Abstract
Multifunctional proteins often appear to result from fusion of smaller proteins and in such cases typically can be separated into their ancestral components simply by cleaving the linker regions that separate the domains. Though possibly guided by sequence alignment, structural evidence, or light proteolysis, determination of the locations of linker regions remains empirical. We have developed an algorithm, named UMA, to predict the locations of linker regions in multifunctional proteins by quantification of the conservation of several properties within protein families, and the results agree well with structurally characterized proteins. This technique has been applied to a family of fungal type I iterative polyketide synthases (PKS), allowing prediction of the locations of all of the standard PKS domains, as well as two previously unidentified domains. Using these predictions, we report the cloning of the first fragment from the PKS norsolorinic acid synthase, responsible for biosynthesis of the first isolatable intermediate in aflatoxin production. The expression, light proteolysis and catalytic abilities of this acyl carrier protein–thioesterase didomain are discussed.
Keywords: polyketide synthase, multifunctional proteins, protein linkers, aflatoxin, domain prediction
Introduction
Many enzymes exist as multifunctional proteins, which are thought to be the product of gene fusion and/or duplication events. Their functions range from signaling,1 DNA polymerization, and primary metabolism,2,3 to the extremely large and complicated modular enzyme systems that synthesize polyketide4 and non-ribosomal peptide5,6 natural products commonly employed by microbes as antibiotics, siderophores or cell pigments. Motivated by intense interest from the natural product community, and the rapid pace of microbial genome sequencing projects, hundreds of such multifunctional genes have been identified. There now exists a great opportunity for the creation of new bioactive compounds through genetic manipulations of biosynthetic proteins.7,8
One of the greatest challenges in the study of the large proteins that these genes encode has been in obtaining crucial tertiary and quaternary structural information necessary for a thorough understanding of an enzyme’s synthetic reactions.9 Though discussed infrequently, the technical limitations of cloning large fragments of DNA in most commercial vectors or hosts are often nearly impossible to overcome. Furthermore, the large sizes of the resulting proteins make over-expression, functional purification and accurate examination of individual reactions difficult, and the protein may require further modification (glycosylation, phosphorylation, etc.) or the presence of co-factors that may not be readily available in the heterologous host. Finally, their crystallization has been said to be technically equivalent to co-crystallization of multiple proteins, and may be impossible with current technology.10
One approach that has produced significant results in recent years has been the “dissection” approach to understanding multifunctional proteins. Guided initially by the discovery that they could be divided into separate catalytic components by mild proteolysis,11,12 several research groups have employed a modern molecular biological equivalent by cloning, expressing and evaluating specific fragments of larger proteins for their catalytic abilities or structural properties.13 Genetic manipulation also allows the possibility of the insertion or deletion of specific catalytic functionalities into the original protein. Such an approach has led to the biosynthesis of “non-natural” polyketide natural products.4,14 The means by which individual protein chains interact to make large multisubunit enzymes has been probed for polyketide synthases (PKS),15 non-ribosomal peptide synthetases (NRPS)16 and animal fatty acid synthase (FAS).17 Cloning and expression of the adenylation domain of an NRPS has often been used to assay for the amino acid it activates.18,19 Recently, a crystal structure of an individually expressed adenylation domain of GrsA20 led to the development of a sequence comparison method to predict the amino acid residues activated by other adenylation domains using primary sequence data alone.21,22 Crystal or NMR structures are available for several PKS and FAS domains, such as the 6-deoxyerythronolide B synthase thioesterase (TE) domain23 and the actinorhodin acyl carrier protein (ACP) domain,24 providing more acute insight into their mode of action.
We report here a simple algorithm, named Udwary–Merski algorithm (UMA), which has greatly improved our ability to locate the flexible linker regions that occur typically between functional domains in multidomainal proteins. Employing widely used bioinformatics techniques, properties of a target protein are compared with other closely related proteins and a numerical value is assigned to each amino acid residue in the primary sequence relating to its tendency to form linker regions. We have observed that regions within the protein with low levels of conservation tend to correspond to experimentally determined linker regions, and that the predictions produced by UMA are broadly applicable and in good agreement with structurally well-characterized primary metabolic proteins. We then apply the algorithm to deduce the locations of linker regions of a structurally uncharacterized secondary metabolic protein currently of interest in our laboratory, norsolorinic acid (NA) synthase, a type I iterative fungal PKS from Aspergillus parasiticus required for aflatoxin biosynthesis.25 Finally, we discuss illustrative preliminary results from dissection of this protein into its constituent parts on the basis of the predictions made by UMA, and discuss the catalytic abilities of the first protein fragment resulting from the application of the algorithm, the ACP–TE didomain.
Results
Basis for UMA
Several assumptions were made about the fundamental nature of multifunctional proteins. According to the simple beads on a string model, one can consider indeterminate-length regions of the protein to be described by one of the two states: compact, independently folding, bioactive globular regions (domains),26 which are separated from one another by unstructured, flexible regions (linkers). Evolution of such an assembly could occur through simple gene fusions, and is commonly employed by molecular biologists in the construction of chimeric proteins with, for example, maltose-binding27 or polyhistidine metal-chelating regions28 which aid purification by affinity chromatography.
If loss of catalytic activity of the multidomain protein is detrimental to the survivability of the organism, then further assumptions about the natural evolution of the modular protein can be made. It can be expected that mutations in domain regions should, over time, be more strongly selected against than those in linker regions. This would be especially true at or near the active-site residues, and there should be fewer mutations that would significantly alter the domain’s secondary or tertiary structure, as both types of mutation would be detrimental to proper functioning of the protein. Furthermore, we could expect to observe localized pockets with high levels of hydrophobicity within domains, which would lead to hydrophobic collapse,29 allowing the domain to fold properly into its globular shape. Linker regions, on the other hand, should be present primarily to hold the domains in place. We can infer then that the linkers, if unnecessary for overall structure, should exist as relatively short chains of undefined structure, and not be required to maintain a conserved sequence (i.e. more tolerant of random mutation, insertion, or deletion without loss of function). They should have a tendency to be external to the globular fold of the domains, extending into solvent and providing some degree of physical separation between domains, and, therefore, should be more likely to maintain hydrophilic amino acid side-chains. Thus, if overall protein structure is expected to be largely conserved following an ancestral fusion event, then, over time, domains will have a tendency toward conserved sequence and secondary structure, and will contain more hydrophobic residues, while linkers will be less conserved in sequence and structure, and contain more hydrophilic residues.
The UMA algorithm was developed to compare the above properties specifically for a given set of homologous protein sequences. The final score generated quantifies the degree of conservation of each amino acid residue in the primary sequence, and is generated by well-established bioinformatics tools. The UMA score of each individual residue is calculated by combining the results of a multiple sequence alignment (MSA) of the protein in question and several closely related homologs with secondary-structure predictions and local hydrophobicity of these individual proteins. The method was designed to be flexible enough to accommodate the results of current and future predictive techniques. In practice, a low UMA score indicates a tendency of the amino acid at that alignment position to have properties consistent with a linker region, while a high score indicates that the region is conserved, and likely to be important for the structure or function of the protein.
Several well-accepted bioinformatics tools have been utilized to generate input data for the three components of the UMA calculation. The BLOSUM family of homology matrices was employed to define primary structure similarity,30 where BLO-SUM62 was used most frequently with good results. We modified secondary structure log-odds matrices31 to define similarity between secondary structures (Table 1). Our modifications to the matrix were made empirically to take into account gaps derived from the MSAs and low-confidence secondary structure predictions. The final matrix values used are given in Table 1. In addition, to lessen the effects of inaccurate structure predictions, the matrix-derived secondary structure score is multiplied by one-tenth of the integer confidence score given by many structure prediction routines, yielding an adjusted structure score. SOAP was employed to quantify hydrophobicity.32 MSAs were calculated using CLUSTAL W.33 It should be noted that CLUSTAL W is expected to be the most robust of the MSA methods, although other algorithms may be more suitable for a given problem.34 Our tests showed little or no difference utilizing slightly different matrices (such as BLOSUM45 and other modifications of the secondary structure matrix).
Table 1.
Modified secondary structure matrix
| Helix | Sheet | Loop | None | Gap | |
|---|---|---|---|---|---|
| Helix | 4 | −15 | −4 | −2 | −1 |
| Sheet | 8 | −4 | −2 | −1 | |
| Loop | 4 | −1 | −1 | ||
| None | −1 | −1 | |||
| Gap | −1 |
Algorithm
As discussed above, the propensity of an alignment position to be a linker or domain is dependent upon three properties: primary sequence similarity (A ), secondary structure similarity (B ), and hydrophobicity (C ). Therefore, an UMA score (S ) for an amino acid in a sequence may be considered:
Calculation of an UMA output requires both a multiple-sequence alignment and a secondary-structure assignment or prediction for each protein in the alignment. Secondary structure predictions are generated using any highly efficient method that can generate a structure, and (optionally) a confidence value for each residue. Every residue or gap (aa) is assigned a position i,j where j is the specific protein (ranging from 1 to N ) and i is the position within the MSA (ranging from 1 to Ω). A two-dimensional secondary structure array is created, on the basis of the positions of gaps in the primary structure alignment. α is the value obtained from (1) the primary structure homology matrix for any given pair of amino acid residues; (2) from specific gap values if a gap is aligned with an amino acid residue or another gap. Similarly, β is the score generated by the modified secondary structure matrix for a given pair of secondary structure elements (sse, which may include gaps or indeterminate structures). A, then, is the sum of all α (and B the sum of all β) for the target protein against all other proteins at position i:
Finally, C is simply a function that gives the hydrophobicity value assigned to each residue by SOAP.
To allow for adjustment and fine tuning of the contributions of each component of the prediction, a noise-dampening averaging window (Kα, Kβ, Kφ), and weighting values (Qα, Qβ, Qφ) for each component were introduced. For our investigations, only small K values and unity Q values were employed. Thus, the raw UMA score for each residue, T(i,j ), can be written as:
Since UMA was developed to detect large-scale structure within proteins, it was necessary to examine the environment around a given residue position, rather than at that position exclusively. Therefore, an averaging window was introduced, and the final UMA score is the calculated mean T(i,j ) over the window γ:
Noise present in the graphs of the final UMA score against alignment position is highly dependent upon homology of the sequences compared, alignment algorithm, quality of secondary structure assignment, and the K, Q, and γ variables, and so we have not yet found a thoroughly reliable means of automating the final selection of linker regions. Currently, linkers must be chosen by the user. In our hands, true linkers typically have minima in S(i,j ) at or below zero, and show up as somewhat broad, deep valleys on a graph of alignment position against S(i,j ). Specific examples are given in the following sections.
Probability model
Assuming that a given position i is unimportant for function (that is, under little or no selective pressure) and subject to natural base drift, after an infinite number of generations the probability of finding any given amino acid residue at that position will be equivalent to the natural frequency of that amino acid:35
Conversely, if that position is important for the structure or function of the protein, after an infinite number of generations there will be a bias in the amino acid distribution in the position that favors the residues that maintain the functional characteristics of the protein:
Finally, in order to fold properly, the globular functional domains will be likely to conserve a hydrophobic core to facilitate folding. Pi,j will reflect this propensity in these regions.
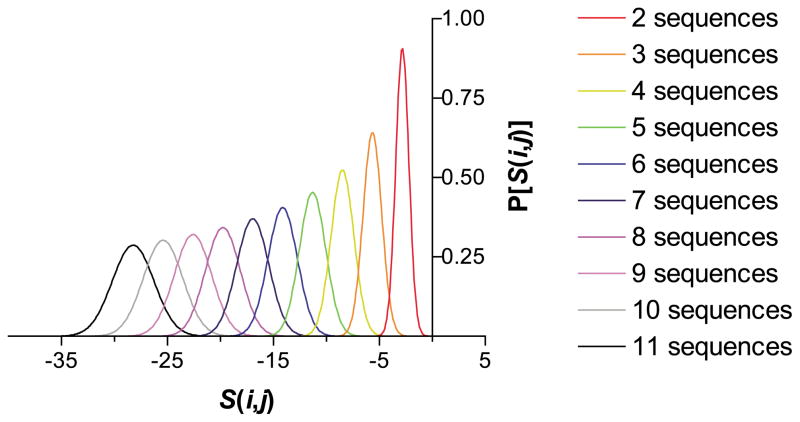
Homology matrices typically disfavor the possibility that a given pair of residues will be similar. UMA utilizes this property to distinguish highly similar domain regions from less similar linker regions. If five or more homologous sequences can be aligned, an S(i,j ) greater than zero will be exceedingly unlikely and therefore significant. Alignments of as few as two or three sequences may be used, but the result will be less reliable. Of course, the more homologous sequences that can be aligned, the better the prediction should be, as this will increase the maximum possible UMA score and decrease the baseline random score (Figure 1). In addition, transitions between low-scoring linker regions and high-scoring domains should be relatively shallow for transitions caused by random noise in the predictions. Transitions due to a string of sequential steps in which S(i,j ) increases continually should be statistically rare and, fortunately, visually apparent on a graph of S(i,j ) versus alignment position. (A mathematical proof of these assertions is available as Supplementary Material.)
Figure 1.
The estimated probability distributions of UMA scores S(i,j ), calculated with the BLO-SUM62 sequence homology matrix, and the secondary structure homology matrix from Table 1. The weighting factors are: Kα = Kβ = KC = 5, Qα = Qβ = QC = 1, and γ = 20. As the number of sequences included in the alignment increases, the most commonly occurring S(i,j ) generated from an alignment of unrelated sequences decreases, demonstrating the lack of conservation between these sequences.
As shown by Figure 1, the distributions for S(i,j ) drop off precipitously as more sequences are added to the alignment, demonstrating that a positive S(i,j ) is extremely unlikely to occur by random chance. Since the score distributions are expected to be Gaussian, the chance of generating a score S(i,j ) or higher can be calculated by the complementary error function.36,37 Thus, UMA should be exquisitely sensitive to differences in the degree of conservation between linker and domain regions present within a family of proteins. Furthermore, the empirically determined window sizes show that the method can determine the environment of an amino acid residue adequately and the window size is small enough to examine different regions within the protein.
Methionine synthase
Bacterial methionine synthases catalyze the methylation of homocysteine, utilizing methyltetrahydrofolate and cobalamin co-factors.38 Many homologous methionine synthases have been identified, most as a result of microbial genome sequencing projects. The Escherichia coli protein is one of the few multidomain proteins for which considerable structural information is available. This monomeric enzyme has been shown to consist of three functional units by trypsin digestion and functions for each domain have been ascribed.39 The 70 kDa N-terminal domain binds substrate homocysteine and methyltetrahydrofolate, and presumably catalyzes methyl transfer via the cobalamin co-factor bound to the second, 28 kDa domain. The third identified domain binds S-adenosylmethionine (S-AdoMet), and is important for the regeneration of oxidized cobalamin via reductive methylation.40 In addition, X-ray crystal structures for the central cobalamin-binding domain,41 the C-terminal S-AdoMet-binding domain,42 and 65 kDa of the C terminus encompassing both domains have been reported.2
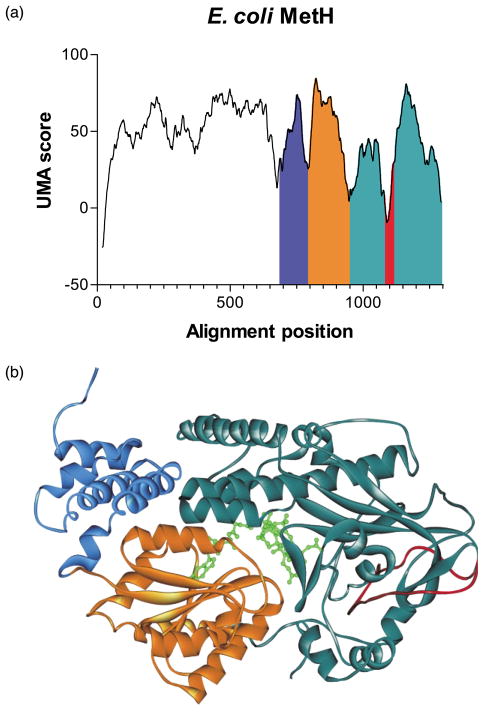
Given the aforementioned structural/functional elements, this enzyme family is ideal for testing the UMA program. Analysis was carried out on 14 prokaryotic sequences with significant homology to the E. coli protein, with all K and Q = 1, and γ = 20. Four separate low-scoring regions can be seen in Figure 2(a). Two of these corresponded perfectly to the locations of the known trypsinolysis cut sites previously used to separate the three domains in E. coli methionine synthase near Y638 (sequence alignment position (a.p.) 676 in Figure 2(a)) and H894 (a.p. 947).39 A third linker was predicted near Q742 (a.p. 794). In the crystal structure of the cobalamin-binding domain, a seven residue loop serves to divide two distinct structures that surround and secure the co-factor in place,41 and should be considered a linker by our definition. A trough in the UMA scores occurs near the middle of this binding domain, with the minimum located at E793 (a.p. 845), which identifies correctly the loop that separates these structural units (see Figure 2(b), border between blue and orange domains). A fourth linker was predicted near V1025 (a.p. 1089), in the middle of the S-AdoMet-binding domain (Figure 2(b), dark green domain). According to the available crystal structures, V1025 is part of a long extended coil (Figure 2(b), red loop) and sequence alignments and structure predictions show considerable variability within this region, properties we have deemed consistent with linkers, even though it appears not to be. This may reflect an ancestral fusion event that is otherwise undetectable by examination of the crystal structure, though further examination of the involvement of this coil in catalytic activity must be examined.
Figure 2.
(a) UMA prediction graph for E. coli MetH. (b) Crystal structure of the C-terminal domains of E. coli MetH.
DNA polymerase I
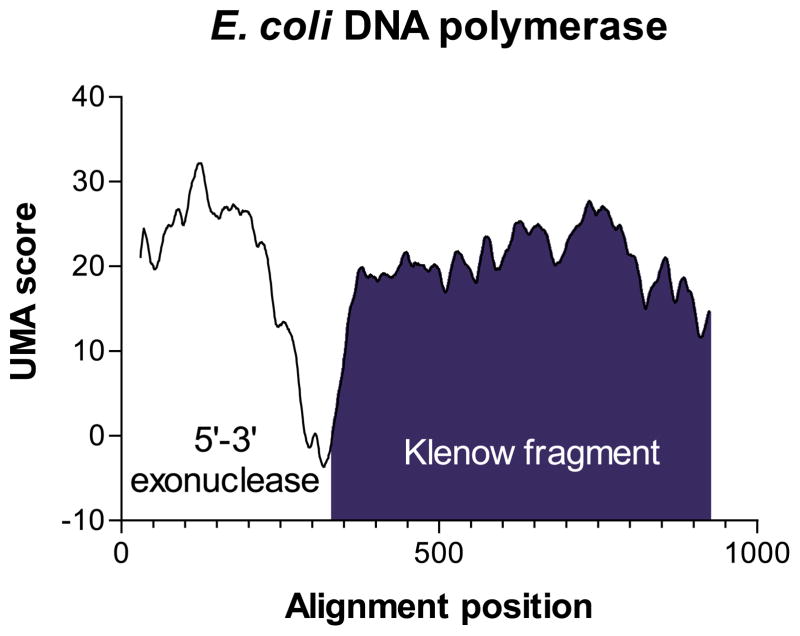
DNA polymerase I is one of the earliest identified and best understood multidomain proteins. Essential for repair of damaged DNA in bacteria, DNA polymerase I consists of two distinct subunits, a 5′–3′ exonuclease region, and a polymerization domain known as the Klenow fragment,12 which has been a workhorse of modern molecular biology.43 No structure of the entire DNA polymerase I molecule has been published, although each fragment has been crystallized separately.44–46 UMA analysis was carried out using five other bacterial sequences homologous to E. coli DNA polymerase I (E. coli AP002567, Acinetobacter calcoaceticus AF038541, Haemophilus influenzae U32767, Neisseria meningitidis AE002546, Salmonella typhimurium AF071212, Vibrio cholerae AE004101). In the calculations, all K = 5, all Q = 1, and γ = 30. From the resulting data, we were able to easily predict a single linker region (all scores <1), corresponding roughly to amino acid residues E283 (a.p. 287) through T323 (a.p. 333) in the E. coli protein. Our prediction is confirmed, as commercially available cloned Klenow fragment begins at position V324 of the wild-type protein (Figure 3, dark blue region), only one amino acid beyond the UMA score minimum of the predicted loop.
Figure 3.
UMA prediction graph for E. coli DNA polymerase I. The region shaded blue corresponds to the Klenow fragment.
ThiI
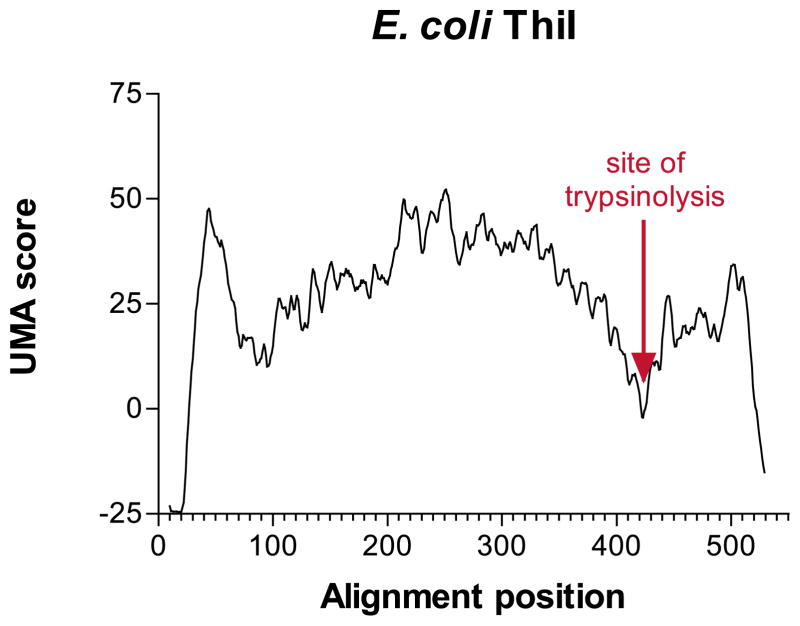
In bacteria, the thiI gene has been identified to be involved in the sulfur transfer step in biosynthesis of thiamin47 and 4-thiouridine,48 both necessary for bacterial tRNA formation. Currently, 23 known thiI homologs exist in GenBank. However, eight of these homologs have an additional C-terminal extension of approximately 100 amino acid residues with similarity to rhodanese,49 an enzyme used in sulfur transfer from thiosulfate to cyanide. Selective mutagenesis of all cysteine residues in E. coli ThiI suggests that this protein has evolved a mechanism by which an active-site cysteine residue in each domain forms a disulfide crosslink, and acts as the reactive species for sulfur transfer from a second enzyme, IscS.50 A protease protection experiment (with ATP) indicates that the dual-domain E. coli ThiI protein is cleaved readily by trypsinolysis to a 45 kDa fragment,51 and N-terminal sequencing of a second, smaller fragment showed that the proteolysis occurs at amino acid residue R385 (E. G. Mueller, personal communication). When the eight available dual-domain ThiI sequences were compared by UMA analysis (all K = Q = 1, and γ = 10), the presence of a linker region is indicated strongly by a sharp region of low UMA scores that can be seen over E. coli ThiI amino acid residues 370–390, with a minimum score at position 384 (Figure 4), only one amino acid residue different from the trypsinolysis result (Figure 4, as indicated). A second, lesser linker region was predicted near the N terminus of the protein, near residues 60–65, although this could not be confirmed.
Figure 4.
UMA prediction graph for E. coli ThiI with the known trypsin-preferred cut site as indicated.
Animal fatty acid synthase
In animal tissues, the fatty acid palmitate is synthesized by FAS, a homodimer consisting of identical 275 kDa subunits. Each polyprotein contains catalytic regions for β-ketoacyl synthase (KS), acyl/malonyl transferase (A/MT), dehydratase (DH), enoyl reductase (ER), ketoreductase (KR), ACP, and TE activities arrayed from the N to C terminus of each peptide. It has long been believed that the subunits are arranged head-to-tail, yielding two simultaneously active regions, with strong physical evidence to indicate that this is the case.17,52 However, other studies have indicated the possibility of a more complicated subunit interaction.53
The seminal work describing the architecture of animal FAS appeared as a series of four papers nearly 20 years ago,11,54–56 which described the results of extensive proteolysis experiments and active-site assays of the fragments retrieved from purified chicken liver FAS. Each peptide subunit is said to consist of three major domains separated by large, extensively proteolyzable regions (linker regions by our definition above). Domain I, 127 kDa in size, contains active-site regions for KS, A/MT, and DH.57 The ~107 kDa domain II contains functional units for KR, ER and ACP, and the 33 kDa TE domain constitutes domain III. In addition, domains II and I can be further proteolyzed. However, many of the proteases used in these experiments have amino acid sequence specificity requirements. It should be further noted that because FAS is a known homodimer, its quaternary structure might inhibit access of proteases to linkers. For these reasons, there have been conflicting reports of the locations of borders between domains, even from within the same research groups.58
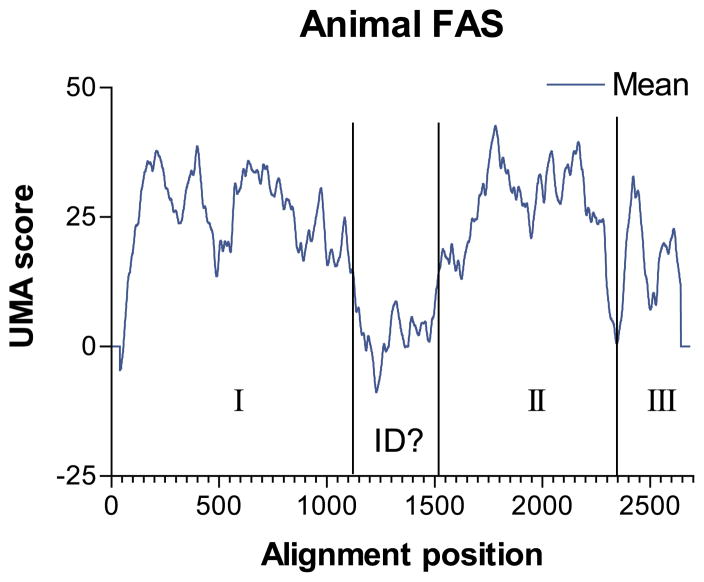
With these experiments in mind, we undertook an in silico dissection of animal FAS. UMA analysis was performed by an alignment of the seven available animal type I FASs: chicken (GenBank accession P12276), human (P49327), rat (P12785), mouse (AAG02285), and the closely related Drosophila melanogaster (AAF51148), Caenorhabditis elegans (NP_492417), and silkworm (T18201) homologs. Calculations were performed with all K = 5, all Q = 1, and γ = 40. The mean result (Figure 5, predictions for each organism are qualitatively identical) indicates the presence of two major linker regions. A C-terminal domain corresponding to the TE domain region (domain III) was separated from the remainder of the FAS by a linker (approximately A2192–P2224 in human FAS), and this domain was predicted to be 33 kDa, identical with that reported following proteolysis. A long stretch (~350 amino acid residues) of low-scoring residues, which falls within a proposed interdomain region separating domains I and II, has been shown to be necessary for dimerization and catalytic activity.17 It is probably unreasonable to expect a ~350 amino acid residue flexible linker region, although the specific structure of the protein here is unclear in Wakil & Chiu’s electron microscopy experiments.52 Instead, the consistently low scores over this part of the protein probably reflect the fact that the interdomain region is not specifically necessary for catalytic activity, and could readily mutate so long as the mutations did not weaken association of the homodimer significantly. Such a lack of conservation may indicate that some of the FASs may have different specific interactions at this domain.
Figure 5.
UMA prediction graph for human FAS. I, II, and III indicate Wakil’s domains I, II, and III, respectively. ID indicates the predicted linker-like interdomain region.
The prediction resulting from use of a smaller averaging value (γ = 15) did not increase the resolution of the prediction significantly, although further linkers can be predicted between the KS and A/MT domains (near R477 in the human FAS), and between the A/MT and DH domains (near P855) within domain I. The translated masses of domains between these predicted linkers appear to be in approximate agreement with the orginal mass determinations made by gel electrophoresis.11 Delineation of subdomains within domain II could not be discerned.
Norsolorinic acid synthase-class PKS
The biosynthesis of the hepatic carcinogen aflatoxin in the fungal genus Aspergillus begins with the construction of the polyketide NA. An unusual FAS-derived hexanoyl starter unit59 is taken up and extended by a type I iterative PKS.60 This is followed by successive oxidations and rearrangements of the NA carbon skeleton,61,62 ultimately yielding aflatoxin B1,25 which draws its toxicity from an easily oxidized bisfuran ring,63 and affinity for double-stranded DNA. While considerable progress has been made in recent years toward the understanding of the mechanisms of interaction of the more common type I modular PKSs,4 the iterative classes of type I PKSs are less well understood, and the means by which the polyketide chain length and cyclization pattern are controlled is a considerable mystery. Previous work in our laboratory has concentrated on the A. parasiticus version of the NA synthase PKS, and more specifically on its interaction with the dedicated hexanoyl-producing FAS subunits HexA and HexB.64,65
Seven other similar PKSs were identified by BLAST homology. The A. nidulans StcA protein is known to produce NA as well.66 Another A. nidulans gene, wA, has been investigated by Ebizuka and co-workers,67,68 and is known to produce the heptaketide naphthopyrone. In A. fumigatus, two nearly identical PKS sequences are present in GenBank, labeled as alb169 and pksP.70 In other fungal species, similar PKSs include the Colletotrichum lagenarium PKS1, known to produce tetrahydroxynaphthalene,71 pks4 of Giberella fujikoroi, identified to be involved in bikaverin biosynthesis (GenBank accessions CAB92399, CAC88775), and a melanin-producing PKS from a Nodulisporium species.72 By MSA, all of these PKSs are believed to have nearly identical domain organization, with identifiable active-site motifs for ketosynthase (KS), acyl transferase (AT), ACP and TE (sometimes referred to as Claisen cyclase, or CC) domains, with the exception that the G. fujikoroi and A. parasiticus PKSs have only one ACP domain motif, while the others have two. Calculations were carried out with all K = Q = 1, and γ = 20.
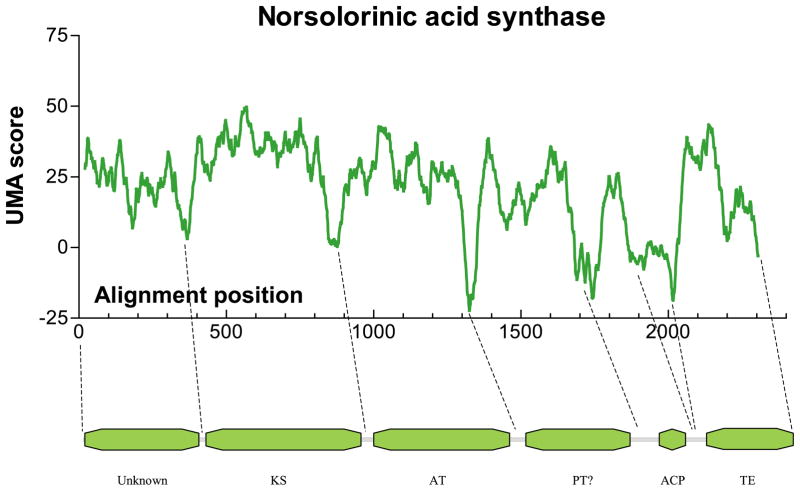
UMA predictions on each protein sequence were highly consistent except (as expected) in the region of the second ACP domain, and each was predicted to contain five (one ACP) or six (two ACP) linkers connecting six or seven domains, respectively. Active-site motifs were identified for the KS in the second domain (~50 kDa), AT in the third domain (~45 kDa), TE in the C-terminal domain (~28 kDa), and ~10 kDa ACP(s) were located adjacent to the TE domain (Figure 6). The UMA score minima (which should correspond to the locations of predicted linker regions) for each protein in this family are given in Table 2.
Figure 6.
UMA prediction graph for A. parasiticus NA synthase PKS with corresponding predicted domains.
Table 2.
UMA score minima for NA synthase family PKSs
| Protein | NT–KS | KS–AT | AT–PT | PT–ACP | ACP1–ACP2 | ACP–TE |
|---|---|---|---|---|---|---|
| A. parasiticus | I341 | A829 | E1287 | K1666 | – | P1841 |
| A. nidulans stcA | P361 | N831 | D1273 | S1669 | R1807 | T1931 |
| A. nidulans wA | L344 | A833 | A1279 | A1626 | P1751 | S1876 |
| A. fumigatus | R345 | A832 | A1279 | G1634 | S1750 | S1872 |
| C. lagenarium | R359 | N843 | L1283 | K1637 | S1769 | P1899 |
| G. fujikoroi | R321 | A804 | V1253 | V1600 | – | S1746 |
| Nodulisporium | G337 | A839 | K1283 | K1631 | S1763 | T1884 |
Two predicted domains could not be identified. The ~38 kDa N-terminal domain contained a low level of overall sequence similarity, with no conserved identifiable motifs or homology, PKS or otherwise. However, secondary structure predictions and local hydrophobicity scores were surprisingly similar. With no apparent catalytic residues or active site, and no previously reported function, yet having clear structural conservation, we speculate that the N-terminal domain may be important for tertiary or quaternary structure, possibly used as a multimerization domain as observed in animal FAS.17 Although this prediction cannot be confirmed, attempts to express this domain in E. coli are underway.
The fourth domain, ~34 kDa in size, we initially attributed to a structural domain as well, due to its central location, as observed in animal FAS. However, upon closer examination, specific motifs can be observed, including VNHGWDS located at V1567, and GHXVXGX5PS at G1344. The domain is not known by BLAST homology to be present in any other protein sequences, and has no significant homology to any known proteins outside of this family of PKSs. Recent work by Ebizuka and co-workers73 may contain a clue as to its function. A chimeric protein, containing portions of the wA gene and the C. lagenarium pks1 produced polyketide products distinct from either original PKS. Although specific primer locations used for the construction of the chimera are not given, a figure shows that the fusion occurs somewhere in this uncharacterized domain. Therefore, we must conclude that this predicted domain is responsible, at least in part, for the chain length of the polyketide product, and may have a role in control of cyclization. We have tentatively labeled this domain as a “product template domain” (PT), to differentiate it from previously known chain length factor (CLF) proteins.74
To evaluate one of our predictions experimentally, we cloned and expressed a fragment of NA synthase containing both the ACP and TE domains. Previous work has confirmed the TE activity of the C-terminal domain in the homologous wA protein,75 and by expressing both domains we anticipated the opportunity to probe for structure by light proteolysis. The TA1 gene fragment, beginning at P1664 and continuing through to the C-terminal end of the gene, was ligated into pET28a and expressed with an N-terminal His6 fusion in E. coli by standard techniques. The resulting 52 kDa peptide was soluble when expressed at 20 °C, and was purified easily by nickel or cobalt resin chromatography.
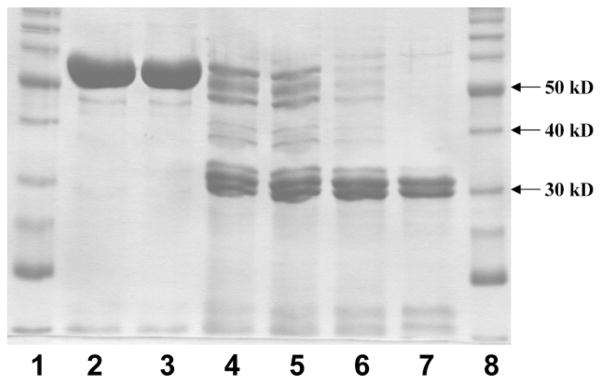
The purified protein was subjected to partial proteolysis with the fungal protease, proteinase K. Analysis by denaturing polyacrylamide gel electrophoresis (SDS-PAGE) showed that after ten minutes at ambient temperature, the peptide was fragmented into three bands appearing at roughly 30 kDa, 31 kDa and 32 kDa (Figure 7). N-terminal sequencing of these bands showed that the cuts had occurred at S1818, E1825, and E1832, all within the predicted linker region between the ACP and TE domains, which extends roughly from D1789 to P1851. A smaller ~15 kDa fragment, which we expect to correspond to the His6 tag and ACP domain, was visible on the gels, but could not be sequenced.
Figure 7.
Light proteolysis of TA1 by proteinase K. Lanes: (1) Benchmark protein ladder (Invitrogen); (2) 0 minute, no proteinase K; (3) 0 minute, added proteinase K; (4) one minute; (5) two minutes; (6) five minutes; (7) ten minutes; (8) Benchmark protein ladder.
In addition, the ACP–TE didomain cleaved thioesters readily. An assay method for the DEBS TE domain was adapted for this purpose.13 While the DEBS TE is involved in macrolactonization in erythromycin biosynthesis,23 and the NA synthase TE likely mediates Claisen cyclization,75 both enzymes catalyze TE reactions. Aliquots of the reaction mixture were removed at fixed time-points, the enzyme denatured with urea, Ellman’s reagent (5,5′-dithio-bis(2-nitrobenzoic acid, or DTNB) added, and the extent of reaction monitored by measuring the absorbance at 412 nm. Under these conditions, TA1 cleaved benzoyl N-acetyl cysteamine (benzSNAC) with KM = 1.9 mM, kcat = 0.84 s−1, with a resulting V/K = 0.44 s−1mM−1. By comparison, the most active substrate used in the DEBS TE assay reportedly hydrolyzed with a similar KM = 0.8 mM, but much lower kcat = 0.012 s−1. Other SNAC thioesters could be cleaved by TA1 with similar efficiency, although rigorous kinetic measurements were not taken. Unfortunately, the inherent difficulty in synthesizing the presumed natural substrate for the NA synthase TE precludes its use in these experiments. However, we anticipate further experimentation to probe for Claisen cyclization activity in this domain.75
It is important to note that the structural predictions given above are applicable to the other members of this family of type I iterative PKSs. The overall conserved sequence and presence of motifs leaves little doubt that domain structures in all of these PKSs are identical (with the exception of the lack of second ACP domain in those discussed above). If a more specific engineered approach can be made to swapping or mutation of PKS domains, it should be possible to tailor these systems to make biosynthetically novel fused aromatic products. It will be interesting to study the specific roles of the unknown domains, and gain a better understanding of the construction and control of the highly reactive poly β-keto chain.
Discussion
The above results have shown the UMA algorithm is able to predict the location of linker regions between folded domains accurately in nearly all of the protein families we examined and yielded insight into the type I iterative PKS NA synthase from A. parasiticus. The prediction has allowed us to identify the borders between all of the known catalytic functional units of this protein, and to propose the existence of two new structural units, including a previously unidentified domain that we believe to be responsible for chain-length determination and/or cyclization. Cloning and expression of the ACP–TE didomain by standard techniques in E. coli demonstrated full catalytic ability. In addition, with the application to other members of this family of proteins, we have opened up the possibility to perform specific bio-synthetic engineering experiments by altering, deleting or swapping domains between members of this family of PKS, much as has been attempted for type I modular PKSs and NRPSs.4,76
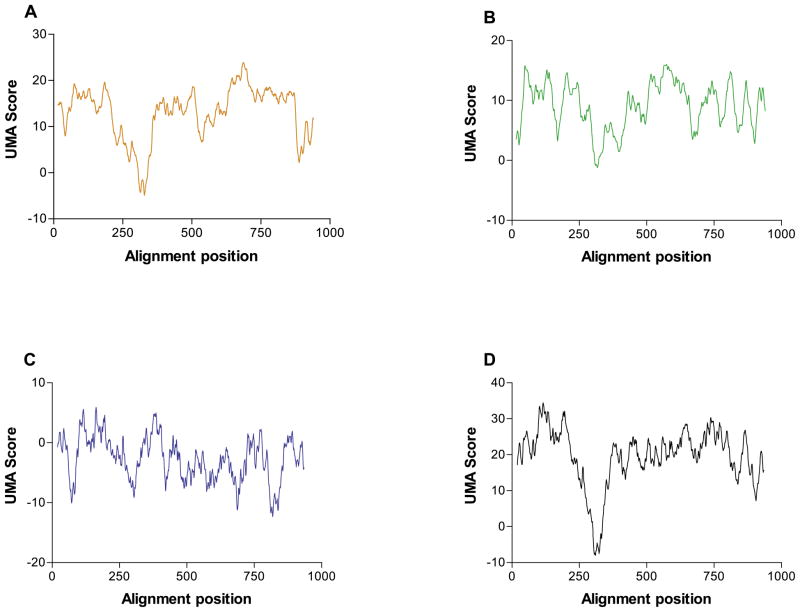
The UMA algorithm provided better, more quantifiable predictions in our hands than sequence alignments alone. When the three individual components of the prediction, sequence similarity, secondary structure prediction similarity, and local hydrophobicity, were examined, we found that the final, combined score was more accurate and consistent than any one or combination of two of the components alone. To illustrate this point, the components of the prediction for E. coli DNA polymerase I are shown in Figure 8. While the major contributor to the final score is clearly the primary sequence alignment (Figure 8(a)), we see that if only it is taken into account, linkers might also be predicted near alignment positions 530 and 880. The secondary structure (Figure 8(b)) and hydrophobicity (Figure 8(c)) profiles also give results individually inconsistent with the structure of the actual protein. It is only once all three components are combined by UMA (Figure 8(d)), however, that a good prediction is achieved; that of a single linker region near alignment position 320 (corresponding to E316).
Figure 8.
Components of the prediction of E. coli DNA polymerase I. (a) The sequence similarity component: Qα = 1, Qβ = QC = 0; (b) secondary structure component: Qα = QC = 0, Qβ = 1; (c) hydrophobicity component: Qα = Qβ = 0, QC = 1; (d) final score Qα = Qβ = QC = 1.
Because the UMA algorithm itself was designed to be independent of the programs used to generate its input, we were able to rapidly test many different alignment methods and structure prediction programs. As new methods are developed or new sources of structural information become available (for example, crystal structures, new alignment techniques, more accurate structure prediction routines),77 they can easily be harnessed to generate better predictions. Finally, because UMA is essentially making a structure prediction for an entire class of proteins, it should be trivial to apply the prediction to any newly discovered members of that class simply by sequence alignment. Because secondary structure (and by extension, tertiary structure) should almost certainly be conserved among members of a protein family,78 the locations of linker regions in the protein chain will be conserved, though they may be shortened, lengthened or otherwise modified because of their susceptibility to mutation.
Despite the successes reported above, we have encountered limitations in the predictive abilities of the algorithm. As with any comparative method, the availability of sequences with significant similarity to the target is of paramount importance. Although we developed the technique primarily for use in engineering of natural product-producing enzymes, availability of sequence is still a major technical hurdle, and for that reason the algorithm may currently be more applicable to examination of gene products involved in prokaryotic primary metabolism. However, one can be confident that more natural product gene sequences and clusters will continue to be explored and, therefore, more and more accurate predictions can be made as homologous sequences are discovered. A second limitation is that the sequences must be aligned accurately to enable the comparative routines of the algorithm to yield reliable results. In some cases, it was necessary to correct misalignments by hand, especially when working with sets of large modular proteins in which a particular domain in a sequence may be missing or inserted. While we have largely utilized the Clustal family of programs for our sequence algorithms with good results, it may be more appropriate in some cases to use a different method for specific alignment tasks.34 The Dialign MSA program79 has been shown to be more useful for modular proteins, though we found that it produced rigid “pockets” of alignment, and grossly misaligned linker regions, unlike Clustal. Ultimately, any sequence alignment should be evaluated by the user for accuracy.80
There has been much discussion of the potential for creation of new bioactive compounds by the manipulation of natural product-producing proteins.4 However, few truly successful examples of such techniques exist. Arguably, the most severe limitation to accomplishing this goal has been an understanding of the structure of these large proteins and how they interact, and thus how to take them apart and put them back together with different catalytic abilities. Requiring only primary sequence data, the UMA algorithm should prove a valuable tool for gaining insight into the structural properties of multifunctional proteins without necessitating the laborious processes of cloning, expression, purification and proteolysis of extremely large proteins.
Materials and Methods
Calculations
The UMA program was written entirely in the Perl 5 programming language. All calculations were performed on standard Windows 98 or Unix-based computers, and computation times were typically less than one minute. All calculations were performed using the following method: sequences homologous to the target protein were retrieved by analysis with BLASTp81 and DART,82 and a PIR-formatted MSA was generated using CLUSTAL W33 or with DIALIGN79 as indicated. A secondary structure prediction for each protein sequence in the MSA was generated with the PHDsec program available through the PredictProtein server.83,84 A custom Perl script was applied to each prediction to extract only the prediction and confidence values, and to generate a standard secondary structure file readable by UMA. Multiple rounds of calculation were performed, adjusting the K, Q, and γ evaluation constants as necessary in order to enhance the signal to noise ratio. A Perl/ Tk-based graphical analysis tool, and the program source code are available from the authors upon request.
Cloning, expression and purification of NA synthase domains
The locations of linker regions were identified in the NA synthase PKS using the UMA algorithm. PCR primers for use against the known sequence (GenBank L42766) were synthesized with overhanging restriction sites suitable for ligation into pET28 vectors (Novagen). The resulting PCR products were initially ligated into a high-copy vector for screening and sequencing, pT7Blue3 from the Perfectly Blunt cloning kit (Novagen) or pCR2.1-TOPO (Invitrogen). Inserts were digested with suitable restriction enzymes and purified with a Qiaquick gel purification kit (Qiagen), followed by ligation into pET28a(+) so as to allow expression of the resulting protein with an N-terminal His6 fusion.
Expression was performed under standard low-temperature conditions. Cultures were inoculated in LB supplemented with 25 μM kanamycin and incubated with shaking at 300 rpm and 37 °C to A600 = 0.25. Cultures were then placed at 20 °C for two hour, after which time expression was induced by the addition of IPTG to 0.5 mM, and shaking for 12 hours. Cells were harvested by centrifugation at 5000 g, sonicated, and soluble His-tagged proteins were purified with a Ni or Co resin by standard chromatographic techniques.28
Proteolytic digest of the ACP–TE didomain
TA1 protein, consisting of the 466 residues of the C terminus of the NA synthase PKS encompassing the predicted ACP and TE domains was expressed with an N-terminal His6-tag and purified as described above. TA1 protein (320 μg) was treated with 6.4 μg of proteinase K (Fisher) at room temperature, in 200 μl of 50 mM phosphate buffer (pH 8). Samples (40 μl) were removed after zero, one, two, five and ten minutes, and precipitated immediately by the addition of 40 μl of 20% (w/v) trichloroacetic acid. The samples were redissolved in SDS-PAGE sample buffer (125 mM Tris–HCl (pH 6.75), 20% (v/v) glycerol, 10% (v/v) β-mercaptoethanol, 4% (w/v) SDS, 0.02% (w/v) bromphenol blue), pH adjusted with 3 μl of 1 M Tris (pH 8.5), and analyzed by SDS-10% PAGE electrophoresis. N-terminal protein sequencing of the resulting fragments was performed by the Synthesis and Sequencing Facility, Department of Biological Chemistry, The Johns Hopkins School of Medicine (Baltimore, MD).
Catalytic activity of the ACP–TE didomain
The catalytic activity of the ACP–TE didomain was evaluated in a manner similar that described:13 50 μl of purified TA1 protein (10 μM) was mixed with 190 μl of 50 mM potassium phosphate (pH 7.5) and 10 μl of benzoyl N-acetylcysteamine (benzSNAC, 2.5–250 mM in DMSO). Aliquots (50 μl) were removed at two, five, ten and 15 minute intervals and mixed immediately with 50 μl of saturated aqueous urea and placed on ice. After centrifugation for five minutes (16,000 g, 4 °C), 75 μl of supernatant was removed and 4 μl of saturated DTNB was added. Absorbance at 412 nm was measured after 15 minutes at room temperature. The kinetic constants were obtained by fitting the data with the program HYPER.85
Supplementary Material
Acknowledgments
Thanks are due to Professor Eugene G. Mueller (University of Delaware) for providing proteolytic data on the ThiI protein, and to Professor Burkhard Rost (Columbia University) for answering questions concerning the PhD program. We thank Professor Douglas C. Poland, Dr Timothy S. Hitchman, Dr Jason D. Galpin, Daniel P. Grilley, Gregory Lopez, and Katherine I. Miller for stimulating discussions, and Professor David E. Draper and Professor Neil D. Clarke for reading the manuscript. We are grateful to the National Institutes of Health for financial support (NIH ES01670).
Abbreviations used
- UMA
Udwary–Merski algorithm
- NRPS
non-ribosomal peptide synthetases
- FAS
fatty acid synthase
- PKS
polyketide synthase
- KS
ketoacyl synthase
- A/MT
acyl/malonyl transferase
- DH
dehydratase
- ER
enoyl reductase
- KR
ketoreductase
- NA
norsolorinic acid
- ACP
acyl carrier protein
- TE
thioesterase
- S-AdoMet
S-adenosylmethionine
- a.p
sequence alignment position
- PT
product template domain
- CLF
chain length factor
- MSA
multiple sequence alignment
Footnotes
Supplementary Material for this paper comprising proof of the validity of scores generated by UMA is available on IDEAL
References
- 1.Cohen GB, Ren R, Baltimore D. Modular binding domains in signal transduction proteins. Cell. 1995;80:237–248. doi: 10.1016/0092-8674(95)90406-9. [DOI] [PubMed] [Google Scholar]
- 2.Bandarian V, Pattridge KA, Lennon BW, Huddler DP, Matthews RG, Ludwig ML. Domain alternation switches B(12)-dependent methionine synthase to the activation conformation. Nature Struct Biol. 2002;9:53–56. doi: 10.1038/nsb738. [DOI] [PubMed] [Google Scholar]
- 3.Thoden JB, Miran SG, Phillips JC, Howard AJ, Raushel FM, Holden HM. Carbamoyl phosphate synthetase: caught in the act of glutamine hydrolysis. Biochemistry. 1998;37:8825–8831. doi: 10.1021/bi9807761. [DOI] [PubMed] [Google Scholar]
- 4.Staunton J, Weissman KJ. Polyketide biosynthesis: a millennium review. Nature Prod Rep. 2001;18:380–416. doi: 10.1039/a909079g. [DOI] [PubMed] [Google Scholar]
- 5.Marahiel MA, Stachelhaus T, Mootz HD. Modular peptide synthetases involved in nonribosomal peptide synthesis. Chem Rev. 1997;97:2651–2674. doi: 10.1021/cr960029e. [DOI] [PubMed] [Google Scholar]
- 6.von Dohren H, Keller U, Vater J, Zocher R. Multifunctional peptide synthetases. Chem Rev. 1997;97:2675–2706. doi: 10.1021/cr9600262. [DOI] [PubMed] [Google Scholar]
- 7.Walsh CT. Combinatorial biosynthesis of antibiotics: challenges and opportunities. ChemBio-Chem. 2002;3:124–134. doi: 10.1002/1439-7633(20020301)3:2/3<124::AID-CBIC124>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 8.Khosla C. Natural product biosynthesis: a new interface between enzymology and medicine. J Org Chem. 2000;65:8127–8133. doi: 10.1021/jo000849y. [DOI] [PubMed] [Google Scholar]
- 9.Townsend CA. Structural studies of natural product biosynthetic proteins. Chem Biol. 1997;4:721–730. doi: 10.1016/s1074-5521(97)90310-0. [DOI] [PubMed] [Google Scholar]
- 10.Gewolb J. Bioengineering. Working outside the protein-synthesis rules. Science. 2002;295:2205–2207. doi: 10.1126/science.295.5563.2205. [DOI] [PubMed] [Google Scholar]
- 11.Mattick JS, Tsukamoto Y, Nickless J, Wakil SJ. The architecture of the animal fatty acid synthetase. I Proteolytic dissection and peptide mapping. J Biol Chem. 1983;258:15291–15299. [PubMed] [Google Scholar]
- 12.Klenow H, Hennigsen I. Selective elimination of the exonuclease activity of the deoxyribonucleic acid polymerase from Escherichia coli B by limited proteolysis. Proc Natl Acad Sci USA. 1970;65:168. doi: 10.1073/pnas.65.1.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gokhale RS, Hunziker D, Cane DE, Khosla C. Mechanism and specificity of the terminal thioesterase domain from the erythromycin polyketide synthase. Chem Biol. 1999;6:117–125. doi: 10.1016/S1074-5521(99)80008-8. [DOI] [PubMed] [Google Scholar]
- 14.Yoon YJ, Beck BJ, Kim BS, Kang HY, Reynolds KA, Sherman DH. Generation of multiple bioactive macrolides by hybrid modular polyketide synthases in Streptomyces venezuelae. Chem Biol. 2002;9:203–214. doi: 10.1016/s1074-5521(02)00095-9. [DOI] [PubMed] [Google Scholar]
- 15.Tsuji SY, Cane DE, Khosla C. Selective protein–protein interactions direct channeling of intermediates between polyketide synthase modules. Biochemistry. 2001;40:2326–2331. doi: 10.1021/bi002463n. [DOI] [PubMed] [Google Scholar]
- 16.Linne U, Marahiel MA. Control of directionality in nonribosomal peptide synthesis: role of the condensation domain in preventing misinitiation and timing of epimerization. Biochemistry. 2000;39:10439–10447. doi: 10.1021/bi000768w. [DOI] [PubMed] [Google Scholar]
- 17.Chirala SS, Jayakumar A, Gu ZW, Wakil SJ. Human fatty acid synthase: role of inter-domain in the formation of catalytically active synthase dimer. Proc Natl Acad Sci USA. 2001;98:3104–3108. doi: 10.1073/pnas.051635998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kleinkauf H, von Dohren H. Nonribosomal biosynthesis of peptide antibiotics. Eur J Biochem. 1990;192:1–15. doi: 10.1111/j.1432-1033.1990.tb19188.x. [DOI] [PubMed] [Google Scholar]
- 19.Weber T, Marahiel MA. Exploring the domain structure of modular nonribosomal peptide synthetases. Structure. 2001;9:R3–R9. doi: 10.1016/s0969-2126(00)00560-8. [DOI] [PubMed] [Google Scholar]
- 20.Conti E, Stachelhaus T, Marahiel MA, Brick P. Structural basis for the activation of phenylalanine in the non-ribosomal biosynthesis of gramicidin S. EMBO J. 1997;16:4174–4183. doi: 10.1093/emboj/16.14.4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Challis GL, Ravel J, Townsend CA. Predictive, structure-based model of amino acid recognition by nonribosomal peptide synthetase adenylation domains. Chem Biol. 2000;7:211–224. doi: 10.1016/s1074-5521(00)00091-0. [DOI] [PubMed] [Google Scholar]
- 22.Stachelhaus T, Mootz HD, Marahiel MA. The specificity-conferring code of adenylation domains in nonribosomal peptide synthetases. Chem Biol. 1999;6:493–505. doi: 10.1016/S1074-5521(99)80082-9. [DOI] [PubMed] [Google Scholar]
- 23.Tsai SC, Miercke LJ, Krucinski J, Gokhale R, Chen JC, Foster PG, et al. Crystal structure of the macrocycle-forming thioesterase domain of the erythromycin polyketide synthase: versatility from a unique substrate channel. Proc Natl Acad Sci USA. 2001;98:14808–14813. doi: 10.1073/pnas.011399198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crump MP, Crosby J, Dempsey CE, Parkinson JA, Murray M, Hopwood DA, Simpson TJ. Solution structure of the actinorhodin polyketide synthase acyl carrier protein from Streptomyces coelicolor A3(2) Biochemistry. 1997;36:6000–6008. doi: 10.1021/bi970006+. [DOI] [PubMed] [Google Scholar]
- 25.Minto RE, Townsend CA. Enzymology and molecular biology of aflatoxin biosynthesis. Chem Rev. 1997;97:2537–2555. doi: 10.1021/cr960032y. [DOI] [PubMed] [Google Scholar]
- 26.Rashin AA. Location of domains in globular proteins. Nature. 1981;291:85–87. doi: 10.1038/291085a0. [DOI] [PubMed] [Google Scholar]
- 27.di Guan C, Li P, Riggs PD, Inouye H. Vectors that facilitate the expression and purification of foreign peptides in Escherichia coli by fusion to maltose-binding protein. Gene. 1988;67:21–30. doi: 10.1016/0378-1119(88)90004-2. [DOI] [PubMed] [Google Scholar]
- 28.Smith MC, Furman TC, Ingolia TD, Pidgeon C. Chelating peptide-immobilized metal ion affinity chromatography. A new concept in affinity chromatography for recombinant proteins. J Biol Chem. 1988;263:7211–7215. [PubMed] [Google Scholar]
- 29.Dill KA. Dominant forces in protein folding. Biochemistry. 1990;29:7133–7155. doi: 10.1021/bi00483a001. [DOI] [PubMed] [Google Scholar]
- 30.Henikoff S, Henikoff JG. Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci USA. 1992;89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wallqvist A, Fukunishi Y, Murphy LR, Fadel A, Levy RM. Iterative sequence/secondary structure search for protein homologs: comparison with amino acid sequence alignments and application to fold recognition in genome databases. Bioinformatics. 2000;16:988–1002. doi: 10.1093/bioinformatics/16.11.988. [DOI] [PubMed] [Google Scholar]
- 32.Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 33.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucl Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thompson JD, Plewniak F, Poch O. A comprehensive comparison of multiple sequence alignment programs. Nucl Acids Res. 1999;27:2682–2690. doi: 10.1093/nar/27.13.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Voet D, Voet J. Biochemistry. Wiley; New York: 1995. [Google Scholar]
- 36.Goldman M. Introduction to Probability and Statistics. Harcourt, Brace & World; New York: 1970. [Google Scholar]
- 37.Maksoudian YL. Probability and Statistics with Applications. International Textbook Company; Scranton, PA: 1969. [Google Scholar]
- 38.Matthews RG. Cobalamin-dependent methionine synthase. In: Banerjee R, editor. Chemistry and Biochemistry of B12. Wiley; New York: 1999. pp. 681–706. [Google Scholar]
- 39.Drummond JT, Loo RR, Matthews RG. Electrospray mass spectrometric analysis of the domains of a large enzyme: observation of the occupied cobalamin-binding domain and redefinition of the carboxyl terminus of methionine synthase. Biochemistry. 1993;32:9282–9289. doi: 10.1021/bi00087a004. [DOI] [PubMed] [Google Scholar]
- 40.Goulding CW, Postigo D, Matthews RG. Cobalamin-dependent methionine synthase is a modular protein with distinct regions for binding homocysteine, methyltetrahydrofolate, cobalamin, and adenosylmethionine. Biochemistry. 1997;36:8082–8091. doi: 10.1021/bi9705164. [DOI] [PubMed] [Google Scholar]
- 41.Drennan CL, Huang S, Drummond JT, Matthews RG, Lidwig ML. How a protein binds B12: a 3.0 Å X-ray structure of B12-binding domains of methionine synthase. Science. 1994;266:1669–1674. doi: 10.1126/science.7992050. [DOI] [PubMed] [Google Scholar]
- 42.Dixon MM, Huang S, Matthews RG, Ludwig M. The structure of the C-terminal domain of methionine synthase: presenting S-adenosylmethionine for reductive methylation of B12. Structure. 1996;4:1263–1275. doi: 10.1016/s0969-2126(96)00135-9. [DOI] [PubMed] [Google Scholar]
- 43.Sambrook J, Fristch E, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1989. [Google Scholar]
- 44.Brautigam CA, Steitz TA. Structural principles for the inhibition of the 3′–5′ exonuclease activity of Escherichia coli DNA polymerase I by phosphorothioates. J Mol Biol. 1998;277:363–377. doi: 10.1006/jmbi.1997.1586. [DOI] [PubMed] [Google Scholar]
- 45.Ollis DL, Brick P, Hamlin R, Xuong NG, Steitz TA. Structure of large fragment of Escherichia coli DNA polymerase I complexed with dTMP. Nature. 1985;313:762–766. doi: 10.1038/313762a0. [DOI] [PubMed] [Google Scholar]
- 46.Beese LS, Derbyshire V, Steitz TA. Structure of DNA polymerase I Klenow fragment bound to duplex DNA. Science. 1993;260:352–355. doi: 10.1126/science.8469987. [DOI] [PubMed] [Google Scholar]
- 47.Mueller EG, Buck CJ, Palenchar PM, Barnhart LE, Paulson JL. Identification of a gene involved in the generation of 4-thiouridine in tRNA. Nucl Acids Res. 1998;26:2606–2610. doi: 10.1093/nar/26.11.2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Webb E, Claas K, Downs DM. Characterization of thiI, a new gene involved in thiazole biosynthesis in Salmonella typhimurium. J Bacteriol. 1997;179:4399–4402. doi: 10.1128/jb.179.13.4399-4402.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Palenchar PM, Buck CJ, Cheng H, Larson TJ, Mueller EG. Evidence that ThiI, an enzyme shared between thiamin and 4-thiouridine biosynthesis, may be a sulfurtransferase that proceeds through a persulfide intermediate. J Biol Chem. 2000;275:8283–8286. doi: 10.1074/jbc.275.12.8283. [DOI] [PubMed] [Google Scholar]
- 50.Mueller EG, Palenchar PM, Buck CJ. The role of the cysteine residues of ThiI in the generation of 4-thiouridine in tRNA. J Biol Chem. 2001;276:33588–33595. doi: 10.1074/jbc.M104067200. [DOI] [PubMed] [Google Scholar]
- 51.Kambampati R, Lauhon CT. Evidence for the transfer of sulfane sulfur from IscS to ThiI during the in vitro biosynthesis of 4-thiouridine in Escherichia coli tRNA. J Biol Chem. 2000;275:10727–10730. doi: 10.1074/jbc.275.15.10727. [DOI] [PubMed] [Google Scholar]
- 52.Brink J, Ludtke SJ, Yang CY, Gu ZW, Wakil SJ, Chiu W. Quaternary structure of human fatty acid synthase by electron cryo-microscopy. Proc Natl Acad Sci USA. 2002;99:138–143. doi: 10.1073/pnas.012589499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rangan VS, Joshi AK, Smith S. Mapping the functional topology of the animal fatty acid synthase by mutant complementation in vitro. Biochemistry. 2001;40:10792–10799. doi: 10.1021/bi015535z. [DOI] [PubMed] [Google Scholar]
- 54.Mattick JS, Nickless J, Mizugaki M, Yang CY, Uchiyama S, Wakil SJ. The architecture of the animal fatty acid synthetase. II Separation of the core and thioesterase functions and determination of the N–C orientation of the subunit. J Biol Chem. 1983;258:15300–15304. [PubMed] [Google Scholar]
- 55.Wong H, Mattick JS, Wakil SJ. The architecture of the animal fatty acid synthetase. III Isolation and characterization of beta-ketoacyl reductase. J Biol Chem. 1983;258:15305–15311. [PubMed] [Google Scholar]
- 56.Tsukamoto Y, Wong H, Mattick JS, Wakil SJ. The architecture of the animal fatty acid synthetase complex. IV Mapping of active centers and model for the mechanism of action. J Biol Chem. 1983;258:15312–15322. [PubMed] [Google Scholar]
- 57.Chirala SS, Huang WY, Jayakumar A, Sakai K, Wakil SJ. Animal fatty acid synthase: functional mapping and cloning and expression of the domain I constituent activities. Proc Natl Acad Sci USA. 1997;94:5588–5593. doi: 10.1073/pnas.94.11.5588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tsukamoto Y, Wakil SJ. Isolation and mapping of the beta-hydroxyacyl dehydratase activity of chicken liver fatty acid synthase. J Biol Chem. 1988;263:16225–16229. [PubMed] [Google Scholar]
- 59.Hitchman TS, Schmidt EW, Trail F, Rarick MD, Linz JE, Townsend CA. Hexanoate synthase, a specialized type I fatty acid synthase in aflatoxib B1 biosynthesis. Bioorg Chem. 2001;29:293–307. doi: 10.1006/bioo.2001.1216. [DOI] [PubMed] [Google Scholar]
- 60.Townsend CA, Christensen SB, Trautwein K. Hexanoate as a starter unit in polyketide biosynthesis. J Am Chem Soc. 1984;106:3868–3869. [Google Scholar]
- 61.Watanabe CMH, Townsend CA. The in vitro conversion of norsolorinic acid to aflatoxin B1. An improved method of cell-free enzyme preparation and stabilization. J Am Chem Soc. 1998;120:6231–6239. [Google Scholar]
- 62.Udwary DW, Casillas LK, Townsend CA. Synthesis of 11-hydroxyl O-methylsterigmatocystin and the role of a cytochrome P-450 in the final step of aflatoxin biosynthesis. J Am Chem Soc. 2002;124:5294–5303. doi: 10.1021/ja012185v. [DOI] [PubMed] [Google Scholar]
- 63.Johnston DS, Stone MP. Refined solution structure of 8,9-dihydro-8-(N7-guanyl)-9-hydroxyaflatoxin B1 opposite CpA in the complementary strand of an oligodeoxynucleotide duplex as determined by 1H NMR. Biochemistry. 1995;34:14037–14050. doi: 10.1021/bi00043a009. [DOI] [PubMed] [Google Scholar]
- 64.Watanabe CMH, Wilson D, Linz JE, Townsend CA. Demonstration of the catalytic roles and evidence for the physical association of type I fatty acid synthases and a polyketide synthase in the biosynthesis of aflatoxin B1. Chem Biol. 1996;3:463–469. doi: 10.1016/s1074-5521(96)90094-0. [DOI] [PubMed] [Google Scholar]
- 65.Watanabe CMH, Townsend CA. Initial characterization of a type I fatty acid synthase and polyketide synthase multienzyme complex NorS in the biosynthesis of aflatoxin B1. Chem Biol. 2002;9:981–988. doi: 10.1016/s1074-5521(02)00213-2. [DOI] [PubMed] [Google Scholar]
- 66.Brown DW, Yu JH, Kelkar HS, Fernandes M, Nesbitt TC, Keller NP, et al. Twenty-five co-regulated transcripts define a secondary metabolite gene cluster in Aspergillus nidulans. Proc Natl Acad Sci USA. 1996;93:1418–1422. doi: 10.1073/pnas.93.4.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Watanabe A, Ono Y, Fujii I, Sankawa U, Mayorga ME, Timberlake WE, Ebizuka Y. Product identification of polyketide synthase coded by Aspergillus nidulans wA gene. Tetrahedron Letters. 1998;39:7733–7736. [Google Scholar]
- 68.Watanabe A, Fujii I, Sankawa U, Mayorga ME, Timberlake WE, Ebizuka Y. Re-identification of Aspergillus nidulans wA gene to code for a polyketide synthase of naphthopyrone. Tetrahedron Letters. 1999;40:91–94. [Google Scholar]
- 69.Tsai HF, Chang YC, Washburn RG, Wheeler MH, Kwon-Chung KJ. The developmentally regulated alb1 gene of Aspergillus fumigatus: its role in modulation of conidial morphology and virulence. J Bacteriol. 1998;180:3031–3038. doi: 10.1128/jb.180.12.3031-3038.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Langfelder K, Jahn B, Gehringer H, Schmidt A, Wanner G, Brakhage AA. Identification of a polyketide synthase gene (pksP) of Aspergillus fumigatus involved in conidial pigment biosynthesis and virulence. Med Microbiol Immunol. 1998;187:79–89. doi: 10.1007/s004300050077. [DOI] [PubMed] [Google Scholar]
- 71.Feng B, Wang X, Hauser M, Kaufmann S, Jentsch S, Haase G, et al. Molecular cloning and characterization of WdPKS1, a gene involved in dihydroxynaphthalene melanin biosynthesis and virulence in Wangiella (Exophiala ) dermatitidis. Infect Immun. 2001;69:1781–1794. doi: 10.1128/IAI.69.3.1781-1794.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fulton TR, Ibrahim N, Losada MC, Grzegorski D, Tkacz JS. A melanin polyketide synthase (PKS) gene from Nodulisporium sp that shows homology to the pks1 gene of Colletotrichum lagenarium. Mol Gen Genet. 1999;262:714–720. doi: 10.1007/s004380051133. [DOI] [PubMed] [Google Scholar]
- 73.Watanabe A, Ebizuka Y. A novel hexaketide naphthalene synthesized by a chimeric polyketide synthase composed of fungal pentaketide and heptaketide synthases. Tetrahedron Letters. 2002;43:843–846. [Google Scholar]
- 74.Burson KK, Khosla C. Dissecting the chain length specificity in bacterial aromatic polyketide synthases using chimeric genes. Tetrahedron. 2000;56:940–9408. [Google Scholar]
- 75.Fujii I, Watanabe A, Sankawa U, Ebizuka Y. Identification of Claisen cyclase domain in fungal polyketide synthase WA, a naphthopyrone synthase of Aspergillus nidulans. Chem Biol. 2001;8:189–197. doi: 10.1016/s1074-5521(00)90068-1. [DOI] [PubMed] [Google Scholar]
- 76.Mootz HD, Schwarzer D, Marahiel MA. Construction of hybrid peptide synthetases by module and domain fusions. Proc Natl Acad Sci USA. 2000;97:5848–5853. doi: 10.1073/pnas.100075897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rost B. Review: protein secondary structure prediction continues to rise. J Struct Biol. 2001;134:204–218. doi: 10.1006/jsbi.2001.4336. [DOI] [PubMed] [Google Scholar]
- 78.Abagyan RA, Batalov S. Do aligned sequences share the same fold? J Mol Biol. 1997;273:355–368. doi: 10.1006/jmbi.1997.1287. [DOI] [PubMed] [Google Scholar]
- 79.Morgenstern B, Frech K, Dress A, Werner T. DIALIGN: finding local similarities by multiple sequence alignment. Bioinformatics. 1998;14:290–294. doi: 10.1093/bioinformatics/14.3.290. [DOI] [PubMed] [Google Scholar]
- 80.Rost B. Twilight zone of protein sequence alignments. Protein Eng. 1999;12:85–94. doi: 10.1093/protein/12.2.85. [DOI] [PubMed] [Google Scholar]
- 81.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 82.Marchler-Bauer A, Panchenko AR, Shoemaker BA, Thiessen PA, Geer LY, Bryant SH. CDD: a database of conserved domain alignments with links to domain three-dimensional structure. Nucl Acids Res. 2002;30:281–283. doi: 10.1093/nar/30.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rost B. Predicting one-dimensional protein structure by profile-based neural networks. Methods Enzymol. 1996;266:525–539. doi: 10.1016/s0076-6879(96)66033-9. [DOI] [PubMed] [Google Scholar]
- 84.Rost B, Sander C. Improved prediction of protein secondary structure by use of sequence profiles and neural networks. Proc Natl Acad Sci USA. 1993;90:7558–7562. doi: 10.1073/pnas.90.16.7558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cleland WW. Statistical analysis of enzyme kinetic data. Methods Enzymol. 1975;63:103–139. doi: 10.1016/0076-6879(79)63008-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.