Abstract
Embryonic stem (ES) cells are derived from blastocysts. They can differentiate into the three embryonic germ layers and essentially any type of somatic cells. Therefore, they hold great potentials in tissue regeneration therapy. The ethical issues associated with the use of human embryonic stem cells are resolved by the technical break-through of generating induced pluripotent stem (iPS) cells from various types of somatic cells. However, how ES and iPS cells self-renew and maintain their pluripotency is still largely unknown in spite of the great progresses that have been made in the last two decades. Integrative genome-wide approaches, such as gene expression microarray, chromatin immunoprecipitation based microarray (ChIP-chip) and chromatin immunoprecipitation followed by massive parallel sequencing (ChIP-seq) offer unprecedented opportunities to elucidate the mechanism of the pluripotency, reprogramming and DNA damage response of ES and iPS cells. This review summarized the fundamental biological questions about ES and iPS cells and reviewed the recent advances in ES and iPS cell research using genome-wide technologies. In the end, we offered our perspectives on the future of genome-wide studies on stem cells.
1. Introduction to the platforms for genome-wide analysis
One of the breakthroughs in modern stem cell biology is the cloning of embryonic stem (ES) cells from mouse and human 1–3. These cells are derived from the inner cell mass of developing blastocysts and can be cultured in vitro indefinitely without losing the ability to develop into all the three germ layers in an embryo. The pluripotency of ES cells is demonstrated by their ability to form a whole organism in a tetraploid complementation assay. ES cells not only serve as a good model system to study developmental processes but also have important therapeutic values. Human embryonic stem (hES) cells hold great potentials in tissue regeneration and personalized therapy. It is critical for ES cells to maintain their genomic stability because mutations generated by exogenous and endogenous DNA damage, and/or by re-activation of endogenous retroviruses in the genome could be detrimental to all the offspring cells. Therefore, ES cells are also endowed with an exquisite ability to cope with insults that cause genomic instability 4. The creation induced pluripotent stem (iPS) cells from somatic cells represents another major advance in stem cell research. It is of great interest to study how iPS cells are formed and how they are different from ES cells.
The availability of genome-wide approaches, such as gene expression microarray, chromatin immunoprecipitation assay based microarray (ChIP-chip), massive parallel sequencing of RNA (RNA sequencing) and chromatin immunoprecipitation assay based massive parallel sequencing (ChIP-seq), offers an unprecedented opportunity to explore the mechanisms underlying these fundamental biological questions of ES and iPS cells.
1) Gene expression microarray and NanoString platforms
Gene expression microarray is probably one of the earliest platforms for genome-wide study. It can measure the expression levels of thousands of genes simultaneously. Generally, reverse transcribed cDNAs are labeled with various fluorescent dyes and then hybridized to the microarray that contains thousands of gene-specific probes. The abundance of each transcript is determined by the relative intensity of fluorescence signal. One of the biggest advantages of this technique is its high throughput compared to quantitative realtime PCR (qPCR). Old versions of gene expression microarrays contain probes that hybridize to regions close to the 3’ end of transcripts and were widely used for numerous studies on ES cells5–8. However, they failed to detect alternative splicing that might play important roles during differentiation. Some latest version of microarrays, such as exon microarrays, can detect alternative splicing of genes. Thus, they offer another level of complexity to study the gene expression and regulation. To our best knowledge, there has been no formal report to link alternative splicing to the “stemness” or differentiation of ES cells. In addition, the mRNA levels do not always correlate well with the protein levels. Indeed, using mass spectrometry, many proteins found over-expressed in ES cells were not detected by gene expression microarray9.
The dynamic range and sensitivity of microarray are normally lower than qPCR. Therefore, gene expression microarray sometimes fails to detect subtle gene expression changes which might have biological meanings. A multiplex system called NanoString nCounter gene expression system filled the gap between gene expression microarray and qPCR 10. NanoString uses a capture probe and a reporter probe to detect a transcript of interest. The abundance of a transcript is determined by an automated system called nCounter System. NanoString is based on hybridization and no reverse transcriptase or other enzymes are used in the process. It has similar sensitivity to qPCR and much better sensitivity than gene expression microarray10. Therefore, for detecting a small set of genes, such as a gene family, or validating the result from gene expression microarray, NanoString could be a good choice.
2) RNA sequencing platforms
RNA sequencing is based on massive parallel sequencing (also called deep sequencing). It is an alternative platform to detect the expression level of thousands of genes. Deep sequencing is a hybridization-free approach to sequence millions of DNA tags. The length of sequenced tags is normally 25–100 base-pair long. Afterwards, the sequenced tags are aligned to a pre-sequenced genome to determine the identity of each DNA fragment. When the number of sequenced tags is big enough, the relative abundance of each tag can be determined and is statistically correlated with the expression levels of its corresponding mRNAs. Because of the alignment step, the genome sequence needs to be determined before performing deep sequencing. When performing RNA sequencing, RNA is reversely transcribed to DNA, which then is subject to deep sequencing. Compared to gene expression microarray, it is more powerful to discover novel RNAs including non-coding RNAs11, 12. The basic principle of RNA sequencing is similar to an earlier platform called Serial Analysis of Gene Expression (SAGE) 13. Although RNA sequencing is still in its infancy and the cost is relatively high, it has the potential to completely replace the gene expression microarray because of its high sensitivity, preciseness and essentially unlimited dynamic range.
Recently, a technology called HeliScope Genetic Analysis system has been introduced 14. HeliScope Genetic Analysis does not involve reverse transcription or ligation/amplification step. Instead, it captures single molecule of RNA on a solid surface and directly sequences a large number of RNA molecules simultaneously. Although the error rate needs to be further improved, this technology has the potential to map the complete transcriptome of a genome at single-nucleotide resolution. Importantly, the strand specificity can be easily determined since no DNA synthesis is required prior to the sequencing. This feature is particularly useful when the strand information about a RNA molecule is not available.
3) ChIP-chip platform
ChIP-chip technology was initially developed for mapping the interaction between a transcription factor and DNA in a genome-wide manner15. Later on, it has been shown to be extremely powerful in epigenetic studies, such as measuring the levels of histone modifications. Briefly, a specific antibody recognizing a transcription factor or a histone modification is used to precipitate its associated DNA. The precipitated DNA and input DNA are then labeled with different fluorescence dyes and subsequently hybridized to microarray to detect the relative abundance of precipitated DNA. Because of the use of hybridization, ChIP-chip inherits most of the technical advantages and disadvantages of gene expression microarray platform. In addition, for large genomes, probe design is extremely time-consuming and costly, which could become a major challenge for some custom designed arrays. Several previous ES cell studies utilized promoter arrays covering the promoter regions of well annotated genes 5, 8. Although several important conclusions were drawn from these studies, the limited coverage (5–7%) of the genome could under-estimate the importance of intergenic regions. Knowledge from studying other transcription factors such as FoxA1 and estrogen receptor suggests that intergenic regions can also be functionally important16. This limitation was overcome later by using genomic tiling array that covers essentially the whole human genome7, 17.
4) ChIP-seq and other alternative platforms
Similar to RNA sequencing, ChIP-seq is also based on deep sequencing. After immunoprecipitation, the precipitated DNA is sequenced and aligned to the genome sequence to determine the location of the sequenced DNA in the genome. Compared to ChIP-chip, ChIP-seq does not require probe design. In general, deep sequencing-based approaches (RNA sequencing and ChIP-seq) are more powerful to generate genome-wide information than hybridization-based approaches (gene expression microarray and ChIP-chip). However, deep sequencing fails to focus on certain regions of the genome. Therefore, for large genomes, such as human genome that contains about 3 billion bases, enough sequencing depth is required to generate statistically significant data, in particular, at single-nucleotide resolution. The assumption is that the whole genome has to be sequenced as least once to make “yes/no” judgement call. This is particularly true for proteins that do not have substantial bias to certain regions of the genome, such as histones. For transcription factors or histone modifications, 10–20 millions of reads are generally sufficient for identifying the binding sites because they tend to concentrate at specific regions of the genome. As the sequencing depth continues to improve, the loci with lower enrichment can be identified.
The large amount of data generated by ChIP-seq is a daunting challenge for the bioinformatic and bio-statistic infrastructure of a lab or even some small institutes. Depending on the nature of the study, ChIP-chip sometimes is a practical choice. ChIP-chip requires relatively less sophisticated bioinformatic and bio-statistic tools and is particularly useful for investigating small genomes, such as yeast genome. A noteworthy new technology called SureSelect Target Enrichment System developed by Agilent is based on both hybridization and subsequent deep sequencing. It uses location-specific probes to enrich the regions of interest via hybridization, and then deep-sequence the enriched regions. Thus, it is especially useful for the studies in which the regions of interest are known. The enrichment step could greatly increase the statistical power of the deep sequencing result. Before the adoption of ChIP-seq, a very similar technique called ChIP Paired-End diTag (ChIP-PET) was developed to study the genomic binding sites of pluripotent transcription factors or other transcription factors 18, 19. ChIP-PET has been discussed in another review20, and therefore it will not be covered in this article.
ChIP-chip and ChIP-seq both involve purification, enzymatic and PCR steps to amplify the immunoprecipitated DNA. Normally, nanogram quantity of DNA is required to perform such studies. The new HeliScope Genetic Analysis system discussed above allows the sequencing of single molecule and does not use PCR-based approach 21. Importantly, only 50 picogram of DNA is needed to perform viral genome sequencing and genome-wide profiling of the binding sites of histone modifications 21, 22. Therefore, the technology of direct sequencing enables genome-wide studies to be performed in cell types that are difficult to obtain large amount, such as adult stem cells.
This review will summarize recent progress of using genome-wide approaches, including gene expression microarray, RNA sequencing, ChIP-chip and ChIP-seq to investigate the fundamental biological questions of ES and iPS cells: 1) How ES cells maintain pluripotency? 2) How iPS cells are formed from somatic cells? 3) How ES cells maintain their genomic stability? 4) How ES cells epigenetically silence the retroviruses? Because this is a rapidly expanding area, we apologize to our colleagues if this review did not cover their research. The unique features of each platform were summarized in Table 1.
Table 1.
Commonly used genome-wide platforms in ES cell studies and their unique features
| Principle | Platforms | Applications | Coverage | Notes |
|---|---|---|---|---|
| Hybridization based | Gene expression microarray | Gene expression Non-coding RNA expression Alternative splicing analysis |
Partial genome or whole genome | Need probe designing High cost for large genome Low dynamic range Less powerful than RNA sequencing in discovering novel coding and non-coding RNAs |
| ChIP-chip | Mapping DNA-protein interaction DNA methylation analysis |
Partial genome or whole genome | Need probe designing High cost for large genome Low dynamic range Low resolution |
|
| Sequencing based | RNA sequencing | Gene expression Non-coding RNA expression Alternative splicing analysis |
Whole genome | Pre-sequenced genome Unlimited dynamic range Easy to discover novel transcripts or non-coding RNAs |
| ChIP-seq | Mapping DNA-protein interaction DNA methylation analysis |
Whole genome | Pre-sequenced genome Unlimited dynamic range High resolution |
|
| DNA sequencing | Genome sequencing DNA methylation analysis |
Whole genome | Pre-sequenced genome Unlimited dynamic range Single nucleotide resolution |
2. Important biological questions about ES cells
ES cells and their “off-spring” cells have the same DNA sequences. However, only ES cells self-renew infinitely, and can develop into the three germ layers, the ability named as pluripotency. How ES cells maintain their pluripotency is an important biological question. Elucidation of the underlying mechanism of pluripotency will greatly facilitate the clinical applications of ES cells. Genome-wide studies undoubtedly provide us unmatched opportunities to address this question.
1) Maintaining the pluripotency of ES cells
a. Mapping the binding sites of pluripotent transcription factors
The pluripotency and self-renewal of embryonic stem cells are maintained by internal transcription network governed by several transcription factors which are referred to as pluripotent transcription factors. The list of these pluripotent factors is still expanding but the major players include Oct4 (Pou5f1), Sox2 and Nanog. Through mapping the binding loci of these transcription factors using ChIP-chip platforms, the transcriptional circuitry of ES cells is delineated5. These pluripotent transcription factors form feedback or feedforward loops to regulate their own expression as well as that of other genes. In the meanwhile, they repress the developmental genes and keep ES cell poised for development signal. The underlying mechanisms as to how these pluripotent factors “decide” when to activate or repress are largely unknown. Using ChIP-seq, the binding sites of 13 pluripotent factors were mapped in mouse ES cells at a genome-wide scale 23. One important observation emerged from this study is that genes bound by several transcription factors generally have higher expression levels in ES cells than those bound by a single factor. An follow-up question is whether the co-occupancy of these transcription factors is determined merely DNA sequence or certain epigenetic events are involved.
The internal transcriptional circuitry is connected to external signals, such as leukemia inhibitory factor (LIF), bone morphogenetic proteins (BMPs) and Wnts, via transcriptional factor Stat3, Smad1 and Tcf3, respectively24–26. In un-differentiation condition, ES cells self-renew. Upon withdrawal of LIF, ES cells initiate a differentiation program characteristic of the loss of Oct4, Sox2 and Nanog, which probably is mediated by mitogen-activated protein kinase (MAPK) pathway 27. The exact events occurring during the transition from un-differentiation to differentiation are currently unclear. Systematical mapping of the binding loci of pluripotent transcription factors and measuring the global gene expression changes will provide the molecular insights.
Recent genome-wide studies on Tcf3 using ChIP-chip in ES cells serve as good examples of how genome-wide studies provide molecular insights into the underlying mechanism of pluripotency 25, 26. Tcf3 occupies a large number of genes that are also bound by Oct4, Sox2 and/or Nanog in ES cells. Therefore, canonical Wnt signaling appears to play critical roles in ES cells. It is worth pointing out that embryos without beta-catenin, an important downstream mediator of canonical Wnt signaling, successfully pass the developmental stage of blastocysts and arrest at gastrulation phase, suggesting that the canonical Wnt signaling is not essential for ES cell maintenance 28. Therefore, it is possible that non-canonical Wnt signaling is also critical for mediating the function of Wnts in ES cells.
b. Unique features of histone modification patterns in ES cells
Epigenetic differences between ES and somatic cells are subjects of extensive studies. A noteworthy epigenetic feature of ES cells is that some genes, in particular developmental genes, are simultaneously marked with bivalent histone modifications, i.e., histone H3 lysine 4 (H3K4me3) and lysine 27 (H3K27me3) trimethylation 29. This novel observation was initially discovered using ChIP-chip 29, which later on was confirmed by ChIP-seq 30. The bivalent marks are featured with large blocks of H3K27me3 and small clusters of H3K4me3. Therefore, ChIP-chip and ChIP-seq are powerful enough to detect this pattern which normally will escape the “radar” of classical ChIP assay. Upon differentiation, these bivalent genes either put on an active mark, H3K4me3, or a repressive mark, H3K27me3. Using gene expression microarray, it was shown that the expression levels of these bivalent genes are normally low in ES cells29. It is postulated that the bivalent marks poise the developmental genes for activation during development. However, it is unclear how these bivalent modifications are established and resolved during the developmental process. Mixed Lineage Leukemia (MLL) and Polycomb group (PRC) complexes are definitely playing critical roles in these events. Precise mapping of the binding sites of each member of MLL and PRC complexes during the differentiation of ES cells should shed light on the regulation of bivalent modification. Answering this question also requires collecting samples from the various developmental stages before blastocyst, which cast technical challenges for genome-wide studies such as ChIP-chip and ChIP-seq.
Another distinct histone modification pattern between ES and differentiated cells is histone H3 lysine 9 dimethylation (H3K9me2). Using ChIP-chip, Wen et al. found that H3K9me2 forms large blocks (designated as LOCKs) in differentiated cells 31. The LOCKs cover up to 4.3 megabase regions in differentiated cells and 31% of the genome. While in ES cells, they only represent about 4% of the genome. Interestingly, the LOCKs appear to lock the differentiated cells in their lineages. For example, genes with non-liver function are highly enriched with LOCKs in liver tissue while those playing roles in liver are not covered by LOCKs. Similar to the bivalent modifications, the mechanisms as to how LOCKs are established during the differentiation and whether the deregulation of LOCKs could contribute to tumorigenesis are not fully appreciated. Because ES and somatic cells have the same genomic DNA, epigenetic mechanism is definitely the major player for the establishment and maintenance of LOCKs.
c. Roles of histone variants in ES cells
In addition to the major histones, H1, H2A, H2B, H3 and H4, there are numerous histone variants such as H3.1, H3.3, H2AX, H2AZ and macroH2A 32. Recently, the role of histone variant H2AZ has been studied in mouse ES cells. H2AZ depletion did not affect the self-renewal of ES cells but the pluripotency was severely affected 17. These observations agree to the in vivo findings very nicely 33. The authors further studied the underlying mechanism by which H2AZ regulates the developmental potentials of ES cells. Using ChIP-chip, H2AZ and SUZ12, a critical component of polycomb group proteins, were found to co-localize and co-regulate numerous developmental genes. However, H2AZ has not been found to co-exist in the polycomb group protein complex, suggesting that the cross-talk between H2AZ and polycomb group protein is a dynamic process, which may be difficult to be detected by conventional biochemical assays. The recruitment of H2AZ is mediated by chaperone protein SWR1 (also designated as SRCAP)34. It remains to be determined whether the deposition of H2AZ to SUZ12 occupied loci is SWR1-dependent.
Recently, the genomic binding sites of histone variant H3.3 have been elegantly documented using ChIP-seq35. Because there is not available ChIP grade antibody available for H3.3, the authors added HA or EYFP tag to endogenous H3.3 protein using the novel zinc finger nuclease technology. This allows them to interrogate the binding of endogenous H3.3. Several important findings were generated from this study. First, the distribution of H3.3 is altered in differentiated cells versus ES cells, suggesting a role of H3.3 in ES cells. Second, the canonical H3.3 chaperone, HIRA, is not the only protein that controls the deposition of H3.3. Atrx is also critical for regulating the recruitment of H3.3, particularly to telomeric regions.
d. DNA methylation in ES cells
Genome-wide DNA methylations in mouse ES and ES-derived neural progenitor cells (NPC) were also mapped using ChIP-seq 36. Results from this study revealed that DNA methylation correlates with histone modifications better than with underlying DNA sequence, suggesting that histone modifications play important roles in shaping the epigenome of ES cells and other cells types. Higher resolution of DNA methylations in human ES cells and fibroblasts were determined at single-nucleotide resolution using deep sequencing 37. This study found that DNA methylation is predominantly CpG methylation in somatic cells. While in human ES cells, a significant portion (about 25%) of DNA methylation is non-CpG methylation. Whether these non-CpG methylations play important roles in maintaining the stemness and regulating the pluripotency of ES cells needs to be further studied.
2) DNA damage response of ES cells
DNA damage is one of the major drivers of DNA mutation. How ES cells deal with DNA damage insults is an important biological question. Intuitively, DNA mutations in embryonic stem cells are more detrimental to an organism than those in adult stem cells and somatic cells. Therefore, ES cells must have their unique way to cope with DNA damage. Indeed, the spontaneous mutation rate in ES cells is 100 times less than differentiated cells, such as fibroblasts4. The tumor suppressor, p53, plays important roles in maintain the genomic stability of ES cells. Upon DNA damage, p53 quickly binds to the promoter of Nanog and represses its expression. The loss of Nanog leads to the differentiation of ES cells and restrict the potential DNA mutations to the damaged cells. Recently, using an integrative genome-wide approach, our group has discovered a novel function of p53 in ES cells38. Damaged ES cells quickly undergo apoptosis, and simultaneously secret Wnt ligands through p53 to act on neighboring ES cells. The secreted Wnt ligands delay the differentiation of neighboring cells, presumably giving them more time to divide and compensate for the loss of damaged ES cells. Therefore the stability of ES cell population is maintained. It is currently unclear why tumor suppressor p53 activates the expression of the Wnt ligands that are thought to be predominantly oncogenic in somatic cells 39. What is the mechanism that determines the ES-specific regulation of Wnts by p53? The possible explanations were discussed in another review article40.
3) Epigenetic silencing of retroviruses in ES cells
Endogenous retroviruses and retrovirus-like elements represent a significant portion of human and mouse genomes. Some of the active ones become a major threat to the genomic stability of ES cells. Epigenetic silencing of endogenous and exogenous retroviruses in ES cells presumably serves as an ES-specific defense mechanism to minimize the impact of these viruses on the genome. ES cells uses KAP1 (also called Trim28)-ESET (also called SETDB1) mediated histone H3 lysine 9 trimethylation (H3K9me3) and DNA methylation to silence the expression of these retroviruses 30, 41, 42. Using RNA sequencing, Rowe et al., found that depletion of KAP1 re-activated the expression of intracisternal A-type particles (IAP) through the loss of H3K9me3 41. Because most commercially available gene expression microarrays focus on the coding regions of genome, RNA sequencing is an ideal platform to study the expression of endogenous retroviruses and retrovirus-like elements. Since some of the retroviral elements are repetitive sequences and deep sequencing requires an alignment step, certain adaption is required before this technique is used in retrovirus study.
3. The biological questions about iPS cells
The generation of iPS cells revolutionized stem cell research 43, 44. Human iPS cells also can circumvent the ethical issue associated with using embryos44, 45. Since the establishment of iPS cells in 2006, our knowledge on iPS cells has significantly expanded. However, before the ultimate clinical applications of iPS cells, we need to know whether and how iPS cells are different from ES cells. Conventional biological assays, such as embryoid body formation, teratoma formation and in vitro differentiation assay, are invaluable to assess iPS cells. Genome-wide analyses, by mapping genetic and epigenetic landscapes of iPS cells, will facilitate the development of better approaches to make iPS cells and accelerate the generation of risk-free iPS cells.
1) Roles of reprogramming factors in making iPS cells
It has drawn a lot of attentions to generate iPS cells with rational design. Although the methods of generating iPS cells vary, the common theme is to introduce pluripotent or reprogramming transcription factors, such as Oct4/Sox2/Klf4/cMyc or Oct4/Sox2/Nanog/Lin28 into somatic or adult stem cells to reprogram them into ES cell status. Several groups are seeking to use chemical approach to make iPS cells 46, 47. So far, pure chemical approach to generate iPS cells has not been successful. Sridharan et al., uses ChIP-chip and gene expression microarray probed the roles of the reprogramming factors during the reprogramming process 48. The study found that c-Myc is involved in the early events of reprogramming while Oct4, Sox2 and Klf4 in the later stages. This result not only shed light on the reprogramming process, but also has ramifications in tumorigenesis. Many types of cancers have gene signature of embryonic stem cells and cMyc is one of the factors that regulate both cancer and stem cells gene expression49. Does this suggest that reprogramming is a natural tumorigenic process or that cMyc simply plays two separate roles during tumorigenesis and reprogramming? Or is there certain c-Myc downstream gene(s) that can replace cMyc during the iPS generation without causing tumorigenesis? Future genome-wide studies need to address these questions in order to harness the reprogramming role of cMyc and, in the meanwhile, minimize its tumorigenic function.
2) DNA methylation and histone modifications of iPS cells compared to ES cells and fibroblasts
DNA methylation, chromatin states and gene expression were systematically documented in mouse and human iPS cells 50, 51. In general, the DNA methylation pattern in iPS cells is similar to that in ES cells, although some local abnormality has been found51, 52. Using an integrative genomic approach, DNA de-methylation was shown to be one of the major barriers to reprogramming. Partially reprogrammed iPS (piPS) cells fail to demethylate the pluripotent genes 53. A notable genome-wide study on difference between human ES (hES) and human iPS (hiPS) cells indicates that hiPS cells at late passage are more similar to hES cells than those at early passage 54. Thus, induced pluripotency is a slow and stochastic event. Interestingly, the hypomethylated (iPS versus fibroblasts) differentially methylated regions (DMRs) are associated with the bivalent domains, which overlap with hypermethylated DMRs in colon cancers 51. Accumulating evidence has suggested that abnormal histone modifications and DNA methylation could contribute to cancers 55–59. Therefore, a careful interrogation of histone modification landscape and DNA methylation needs to be performed with genome-wide studies before the clinical application of iPS technology. Results from whole genome sequencing will be extremely useful to address this demand.
4. Future perspective and challenges
Genome-wide approaches have provided important insights into the maintenance of pluripotency, reprogramming, DNA damage response and epigenetic silencing of retroviruses. As we are transiting from genomic to epigenomic era, how epigenetic and chromatic events regulate these biological processes is a fascinating question60. There are more than thirty histone modifications and 10–20 pluripotent transcription factors that have been discovered. This list is rapidly expanding. The genome-wide bindings of some of these histone modifications and transcription factors have already been mapped in ES cells or somatic cells 8, 61, 62. A major challenge of genome-wide studies, in general, is to extract biologically meaningful information from the ocean of data generated from these studies. Obviously, it requires more powerful bioinformatic and bio-statistic tools and the close communication between biologists, bioinformaticists and/or bio-statisticians. Data sharing and mining will undoubtedly expedite the process of scientific discovery.
Because ES cells divide symmetrically, it is easy to obtain a large amount of materials to perform genome-wide study. For adult stem cells, such as hematopoietic stem cells and neural stem cells, they tend to divide asymmetrically in vivo and it is difficult to obtain a sufficient amount of cells to perform genome-wide studies. Therefore, novel sample isolation and preparation approaches for the rare cell populations are particularly useful for addressing their biological function 63, 64. Integrative genome-wide studies on ES cells will pave the road for the future studies on adult stem cells.
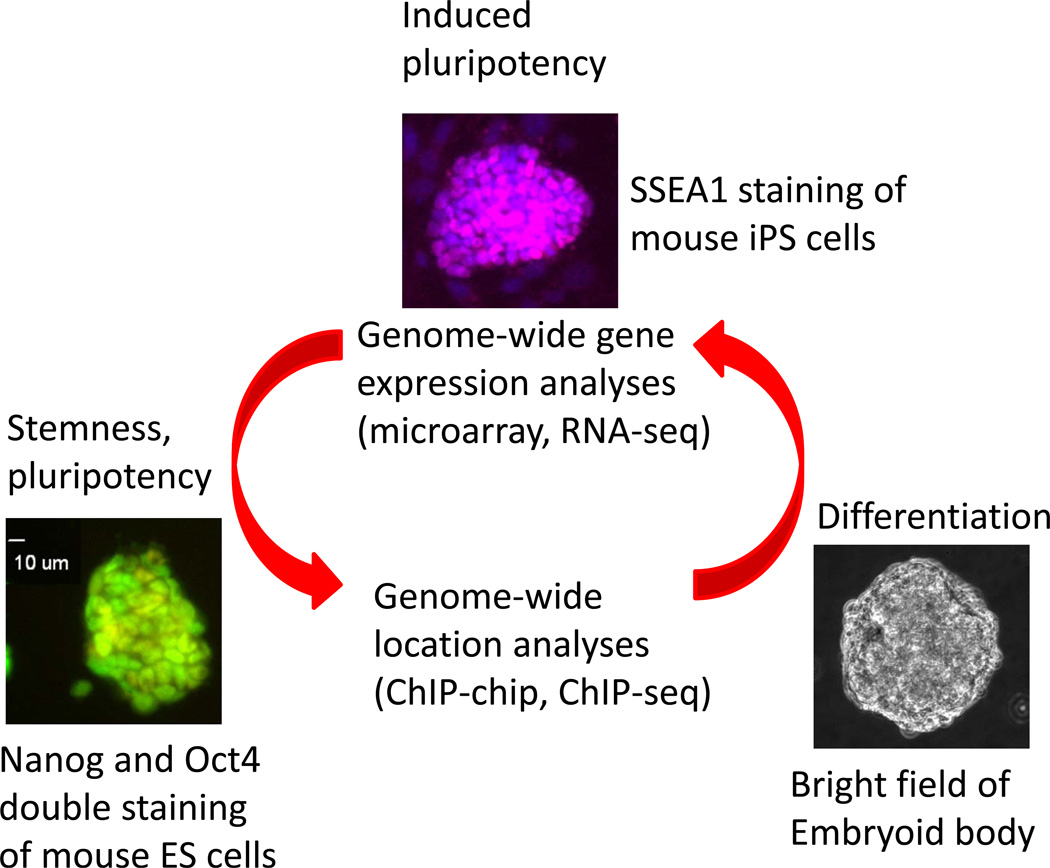
Figure 1.
Acknowledgements
This research was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute and Center for Cancer Research. We thank Dr. Nan Roche for critically reading the manuscript.
Footnotes
Conflict of Interests
The authors declare no conflict of interests.
References
- 1.Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 2.Martin GR. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci U S A. 1981;78:7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thomson JA, Itskovitz-Eldor J, Shapiro SS, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 4.Cervantes RB, Stringer JR, Shao C, et al. Embryonic stem cells and somatic cells differ in mutation frequency and type. Proc Natl Acad Sci U S A. 2002;99:3586–3590. doi: 10.1073/pnas.062527199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyer LA, Lee TI, Cole MF, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyer LA, Plath K, Zeitlinger J, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 7.Lee TI, Jenner RG, Boyer LA, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim J, Chu J, Shen X, et al. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van Hoof D, Passier R, Ward-Van Oostwaard D, et al. A quest for human and mouse embryonic stem cell-specific proteins. Mol Cell Proteomics. 2006;5:1261–1273. doi: 10.1074/mcp.M500405-MCP200. [DOI] [PubMed] [Google Scholar]
- 10.Geiss GK, Bumgarner RE, Birditt B, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol. 2008;26:317–325. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- 11.Babiarz JE, Ruby JG, Wang Y, et al. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev. 2008;22:2773–2785. doi: 10.1101/gad.1705308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marson A, Levine SS, Cole MF, et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Velculescu VE, Zhang L, Vogelstein B, et al. Serial analysis of gene expression. Science. 1995;270:484–487. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- 14.Ozsolak F, Platt AR, Jones DR, et al. Direct RNA sequencing. Nature. 2009;461:814–818. doi: 10.1038/nature08390. [DOI] [PubMed] [Google Scholar]
- 15.Ren B, Robert F, Wyrick JJ, et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- 16.Lupien M, Eeckhoute J, Meyer CA, et al. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell. 2008;132:958–970. doi: 10.1016/j.cell.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Creyghton MP, Markoulaki S, Levine SS, et al. H2AZ is enriched at polycomb complex target genes in ES cells and is necessary for lineage commitment. Cell. 2008;135:649–661. doi: 10.1016/j.cell.2008.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loh YH, Wu Q, Chew JL, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- 19.Wei CL, Wu Q, Vega VB, et al. A global map of p53 transcription-factor binding sites in the human genome. Cell. 2006;124:207–219. doi: 10.1016/j.cell.2005.10.043. [DOI] [PubMed] [Google Scholar]
- 20.Mathur D, Danford TW, Boyer LA, et al. Analysis of the mouse embryonic stem cell regulatory networks obtained by ChIP-chip and ChIP-PET. Genome Biol. 2008;9:R126. doi: 10.1186/gb-2008-9-8-r126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harris TD, Buzby PR, Babcock H, et al. Single-molecule DNA sequencing of a viral genome. Science. 2008;320:106–109. doi: 10.1126/science.1150427. [DOI] [PubMed] [Google Scholar]
- 22.Goren A, Ozsolak F, Shoresh N, et al. Chromatin profiling by directly sequencing small quantities of immunoprecipitated DNA. Nat Methods. 7:47–49. doi: 10.1038/nmeth.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen X, Xu H, Yuan P, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 24.Niwa H, Burdon T, Chambers I, et al. Self-renewal of pluripotent embryonic stem cells is mediated via activation of STAT3. Genes Dev. 1998;12:2048–2060. doi: 10.1101/gad.12.13.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cole MF, Johnstone SE, Newman JJ, et al. Tcf3 is an integral component of the core regulatory circuitry of embryonic stem cells. Genes Dev. 2008;22:746–755. doi: 10.1101/gad.1642408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tam WL, Lim CY, Han J, et al. T-cell factor 3 regulates embryonic stem cell pluripotency and self-renewal by the transcriptional control of multiple lineage pathways. Stem Cells. 2008;26:2019–2031. doi: 10.1634/stemcells.2007-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ying QL, Wray J, Nichols J, et al. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haegel H, Larue L, Ohsugi M, et al. Lack of beta-catenin affects mouse development at gastrulation. Development. 1995;121:3529–3537. doi: 10.1242/dev.121.11.3529. [DOI] [PubMed] [Google Scholar]
- 29.Bernstein BE, Mikkelsen TS, Xie X, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 30.Mikkelsen TS, Ku M, Jaffe DB, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wen B, Wu H, Shinkai Y, et al. Large histone H3 lysine 9 dimethylated chromatin blocks distinguish differentiated from embryonic stem cells. Nat Genet. 2009;41:246–250. doi: 10.1038/ng.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Henikoff S, Ahmad K. Assembly of variant histones into chromatin. Annu Rev Cell Dev Biol. 2005;21:133–153. doi: 10.1146/annurev.cellbio.21.012704.133518. [DOI] [PubMed] [Google Scholar]
- 33.Faast R, Thonglairoam V, Schulz TC, et al. Histone variant H2A.Z is required for early mammalian development. Curr Biol. 2001;11:1183–1187. doi: 10.1016/s0960-9822(01)00329-3. [DOI] [PubMed] [Google Scholar]
- 34.Mizuguchi G, Shen X, Landry J, et al. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science. 2004;303:343–348. doi: 10.1126/science.1090701. [DOI] [PubMed] [Google Scholar]
- 35.Goldberg AD, Banaszynski LA, Noh KM, et al. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell. 140:678–691. doi: 10.1016/j.cell.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meissner A, Mikkelsen TS, Gu H, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lister R, Pelizzola M, Dowen RH, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee KH, Li M, Michalowski AM, et al. A genomewide study identifies the Wnt signaling pathway as a major target of p53 in murine embryonic stem cells. Proc Natl Acad Sci U S A. 107:69–74. doi: 10.1073/pnas.0909734107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Polakis P. Wnt signaling and cancer. Genes Dev. 2000;14:1837–1851. [PubMed] [Google Scholar]
- 40.Li M, Huang J. A new puzzling role of p53 in mouse embryonic stem cells. Cell Cycle. 9:1669–1670. doi: 10.4161/cc.9.9.11596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rowe HM, Jakobsson J, Mesnard D, et al. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature. 463:237–240. doi: 10.1038/nature08674. [DOI] [PubMed] [Google Scholar]
- 42.Matsui T, Leung D, Miyashita H, et al. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature. 464:927–931. doi: 10.1038/nature08858. [DOI] [PubMed] [Google Scholar]
- 43.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 44.Yu J, Vodyanik MA, Smuga-Otto K, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 45.Takahashi K, Tanabe K, Ohnuki M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 46.Shi Y, Desponts C, Do JT, et al. Induction of pluripotent stem cells from mouse embryonic fibroblasts by Oct4 and Klf4 with small-molecule compounds. Cell Stem Cell. 2008;3:568–574. doi: 10.1016/j.stem.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 47.Huangfu D, Maehr R, Guo W, et al. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat Biotechnol. 2008;26:795–797. doi: 10.1038/nbt1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sridharan R, Tchieu J, Mason MJ, et al. Role of the murine reprogramming factors in the induction of pluripotency. Cell. 2009;136:364–377. doi: 10.1016/j.cell.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wong DJ, Liu H, Ridky TW, et al. Module map of stem cell genes guides creation of epithelial cancer stem cells. Cell Stem Cell. 2008;2:333–344. doi: 10.1016/j.stem.2008.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wernig M, Meissner A, Foreman R, et al. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- 51.Doi A, Park IH, Wen B, et al. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat Genet. 2009;41:1350–1353. doi: 10.1038/ng.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stadtfeld M, Apostolou E, Akutsu H, et al. Aberrant silencing of imprinted genes on chromosome 12qF1 in mouse induced pluripotent stem cells. Nature. 465:175–181. doi: 10.1038/nature09017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mikkelsen TS, Hanna J, Zhang X, et al. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chin MH, Mason MJ, Xie W, et al. Induced pluripotent stem cells and embryonic stem cells are distinguished by gene expression signatures. Cell Stem Cell. 2009;5:111–123. doi: 10.1016/j.stem.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chi P, Allis CD, Wang GG. Covalent histone modifications--miswritten, misinterpreted and mis-erased in human cancers. Nat Rev Cancer. 10:457–469. doi: 10.1038/nrc2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Villa R, Pasini D, Gutierrez A, et al. Role of the polycomb repressive complex 2 in acute promyelocytic leukemia. Cancer Cell. 2007;11:513–525. doi: 10.1016/j.ccr.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 57.Martinez-Garcia E, Licht JD. Deregulation of H3K27 methylation in cancer. Nat Genet. 42:100–101. doi: 10.1038/ng0210-100. [DOI] [PubMed] [Google Scholar]
- 58.Schlesinger Y, Straussman R, Keshet I, et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet. 2007;39:232–236. doi: 10.1038/ng1950. [DOI] [PubMed] [Google Scholar]
- 59.Keshet I, Schlesinger Y, Farkash S, et al. Evidence for an instructive mechanism of de novo methylation in cancer cells. Nat Genet. 2006;38:149–153. doi: 10.1038/ng1719. [DOI] [PubMed] [Google Scholar]
- 60.Wang Z, Schones DE, Zhao K. Characterization of human epigenomes. Curr Opin Genet Dev. 2009;19:127–134. doi: 10.1016/j.gde.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li G, Margueron R, Ku M, et al. Jarid2 and PRC2, partners in regulating gene expression. Genes Dev. 24:368–380. doi: 10.1101/gad.1886410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Barski A, Cuddapah S, Cui K, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 63.Irimia D, Mindrinos M, Russom A, et al. Genome-wide transcriptome analysis of 150 cell samples. Integr Biol (Camb) 2009;1:99–107. doi: 10.1039/b814329c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gibson JD, Jakuba CM, Boucher N, et al. Single-cell transcript analysis of human embryonic stem cells. Integr Biol (Camb) 2009;1:540–551. doi: 10.1039/b908276j. [DOI] [PubMed] [Google Scholar]