Abstract
Background
Chronic inflammation is etiologically-related to several cancers. We evaluated the performance (ability to detect concentrations above the assay’s lower limit of detection, coefficients-of-variation [CVs], and intraclass correlation coefficients [ICCs]) of 116 inflammation, immune, and metabolic markers across two luminex bead-based commercial kits and three specimen types.
Methods
From 100 cancer-free participants in the Prostate, Lung, Colorectal, and Ovarian Cancer Trial, serum, heparin plasma, and EDTA plasma samples were utilized. We measured levels of 67 and 97 markers using Bio-Rad and Millipore kits, respectively. Reproducibility was assessed using 40 blinded duplicates (20 within-batches and 20 across-batches) for each specimen type.
Results
A majority of markers were detectable in >25% of individuals on all specimen types/kits. Of the 67 Bio-Rad markers, 51, 52, and 47 markers in serum, heparin plasma, and EDTA plasma, respectively, had across-batch CVs <20%. Likewise, of 97 Millipore markers, 75, 69, and 78 markers in serum, heparin plasma, and EDTA plasma, respectively, had across-batch CVs <20%. When results were combined across specimen types, 45 Bio-Rad and 71 Millipore markers had acceptable performance (>25% detectability on all 3 specimen types and across-batch CVs <20% on at least 2 of 3 specimen types). Median concentrations and ICCs differed to a small extent across specimen types and to a large extent between Bio-Rad and Millipore.
Conclusions
Inflammation and immune markers can be measured reliably in serum and plasma samples using multiplexed luminex-based methods.
Impact
Multiplexed assays can be utilized for epidemiologic investigations into the role of inflammation in cancer etiology.
Background
Chronic inflammation is now recognized as a major etiologic factor for a range of malignancies, including cancers of the esophagus, stomach, gall bladder, liver, pancreas, colon and rectum, prostate, urinary bladder, and lung (1–3). Chronic inflammation in tissues arises from sustained activation of the innate immune system (neutrophils, macrophages, and fibroblasts) as well as the adaptive immune system (B and T cells) (4). This chronic inflammatory response to persistent infections or environmental insults increases cancer risk both directly, through DNA damage, and indirectly, through tissue remodeling and fibrosis (4).
One strategy to evaluate the relationship of cancer with chronic inflammation is to measure circulating levels of inflammatory markers. Most previous epidemiologic investigations of circulating inflammatory markers and cancer have included a narrow range of markers (e.g., CRP, IL-6, IL10, TNF-alpha etc.) (5). The process of inflammation is complex and involves multiple key mediators (3), including chemokines, pro-inflammatory cytokines, anti-inflammatory cytokines, growth factors, angiogenesis factors, and metabolic markers. Therefore, a thorough epidemiologic characterization of inflammatory biomarkers and pathways involved in carcinogenesis requires a comprehensive evaluation of a wide range of markers.
Emerging multiplex technologies allow for the simultaneous quantification of over 100 analytes in low specimen volumes (6;7), underscoring their potential utility for large-scale epidemiologic investigations. While the obvious benefits of multiplexed assays include reductions in time and specimen volume, several aspects of these assays warrant thorough evaluation and standardization, including assay validity, reproducibility, stability, and appropriateness of specimen types (e.g. serum vs. plasma) (6). A majority of the previous studies that have formally assessed the performance of multiplexed assays were small in size and limited in the number of markers (8–15).
In the current study, we evaluated the performance of 116 inflammation, immune, and metabolic markers across two luminex bead-based commercial kits (Millipore and Bio-Rad) and three specimen types (serum, heparin plasma, and EDTA plasma). We specifically addressed the epidemiologic utility of these assays, as measured by their detectability in specimens from cancer-free individuals (i.e. values above the assay’s lower limit of detection) and reproducibility, as measured by coefficients-of-variation and intraclass correlation coefficients. Our primary aim was to evaluate the performance of each marker within a specimen type and kit type. Our secondary aims were to compare assay performance across specimen types within each kit and across kits within each specimen type.
Materials and Methods
Study design
We conducted this study among 100 cancer-free individuals who participated in the screening arm of the Prostate, Lung, Colorectal, and Ovarian (PLCO) Cancer Screening Trial. Briefly, between 1993 and 2002, the PLCO trial recruited approximately 155,000 men and women into either the screening arm or the control arm (16). Screening arm participants provided blood specimens at the baseline visit (T0) and annually during follow-up (T1 through T5) (16). All samples were processed by centrifugation at 2400–3000 RPM for 15 minutes. Specimens were frozen within 2 hours of collection and stored at −80° Centigrade until further use. Specimens used for the current study underwent two thaw cycles—one for aliquoting and one for laboratory testing.
We selected 100 participants with available T0 serum and T0 heparin plasma as well as EDTA plasma samples collected at the third annual visit (T3). To ensure comparability of the T3 EDTA plasma samples with another specimen type, we also included T3 heparin plasma samples from 50 of the 100 individuals. This design allowed us to compare assay performance between T0 serum versus T0 heparin plasma samples (n=100) as well as between T3 EDTA plasma samples versus T3 heparin plasma samples (n=50). The T0 and T3 heparin plasma samples were analyzed separately. To evaluate reproducibility of marker measurements, from each of the three specimen types (T0 serum, T0 heparin plasma, and T3 EDTA plasma), we selected 40 individuals as blinded duplicates, and placed 20 pairs as within-batch duplicates and 20 as across-batch duplicates. A batch denotes one plate of 37 unique samples including blinded duplicates. The subjects selected for blinded duplicates varied by specimen type, but were the same across the two kits given a specimen type.
Laboratory methods
We evaluated the performance of 116 inflammation, immune and metabolic markers—67 on Bio-Rad and 97 on Millipore, with 48 markers measured on both kits. Using magnetic bead-based assays, the Bio-Rad markers were measured in 150 ul of specimen across 4 panels: cytokine panel 1 (27 markers), cytokine panel 2 (21 markers), acute-phase protein panel (9 markers), and diabetes panel (12 markers). The Millipore kits utilized polystyrene bead-based assays to measure 97 markers in 400 ul of specimen across 6 panels: cytokine panel 1 (39 markers), cytokine panel 2 (23 markers), cytokine panel 3 (9 markers), soluble receptor panel (13 markers), metabolic panel (10 markers), and acute-phase protein panel (3 markers). On both kits, specimens were assayed in duplicate and the duplicate measurements were averaged. Based on the measurements of 7 standard concentrations provided by the manufacturer, a five parameter standard curve was utilized to convert optical density values into concentrations (pg/mL). Using the curve-fit measurements for each standard, we also estimated coefficients of variation across unblinded duplicates as well as recovery—calculated as the ratio of the observed and expected concentrations. We note that these recoveries indicate the goodness-of-fit of the standard curve rather than recoveries based on known, spiked concentrations. All assays were performed according to the manufacturer’s instructions.
Statistical analyses
For each marker, separately within each kit and specimen type, we evaluated assay performance using three measures: 1) detectability—the proportion of samples with values above the assay’s lower limit of detection (based on the 100 unique measurements for T0 serum, T0 heparin plasma, and T3 EDTA plasma); 2) coefficients-of-variation (CVs) for within-batch and across-batch duplicates (based on 20 pairs each for each specimen type); and 3) intraclass correlation coefficients (ICCs), which capture the proportion of total variability in measurements that arises from inter-individual variability (based on 20 pairs each of within-batch and across-batch duplicates for each specimen type). Observed concentrations of each marker were log-transformed to achieve approximate normality. CVs and ICCs were estimated using the ANOVA procedure. We considered detectability greater than 25% as acceptable, given the common use of quartiles in epidemiologic studies. CVs less than 20% were deemed acceptable.
To generalize marker performance across the three specimen types (T0 serum, T0 heparin plasma, and T3 EDTA plasma), we defined acceptable performance for a marker as: 1) being detectable in greater than 25% of the 100 samples on all three specimen types and 2) across-batch CVs of <20% on at least 2 of the 3 specimen types. These criteria allowed us to identify markers with acceptable performance within each kit across different specimen types.
We compared detectability across specimen types given a kit (T0 serum vs. T0 heparin plasma and T3 EDTA plasma vs. T3 heparin plasma) and across kits (Bio-Rad vs. Millipore) given a specimen type using the McNemar’s test. Median concentrations of each marker across specimen types and kits were compared using the Wilcoxon signed-rank test. Correlations of marker measurements across specimen types and kits were estimated using the Spearman’s rank correlation coefficient. Analyses for comparisons of detectability and medians and for correlation coefficients were based on the 100 unique measurements for T0 serum, T0 heparin plasma, and T3 EDTA plasma.
Head-to-head comparisons of ICCs across specimen types (given a kit) and between kits (given a specimen type) were conducted using Variance Components analyses. For each specimen type and kit, we estimated the ICC as the proportion of variation attributed to inter-individual variation using all observations (including blinded duplicates) in mixed effects models that included the batch number and study subjects nested within batches.
Results
Given the large number of markers as well as the different specimen types and kit types, we present assay performance and type/kit comparisons as the number of markers with acceptable or poor performance. Detailed results for each marker, including median observed concentrations, % detectability, CVs, and ICCs separately for each kit and specimen type as well as correlations of marker measurements across specimen types and kits are presented as supplemental material (Supplemental Tables 1 and 2).
Bio-Rad markers
For the 67 markers measured on Bio-Rad, we initially evaluated CVs as well as recoveries on unblinded duplicates across the 7 known standard concentrations used for curve-fit. Across the markers, CVs ranged from 4.3% to 27%, with only 2 markers (PCT and Ferritin) having CVs > 20%. Likewise, recoveries ranged from 90% to 670%, with a majority of markers (49 of 67 markers) having recoveries in the 80%–120% range.
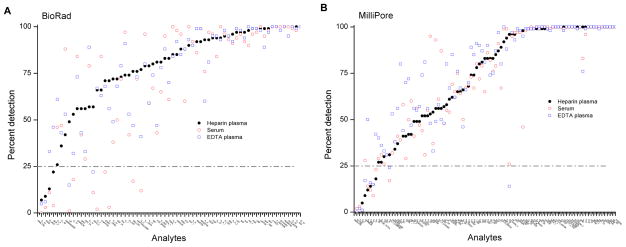
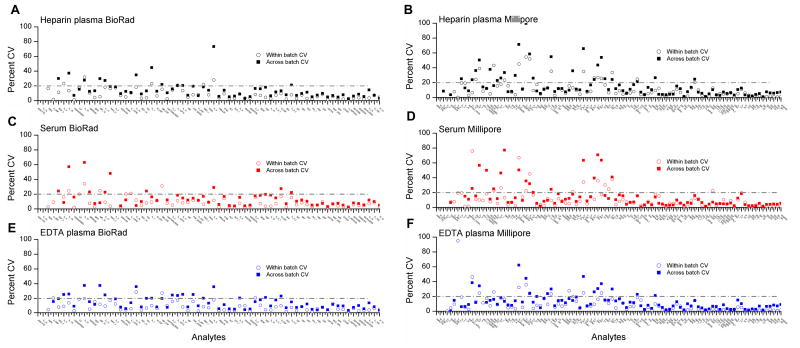
Using a criterion of detectable values in greater than 25 of the 100 individuals for each specimen type, a high proportion of markers were detectable (56 markers on serum, 63 markers on heparin plasma, and 64 markers on EDTA plasma) (Figure 1A and Table 1). Likewise, a high proportion of markers had CVs for across-batch duplicates less than 20% (51, 52, and 47, respectively, on serum, heparin plasma, and EDTA plasma) (Figure 2A, C, E and Table 1). Additionally, for a majority of markers, within-batch CVs were lower than across-batch CVs on each specimen type (Figure 2A, C, E).
Figure 1. Detectability on Bio-Rad and Millipore assays.
Figure 1 shows the proportion of samples with detectable levels for 67 markers on Bio-Rad (panel A) and 97 markers on Millipore (Panel B) across three specimen types—serum (open circles), heparin plasma (filled circles), and EDTA plasma (open squares).
Table 1.
Summary of results for evaluation of multiplexed inflammation marker assays
| Bio-Rad number of markers (%) | Millipore number of markers (%) | |
|---|---|---|
|
| ||
| Total | 67 (100.0) | 97 (100.0) |
|
| ||
| Markers with > 25% detection | ||
| Serum | 56 (83.5) | 89 (91.7) |
| Heparin plasma | 63 (94.0) | 89 (91.7) |
| EDTA plasma | 64 (95.5) | 89 (91.7) |
|
| ||
| Markers with < 20% CV for across-batch duplicatesa | ||
| Serum | 51 (76.2) | 75 (77.3) |
| Heparin plasma | 52 (77.6) | 69 (71.1) |
| EDTA plasma | 47 (70.1) | 78 (80.4) |
|
| ||
| ICCs for across-batch duplicates | ||
| Serum | ||
| <0.50 | 6 (8.9) | 5 (5.1) |
| 0.50–0.80 | 29 (43.3) | 32 (33.0) |
| 0.80–0.90 | 12 (17.9) | 19 (19.5) |
| ≥0.90 | 11 (16.4) | 34 (35.1) |
| Heparin plasma | ||
| <0.50 | 3 (4.5) | 9 (9.3) |
| 0.50–0.80 | 38 (56.7) | 31 (31.9) |
| 0.80–0.90 | 15 (22.3) | 25 (25.8) |
| ≥0.90 | 7 (10.4) | 28 (28.9) |
| EDTA plasma | ||
| <0.50 | 5 (7.5) | 7 (7.2) |
| 0.50–0.80 | 49 (73.1) | 25 (25.8) |
| 0.80–0.90 | 5 (7.5) | 24 (24.7) |
| ≥0.90 | 5 (7.5) | 37 (38.1) |
|
| ||
| Markers with acceptable performanceb | 45 (67.2) | 71 (73.2) |
Abbreviations: CV= coefficient of variation; ICC=intraclass correlation coefficient.
CVs and ICCs were calculated for 20 blinded duplicate samples for each specimen type that were placed across different batches.
Acceptable performance was defined as: 1) being detectable in greater than 25% of the 100 samples on all three specimen types and 2) across-batch CVs of <20% for blinded duplicates placed on at least 2 of the three specimen types.
Figure 2. Coefficients of variation in blinded duplicate samples stratified by specimen type and kit.
Figures 2 shows the coefficient of variation (CV) for 40 blinded duplicates, 20 placed within the same batch and 20 placed across different batches. Results are shown separately for 67 Bio-Rad markers (panels A, C, and E) and 97 Millipore markers (panels B, D, and F) across three specimen types—serum, heparin plasma, and EDTA plasma.
When the performance across the three specimen types was combined, 45 of 67 markers had acceptable performance in terms of detectability and across-batch CVs (Table 1). Markers with poor performance (< 25% detection on at least one specimen type or across-batch CVs > 20% on at least 2 specimen types) included: B-NGF, GM-CSF, G-CSF, IFNA2, IL-1A, IL-1B, IL-2, IL-3, IL-4, IL-5, IL-7, IL-10, IL-12 p40, IL-12 p70, IL-13, IL-15, IL-17, LIF, MCP-3, MIP-1A, TNFB, and TPA (Table 2). Across the four panels, 12 of 27 markers on cytokine panel 1, 13 of 21 markers on cytokine panel 2, 8 of 9 markers on the acute-phase panel, and all 12 markers on the diabetes panel had acceptable performance.
Table 2.
Summary of performance of multiplexed markers on Bio-Rad and Millipore
| Bio-Rad markers with acceptable performance a | Bio-Rad markers with <25% detectability b | Millipore markers with acceptable performance | Millipore markers with acceptable performance a | Millipore markers with < 25% detectability b |
|---|---|---|---|---|
| A2M CRP CTACK C_PEPTIDE EOTAXIN FERRITIN FGF_BASIC FIBRINOGEN GHRELIN GIP GLP_1 GLUCAGON GRO HAPTOGLOBIN HGF IFNG IL_16 IL_18 IL_1RA IL_2RA IL_6 IL_8 IL_9 INSULIN IP_10 LEPTIN MCP_1MC MIF MIG MIP_1B M_CSF PAI_1 PCT PDGF_BB RANTES RESISTIN SAA SAP SCF SCGF_B SDF_1A TNFA TRAIL VEGF VISFATIN |
B_NGF GM_CSF IFNA2 IL_12P40 IL_15 IL_1A IL_2 IL_3 LIF MCP_3 TNFB Bio-Rad markers with >20% CV c B_NGF GM_CSF G_CSF IL_10 IL_12P70 IL_13 IL_15 IL_17 IL_1A IL_1B IL_2 IL_4 IL_5 IL_7 LIF MCP_3 MIP_1A TNFB TPA |
AMYLIN_TOTAL BCA_1 CCL19_MIP3B CCL20_MIP3A CKINE CRP CTACK CXCL11_I_TAC CXCL6_GCP2 CXCL9_MIG C_PEPTIDE EGF ENA_78 EOTAXIN EOTAXIN_2 FGF_BASIC FIT_3_LIGAND FRACTALKINE GIP GLP_1 GLUCAGON GRO G_CSF IFNA2 IFNG IL_11 IL_12P40 IL_16 IL_1A IL_29_IFNG1 IL_33 IL_8 INSULIN IP_10 LEPTIN LIF MCP_1 MCP_2 MCP_3 MCP_4 MDC MIP_1A MIP_1B |
MIP_1D PP PYY SAA SAP SCD30 SCD40L SCF SDF_1A SEGFR SGP130 SILRII SIL_1RI SIL_2RA SIL_4R SIL_6R SRAGE STNFRI STNFRII SVEGFR1 SVEGFR2 SVEGFR3 TARC TNF-A TPO TRAIL TSLP VEGF |
EOTAXIN_3 GHRELIN IL_20 IL_21 IL_28A IL_3 IL_4 IL_5 M_CSF XCL1_LYMPHO Millipore markers with >20% CV c GM_CSF IL_10 IL_12P70 IL_13 IL_15 IL_17 IL_1B IL_1RA IL_2 IL_21 IL_23 IL_28A IL_3 IL_4 IL_5 IL_6_1 IL_7 IL_9 I_309 M_CSF TGFA TNFB XCL1_LYMPHO |
Acceptable performance was defined as: 1) being detectable in greater than 25% of the 100 samples on all three specimen types and 2) across-batch CVs of <20% for blinded duplicates placed on at least 2 of the three specimen types.
Markers with <25% detectability on at least one of three (serum, heparin plasma, and EDTA plasma) specimen types.
Markers with coefficients of variation (CVs) for across-batch duplicates >20% on 2 or more of 3 specimen types. CVs were calculated for 20 blinded duplicate samples for each specimen type that were placed across different batches.
On all three specimen types, ICCs for across-batch duplicates ranged from 0.31 to 0.99, with 23 markers on serum, 22 on heparin plasma, and 10 on EDTA plasma having ICCs greater than 0.8 (Table 1).
Millipore markers
Across the 97 Millipore markers, CVs for the 7 standard concentrations ranged from 3.4% to 14.7% and recoveries ranged from 72% to 319%. A majority of markers (82 of 97 markers) had recoveries in the 80%–120% range.
On serum, heparin plasma, and EDTA plasma samples, 89 markers each had detectable concentrations in greater than 25% of the 100 individuals (Table 1 and Figure 1B). A high proportion of markers (75 on serum, 69 on heparin plasma, and 78 on EDTA plasma) had across-batch CVs < 20% (Table 1 and Figure 2B, D, F). Similar to the Bio-Rad results, on each specimen type, within-batch CVs were generally less than across-batch CVs.
Combining detectability and across-batch CVs for the 3 specimen types, 71 of 97 markers had acceptable performance (Table 1). Markers with poor performance (< 25% detection on at least one specimen type or across-batch CVs > 20% on at least 2 specimen types) included: Eotaxin-3, Ghrelin, GM-CSF, IL-1B, IL-1 RA, IL-2, IL-3, IL-4, IL-5, IL-6, IL-7, IL-9, IL-10, IL-12 p70, Il-13, IL-15, IL-17, IL-20, IL-21, IL-23, IL-28A, I-309, M-CSF, TGF-A, TNF-B, and CXCL1 (Table 2). Across the different Millipore panels, 22 of 39 markers on cytokine panel 1, 17 of 23 markers on cytokine panel 2, 7 of 9 markers on cytokine panel 3, 9 of 10 markers on the metabolic panel, and all 3 markers on the acute-phase and all 13 markers soluble receptors panel had acceptable performance.
ICCs for across-batch duplicates ranged from 0.08–0.99, with 53, 53, and 61 markers on serum, heparin plasma, and EDTA plasma, respectively, having ICCs greater than 0.8 (Table 1).
Comparison of assay performance across specimen types and kits
We conducted comparisons of assay performance across the 3 specimen types and 2 kits for markers with acceptable performance (Table 3; 45 Bio-Rad markers, 71 Millipore markers, and 23 markers measured on both Bio-Rad and Millipore). On both Bio-Rad and Millipore, a majority of markers had similar % detectability for T0 serum vs. T0 heparin plasma as well as for T3 EDTA plasma vs. T3 heparin plasma. In contrast, for both Bio-Rad and Millipore, for a considerable number of markers, median cytokine concentrations differed between T0 serum vs. T0 heparin plasma and between T3 EDTA plasma vs. T3 heparin plasma (Table 3).
Table 3.
Comparison of assay performance across specimen types and kitsa
| Comparison | % Detectability # of markers b | Median concentration # of markers c |
|---|---|---|
|
| ||
| Bio-Rad (45 markers) | ||
|
| ||
| T0 serum vs. T0 heparin plasma | ||
| Significantly higher in serum | 7 | 16 |
| Significantly higher in plasma | 10 | 19 |
| Similar | 21 | 10 |
| Not Evaluable d | 7 | |
|
| ||
| T3 EDTA plasma vs. T3 heparin plasma | ||
| Significantly higher in EDTA | 2 | 7 |
| Significantly higher in heparin | 2 | 12 |
| Similar | 32 | 26 |
| Not Evaluable | 9 | |
|
| ||
| Millipore (71 markers) | ||
|
| ||
| T0 serum vs. T0 heparin plasma | ||
| Significantly higher in serum | 4 | 18 |
| Significantly higher in plasma | 5 | 13 |
| Similar | 51 | 40 |
| Not Evaluable | 11 | |
|
| ||
| T3 EDTA plasma vs. T3 heparin plasma | ||
| Significantly higher in EDTA | 5 | 3 |
| Significantly higher in heparin | 1 | 30 |
| Similar | 49 | 38 |
| Not Evaluable | 16 | |
|
| ||
| Bio-Rad vs. Millipore (23 markers) | ||
|
| ||
| T0 Serum | ||
| Significantly higher in Bio-Rad | 2 | 15 |
| Significantly higher in Millipore | 2 | 7 |
| Similar | 10 | 1 |
| Not Evaluable | 9 | |
| T0 heparin plasma | ||
| Significantly higher in Bio-Rad | 4 | 16 |
| Significantly higher in Millipore | 4 | 7 |
| Similar | 6 | 0 |
| Not Evaluable | 9 | |
|
| ||
| T3 EDTA plasma | ||
| Significantly higher in Bio-Rad | 3 | 15 |
| Significantly higher in Millipore | 3 | 8 |
| Similar | 7 | 0 |
| Not Evaluable | 10 | |
Comparisons were restricted to markers with acceptable performance—45 Bio-Rad markers, 71 Millipore markers, and 23 markers measured using both kits.
Comparisons of % detectability across specimen types and kits were performed using the McNemar’s test.
Comparisons of median observed concentrations across specimen types and kits were performed using the Wilcoxon signed-rank test. Observations below the assay’s lower limit were excluded.
McNemar’s p-value could not be evaluated because one of the groups had 100% detectability.
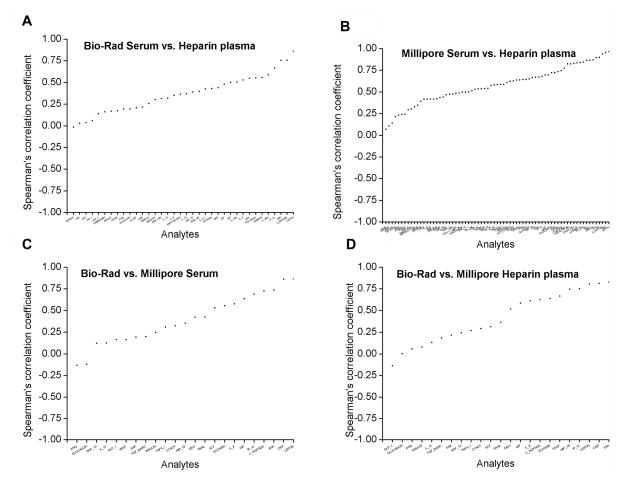
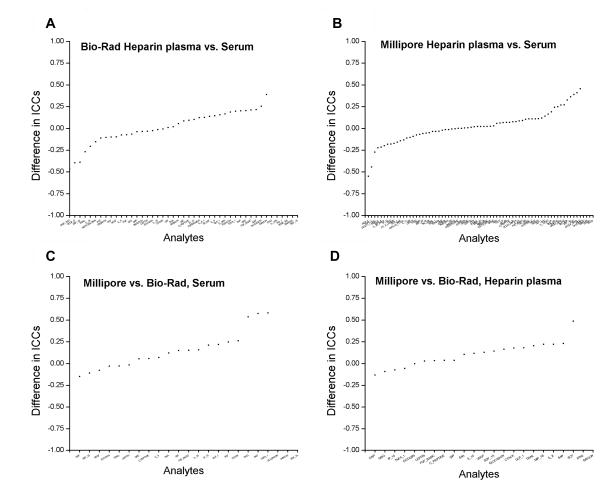
For 45 Bio-Rad markers with acceptable performance (Figure 3A), correlation coefficients between T0 serum and T0 heparin plasma were < 0.5 for 33 markers, 0.5–0.75 for 9 markers and ≥0.75 for 3 markers. For 71 Millipore markers with acceptable performance (Figure 3B), correlation coefficients between T0 serum and T0 heparin plasma were <0.5 for 25 markers, 0.5–0.75 for 31 markers, and ≥0.75 for 15 markers. In variance components analyses, <10% difference in ICCs between T0 serum and T0 heparin plasma was observed for 16 of 39 evaluable Bio-Rad markers with acceptable performance and for 35 of 67 evaluable Millipore markers with acceptable performance (Figure 4A and B).
Figure 3. Correlation of marker measurements across specimen types and kits.
Spearman’s correlation coefficients are shown for comparisons of the rank order of marker concentrations between T0 serum and T0 heparin plasma on Bio-Rad (panel A) and on Millipore (panel B). Comparisons between Bio-Rad and Millipore for T0 serum (panel C) and T0 heparin plasma (panel D) are also shown. T0 denotes baseline visit in the PLCO study.
Figure 4. Comparison of intraclass correlation coefficients by specimen type and kit.
Differences in intraclass correlations coefficients between T0 heparin plasma vs. T0 Serum for Bio-Rad (panel A) and for Millipore (panel B). Differences in ICCs between Millipore vs. Bio-Rad for T0 serum (panel C) and T0 heparin plasma (panel D) are also shown. ICCs were estimated using variance components analyses.
Across the three specimen types, % detectability and median concentrations were significantly different between Bio-Rad and Millipore for a majority of the 23 markers with acceptable performance. Likewise, for all three specimen types (Figure 3C and D), correlation coefficients between Bio-Rad and Millipore were low (for T0 serum: <0.5 for 12 markers, 0.5–0.75 for 7 markers, and ≥0.75 for 4 markers; for T0 heparin plasma: <0.5 for 14 markers, 0.5–0.75 for 7 markers, and ≥0.75 for 2 markers; for T3 EDTA plasma: <0.5 for 15 markers, 0.5–0.75 for 4 markers, and ≥0.75 for 4 markers). In variance components analyses, ICCs differed between Bio-Rad and Millipore for a majority of markers (of 23 acceptable markers on both kits, 7 of 20 evaluable markers on T0 serum, 8 of 21 evaluable markers on T0 heparin plasma, and 7 of 19 evaluable markers on T3 EDTA plasma had < 10% difference in ICCs) (Figure 4C and D).
Discussion
In this large methodologic study, we show that a majority of multiplexed inflammation, immune, and metabolic markers can be measured reliably in serum and plasma specimens, as evidenced by low CVs and high ICCs, on both Bio-Rad and Millipore. Median analyte concentrations and ICCs differed to a small extent across specimen types and to a large extent between Bio-Rad and Millipore. Likewise, correlations in analyte levels were moderate to high across specimen types, but were low between the 2 commercial kits.
Our results underscore the utility of multiplexed technologies for large-scale investigations into the role of inflammation and immune dysregulation in the etiology of cancer and other diseases. Notably, the 45 markers on Bio-Rad and 71 markers on Millipore with good detectability and reproducibility include several components of the inflammation and immune response such as pro-inflammatory markers (e.g. IL-8, TNF-A, IFN-G, GRO), anti-inflammatory markers (e.g. IL-16), acute-phase proteins (e.g. CRP, serum amyloid A [SAA]), and growth and angiogenesis factors (e.g. FGF, VEGF). Reliable detection of these markers in serum and plasma samples provides the opportunity to comprehensively evaluate the role of immunity and inflammation in cancer etiology in cohort and case-control studies. Furthermore, the redundant and pleiotropic nature of most inflammation markers provides the opportunity to evaluate the association of groups of markers (defined through principal components or factor analyses) with cancer risk (9).
Despite the large number of markers with acceptable performance, classic Th1-type markers such as IL-2, IL-12, and IL-15 and Th2-type markers such as IL-4, IL-10, and IL-13 had a low proportion of samples with detectable levels, unacceptably high CVs, or low ICCs. Notably, a majority of these markers were included in panels with higher numbers of markers, and we found that assay performance decreased with increasing number of markers on a panel. For example, 17 markers (43%) on Millipore’s 39-plex panel and 15 markers (55%) on Bio-Rad’s 27-plex panel had poor detectability and/or reproducibility. Because markers such as IL-2 and IL-10 from the same vendors had acceptable performance on previous studies which simultaneously measured a limited number of markers (9;17), it is likely that interference from other markers affected the performance of these markers.
Measurement of circulating inflammation markers is potentially sensitive to several factors such as specimen types, sample handling, and processing methods (18;19). Previous studies have reported that marker measurements are not interchangeable between serum and plasma samples (9), and these differences are believed to arise from factors such as degradation of markers during the process of clotting and degranulation of granulocytes (9). Consistent with these studies, we found that on both Bio-Rad and Millipore, for a considerable number of markers, median analyte concentrations and ICCs differed between serum vs. heparin plasma and EDTA plasma vs. heparin plasma. Additionally, irrespective of the specimen type, for a majority of markers, % detectability, median concentrations, and ICCs differed between Bio-Rad and Millipore. Therefore, our observations indicate that results from different studies utilizing different specimen types and different multiplexed kits may not be directly comparable (20).
Circulating levels of inflammation, immune, and metabolic markers are also influenced by several demographic and behavioral characteristics such as age, sex, race, smoking, body mass index, and diet (21). Therefore, in separate studies, we are currently evaluating predictors of an inflammatory response for single markers as well as empirical groupings of markers, and the temporal stability of markers with acceptable performance. The temporal stability of circulating markers is largely unknown, and single time-point measurements in prospective cohort studies could bias results to the null for unstable markers (22).
Our study has several strengths, including the standardized collection, processing, and storage of specimens in the PLCO study (16) and comprehensive evaluation of over 100 multiplexed markers on different specimen types. We also note the limitations of our study. Importantly, our study focused on reliability, but not validity, of marker measurements. Nevertheless, previous studies comparing the performance of multiplexed marker measurements with ELISA assays show high validity (13–15). Finally, we defined less than 25% detectability as poor performance, in part, because samples with low detection levels are generally accompanied by unacceptably high CVs and low ICCs. However, we note the possibility that some markers could be expressed only in disease conditions and therefore could be informative for disease associations.
In conclusion, our key observation was that Bio-Rad and Millipore multiplexed markers are broadly reproducible and can therefore be utilized for large-scale epidemiologic studies. Our results highlight the opportunity to comprehensively evaluate the role of a large number of circulating inflammatory markers representative of a range of immune-related processes and pathways in cancer etiology and prognosis.
Supplementary Material
Reference List
- 1.Moss SF, Blaser MJ. Mechanisms of disease: Inflammation and the origins of cancer. Nat Clin Pract Oncol. 2005 Feb;2(2):90–7. doi: 10.1038/ncponc0081. [DOI] [PubMed] [Google Scholar]
- 2.Thun MJ, Henley SJ, Gansler T. Inflammation and cancer: an epidemiological perspective. Novartis Found Symp. 2004;256:6–21. [PubMed] [Google Scholar]
- 3.Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002 Dec 19;420(6917):860–7. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Visser KE, Coussens LM. The inflammatory tumor microenvironment and its impact on cancer development. Contrib Microbiol. 2006;13:118–37. doi: 10.1159/000092969. [DOI] [PubMed] [Google Scholar]
- 5.Heikkila K, Harris R, Lowe G, Rumley A, Yarnell J, Gallacher J, et al. Associations of circulating C-reactive protein and interleukin-6 with cancer risk: findings from two prospective cohorts and a meta-analysis. Cancer Causes Control. 2009 Feb;20(1):15–26. doi: 10.1007/s10552-008-9212-z. [DOI] [PubMed] [Google Scholar]
- 6.Elshal MF, McCoy JP. Multiplex bead array assays: performance evaluation and comparison of sensitivity to ELISA. Methods. 2006 Apr;38(4):317–23. doi: 10.1016/j.ymeth.2005.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de JW, Rijkers GT. Solid-phase and bead-based cytokine immunoassay: a comparison. Methods. 2006 Apr;38(4):294–303. doi: 10.1016/j.ymeth.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 8.Hosnijeh FS, Krop EJ, Portengen L, Rabkin CS, Linseisen J, Vineis P, et al. Stability and reproducibility of simultaneously detected plasma and serum cytokine levels in asymptomatic subjects. Biomarkers. 2010 Mar;15(2):140–8. doi: 10.3109/13547500903340570. [DOI] [PubMed] [Google Scholar]
- 9.Wong HL, Pfeiffer RM, Fears TR, Vermeulen R, Ji S, Rabkin CS. Reproducibility and correlations of multiplex cytokine levels in asymptomatic persons. Cancer Epidemiol Biomarkers Prev. 2008 Dec;17(12):3450–6. doi: 10.1158/1055-9965.EPI-08-0311. [DOI] [PubMed] [Google Scholar]
- 10.Gu Y, Zeleniuch-Jacquotte A, Linkov F, Koenig KL, Liu M, Velikokhatnaya L, et al. Reproducibility of serum cytokines and growth factors. Cytokine. 2009 Jan;45(1):44–9. doi: 10.1016/j.cyto.2008.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chowdhury F, Williams A, Johnson P. Validation and comparison of two multiplex technologies, Luminex((R)) and Mesoscale Discovery, for human cytokine profiling. J Immunol Methods. 2009 Jan 1;340(1):55–64. doi: 10.1016/j.jim.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 12.Khan SS, Smith MS, Reda D, Suffredini AF, McCoy JP., Jr Multiplex bead array assays for detection of soluble cytokines: comparisons of sensitivity and quantitative values among kits from multiple manufacturers. Cytometry B Clin Cytom. 2004 Sep;61(1):35–9. doi: 10.1002/cyto.b.20021. [DOI] [PubMed] [Google Scholar]
- 13.Djoba Siawaya JF, Roberts T, Babb C, Black G, Golakai HJ, Stanley K, et al. An evaluation of commercial fluorescent bead-based luminex cytokine assays. PLoS ONE. 2008;3(7):e2535. doi: 10.1371/journal.pone.0002535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.dupont NC, Wang K, Wadhwa PD, Culhane JF, Nelson EL. Validation and comparison of luminex multiplex cytokine analysis kits with ELISA: determinations of a panel of nine cytokines in clinical sample culture supernatants. J Reprod Immunol. 2005 Aug;66(2):175–91. doi: 10.1016/j.jri.2005.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fu Q, Zhu J, Van Eyk JE. Comparison of multiplex immunoassay platforms. Clin Chem. 2010 Feb;56(2):314–8. doi: 10.1373/clinchem.2009.135087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hayes RB, Sigurdson A, Moore L, Peters U, Huang WY, Pinsky P, et al. Methods for etiologic and early marker investigations in the PLCO trial. Mutat Res. 2005 Dec 30;592(1–2):147–54. doi: 10.1016/j.mrfmmm.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 17.Kemp TJ, Hildesheim A, Garcia-Pineres A, Williams MC, Shearer GM, Rodriguez AC, et al. Elevated systemic levels of inflammatory cytokines in older women with persistent cervical human papillomavirus infection. Cancer Epidemiol Biomarkers Prev. 2010 Aug;19(8):1954–9. doi: 10.1158/1055-9965.EPI-10-0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aziz N, Nishanian P, Mitsuyasu R, Detels R, Fahey JL. Variables that affect assays for plasma cytokines and soluble activation markers. Clin Diagn Lab Immunol. 1999 Jan;6(1):89–95. doi: 10.1128/cdli.6.1.89-95.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tworoger SS, Hankinson SE. Use of biomarkers in epidemiologic studies: minimizing the influence of measurement error in the study design and analysis. Cancer Causes Control. 2006 Sep;17(7):889–99. doi: 10.1007/s10552-006-0035-5. [DOI] [PubMed] [Google Scholar]
- 20.Toedter G, Hayden K, Wagner C, Brodmerkel C. Simultaneous detection of eight analytes in human serum by two commercially available platforms for multiplex cytokine analysis. Clin Vaccine Immunol. 2008 Jan;15(1):42–8. doi: 10.1128/CVI.00211-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O’Connor MF, Irwin MR. Links between behavioral factors and inflammation. Clin Pharmacol Ther. 2010 Apr;87(4):479–82. doi: 10.1038/clpt.2009.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Platz EA, Sutcliffe S, De Marzo AM, Drake CG, Rifai N, Hsing AW, et al. Intra-individual variation in serum C-reactive protein over 4 years: an implication for epidemiologic studies. Cancer Causes Control. 2010 Jun;21(6):847–51. doi: 10.1007/s10552-010-9511-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.