells were imaged in total internal reflection fluorescence (TIRF) mode using activation and excitation lasers with wavelengths 405 nm (50 mW Cube, Coherent), 488 nm (50-mW Sapphire, Coherent) and 561 nm (40 mW Compass, Coherent). Fluorescence emission was detected with an electron-multiplying charge-coupled device (EM-CCD) camera (Andor Technology, DV887ECS-BV). Images of a 43 μm × 43 μm area were collected with an exposure time of 100 ms. PAGFP was simultaneously activated and excited with 488-nm laser with the intensity set to 400 μW (as measured at rear aperture of the objective). Activation power was commonly increased toward the end of the experiment once most of the molecules were depleted. Activated fluorescent molecules were typically depleted after 10,000–50,000 frames. For two-color imaging, PAGFP fluorescence was collected first by simultaneously activating and exciting with 488-nm laser until PAGFP was completely exhausted. Next PAmCh fluorescence was collected using 405 nm (1–15 μW), and 561 nm (800 μW) lasers for activation and excitation, respectively. Tetraspec beads (Invitrogen) were used as fiducial markers to correct for drift during image acquisition and to overlay two-color images. Confocal imaging was performed with a Marianas spinning disc (Intelligent Imaging Innovations) attached to a Zeiss Observer.Z1 microscope (Carl Zeiss) equipped with a 63× Plan Apochromat 1.4 NA (Carl Zeiss) objective lens and a Leica TCS SP2 spectral confocal system (Leica) with a 63× oil-immersion objective. Maximum-intensity projection images were constructed using Slidebook 5.0 software (Intelligent Imaging Innovations).
Data analysis
Peaks were localized using a previously described algorithm written in IDL (Research Systems)11. Peaks identified in each frame were fit using a cylindrically symmetric Gaussian point spread function. The average localization precision (σraw) of the peaks ranged from 20–35 nm. PALM images were collected under conditions where the average distance between peaks in a single frame was substantially larger than the precision (σraw) of the peak positions. All the detected peaks were used for subsequent analysis using code custom written in MATLAB (MathWorks).
Correlation analysis
Peaks appearing in consecutive frames within a radius of 2.5 × σraw were considered to belong to the same molecule and were replaced by a single peak whose sigma (σpeaks) and position coordinates were estimated as a weighted average of the sigma values and position coordinates of the contributing peaks. Average localization precision of resulting peaks ranged between 16 and 20 nm (Supplementary Fig. 6). In a PALM image assembled by combining all the frames, an individual protein is represented by multiple peaks, which are spatially separated owing to uncertainty of their localization and are temporally separated owing to blinking of the fluorophores. However, the peaks belonging to the same protein have a specific spatial dispersion and they are distributed approximately over a 2D Gaussian surface centered at the actual position of the protein. We took advantage of this defined spatial distribution of peaks belonging to a single molecule in our analysis, as described below.
Binary PALM images of entire cells were constructed using the best fit coordinates of the peak centers, such that the pixels have a value of 1 at peak centers and a value of 0 elsewhere. Auto- and cross-correlation was performed on randomly selected sections (4–16 μm2 squares) of these images. Pair correlation functions were computed using Fast Fourier Transforms in MATLAB (Supplementary Note 1). Sections of cells were selected in such a way that visible gaps (holes) in the zoomed out image (of distribution of peaks) were avoided. As we obtained distinct membrane organization for the five proteins tested, we are confident that membrane topology does not influence our results substantially. This is also supported by the fact that we see changes in the distribution of proteins in response to membrane perturbations and cross-linking of proteins. Both auto- and cross-correlation were normalized to the average density of peaks over the entire image. By this definition, correlation functions quantify the probability of finding a second particle at a distance r away from a given particle, and a value of 1 indicates that particles are randomly distributed at that distance. Correlation function with a value greater than 1 indicates a clustered distribution of molecules. Mathematical terms are defined in Supplementary Note 2.
Autocorrelation model
The measured autocorrelation of the peaks, g(r)peaks, can be separated out into two contributions from (i) the protein correlation at r = 0 which we describe as correlation function of the centroids of the proteins, viz. g(r)centroid, and, (ii) the protein correlation at (r > 0), g(r)protein, which describes the spatial organization of the proteins. The two components of the correlation function, g(r)peaks and g(r)centroid, can be treated as statistically independent functions. Owing to the uncertainty in localization precision imposed by the experimental technique, both these correlation functions are convolved with the effective point spread function of the uncertainty in position determination, g(r)PSF. The total measured autocorrelation is the sum of these two correlation functions:
| (3) |
The defined spatial signature of peaks belonging to the same protein was used to derive a functional form of g(r)PSF. The centers and localization precision of each of the peaks was calculated by fitting a 2D Gaussian function. The spatial distribution of the cluster of peaks belonging to a single protein can be approximated by a 2D Gaussian surface (centered at the actual position of the protein), whose sigma is represented as σs. The effective point spread function of the uncertainty in position determination will be given by this 2D Gaussian function. The autocorrelation of this point spread function is also a Gaussian with increased sigma of √2 × σs. Thus, the functional form of g(r)PSF is given by:
| (4) |
The total correlation, g(r)peaks, takes the form:
| (5) |
As described before, the stochastic uncertainty in position determination leads to each molecule being represented as multiple peaks spread across the effective point spread function. The convolution of g(r)centroid with g(r)PSF represents the contribution of this cluster of peaks (corresponding to a single molecule) to the measured autocorrelation function. The correlation of the centroids, g(r)centroid, is a delta function with amplitude equal to inverse of the average density of the protein, ρaverage.
Thus, the total correlation of peaks can be represented by:
| (6) |
The first term of equation (6) represents the correlation arising from the multiple appearance of each protein and associated stochastic uncertainty in position determination. The second term of equation (6) (that is, g(r)protein convolved with g(r)PSF) represents the contribution from relative spatial position of the protein molecules.
If we define g(r)stoch as the stochastic correlation arising from multiple appearances of single protein, then:
| (7) |
The same underlying principle is used for detailed analysis of individual molecules in electron microscopy and STORM data (S.L. Veatch, B. Machta, S. Shelby, E. Chiang, D. Holowka and B. Baird, arXiv:1106.6068).
Randomly distributed proteins
Protein correlation function quantifies the increased probability of finding a protein at a distance r from a given protein compared to that expected for random distribution of proteins. Thus, in the special case where the proteins are randomly distributed, the protein correlation function g(r)protein (r > 0) ~ 1, and the second term of equation (6) becomes unity. Thus, the measured correlation now takes the following form:
| (8) |
Therefore, if equation (8) can describe the measured correlation function, then it indicates that the proteins are randomly organized. The mean localization precision (σpeaks) of all the peaks determines the s.d. (σs) of the 2D Gaussian surface describing the distribution of peaks belonging to the same protein. We obtained an estimate of σs by fitting the distribution of σpeaks from each PALM dataset to a skewed Gaussian function (Supplementary Fig. 7). If ρpeaks represent the average density of peaks, and α represents the average number of discrete appearances of a protein owing to blinking, the average density of proteins, ρaverage, will be given by ρavearge = ρpeaks/α. ρpeaks was calculated from the number of peaks present in the image. We could obtain an estimate of α, the average number of appearances of individual molecules, by analyzing images where PA-FP proteins are present at a very sparse density. α was 4–5 for all the proteins examined under our imaging conditions. The measured autocorrelation was fit to equation (8) using nonlinear least square fitting algorithm while adjusting the two free parameters σs and α. The distribution was considered random if the values of fit parameters, σs and α, were reasonable and consistent with the estimated values (±25% of the estimated values) and g(r)protein (r > 0) was less than 1.5.
We evaluated g(r)stoch by using the fit parameters σs and α, and subsequently calculated g(r)protein by subtracting g(r)stoch from the measured autocorrelation g(r)peaks. For random distribution of molecules, g(r)protein ~1 for all values of (r > 0).
Clustered distribution
In our model, deviations of experimental correlation functions from equation (8) indicate a non-random distribution of the proteins on the plasma membrane. If proteins are organized into clusters, the computed protein correlation will be greater than 1 along the length scales of the clusters, and approach the value expected from random distribution (that is, 1) at longer length scales. Under such conditions, we need to define a functional form of g(r)protein. We used two different criteria to choose a model for g(r)protein. If a protein is part of a cluster, the probability of finding another protein in the neighborhood is dependent on the density and distance, and in a 2D-system of clusters with no predefined shape, this probability can be approximated by an exponentially decaying function (Supplementary Fig. 1). We used an additional criterion to define g(r)protein. The correlation function g(r)protein quantifies the increased probability of finding a protein at distance r from a given protein relative to the expected probability for random distribution of proteins. Thus g(r)protein should decay to 1 at longer distances where the measured probability density approaches the value expected for random distribution of proteins. We chose a functional form of g(r)protein that satisfies both the above physical requirements:
| (9) |
Here the correlation length ξ gives an estimate of the radius of the domain and A is a measure of the protein density in the domains. Thus, the measured correlation function of all the peaks will take the following form:
| (10) |
The correlation function is fit to equation (10) by adjusting the free parameters ξ and A. σs and α are constrained to be consistent with the estimated values, as described before for random model fitting.
The cluster size, average number of proteins per cluster, and the magnitude of clustering can be evaluated from the fit parameters. The radius of the cluster of proteins is given by the correlation length ξ. The average number of proteins per cluster, Ncluster, is given by:
| (11) |
As described above, we obtained an estimate of α by analyzing the average number of appearances of individual proteins when they are expressed sparsely, conditions under which proteins can be grouped with more confidence. Thus, it should be noted that any uncertainty in estimation of α values would affect our calculation of Ncluster. This effect will be more severe for situations where the correlation lengths of the clusters are similar in magnitude to σs, as in the cases for PAGFP-GPI and VSVG-PAGFP. However, as we obtained similar values of α for the PAGFP-tagged proteins at low density, it is reasonable to assume that our estimate is close to the actual values when high density of proteins are imaged under similar conditions.
We defined ψcluster as the increased density of proteins in clusters relative to the overall average density of proteins across the whole image, and it is given by:
| (12) |
Although we optimized our imaging scheme for activation and detection of PA-FPs, we likely are not detecting the entire population of PA-FPs. This can lead to an underestimation of the number of proteins present in cluster. However, estimates of cluster sizes should not be affected.
Supplementary Material
Acknowledgments
We thank G. Patterson (US National Institute of Biomedical Imaging and Bioengineering) for providing plasmid constructs; E. Ambroggio (US National Institute of Child Health and Development) for providing purified PAGFP protein; H. Hess and G. Stengel for providing the Peak Selector software and valuable discussion; S. Manley for help in setting up the PALM microscope; B. Baird and D. Holowka for valuable discussions. S.L.V. was supported by US National Institutes of Health grant R00GM87810.
Footnotes
Note: Supplementary information is available on the Nature Methods website.
AUTHOR CONTRIBUTIONS
P.S. and T.J-T. conceived and designed experiments, developed experimental techniques, performed experiments, developed the analytical method, analyzed data and wrote the paper; D.S. helped develop the analytical method; M.R. contributed to characterization of PA-FP and imaging regime; S.L.V. contributed equations used to analyze data; J.L-S. conceived and designed the experiments, and wrote the paper.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Reprints and permissions information is available online at http://www.nature.com/reprints/index.html.
References
- 1.Kusumi A, et al. Paradigm shift of the plasma membrane concept from the two-dimensional continuum fluid to the partitioned fluid: High-speed single-molecule tracking of membrane molecules. Annu Rev Biophys Biomol Struct. 2005;34:351–378. doi: 10.1146/annurev.biophys.34.040204.144637. [DOI] [PubMed] [Google Scholar]
- 2.Eggeling C, et al. Direct observation of the nanoscale dynamics of membrane lipids in a living cell. Nature. 2009;457:1159–1162. doi: 10.1038/nature07596. [DOI] [PubMed] [Google Scholar]
- 3.Simons K, Gerl MJ. Revitalizing membrane rafts: new tools and insights. Nat Rev Mol Cell Biol. 2010;11:688–699. doi: 10.1038/nrm2977. [DOI] [PubMed] [Google Scholar]
- 4.Lingwood D, Simons K. Lipid rafts as a membrane-organizing principle. Science. 2010;327:46–50. doi: 10.1126/science.1174621. [DOI] [PubMed] [Google Scholar]
- 5.Lillemeier BF, Pfeiffer JR, Surviladze Z, Wilson BS, Davis MM. Plasma membrane-associated proteins are clustered into islands attached to the cytoskeleton. Proc Natl Acad Sci USA. 2006;103:18992–18997. doi: 10.1073/pnas.0609009103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prior IA, Muncke C, Parton RG, Hancock JF. Direct visualization of Ras proteins in spatially distinct cell surface microdomains. J Cell Biol. 2003;160:165–170. doi: 10.1083/jcb.200209091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Zanten TS, et al. Hotspots of GPI-anchored proteins and integrin nanoclusters function as nucleation sites for cell adhesion. Proc Natl Acad Sci USA. 2009;106:18557–18562. doi: 10.1073/pnas.0905217106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goswami D, et al. Nanoclusters of GPI-anchored proteins are formed by cortical actin-driven activity. Cell. 2008;135:1085–1097. doi: 10.1016/j.cell.2008.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Glebov OO, Nichols BJ. Lipid raft proteins have a random distribution during localized activation of the T-cell receptor. Nat Cell Biol. 2004;6:238–243. doi: 10.1038/ncb1103. [DOI] [PubMed] [Google Scholar]
- 10.Tanaka KAK, et al. Membrane molecules mobile even after chemical fixation. Nat Methods. 2010;7:865–866. doi: 10.1038/nmeth.f.314. [DOI] [PubMed] [Google Scholar]
- 11.Betzig E, et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 12.Hess ST, Girirajan TPK, Mason MD. Ultra-high resolution imaging by fluorescence photoactivation localization microscopy. Biophys J. 2006;91:4258–4272. doi: 10.1529/biophysj.106.091116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rust MJ, Bates M, Zhuang XW. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nat Methods. 2006;3:793–795. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Folling J, et al. Fluorescence nanoscopy by ground-state depletion and single-molecule return. Nat Methods. 2008;5:943–945. doi: 10.1038/nmeth.1257. [DOI] [PubMed] [Google Scholar]
- 15.Wombacher R, et al. Live-cell super-resolution imaging with trimethoprim conjugates. Nat Methods. 2010;7:717–719. doi: 10.1038/nmeth.1489. [DOI] [PubMed] [Google Scholar]
- 16.Lillemeier BF, et al. TCR and Lat are expressed on separate protein islands on T cell membranes and concatenate during activation. Nat Immunol. 2010;11:90–96. doi: 10.1038/ni.1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hess ST, et al. Dynamic clustered distribution of hemagglutinin resolved at 40 nm in living cell membranes discriminates between raft theories. Proc Natl Acad Sci USA. 2007;104:17370–17375. doi: 10.1073/pnas.0708066104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fuchs J, et al. A photoactivatable marker protein for pulse-chase imaging with superresolution. Nat Methods. 2010;7:627–630. doi: 10.1038/nmeth.1477. [DOI] [PubMed] [Google Scholar]
- 19.Manley S, et al. High-density mapping of single-molecule trajectories with photoactivated localization microscopy. Nat Methods. 2008;5:155–157. doi: 10.1038/nmeth.1176. [DOI] [PubMed] [Google Scholar]
- 20.Owen DM, et al. PALM imaging and cluster analysis of protein heterogeneity at the cell surface. J Biophotonics. 2010;3:446–454. doi: 10.1002/jbio.200900089. [DOI] [PubMed] [Google Scholar]
- 21.Veatch SL, et al. Critical fluctuations in plasma membrane vesicles. ACS Chem Biol. 2008;3:287–293. doi: 10.1021/cb800012x. [DOI] [PubMed] [Google Scholar]
- 22.Balagopalan L, Coussens NP, Sherman E, Samelson LE, Sommers CL. The LAT story: a tale of cooperativity, coordination, and choreography. Cold Spring Harbor Perspect Biol. 2010;2:a005512. doi: 10.1101/cshperspect.a005512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ingley E. Src family kinases: regulation of their activities, levels and identification of new pathways. BBA Proteins Proteomics. 2008;1784:56–65. doi: 10.1016/j.bbapap.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 24.Zagouras P, Rose JK. Dynamic equilibrium between vesicular stomatitis-virus glycoprotein monomers and trimers in the Golgi and at the cell-surface. J Virol. 1993;67:7533–7538. doi: 10.1128/jvi.67.12.7533-7538.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baumgart T, et al. Large-scale fluid/fluid phase separation of proteins and lipids in giant plasma membrane vesicles. Proc Natl Acad Sci USA. 2007;104:3165–3170. doi: 10.1073/pnas.0611357104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schutze S, Tchikov V, Schneider-Brachert W. Regulation of TNFR1 and CD95 signalling by receptor compartmentalization. Nat Rev Mol Cell Biol. 2008;9:655–662. doi: 10.1038/nrm2430. [DOI] [PubMed] [Google Scholar]
- 27.London M, London E. Ceramide selectively displaces cholesterol from ordered lipid domains (rafts)—implications for lipid raft structure and function. J Biol Chem. 2004;279:9997–10004. doi: 10.1074/jbc.M309992200. [DOI] [PubMed] [Google Scholar]
- 28.Romer W, et al. Shiga toxin induces tubular membrane invaginations for its uptake into cells. Nature. 2007;450:670–675. doi: 10.1038/nature05996. [DOI] [PubMed] [Google Scholar]
- 29.Lagerholm BC, Weinreb GE, Jacobson K, Thompson NL. Detecting microdomains in intact cell membranes. Annu Rev Phys Chem. 2005;56:309–336. doi: 10.1146/annurev.physchem.56.092503.141211. [DOI] [PubMed] [Google Scholar]
- 30.Batista FD, Treanor B, Harwood NE. Visualizing a role for the actin cytoskeleton in the regulation of B-cell activation. Immunol Rev. 2010;237:191–204. doi: 10.1111/j.1600-065X.2010.00943.x. [DOI] [PubMed] [Google Scholar]
- 31.Kenworthy AK, et al. Dynamics of putative raft-associated proteins at the cell surface. J Cell Biol. 2004;165:735–746. doi: 10.1083/jcb.200312170. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.