Abstract
Fulvestrant (Faslodex™) is a pure antiestrogen that is approved to treat hormone receptor-positive metastatic breast cancer in postmenopausal women. Previous studies have demonstrated that fulvestrant metabolism in humans involves cytochromes P450 and UDP-glucuronosyltransferases (UGTs). To date, fulvestrant sulfation has not been characterized. This study examined fulvestrant sulfation with nine recombinant sulfotransferases and found that only SULT1A1 and SULT1E1 displayed catalytic activity toward this substrate, with Km of 4.2 ± 0.99 and 0.2 ± 0.16 μM, respectively. In vitro assays of 104 human liver cytosols revealed marked individual variability that was highly correlated with β-naphthol sulfation (SULT1A1 diagnostic substrate; r = 0.98, P < 0.0001), but not with 17β-estradiol sulfation (SULT1E1 diagnostic substrate; r = 0.16, P = 0.10). Fulvestrant sulfation was correlated with both SULT1A1*1/2 genotype (P value = 0.023) and copy number (P < 0.0001). These studies suggest that factors influencing SULT1A1/1E1 tissue expression and/or enzymatic activity could influence the efficacy of fulvestrant therapy.
Keywords: fulvestrant, sulfotransferase, genotype, copy number
Introduction
While tamoxifen has been the gold standard of treatment for estrogen receptor-positive breast cancer, the untoward effects associated with the pharmacological profile of this drug (namely, its partial agonism) and the frequent occurrence of drug resistance has prompted the search for antiestrogens devoid of estrogenicity (ie, pure antiestrogens). Several studies have shown that estradiol (E2) derivatives bearing a functionalized side chain in position 7R satisfy this criterion. Among these compounds, fulvestrant (ICI 182780) was selected for clinical trials because of its high in vivo antitumor activity, notably in animal models of tamoxifen-resistant breast cancer, and is currently in clinical use.1
Fulvestrant (Faslodex™) is the first of a new type of endocrine treatment – an estrogen receptor (ER) antagonist that downregulates the ER and has no agonist effects. Fulvestrant is a 7α-alkylsulphinyl analog of 17β-estradiol, which is distinctly different in chemical structure from the nonsteroidal structures of tamoxifen, raloxifene, and other selective estrogen receptor modulators.2 Fulvestrant competitively inhibits binding of estradiol to the ER, with a binding affinity that is 89% that of estradiol.3 Studies in the MCF-7 human breast cancer cell line have shown that fulvestrant significantly suppresses cellular levels of ER protein4 and inhibits ER-induced expression of the progesterone receptor (PgR), the estrogen-regulated protein pS2, and cathepsin D more strongly than tamoxifen.5 Importantly, fulvestrant has also demonstrated antitumor activity in tamoxifen-resistant MCF-7/TAMR-1 cell lines, confirming a lack of cross-resistance between tamoxifen and fulvestrant.6,7
Previous studies of fulvestrant metabolism in humans indicate the involvement of multiple drug metabolizing enzyme families, including the cytochromes P450, UDP-glucuronosyltransferases (UGTs), and sulfotransferases (SULTs).8 While specific P450 s and UGTs involved in fulvestrant metabolism have been identified,8,9 specific SULT isoforms involved in fulvestrant sulfation have not been described, although fulvestrant sulfates are major metabolites detected in human urine.
The polymorphic nature of SULTs has long been recognized.10–12 These studies have demonstrated large individual variation (about 50-fold in some studies) in the activity of platelet SULT1A1 in humans.13–15 Some of this variability can be explained by a common single-nucleotide polymorphism (SNP; a G to A transition) in the coding region (nucleotide 638) of the SULT1A1 gene.16 Another factor that may also affect enzymatic activity is gene deletion and duplication. Hebbring et al observed that SULT1A1 enzymatic activity is also correlated with SULT1A1 gene copy numbers in vitro.17 Recently, the laboratory of the current study reported that SNPs located in the 3′-UTR and 3′-flanking region of SULT1A1 are significantly associated with SULT1A1 activity, and this effect remains when stratified by SULT1A1 copy number.18 Functional SNPs in SULT1E1, however, are not as well described. The present study examines interindividual variability in fulvestrant sulfation, SULT isoforms involved, and the influence of SULT1A1 genetic variants on fulvestrant sulfation.
Material and methods
Materials
Fulvestrant was provided by AstraZeneca Pharmaceuticals (Macclesfield, Cheshire, UK). 3′-phosphoadenosine-5′-phosphosulfate (PAPS) was obtained from the Department of Chemistry, University of Dayton (Dayton, OH). Sequencing and PCR Primers were purchased from Invitrogen (Grand Island, NY). All other chemicals used were of reagent grade from Fisher HealthCare (Houston, TX).
Mass spectrophotometric analysis of fulvestrant sulfation
Fulvestrant-3-O-sulfate was synthesized according to published methods.19 Fulvestrant and its sulfated metabolite were analyzed using a Waters Acquity Ultra-High Pressure Liquid Chromatography™ (UPLC™) (Milford, MA) connected to a Thermo Scientific Quantum (TSQ®) Ultra™ mass spectrometer (Waltham, MA). Chromatography was performed using a Waters BEH C18 column (2.1 × 50 mm, 1.7 μm particle size) with a gradient of water and methanol starting at 50% methanol; up to 77% methanol over 1.5 minutes; 77% methanol for 1.3 minutes; followed by 100% methanol for 1 minute. Both solvents contained 20 mM ammonium formate. The injection volume was 3 μL. Retention times were found to be 2.15 and 2.80 minutes for fulvestrant-sulfate and fulvestrant, respectively. The mass spectrometer was operated in negative ion mode using an H-ESI probe (Thermo-Fisher Scientific, Waltham, MA). The heated probe was set at 300°C; the heated capillary was held at 350°C; the spray voltage was set to 2300 V; and the sheath and auxiliary gasses were at 45 and 20 arbitrary units, respectively. Detection was by SRM monitoring the transition m/z = 605.3 [M-H]− → 427.3 [M-H-178]− for fulvestrant and m/z = 685.1 [M-H]− → 525.1 [M-H-160]−. External calibration curves were constructed using 1, 5, 10, 25, 100, and 200 nM concentrations of fulvestrant-sulfate, and 5, 10, 25, 100, 200, and 500 μM fulvestrant. The limit of detection was 3 pM.
Sulfation by human liver cytosols
Human liver specimens (n = 104) were obtained from the Cooperative Human Tissue Network (CHTN; Mentor, OH). All liver specimens were from Caucasian donors ranging in age from 10–85 years, with 56 male and 44 female donors. African Americans were excluded from this study due to low numbers that precluded racial comparisons. All liver specimens were snap frozen upon harvest and were confirmed as histologically normal tissue by CHTN. Tissue specimens that exhibited abnormalities were excluded from this study. Cytosols were prepared from human liver tissue as previously described,20 and stored frozen at −80°C until assayed. Cytosolic protein levels were determined using the Bradford method with bovine serum albumin as a standard. Incubations to determine activity toward fulvestrant contained 500 μM fulvestrant (dissolved in dimethyl sulfoxide, DMSO: H2O, 1:3), 50 mM tris-HCl buffer, pH 7.5 and 20 μM PAPS, and 100 μg cytosolic protein in a final volume of 100 μL. The final DMSO concentration in the reactions was <1%. Control reactions were run with no substrate but contained the appropriate volume of the DMSO vehicle. Reactions were incubated for 15 minutes at 37°C and then terminated by adding 50 μL acetonitrile: acetic acid (96:4), then analyzed using a Waters Acquity UPLC connected to a TSQ Ultra mass spectrometer. Activity toward β-naphthol was determined using a colorimetric assay, as previously described.21 Activity assay toward 17β-estradiol used radioactively labeled E2, the sulfate acceptor cosubstrate, rather than radioactively labeled PAPS.22
Sulfation by recombinant SULTs
Sulfation activity was determined using fulvestrant as substrate with each of nine different bacterially expressed human SULT isoforms. All SULTs were expressed in Escherichia coli using the pET vector to generate the native form of the enzyme and then purified by DEAE-Sepharose™ (Fisher Scientific, Houston, TX) chromatography to obtain a preparation suitable for enzymatic characterization.23–26 The resulting preparations were approximately 80% pure, and activities were calculated based on total protein. Assays were performed with each of the expressed human SULTs (SULT2A1, 1E1, 2B1a, 2B1b, 1A1, 1A3, 1B1, 1C1, and 1C2). Fulvestrant and its sulfated metabolite were analyzed using a Waters Acquity UPLC connected to a TSQ Ultra mass spectrometer.
Purification of human SULT1A1 and SULT1E1 for kinetic studies
SULT1A1 and SULT1E1 were expressed using the pMAL-c2 vector in E. coli XL1-Blue cells, as described previously.27 The pMAL-c2 vector generates a maltose binding protein (MBP) tag at the amino-terminal end of the SULTs, allowing for affinity purification with an amylose affinity column. The MBP is cleaved with Factor Xa protease immediately before the initial methionine residue in the SULT. The SULTs are then purified from the MBP and Factor Xa using DEAE-Sepharose CL-6B chromatography and passage through a second small amylose affinity column. The resulting proteins were greater than 90% pure, as determined by SDS-polyacrylamide gel electrophoresis.
Kinetic analysis of fulvestrant sulfation
Fulvestrant sulfation activity was measured using radio-labeled [35 S]-PAPS as the sulfonate donor, as described previously.28 Reactions were performed in 10 mM sodium phosphate (pH 7.4) and 5 mM MgCl2. PAPS concentrations ranged from 0.05 μM to 5 μM, and fulvestrant concentrations ranged from 0.1 μM to 20 μM for SULT1A1, and 0.1 μM to 5 μM for SULT1E1 reactions. Reactions were incubated for 5 minutes at 37°C and stopped with the addition of 200 μM of chloroform. The reactions were then vortexed, centrifuged at 2000 g, and an aliquot spotted onto a Whatman thin layer chromotography plate (Maidstone, Kent, UK), and resolved with a solution of 85 mL of methylene chloride, 15 mL of methanol, and 5 mL of ammonium hydroxide. Radioactive fulvestrant-sulfate was detected by exposure to autoradiograph film, and the radioactive spots scrapped into scintillation vials and quantified by scintillation spectroscopy. Km and Vmax values were determined using two substrate kinetic analysis based on the replots of the Lineweaver–Burk plots.29
Kd determinations of substrate binding by intrinsic fluorescence
The affinity constants for fulvestrant and PAPS/PAP binding to the pure SULTs were calculated using intrinsic fluorescence changes in the enzymes, as described previously.27 Pure enzyme (100 nM) was equilibrated at room temperature in a quartz cuvette in a PerkinElmer® LS-5 fluorescence spectrometer (Waltham, MA). The intrinsic fluorescence was measured using excitation at 280 nm and emission at 345 nm. Fulvestrant or PAPS/PAP was titrated into the solution and the change in fluorescence measured until no additional change in fluorescence was observed. The change in fluorescence was plotted against the concentration of fulvestrant and the Kd was determined from the Lineweaver–Burk plots.29
Chemical inhibition of SULT1A1 activities
2,6-dichloro-4-nitrophenol (DCNP) is a selective inhibitor of SULT1A1 enzymes.30 DCNP was dissolved in ethanol. The final concentrations of DCNP in the SULT assay ranged from 0.1 to 10 μM. After an incubation period (15 minutes), the reactions were halted using 50 μL acetonitrile: acetic acid (96:4).
SULT1A1 genotyping
Genotyping for SULT1A1*1/2 was performed as previously described.31 Genotype was determined by direct sequencing using the CEQ™ DTCS Quick Start Kit and the CEQ™ 8800 Genetic Analysis System (Beckman Coulter, Fullerton, CA). Genotyping for SULT1A1 3′-SNPs was performed as previously described.18
SULT1A1 copy number assay
SULT1A1 copy number determination was performed by real-time PCR in an ABI® PRISM 7900HT Sequence Detection System using the TaqMan® Gene Expression Absolute Quantification Assay (Applied Biosystems, Foster City, CA). A pair of unlabeled PCR primers, 5′TGCCCGCAACGCAAA3′ and 5′GGCCATGTGGTAGAAGTGGTAGT3′, and a FAM dye-labeled TaqMan minor groove binder probe, 5′ATGTGGCAGTTTCC3′, were designed to specifically amplify SULT1A1. VIC dye-labeled TaqMan RNaseP, which has two copies per haploid human genome, was used as a control. Amplification was 10 minutes of initial setup at 95°C, followed by 40 amplification cycles (15 seconds of denaturation at 95°C and 60 seconds of annealing/extension at 60°C). Each sample was examined in quadruplicate and copy number was determined using CopyCaller™ software (Applied Biosystems, Foster City, CA).
Statistical analysis
Parametric (one-way ANOVA) and nonparametric (Spearman’s rank correlation) tests were used to examine the bivariate correlation between fulvestrant sulfation, β-naphthol sulfation, 17β-estradiol sulfation, SULT1A1 SNPs in coding region, and 3′ UTR and SULT1A1 copy number as appropriate. Log transformation was applied for non-Gaussian distributed variables to carry out parametric tests. Haplotypes were constructed, as described previously,18 to assess the collective effects of 3′-UTR SNPs on fulvestrant activity. Statistical significance level was set at α = 0.05 (two-sided) and all analyses were performed using SAS® software (v 9.2, 2008; SAS Institute, Cary, NC).
Results
Sulfation of fulvestrant by human liver cytosols
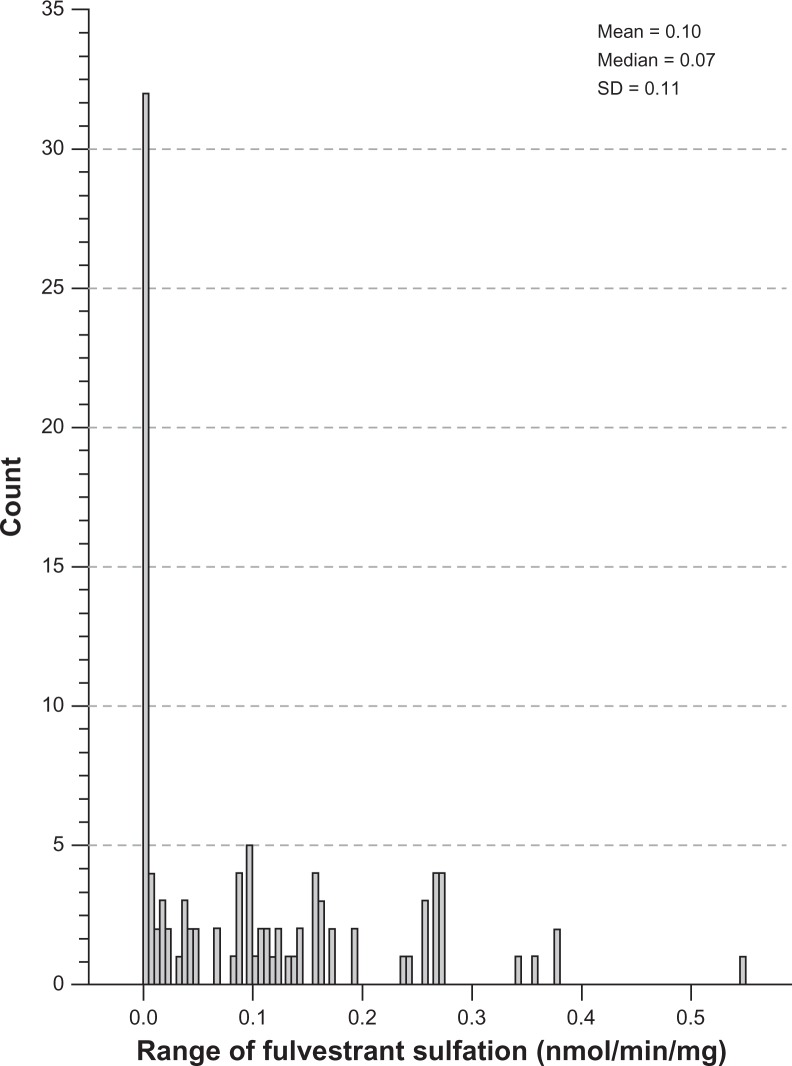
Interindividual variability in fulvestrant sulfation was evaluated in liver cytosols collected from 104 individuals by LC-MS/MS. The histogram in Figure 1 demonstrates the distribution of fulvestrant sulfation ranging from 0.003 to 0.55 nmol/min/mg (0.10 ± 0.11, n = 104). Sulfation activity was significantly higher in females than males (0.14 ± 0.12 vs 0.08 ± 0.10, P < 0.02), with a comparable range of activity within the male and female populations.
Figure 1.
Distribution of fulvestrant sulfation activity in human liver cytosols. Interindividual variability in fulvestrant sulfation was evaluated in liver cytosols collected from 104 individuals. This histogram demonstrates the distribution of fulvestrant ranging from 0.003 to 0.55 nmol/min/mg (0.10 ± 0.11, n = 104).
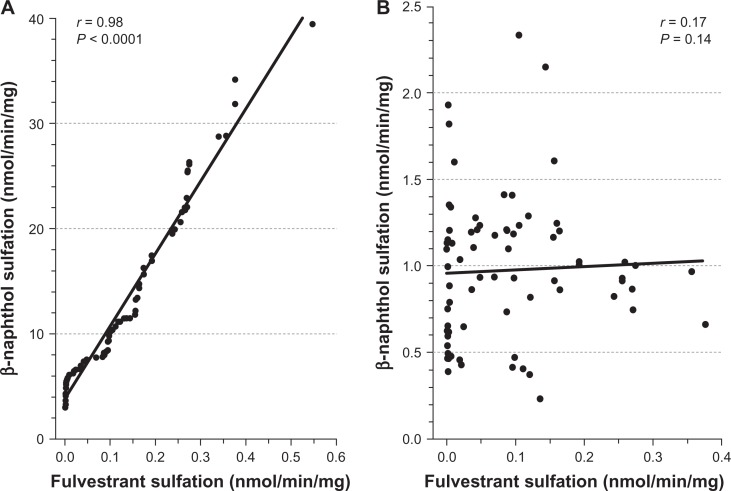
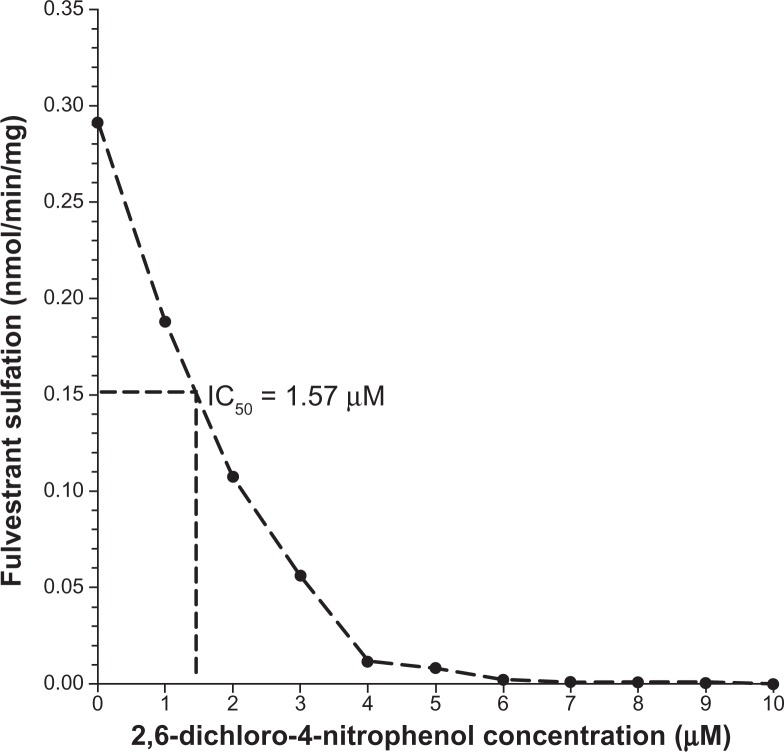
Additional analysis revealed positive and statistically significant correlations between the rates of formation of fulvestrant sulfate and β-naphthol sulfate (a substrate diagnostic for SULT1A1) in the 104 livers studied (Figure 2A; r = 0.98, P < 0.0001) but there was no significant correlation found between fulvestrant sulfate and 17β-estradiol sulfate, a substrate diagnostic for SULT1E1 (Figure 2B; r = 0.17, P = 0.14). The findings of the study also indicated that fulvestrant sulfation in human liver cytosol was potently inhibited by DCNP, a specific inhibitor of SULT1A1 (Figure 3). Preincubation of human liver cytosol with 0.1 to 10 μM DCNP reduced generation of sulfated fulvestrant with an IC50 value of 1.57 μM. When 5 μM (final concentration) DCNP was added to the reaction mixture, the production of sulfated fulvestrant was also significantly inhibited (>95%).
Figure 2.
Correlation between fulvestrant sulfation and β-naphthol sulfation.
Notes: A statistically significant correlation between the rates of fulvestrant sulfate formation and β-naphthol sulfate formation (r = 0.98, P < 0.0001) was demonstrated in 104 human liver cytosols (2A). The correlation between 17β-estradiol sulfation and fulvestrant sulfation was not statistically significant (r = 0.17, P = 0.14, n = 104) (2B).
Figure 3.
Inhibition of fulvestrant sulfation by DCNP. DCNP was dissolved in ethanol. The final concentrations of DCNP in the SULT assay ranged from 0.1 to 10 μM. After an incubation period (15 minutes), the reactions were halted using 50 μL acetonitrile: acetic acid (96:4). Activity was analyzed and IC50 values calculated.
Abbreviation: DCNP, 2,6-dichloro-4-nitrophenol.
Sulfation of fulvestrant by expressed human SULTs
The ability of nine SULT isoforms (SULT1A1, 1A3, 1B1, 1C1, 1C2, 1E1, 2A1, 2B1a, and 2B1b) to conjugate fulvestrant was then investigated. SULT1E1 (1.09 nmol/min/mg) displayed the highest enzymatic activity toward fulvestrant, followed by SULT1A1 (0.628 nmol/min/mg). These differences in sulfation activity were statistically significant (P = 0.045). Other isoforms tested did not exhibit detectable activity.
To further characterize SULT1E1- and SULT1A1-mediated fulvestrant sulfation activity, kinetic analyses were performed using recombinant enzyme in the presence of substrate concentrations varying from 0.1 to 20 μM. Examination of SULT1A1 and SULT1E1 kinetics revealed a calculated Km value of 4.2 ± 0.99 μM and 0.2 ± 0.02 μM, respectively. The Vmax values for SULT1A1- and SULT1E1-catalyzed fulvestrant sulfation were 7.8 ± 0.10 and 62.5 ± 2.57 nmol/min/mg of protein. The normalized Vmax was used to determine the efficiency of sulfation (ratio Vmax/Km) (Table 1). The dissociation constants (Kd) for binding of fulvestrant were also determined to be 2.3 ± 0.4 μM for SULT1A1 and 0.2 ± 0.02 μM for SULT1E1 (Table 1).
Table 1.
Kinetic parameters for fulvestrant sulfation by SULT1A1 and SULT1E1
| Kd | Km | Vmax | |
|---|---|---|---|
| SULT1A1 | 2.3 ± 0.4 μM | 4.2 ± 1.0 μM | 7.8 ± 0.1 pmol/min/μg |
| SULT1E1 | 0.2 ± 0.02 μM | 0.2 ± 0.2 μM | 62.5 ± 2.5 pmol/min/μg |
Note:
error bars represent 95% confidence intervals.
Association of SULT1A1 genotype and copy number with fulvestrant sulfation
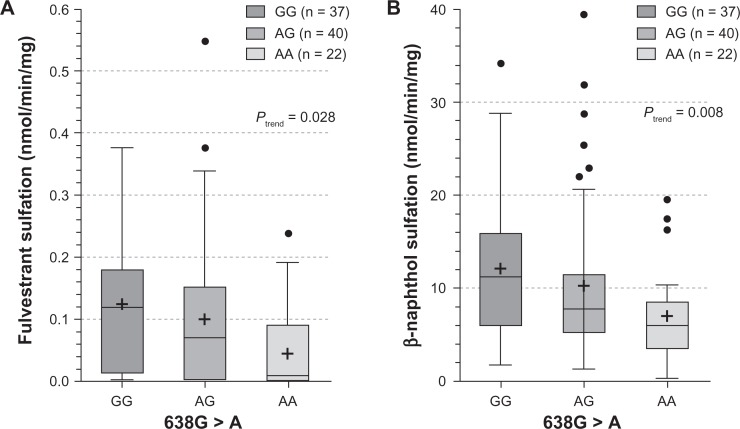
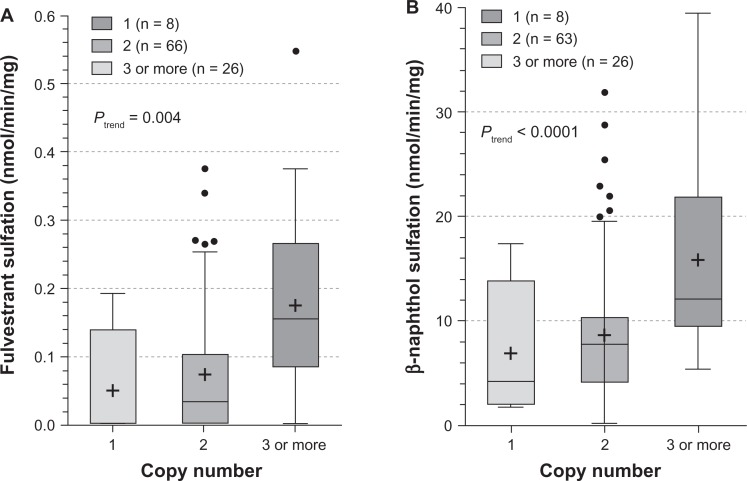
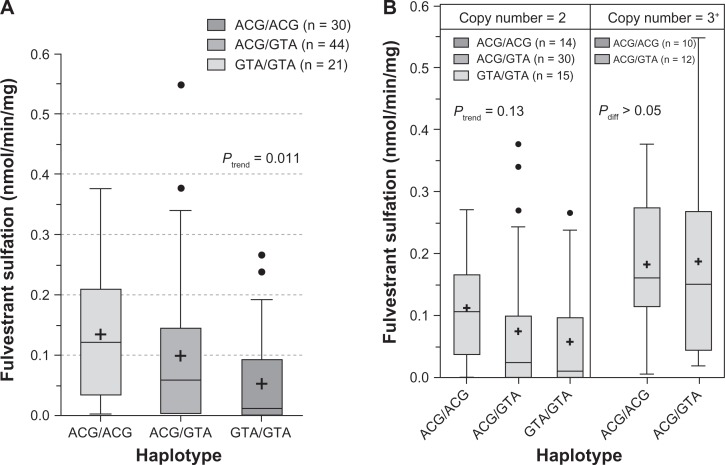
The influence of SULT1A1 genotypes on fulvestrant sulfation in human liver cytosols was then explored. When SULT1A1*1/*2 (638G > A) was examined, both fulvestrant (Figure 4A; P = 0.028) and β-naphthol (Figure 4B; P = 0.008) sulfation were significantly associated with SULT1A1 genotype. When the relationship of SULT1A1 copy number to fulvestrant (Figure 5A, P = 0.004) and β-naphthol (Figure 5B; P < 0.0001) sulfation was assessed, there was found to be a significant correlation between copy number and SULT1A1 activity. The study also examined the association between haplotypes of newly reported 3′ UTR variants18 and fulvestrant sulfation. These variants are in linkage disequilibrium with SULT1A1*1/*2 and have been demonstrated to exert more of an effect on SULT1A1 phenotype than the SULT1A1*1/*2 variant. As shown in Figure 6A, these haplotypes were significantly associated with fulvestrant sulfation. When stratified by copy number, however, the trend was no longer significant (Figure 6B).
Figure 4.
SULT1A1*1/*2 (638G > A) influence on the sulfation of fulvestrant (4A) and β-naphthol (4B). Enzymatic activity was determined either colorimetrically (for β-naphthol) or by LC-MS/MS. Genotype–phenotype relationships were assessed by analysis of variance with phenotype as the dependent variable.
Abbreviation: LC-MS/MS, liquid chromatography–mass spectrometry/tandem mass spectrometry.
Figure 5.
Influence of SULT1A1 copy number on fulvestrant (5A) and β-naphthol (5B) sulfation. Enzymatic activity was determined either colorimetrically (for β-naphthol) or by LC-MS/MS. Copy number–phenotype relationships were assessed by analysis of variance with phenotype as the dependent variable.
Abbreviation: LC-MS/MS, liquid chromatography–mass spectrometry/tandem mass spectrometry.
Figure 6.
Influence of SULT1A1 3′-UTR haplotypes on fulvestrant (A) and β-naphthol (B) sulfation. Enzymatic activity was determined either colorimetrically (for β-naphthol) or by LC-MS/MS. Haplotype–phenotype relationships were assessed by analysis of variance with phenotype as the dependent variable.
Abbreviation: LC-MS/MS, liquid chromatography–mass spectrometry/tandem mass spectrometry.
Discussion
Fulvestrant metabolism involves oxidation, aromatic hydroxylation, and conjugation reactions at the 2, 3, and 17 positions of the steroid nucleus. Identified metabolites are either less active or display similar activity as the parent compound. Sulfates and glucuronides of the parent compound are found in similar proportions to conjugated forms of the individual phase I metabolites (all < 10%) but, overall, the total conjugated metabolites make up a much larger proportion, with sulfation playing a predominant role.8 Because each SULT enzyme displays a distinct pattern of tissue distribution, identifying isoforms involved in the sulfation of a given molecule is required for a better understanding of its pharmacokinetic properties.
Previous studies have identified both fulvestrant-3-glucuronide and fulvestrant-17-glucuronide, but in the present study, only the formation of fulvestrant-3-sulfate was detected. Since an authentic standard for the fulvestrant-17-sulfate was not available, it is possible that low levels of this metabolite could have also been produced. Of the nine SULT isoforms examined, only SULT1A1 and SULT1E1 exhibited enzymatic activity toward fulvestrant. Even though SULT1E1 is more efficient in catalyzing fulvestrant than SULT1A1, correlation analysis of fulvestrant sulfation in human liver showed a significant correlation with β-naphthol sulfation (diagnostic substrate for SULT1A1) but not with 17β-estradiol (diagnostic substrate for SULT1E1). This may be due to the fact that SULT1A1 is the most highly expressed hepatic sulfotransferase, while expression levels of SULT1E1 are relatively low in liver. When enzyme kinetic characteristics were compared between human liver cytosol and recombinant SULT1A1, virtually identical kinetic constants were obtained, suggesting that SULT1A1 is the primary hepatic SULT participating in fulvestrant sulfation.
Additional support for the importance of SULT1A1 in fulvestrant metabolism is the effect of SULT1A1 genotype on fulvestrant sulfation. The SULT1A1*1/*2 SNP, where the *2 variant is associated with decreased enzymatic activity, was significantly associated with both fulvestrant and β-naphthol sulfation. Likewise, the 3′-SNPs, which are in strong linkage disequilibrium with the SULT1A1*1/*2 SNP, were strongly associated with sulfation activity. There was also a significant correlation found between SULT1A1 copy number variants with activity toward either substrate. However, when SNPs were stratified by copy number, the trend became insignificant, most likely due to small numbers in each category. Larger studies are needed to truly define these relationships. Likewise, functional SNPs have been reported in the SULT1E1 coding region,32 but these SNPs occur at a frequency of less than 1% and would therefore require a substantially larger study population to examine their contribution.
In addition to the role of SULT1A1 in the hepatic metabolism of fulvestrant, expression of SULT1A1 in breast tumors could influence tumor response to this therapy. Numerous studies have shown that SULT1A1 expression in normal breast tissue is low, but expression is upregulated in breast tumors, while expression of SULT1E1 is evident in normal breast epithelia, but low in breast tumors.33–42 For this reason, genetic and epigenetic factors influencing the tumor expression levels of SULT1A1 could predict response to fulvestrant therapy. However, SULT1E1 exhibited significantly higher activity towards fulvestrant, and its contribution to the overall metabolism in tissues expressing SULT1E1 may be substantial. Further studies are needed to fully define the role of sulfation in fulvestrant metabolism.
Limitations of the study include the exclusion of all races except Caucasians. Since allele frequencies of SULTs vary across ethnic groups, it will be necessary to accrue more samples before examining fulvestrant sulfation in other ethnic groups. Future studies will include the search for other genetic variants that could influence sulfation in order to more fully elaborate the contribution of genetic variants to fulvestrant sulfation. Studies examining SULT1A1 SNPs in a clinical trial population who receive fulvestrant for metastatic breast cancer are currently underway.
In summary, the formation of sulfated fulvestrant from fulvestrant in vitro is mediated predominantly by SULT1A1 in liver, the expression and activity of which varies substantially between individuals. A similar degree of variability can be expected in the formation of sulfated fulvestrant in vivo. Activity toward fulvestrant in human liver cytosols was significantly associated with SULT1A1*1/*2 genotype, SULT1A1 3′-UTR genotype, and SULT1A1 copy number. Future studies of fulvestrant pharmacogenomics should include functional genetic variants of both SULT1A1 and SULT1E1, which could contribute to treatment decisions for those with unfavorable SULT genotypes in the future.
Acknowledgments
This work was supported by the National Cancer Institute (R01CA118981 to SK) and by AstraZeneca Pharmaceuticals.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Dhingra K. Selective estrogen receptor modulation: the search for an ideal hormonal therapy for breast cancer. Cancer Invest. 2001;19(6):649–659. doi: 10.1081/cnv-100104293. [DOI] [PubMed] [Google Scholar]
- 2.Curran M, Wiseman L. Fulvestrant. Drugs. 2001;61(6):807–813. 814. doi: 10.2165/00003495-200161060-00013. discussion. [DOI] [PubMed] [Google Scholar]
- 3.Wakeling AE, Bowler J. Steroidal pure antioestrogens. J Endocrinol. 1987;112(3):R7–R10. doi: 10.1677/joe.0.112r007. [DOI] [PubMed] [Google Scholar]
- 4.McClelland RA, Gee JM, Francis AB, et al. Short-term effects of pure antioestrogen ICI 182780 treatment on oestrogen receptor, epidermal growth factor receptor and transforming growth factor-alpha protein expression in human breast cancer. Eur J Cancer. 1996;32A(3):413–416. doi: 10.1016/0959-8049(95)00517-x. [DOI] [PubMed] [Google Scholar]
- 5.Nicholson RI, Gee JM, Manning DL, et al. Responses to pure antiestrogens (ICI 164384, ICI 182780) in estrogen-sensitive and -resistant experimental and clinical breast cancer. Ann N Y Acad Sci. 1995;761:148–163. doi: 10.1111/j.1749-6632.1995.tb31376.x. [DOI] [PubMed] [Google Scholar]
- 6.Hu XF, Veroni M, De Luise M, et al. Circumvention of tamoxifen resistance by the pure anti-estrogen ICI 182,780. Int J Cancer. 1993;55(5):873–876. doi: 10.1002/ijc.2910550529. [DOI] [PubMed] [Google Scholar]
- 7.Lykkesfeldt AE, Madsen MW, Briand P. Altered expression of estrogen-regulated genes in a tamoxifen-resistant and ICI 164,384 and ICI 182,780 sensitive human breast cancer cell line, MCF-7/TAMR-1. Cancer Res. 1994;54(6):1587–1595. [PubMed] [Google Scholar]
- 8.Robertson JF, Harrison M. Fulvestrant: pharmacokinetics and pharmacology. Br J Cancer. 2004;90(Suppl 1):S7–S10. doi: 10.1038/sj.bjc.6601630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chouinard S, Tessier M, Vernouillet G, et al. Inactivation of the pure antiestrogen fulvestrant and other synthetic estrogen molecules by UDP-glucuronosyltransferase 1A enzymes expressed in breast tissue. Mol Pharmacol. 2006;69(3):908–920. doi: 10.1124/mol.105.015891. [DOI] [PubMed] [Google Scholar]
- 10.Falany JL, Falany CN. Regulation of estrogen activity by sulfation in human MCF-7 breast cancer cells. Oncol Res. 1997;9(11–12):589–596. [PubMed] [Google Scholar]
- 11.Glatt H. Sulfation and sulfotransferases 4: bioactivation of mutagens via sulfation. FASEB J. 1997;11(5):314–321. doi: 10.1096/fasebj.11.5.9141497. [DOI] [PubMed] [Google Scholar]
- 12.Weinshilboum RM, Otterness DM, Aksoy IA, et al. Sulfation and sulfotransferases 1: sulfotransferase molecular biology: cDNAs and genes. FASEB J. 1997;11(1):3–14. [PubMed] [Google Scholar]
- 13.Weinshilboum R, Aksoy I. Sulfation pharmacogenetics in humans. Chem Biol Interact. 1994;92(1–3):233–246. doi: 10.1016/0009-2797(94)90066-3. [DOI] [PubMed] [Google Scholar]
- 14.Van Loon J, Weinshilboum RM. Human platelet phenol sulfotransferase: familial variation in thermal stability of the TS form. Biochem Genet. 1984;22(11–12):997–1014. doi: 10.1007/BF00499627. [DOI] [PubMed] [Google Scholar]
- 15.Anderson RJ, Weinshilboum RM, Phillips SF, et al. Human platelet phenol sulphotransferase: assay procedure, substrate and tissue correlations. Clin Chim Acta. 1981;110(2–3):157–167. doi: 10.1016/0009-8981(81)90345-4. [DOI] [PubMed] [Google Scholar]
- 16.Raftogianis RB, Wood TC, Otterness DM, et al. Phenol sulfotransferase pharmacogenetics in humans: association of common SULT1A1 alleles with TS PST phenotype. Biochem Biophys Res Commun. 1997;239(1):298–304. doi: 10.1006/bbrc.1997.7466. [DOI] [PubMed] [Google Scholar]
- 17.Hebbring SJ, Adjei AA, Baer JL, et al. Human SULT1A1 gene: copy number differences and functional implications. Hum Mol Genet. 2007;16(5):463–470. doi: 10.1093/hmg/ddl468. [DOI] [PubMed] [Google Scholar]
- 18.Yu X, Dhakal IB, Beggs M, et al. Functional genetic variants in the 3′-untranslated region of sulfotransferase isoform 1A1 (SULT1A1) and their effect on enzymatic activity. Toxicol Sci. 2010;118(2):391–403. doi: 10.1093/toxsci/kfq296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferguson JR, Harding JR, Killick DA, et al. Putative metabolites of fulvestrant, an estrogen receptor downregulator. Improved glucuronidation using trichloroacetimidates. J Chem Soc Perkin Trans. 2001;1:3037–3041. [Google Scholar]
- 20.King RS, Teitel CH, Kadlubar FF. In vitro bioactivation of N-hydroxy-2-amino-alpha-carboline. Carcinogenesis. 2000;21(7):1347–1354. [PubMed] [Google Scholar]
- 21.Frame LT, Ozawa S, Nowell SA, et al. A simple colorimetric assay for phenotyping the major human thermostable phenol sulfotransferase (SULT1A1) using platelet cytosols. Drug Metab Dispos. 2000;28(9):1063–1068. [PubMed] [Google Scholar]
- 22.Falany JL, Krasnykh V, Mikheeva G, et al. Isolation and expression of an isoform of rat estrogen sulfotransferase. J Steroid Biochem Mol Biol. 1995;52(1):35–44. doi: 10.1016/0960-0760(94)00147-e. [DOI] [PubMed] [Google Scholar]
- 23.Falany CN, Krasnykh V, Falany JL. Bacterial expression and characterization of a cDNA for human liver estrogen sulfotransferase. J Steroid Biochem Mol Biol. 1995;52(6):529–539. doi: 10.1016/0960-0760(95)00015-r. [DOI] [PubMed] [Google Scholar]
- 24.Falany CN, Vazquez ME, Kalb JM. Purification and characterization of human liver dehydroepiandrosterone sulphotransferase. Biochem J. 1989;260(3):641–646. doi: 10.1042/bj2600641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang J, Falany JL, Falany CN. Expression and characterization of a novel thyroid hormone-sulfating form of cytosolic sulfotransferase from human liver. Mol Pharmacol. 1998;53(2):274–282. doi: 10.1124/mol.53.2.274. [DOI] [PubMed] [Google Scholar]
- 26.Wilborn TW, Comer KA, Dooley TP, et al. Sequence analysis and expression of the cDNA for the phenol-sulfating form of human liver phenol sulfotransferase. Mol Pharmacol. 1993;43(1):70–77. [PubMed] [Google Scholar]
- 27.Cook IT, Leyh TS, Kadlubar SA, Falany CN. Structural rearrangement of SULT2A1: effects on dehydroepiandrosterone and raloxifene sulfation. Horm Mol Biol Clin Investig. 2010;1(2):81–87. doi: 10.1515/HMBCI.2010.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cook IT, Duniec-Dmuchowski Z, Kocarek TA, et al. 24-hydroxycholesterol sulfation by human cytosolic sulfotransferases: formation of monosulfates and disulfates, molecular modeling, sulfatase sensitivity, and inhibition of liver x receptor activation. Drug Metab Dispos. 2009;37(10):2069–2078. doi: 10.1124/dmd.108.025759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bell JE, Bell ET. Proteins and Enzymes. Englewood Cliffs, NJ: Prentice-Hall; 1988. [Google Scholar]
- 30.Weinshilboum RM. Phenol sulfotransferase in humans: properties, regulation, and function. Fed Proc. 1986;45(8):2223–2228. [PubMed] [Google Scholar]
- 31.Nowell S, Ambrosone CB, Ozawa S, et al. Relationship of phenol sulfotransferase activity (SULT1A1) genotype to sulfotransferase phenotype in platelet cytosol. Pharmacogenetics. 2000;10(9):789–797. doi: 10.1097/00008571-200012000-00004. [DOI] [PubMed] [Google Scholar]
- 32.Adjei AA, Thomae BA, Prondzinski JL, et al. Human estrogen sulfotransferase (SULT1E1) pharmacogenomics: gene resequencing and functional genomics. Br J Pharmacol. 2003;139(8):1373–1382. doi: 10.1038/sj.bjp.0705369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pasqualini JR. Estrogen sulfotransferases in breast and endometrial cancers. Ann N Y Acad Sci. 2009;1155:88–98. doi: 10.1111/j.1749-6632.2009.04113.x. [DOI] [PubMed] [Google Scholar]
- 34.Smuc T, Rizner TL. Expression of 17beta-hydroxysteroid dehydrogenases and other estrogen-metabolizing enzymes in different cancer cell lines. Chem Biol Interact. 2009;178(1–3):228–233. doi: 10.1016/j.cbi.2008.10.038. [DOI] [PubMed] [Google Scholar]
- 35.Singer CF, Fink-Retter A, Gschwantler-Kaulich D, et al. Selective spatial upregulation of intratumoral stromal aromatase in breast cancer patients: evidence for imbalance of local estrogen metabolism. Endocr Relat Cancer. 2006;13(4):1101–1107. doi: 10.1677/erc.1.01230. [DOI] [PubMed] [Google Scholar]
- 36.Aust S, Jaeger W, Klimpfinger M, et al. Biotransformation of melatonin in human breast cancer cell lines: role of sulfotransferase 1A1. J Pineal Res. 2005;39(3):276–282. doi: 10.1111/j.1600-079X.2005.00246.x. [DOI] [PubMed] [Google Scholar]
- 37.Aust S, Obrist P, Klimpfinger M, et al. Altered expression of the hormone- and xenobiotic-metabolizing sulfotransferase enzymes 1A2 and 1C1 in malignant breast tissue. Int J Oncol. 2005;26(4):1079–1085. [PubMed] [Google Scholar]
- 38.Suzuki T, Nakata T, Miki Y, et al. Estrogen sulfotransferase and steroid sulfatase in human breast carcinoma. Cancer Res. 2003;63(11):2762–2770. [PubMed] [Google Scholar]
- 39.Suzuki T, Moriya T, Ishida T, et al. In situ production of estrogens in human breast carcinoma. Breast Cancer. 2002;9(4):296–302. doi: 10.1007/BF02967607. [DOI] [PubMed] [Google Scholar]
- 40.Pasqualini JR, Chetrite GS. Estrone sulfatase versus estrone sulfotransferase in human breast cancer: potential clinical applications. J Steroid Biochem Mol Biol. 1999;69(1–6):287–292. doi: 10.1016/s0960-0760(99)00082-5. [DOI] [PubMed] [Google Scholar]
- 41.Falany JL, Falany CN. Expression of cytosolic sulfotransferases in normal mammary epithelial cells and breast cancer cell lines. Cancer Res. 1996;56(7):1551–1555. [PubMed] [Google Scholar]
- 42.Falany JL, Lawing L, Falany CN. Identification and characterization of cytosolic sulfotransferase activities in MCF-7 human breast carcinoma cells. J Steroid Biochem Mol Biol. 1993;46(4):481–487. doi: 10.1016/0960-0760(93)90102-3. [DOI] [PubMed] [Google Scholar]