Abstract
We present an expansion of the classification of the family Papillomaviridae, which now contains 29 genera formed by 189 papillomavirus (PV) types isolated from humans (120 types), non-human mammals, birds and reptiles (64, 3 and 2 types, respectively). To accommodate the number of PV genera exceeding the Greek alphabet, the prefix “dyo” is used, continuing after the Omega-PVs with Dyodelta-PVs. The current set of human PVs are contained within five genera, whereas mammalian, avian and reptile PVs are contained within 20, 3 and 1 genera, respectively. We propose standardizations to the names of a number of animal PVs. As prerequisite for a coherent nomenclature of animal PVs, we propose founding a Reference Center for Animal PVs. We discuss that based on emerging species concepts derived from genome sequences, PV types could be promoted to the taxonomic level of species, but do not recommend implementing this change at the current time.
Introduction
Papillomaviruses (PVs) infect the epithelia of vertebrates, where they can cause neoplasias or persist asymptomatically. PVs have circular double-stranded DNA genomes approximately 8 kb in size and typically contain eight genes. One of these genes, the L1, encodes the principal capsid protein and is necessary and sufficient to produce the virus-like particles used for the current vaccines. PVs have traditionally been referred to as “types”, a type being a cloned full-length PV genome, whose L1 nucleotide sequence is at least 10% dissimilar from that of any other PV type. The L1 gene is useful for classification and construction of phylogenetic trees, as it is reasonably well conserved and can be aligned for all known PVs. This allows a genome-based approach to PV nomenclature, since PVs are not amenable to classical culture techniques. This technical problem was also a reason the term “strain” was not initially employed, however, it is a taxonomic term used in the publications of the International Committee on Taxonomy of Viruses (ICTV) (Fauquet et al., 2005). Furthermore, PVs do not elicit robust antibody responses, which impeded a classification based on “serotype” designations. As a consequence, classification of PV types based predominantly on nucleotide sequence similarities with some biological and medical properties (Chan et al., 1992; van Ranst et al., 1992b; de Villiers, 1994; Myers et al., 1994; Chan et al., 1995) served as the foundation for a formal nomenclature (de Villiers et al., 2004; Fauquet et al., 2005).
Papillomaviruses were designated as a distinct family, Papillomaviridae, in the 7th Report of the ICTV (van Regenmortel et al., 2002). Within the Papillomaviridae, the publication by de Villiers et al. (2004), whose authors constituted the Study Group of Papillomaviruses of the ICTV, proposed a classification of 92 human papillomavirus (HPV) and 24 animal PV types that consolidated guidelines established by the ICTV and the PV research community. This classification became formalized in the 8th Report of the International Committee on Taxonomy of Viruses (Fauquet et al., 2005). In these two publications, PVs were assigned to genera designated by Greek letters and to species within these genera according to set rules not familiar at that time to the majority of PV researchers. The concept of what constitutes a PV species has been discussed over the years (Van Ranst et al., 1993; Chan et al., 1995), and it was a decision of the ICTV to allocate its placement in the taxonomic hierarchy of PVs (de Villiers et al., 2004; Fauquet et al., 2005), since the “type” concept is not recognized by the ICTV. The adaptation of an official nomenclature that can be utilized by both PV researchers, healthcare workers, scientists and the general public requires compromise and it is the purpose of the current manuscript to expand an effective nomenclature that will serve the broad community for the near future and that is reconciled with the fixed official ICTV taxonomic structure. Table 1 compares commonly used terms referring to PV taxa with the terms defined by the ICTV. The table reflects an inconsistency regarding designation of species, that will be discussed later in this paper: While de Villiers et al. (2004) had given phylogenetic groups of HPVs at the level of species a name consisting of a Greek letter combined with a number, e.g. alpha-9 PV in the case HPV16 and several related HPV types, the ICTV named the “species” after one HPV type (e.g. HPV16) and considered all HPV types within that “species” as “strains”, including HPV16 (Fauquet et al., 2005).
Table 1. Comparison of commonly used and ICTV Papillomavirus nomenclature.
Designation of columns: Scient. abbrev., scientific abbrevation; prev. used abbreviation, previously used abbreviation. For details, see text in article.
| Commonly used names | ICTV term |
|---|---|
| Taxonomic level | |
| Family: Papillomavirus | Family: Papillomaviridae |
| Genus: alpha papillomavirus | Genus: Alphapapillomavirus |
| Species: alpha papillomavirus-9 | Species: Human papillomavirus 16 |
| Types: Human papillomavirus 16, 31, 33, etc. | Strains: Human papillomavirus 16, 31, 33, etc. |
| Type-species: n/a | Type-species: A term that identifies a papillomavirus typical of a genus (Fauquet et al., 2005) or of a species (de Villiers et al., 2004). |
This manuscript addresses the following objectives: (1) to update the nomenclature of genera in order to incorporate 28 novel HPV and 45 novel animal PV types that were described since 2004; (2) to refine the rules to maintain coherence of animal PV nomenclature based on the scientific name of the host species and to describe the foundation of a reference center for animal PVs; (3) to reconcile differences between some commonly used taxonomic names with terms used by the ICTV; (4) to discuss – but not to implement - a phylogenetic species concept for PVs that may lead to raising the taxonomic level of the “type” to the level of a “species”. In order to improve readability of this paper, we use the abbreviation “PV” in isolation or in the context of Greek letters, and it should be pointed out that hyphenated abbreviations like Alpha-PVs identify, strictly speaking, genera like Alphapapillomaviruses.
Results and Discussion
The family Papillomaviridae
The split of the Papovaviridae into two families, Papillomaviridae and Polyomaviridae was accepted by ICTV nearly a decade ago (van Regenmortel et al., 2002). The genomes of papilloma- and polyomaviruses share only a homologous segment within the papillomavirus E1 genes and the polyomavirus T-antigens that corresponds to a helicase, suggesting an ancient common origin of the replication proteins of these viruses (Clertant and Seif, 1984; Rebrikov et al., 2002). This finding has so far no taxonomic ramifications.
Recently, two viruses of marsupials were published to contain a surprising genome organization, early genes resembling the polyomaviruses, and late genes resembling the papillomaviruses (Woolford et al., 2007, Bennet et al., 2008). These polyoma-papilloma “hybrid” viruses are not evaluated in this paper, as it seems more likely that they represent a recombination event rather than share a common ancestor and are not classified within the Papillomaviridae family.
Empiric evidence of PV taxon groupings: Pairwise comparisons of PV types
PV taxa are defined based on L1 nucleotide sequence identities and their topological position within PV phylogenetic trees. To evaluate the natural distribution of PV L1 identities, the L1 genes from the entire set of 189 currently characterized PVs were aligned by global multiple sequence alignment and a matrix of pairwise comparisons calculated and plotted (Fig. 1). As previously demonstrated, the distribution of L1 identities shows a bimodal pattern consistent with the genus and species nomenclature. Based on this classification, three histograms created by separate matrix analyses are displayed (Fig. 2) to evaluate the specific distribution of intraspecies, interspecies (within a genus) and intergeneric identities. (Examples: intraspecies comparison: HPV16 vs. HPV31 in the species alpha papillomavirus 9; interspecies comparison: HPV16 vs. 18, members of the species alpha papillomavirus 9 and 7, respectively; intergeneric comparison HPV16 vs. HPV41, members of the genera alpha and nu papillomavirus, respectively). Most types within a PV genus show less than 60% sequence identity to types of other genera based on global multiple sequence or pairwise alignments of the L1 genes. Nevertheless, the suggested percentage identities that define PV genera have to be taken as general, but not absolute criteria for a number of reasons. For instance, there is overlap between the intergeneric and interspecies PV identities seen at the tails of each histogram. Thus, assignment of PV types to species and genera cannot be relegated to a computer algorithm, but requires curation (i.e. interpretation based on phylogeny, genome organization, biology and pathogenicity).
Fig. 1. Distribution of pairwise L1 nucleotide sequence comparisons of 189 Papillomaviruses.
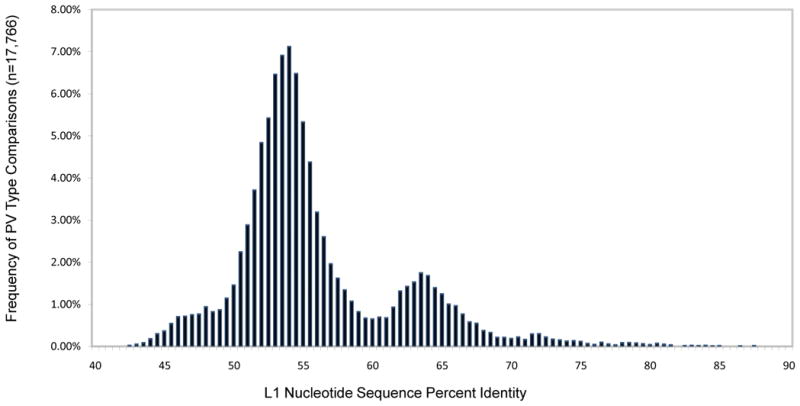
L1 nucleotide global multiple sequence alignments were guided by amino acid alignment using MUSCLE v3.7 (Edgar, 2004) and Seaview v4.1 (Galtier et al., 1996) software. A matrix of 189 L1 regions compared to each other, resulted in a total of 17766 percent identity values. Gaps were included and counted as one position. The Y-axis represents the percent of the total number of comparisons. The X-axis shows the L1 nucleotide sequence percent identity. The figure has a predominantly bimodal distribution with overlap at around 60% nucleotide sequence identity.
Fig. 2. Specific Intergeneric, Interspecies and Intraspecies L1 nucleotide sequence comparisons based on the multiple sequence alignment matrix.
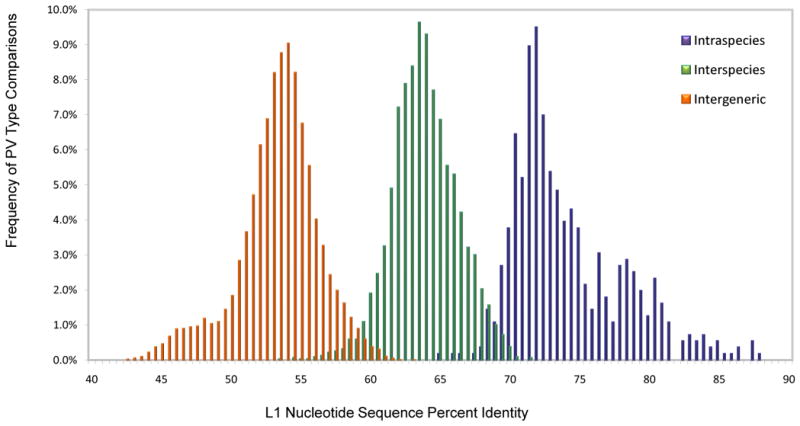
L1 nucleotide global multiple sequence alignment was created as described in the legend to Figure 1. The same matrix was used to evaluate the distribution of- Intraspecies: comparisons of PV types within the same species (161 PV types within 49 species, 558 comparisons); Interspecies: comparisons of PV types within the same genus (161 PV types within 10 genera, 3207 comparisons); Intergeneric: comparisons of all PV types within different genera (189 PVs in 30 genera, 14001 comparison). The Y-axis represents the percent of the total number of comparisons. The X-axis shows the L1 nucleotide sequence percent identity.
Taxonomy of PVs on the level of genera
De Villiers et al. (2004) described the topology of phylogenetic trees, quantitative thresholds in nucleotide sequence comparisons and biologically distinguishing features (host species, target tissues, pathogenicity, genome organization) that determine the classification of PVs on the level of genera. A nomenclature of these genera based on the Greek alphabet was introduced and has rapidly become accepted and widely used by the ICTV and community of PV researchers.
In 2004, sixteen groups of PVs or individual PVs fulfilled the criterion of genera, and the Greek alphabet from the letters alpha to pi was employed to create their nomenclature. Human PVs were members of five genera (Alpha-, Beta-, Gamma-, Mu- and Nu-PVs) and two genera (Eta- and Theta-PVs) were each comprised of a single bird PV. The remaining nine genera contained one or several PVs isolated from various mammals. Research over the ensuing five years has confirmed the notion that phylogenetic congruence of virus lineages with those of the host species is an important, although not the only mechanism of PV evolution. Consequently, search for PVs in previously understudied and remotely related hosts led to the identification of PVs, whose distant relationship with one another and with all previously published PVs fulfilled the criterion to establish 13 additional genera. These include the first two PVs found in reptiles (marine turtles). All established animal PVs are listed in alphabetical order of their abbreviated name in Table 2.
Table 2.
Alphabetic listing of the 69 known animal papillomaviruses.
| Scient. abbrev. | Prev.used abbreviatio | Papillomavirus name | Host scientific name | Host common name | Phylogeny | NCBI # | Reference (* direct submission) |
|---|---|---|---|---|---|---|---|
| AaPV1 | EEPV | Alces alces Papillomavirus 1 | Alces Alces | European elk | delta-1 | M15953 | Ahola et al., 1986 |
| BPV1 | BPV1 | Bos taurus Papillomavirus 1 | Bos taurus | domestic cow | delta-4 | X02346 | Chen et al., 1982 |
| BPV2 | BPV2 | Bos taurus Papillomavirus 2 | Bos taurus | domestic cow | delta-4 | M20219 | Groff et al., 1986 * |
| BPV3 | BPV3 | Bos taurus Papillomavirus 3 | Bos taurus | domestic cow | xi | AF486184 | Terai et al., 2002 |
| BPV4 | BPV4 | Bos taurus Papillomavirus 4 | Bos taurus | domestic cow | xi | X05817 | Patel et al., 1987 |
| BPV5 | BPV5 | Bos taurus Papillomavirus 5 | Bos taurus | domestic cow | epsilon | AF457465 | Pecenkova et al., 1997 |
| BPV6 | BPV6 | Bos taurus Papillomavirus 6 | Bos taurus | domestic cow | xi | AJ620208 | Coggins et al., 1985 |
| BPV8 | BPV8 | Bos taurus Papillomavirus 8 | Bos taurus | domestic cow | epsilon | DQ098913 | Tomita et al., 2007 |
| BPV9 | BPV9 | Bos taurus Papillomavirus 9 | Bos taurus | domestic cow | xi | AB331650 | Hatama et al., 2008 |
| BPV10 | BPV10, AA5 | Bos taurus Papillomavirus 10 | Bos taurus | domestic cow | xi | AB331651 | Hatama et al., 2008 |
| CcaPV1 | RdPV1, CcPV1 | Capreolus capreolus Papillomavirus 1 | Capreolus capreolus | Western Roe deer | delta-5 | EF680235 | Erdelyi et al., 2008 |
| CcPV1 | CcPV1 | Caretta caretta Papillomavirus 1 | Caretta caretta | Loggerhead turtle | dyozeta | EU493092 | Herbst et al., 2009 |
| CgPV1 | CgPV1 | Colobus guereza Papillomavirus 1 | Colobus guereza | Colobus monkey | alpha-14 | GU014532 | Kloster et al., 1988, Reszka et al., 1991 * |
| CgPV2 | CgPV2 | Colobus guereza Papillomavirus 2 | Colobus guereza | Colobus monkey | beta-1 | GU014533 | Kloster et al., 1988 * |
| ChPV1 | ChPV1 | Capra hircus Papillomavirus 1 | Capra hircus | domestic goat | phi | DQ091200 | Van Doorslaer et al., 2006 |
| CmPV1 | CmPV1 | Chelonia mydas Papillomavirus 1 | Chelonia mydas | green seaturtle | dyozeta | EU493091 | Herbst et al., 2009 |
| CPV1 | CoPV, CPV1 | Canis familiaris oral Papillomavirus | Canis familiaris | domestic dog | lambda-2 | D55633 | Isegawa et al., 1995 |
| CPV2 | CPV2, CfPV2 | Canis familiaris Papillomavirus 2 | Canis familiaris | domestic dog | tau | AY722648 | Yuan et al., 2007 |
| CPV3 | CPV3 | Canis familiaris Papillomavirus 3 | Canis familiaris | domestic dog | chi-1 | DQ295066 | Tobler et al., 2006 |
| CPV4 | CPV4 | Canis familiaris Papillomavirus 4 | Canis familiaris | domestic dog | chi-2 | EF584537 | Tobler et al., 2007 * |
| CPV5 | CPV5 | Canis familiaris Papillomavirus 5 | Canis familiaris | domestic dog | chi-1 | FJ492742 | Lange et al., 2009 |
| CPV6 | CPV6 | Canis familiaris Papillomavirus 6 | Canis familiaris | domestic dog | lambda-3 | FJ492743 | Lange et al., 2009 |
| CPV7 | CPV7 | Canis familiaris Papillomavirus 7 | Canis familiaris | domestic dog | tau | FJ492744 | Lange et al., 2009 |
| EcPV1 | EcPV1 | Equus caballus Papillomavirus 1 | Equus ferus caballus | domestic horse | zeta | AF498323 | Ghim et al., 2004 |
| EcPV2 | EqPV2 | Equus caballus Papillomavirus 2 | Equus ferus caballus | domestic horse | dyoiota | EU503122 | Scase et al., 2009 * |
| EdPV1 | EdPV1 | Erethizon dorsatum Papillomavirus 1 | Erethizon dorsatum | North American porcupine | sigma | AY684126 | Rector et al., 2005 |
| EePV1 | EhPV, HhPV1, EEPV | Erinaceus europaeus Papillomavirus 1 | Erinaceus europaeus | European hedgehog | dyoeta | FJ379293 | Schulz et al., 2009 |
| FcPV1 | FcPV | Fringilla coelebs Papillomavirus | Fringilla coelebs | Chaffinch (bird) | eta | AY057109 | Terai et al., 2002 |
| FdPV1 | FdPV1 | Felis domesticus Papillomavirus 1 | Felis domesticus | domestic cat | lambda-1 | AF480454 | Tachezy et al., 2002 |
| FdPV2 | FdPV2 | Felis domesticus Papillomavirus 2 | Felis domesticus | domestic cat | dyotheta | EU796884 | Lange et al., 2009 |
| FlPV1 | FLPV | Francolinus leucoscepus Papillomavirus 1 | Francolinus leucoscepus | Yellownecked Francolin (bird) | dyoepsilon | EU188799 | Van Doorslaer et al., 2009 |
| LrPV1 | LrPV1 | Lynx rufus Papillomavirus 1 | Lynx rufus | Bobcat | lambda-1 | AY904722 | Rector et al., 2007 |
| MaPV1 | HaOPV | Mesocricetus auratus Papillomavirus 1 | Mesocricetus auratus | Syrian Golden hamster | pi-2 | E15111 | Iwasaki et al., 1998 (patent) |
| McPV2 | McPV2 | Mastomys coucha Papillomavirus 2 | Mastomys coucha | Multimammate Mouse | pi-1 | DQ664501 | Nafz et al., 2006 |
| MfPV1 | MfPV1 | Macaca fascicularis Papillomavirus 1 | Macaca fascicularis | Cynomolgus macaque | beta-1 | EF028290 | Joh et al., 2009 |
| MfPV2 | MfPV2 | Macaca fascicularis Papillomavirus 2 | Macaca fascicularis | Cynomolgus macaque | beta-6 | GU014531 | Chen et al., unpub * |
| MfPV3 | MfPV3, RhPV-d | Macaca fascicularis Papillomavirus 3 | Macaca fascicularis | Cynomolgus macaque | alpha-12 | EF558839 | Chen et al., 2009 |
| MfPV4 | MfPV4 | Macaca fascicularis Papillomavirus 4 | Macaca fascicularis | Cynomolgus macaque | alpha-12 | EF558841 | Chen et al., 2009 |
| MfPV5 | MfPV5, MfPV-a | Macaca fascicularis Papillomavirus 5 | Macaca fascicularis | Cynomolgus macaque | alpha-12 | EF558843 | Chen et al., 2009 |
| MfPV6 | MfPV6 | Macaca fascicularis Papillomavirus 6 | Macaca fascicularis | Cynomolgus macaque | alpha-12 | EF558840 | Chen et al., 2009 |
| MfPV7 | MfPV7 | Macaca fascicularis Papillomavirus 7 | Macaca fascicularis | Cynomolgus macaque | alpha-12 | EF558838 | Chen et al., 2009 |
| MfPV8 | MfPV8, RhPV-a | Macaca fascicularis Papillomavirus 8 | Macaca fascicularis | Cynomolgus macaque | alpha-12 | EF558842 | Chen et al., 2009 |
| MfPV9 | MfPV9 | Macaca fascicularis Papillomavirus 9 | Macaca fascicularis | Cynomolgus macaque | alpha-12 | EU490516 | Chen et al., 2009 |
| MfPV10 | MfPV10 | Macaca fascicularis Papillomavirus 10 | Macaca fascicularis | Cynomolgus macaque | alpha-12 | EU490515 | Chen et al., 2009 |
| MfPV11 | MfPV11, RhPV-b | Macaca fascicularis Papillomavirus 11 | Macaca fascicularis | Cynomolgus macaque | alpha-12 | GQ227670 | Chen et al., 2009 |
| MmiPV1 | MmPV1 | Micromys minutus Papillomavirus 1 | Micromys minutus | Old World harvest mouse | pi-1 | DQ269468 | Van Doorslaer et al., 2007 |
| MmPV1 | RhPV-1 | Macaca mulata Papillomavirus 1 | Macacca mulata | Rhesus macaque | alpha-12 | M60184 | Ostrow et al., 1991 |
| MnPV1 | MnPV1, MrPV, MmPV | Mastomys natalensis Papillomavirus 1 | Mastomys natalensis | Multimammate Mouse | iota | U01834 | Tan et al., 1994 |
| OaPV1 | OvPV1 | Ovis aries Papillomavirus 1 | Ovis aries | domestic sheep | delta-3 | U83594 | Karlis et al. 2000 * |
| OaPV2 | OvPV2 | Ovis aries Papillomavirus 2 | Ovis aries | domestic sheep | delta-3 | U83595 | Karlis et al. 2000 * |
| OcPV1 | ROPV | Oryctolagus cuniculus Papillomavirus 2 | Oryctolagus cuniculus | New Zealand white rabbit | kappa-1 | AF227240 | Christensen et al., 2000 |
| OvPV1 | DPV | Odocoileus virginianus Papillomavirus 1 | Odocoileus virginianus | American White-tailed Deer | delta-2 | M11910 | Groff et al., 1986 |
| PcPV1 | PcPV1 | Puma concolor Papillomavirus 1 | Puma concolor | puma | lambda-1 | AY904723 | Rector et al., 2007 |
| PePV1 | PePV | Psittacus erithacus Papillomavirus | Psittacus erithacus | grey parrot (bird) | theta | AF420235 | Tachezy et al., 2002 |
| PlpPV1 | PlpPV1 | Panthera leo persica Papillomavirus 1 | Panthera leo persica | Asiatic lion | lambda-1 | AY904724 | Rector et al., 2007 |
| PlPV1 | PIPV1 | Procyon lotor Papillomavirus 1 | Procyon lotor | raccoon | lambda-4 | AY763115 | Rector et al., 2005 |
| PpPV1 | PpPV1, PCPV | Pan paniscus Papillomavirus 1 | Pan paniscus | pygmy chimpanzee | alpha-10 | X62844 | Van Ranst et al., 1992 |
| PsPV1 | PsPV | Phocoena spinipinnis Papillomavirus | Phocoena spinipinnis | Burmeister’s porpoise | omikron | AJ238373 | Cassonnet et al., 2007* |
| RaPV1 | RaPV1 | Rousettus aegyptiacus Papillomavirus 1 | Rousettus aegyptiacus | Egyptian rousette (fruitbat) | psi | DQ366842 | Rector et al., 2006 |
| RnPV1 | RnPV1 | Rattus norvegicus Papillomavirus 1 | Rattus norvegicus | Norweyian rat | pi-1 | GQ180114 | Schulz et al., 2009 |
| RtPV1 | RtPV, RPV | Rangifer tarandus Papillomavirus 1 | Rangifer tarandus | Reindeer | delta-1 | AF443292 | Burk et al., 2001 |
| SfPV1 | CRPV, SPV | Sylvilagus floridanus Papillomavirus 1 | Sylvilagus floridanus | Cottontail rabbit | kappa-2 | K02708 | Wellstein & Stevens, 1980, Giri et al., 1985 |
| SsPV1 | SsPV1 | Sus scrofa Papillomavirus 1 | Sus scrofa | domestic pig | dyodelta | EF395818 | Stevens et al., 2008 |
| TmPV1 | TmPV1 | Trichechus manatus latirostris Papillomav latirostris | Trichechus manatus | Caribbean manatee | rho | AY609301 | Rector et al., 2004 |
| TtPV1 | TtPV1 | Tursiops truncatus Papillomavirus 1 | Tursiops truncatus | bottlenosed dolphin | upsilon-1 | EU240894 | Rector et al., 2008 |
| TtPV2 | TtPV2 | Tursiops truncatus Papillomavirus 2 | Tursiops truncatus | bottlenosed dolphin | upsilon-2 | AY956402 | Rehtanz et al., 2006 |
| TtPV3 | TtPV3 | Tursiops truncatus Papillomavirus 3 | Tursiops truncatus | bottlenosed dolphin | upsilon-1 | EU240895 | Rector et al., 2008 |
| UmPV1 | UmPV1 | Ursus maritimus Papillomavirus 1 | Ursus maritimus | polar bear | omega | EF536349 | Stevens et al., 2008 |
| UuPV1 | UuPV1 | Uncia uncia Papillomavirus 1 | Uncia uncia | snow leopard | lambda-1 | DQ180494 | Rector et al., 2007 |
Footnote: see supplemental table for ICTV animal papillomavirus nomenclature.
The last official classification of PV genera ended with the genus Pi-PVs. The description of 13 new PV genera however, exhausts the Greek alphabet. In order to create a system that continues with the Greek alphabet, we propose to use the Greek alphabet a second time, employing the prefix “dyo”, (i.e., Greek “a second time”). In addition, we propose to omit the designations Dyoalpha, Dyobeta and Dyogamma, since the Alpha-, Beta- and Gamma-PV genera include the most common and medically important HPVs. To designate specific genera, we have followed the temporal order of publication and/or GenBank submissions of all PV nucleotide sequences and named the new genera Rho- to Omega-PV, and continued with the terms e.g., “Dyodelta-PVs”, “Dyoepsilon-PVs”. Table 3 contains a list of all PV genera and species, particularly the name of those PVs that led to the establishment of the new genera. A phylogenetic tree using the L1 nucleotide sequences of 189 PVs was generated using a Bayesian algorithm (Fig. 3) and visualizes the relationship between all previously and newly described genera highlighting the three major genera containing the majority of HPV types.
Table 3.
Papillomavirus genera and species.
| genus | species (common use species (ICTV) | |
|---|---|---|
| alphapapillomavirus | alpha-1 | Human Papillomavirus 32 |
| alpha-2 | Human Papillomavirus 10 | |
| alpha-3 | Human Papillomavirus 61 | |
| alpha-4 | Human Papillomavirus 2 | |
| alpha-5 | Human Papillomavirus 26 | |
| alpha-6 | Human Papillomavirus 53 | |
| alpha-7 | Human Papillomavirus 18 | |
| alpha-8 | Human Papillomavirus 7 | |
| alpha-9 | Human Papillomavirus 16 | |
| alpha-10 | Human Papillomavirus 6 | |
| alpha-11 | Human Papillomavirus 34 | |
| alpha-12 | Macaca mulata Papillomavirus 1 | |
| alpha-13 | Human Papillomavirus 54 | |
| alpha-14 | Human Papillomavirus 90 | |
| betapapillomavirus | beta-1 | Human Papillomavirus 5 |
| beta-2 | Human Papillomavirus 9 | |
| beta-3 | Human Papillomavirus 49 | |
| beta-4 | Human Papillomavirus 92 | |
| beta-5 | Human Papillomavirus 96 | |
| beta-6 | Macaca fascicularis Papillomavirus 2 | |
| gammapapillomavirus | gamma-1 | Human Papillomavirus 4 |
| gamma-2 | Human Papillomavirus 48 | |
| gamma-3 | Human Papillomavirus 50 | |
| gamma-4 | Human Papillomavirus 60 | |
| gamma-5 | Human Papillomavirus 88 | |
| gamma-6 | Human Papillomavirus 101 | |
| gamma-7 | Human Papillomavirus 109 | |
| gamma-8 | Human Papillomavirus 112 | |
| gamma-9 | Human Papillomavirus 116 | |
| gamma-10 | Human Papillomavirus 121 | |
| Deltapapillomavirus | delta-1 | Alces alces Papillomavirus 1 |
| delta-2 | Odocoileus virginianus Papillomavirus 1 | |
| delta-3 | Ovis aries Papillomavirus 1 | |
| delta-4 | Bos taurus Papillomavirus 1 | |
| delta-5 | Capreolus capreolus Papillomavirus 1 | |
| Epsilonpapillomavirus | epsilon-1 | Bos taurus Papillomavirus 5 |
| Zetapapillomavirus | Zeta-1 | Equus caballus Papillomavirus 1 |
| Etapapillomavirus | Eta-1 | Fringilla coelebs Papillomavirus |
| Thetapapillomavirus | Theta-1 | Psittacus erithacus Papillomavirus 1 |
| Iotapapillomavirus | Iota-1 | Mastomys natalensis Papillomavirus 1 |
| Kappapapillomavirus | Kappa-1 | Oryctolagus cuniculus Papillomavirus 1 |
| Kappa-2 | Sylvilagus floridanus Papillomavirus 1 | |
| Lambdapapillomavirus | Lambda-1 | Felis domesticus Papillomavirus 1 |
| Lambda-2 | Canis familiaris Papillomavirus 1 | |
| Lambda-3 | Canis familiaris Papillomavirus 6 | |
| Lambda-4 | Procyon lotor Papillomavirus 1 | |
| Mupapillomavirus | Mu-1 | Human Papillomavirus 1 |
| Mu-2 | Human papillomavirus 63 | |
| Nupapillomavirus | Nu-1 | Human papillomavirus 41 |
| Xipapillomavirus | Xi-1 | Bos taurus Papillomavirus 3 |
| Pipapillomavirus | Pi-1 | Mesocricetus auratus Papillomavirus 1 |
| Pi-2 | Micromys minutus Papillomavirus 1 | |
| Rhopapillomavirus | Rho-1 | Trichechus manatus latirostris Papillomavirus |
| Sigmapapillomavirus | Sigma-1 | Erethizon dorsatum Papillomavirus 1 |
| Taupapillomavirus | Tau-1 | Canis familiaris Papillomavirus 2 |
| Upsilonpapillomavirus | Upsilon-1 | Tursiops truncatus Papillomavirus 1 |
| Upsilon-2 | Tursiops truncatus Papillomavirus 2 | |
| Phipapillomavirus | Phi-1 | Capra hircus Papillomavirus 1 |
| Chipapillomavirus | Chi-1 | Canis familiaris Papillomavirus 3 |
| Chi-2 | Canis familiaris Papillomavirus 4 | |
| Psipapillomavirus | Psi-1 | Rousettus aegyptiacus Papillomavirus 1 |
| Omegapapillomavirus | Omega-1 | Ursus maritimus Papillomavirus 1 |
| Dyodeltapapillomavirus | Dyodelta-1 | Sus scrofa Papillomavirus 1 |
| Dyoepsilonpapillomavirus | Dyoepsilon-1 | Francolinus leucoscepus Papillomavirus 1 |
| Dyozetapapillomavirus | Dyozeta-1 | Caretta caretta Papillomavirus 1 |
| Dyoetapapillomavirus | Dyoeta-1 | Erinaceus europaeus Papillomavirus 1 |
| Dyothetapapillomavirus | Dyotheta-1 | Felis domesticus Papillomavirus 2 |
| Dyoiotapapillomavirus | Dyoiota-1 | Equus caballus Papillomavirus 2 |
Fig. 3. Phylogenetic tree inferred from the L1 nucleotide sequences of 189 papillomaviruses.
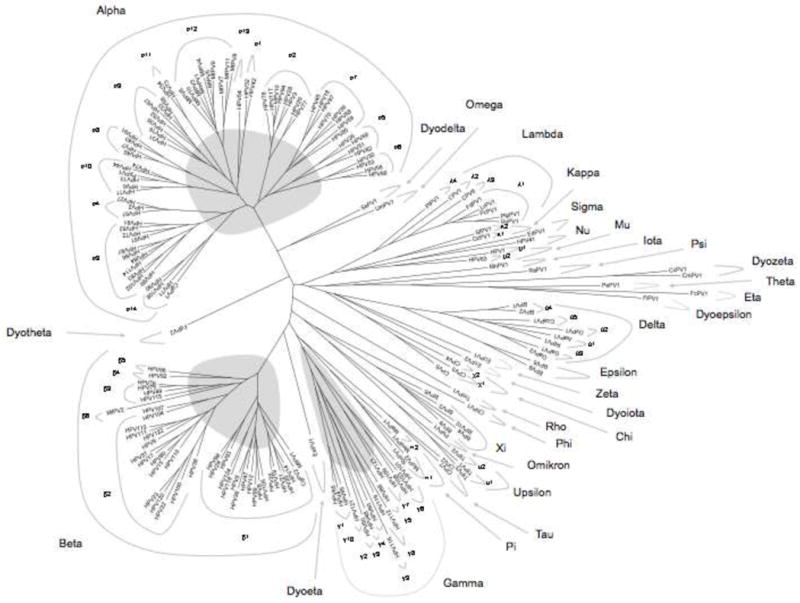
The phylogeny analysis is based on the multiple L1 nucleotide sequence alignment of 189 PV types that was used in Figures 1 and 2. MrBayes v3.1.2 (Huelsenbeck and Ronquist, 2001; Ronquist and Huelsenbeck, 2003) with 10,000,000 cycles for the Markov chain Monte Carlo (MCMC) algorithm was used to generate a phylogenetic tree. For Bayesian tree construction, the computer program ModelTest v3.7 (Posada and Crandall, 1998) identified the best evolutionary model. The identified gamma model was set for among-site rate variation and allowed all substitution rates of aligned sequence to be different.
Nomenclature of animal PV types
Table 2 lists the abbreviated scientific name of all animal PV types, the previously used names, the proposed phylogenetic genus, database accession numbers and references. The abbreviations have been edited to assure a coherent nomenclature of all animal PVs. Consensus within the community of papillomavirus researchers established that the name of an animal PV should be based on the scientific name of the host, using the host genus and species designation, for example FdPV1 for Felis domesticus PV type 1. We have applied this rule systematically, since an uncurated nomenclature of animal PVs resulted in confusion and multiple use of the same or similar abbreviations for a single type of PV. For example, a PV isolated from the European Elk had been named EEPV (Ahola et al., 1986), and subsequently a different PV isolated from Erinaceus europaeus (hedgehog) received a similar designation, EePV (Schulz et al., 2009). Moreover, ChPV1 has been used to describe Capra hircus PV type 1 (van Doorslaer et al., 2006) and chimpanzee PV type 1 (van Ranst et al., 1992a). To facilitate the isolation, characterisation and publication of novel animal PVs based on a standardised evaluation and nomenclature, an Animal Papillomavirus Reference Center is proposed (see below). While this paper was compiled using publication data and database acceptance dates, deposit of the cloned viral genome will be a future requirement to recognize new animal PV names and inclusion in the official PV taxonomy, just as it has been practiced for many years in the case of human PVs.
To enhance a robust and workable nomenclature, we have sustained the historical use of the abbreviation “HPV” (with H standing for human or Homo, but avoiding the species designation “sapiens”) as well as “BPV” (with B standing for bovine or Bos, but avoiding the species designation “taurus”). However, the name of the cottontail rabbit PV was modified from CRPV1 to SfPV1 (for Sylvilagus floridanus), as it had been originally named SPV by one group at the time it was cloned (Wettstein and Stevens, 1980). This nomenclature affects the taxonomic designation, but not necessarily the working terminology as listed under “previously used abbreviations” in Table 2. With the revised nomenclature set forth here, all PV names are unique. Where renaming was necessary, we kept the name of the PV type with publication priority or, as in the example above (EePV1) the one that adapted the name adherent to these rules. In case of overlaps, a third letter was and will be added (e.g., the Caretta caretta PV became CcPV1, while the Capreolus capreolus PV became CcaPV1). All prototype PVs for a genus were consistently given the number “1”, even when there was no second PV yet known from the same host. Subsequent types were and will be numbered sequentially following the publication date of the virus and/or the submission of the sequence to a public database and deposit of the cloned viral genome. The numbers are added without hyphens to facilitate electronic searches.
The taxonomic status of new HPV types
De Villiers et al. (2004) described the taxonomy of human papillomaviruses HPV1 to HPV96. Since HPV46, 55, 64 and 79 did not meet the criteria as a unique HPV type, they were omitted and their numbers left vacant (de Villiers 2004). In addition, PV types cloned from PCR products are now accepted for full classification and the term “candidate” has been eliminated. Table 4 lists 28 HPV types (ICTV strains, see Table 1) described since 2004 and their placement within genera. Five PVs (HPV97, 102, 106, 114, 117) belong to the Alpha-PVs, fourteen PVs (HPV98, 99, 100, 104, 105, 107, 110, 111, 113, 115, 118, 120, 122, 124) to the Beta-PVs, and nine PVs (HPV101, 103, 108, 109, 112, 116, 119, 121, 123) to the Gamma-PVs. Among these nine PVs, HPV101, 103, and 108 diverge convincingly from all other HPV types in that they lack an E6 ORF (Chen et al., 2007a; Nobre et al., 2009). In spite of this distinction, these three types are included in the genus Gamma-PV based on the present rules of sequence similarities in the L1 ORF and the resulting topology of the phylogenetic tree.
Table 4.
Human papillomaviruses characterized since 2004.
| HPV type | PV genus | PV species (common use) | PV species (ICTV) | GenBank No. | Reference |
|---|---|---|---|---|---|
| HPV 97 | Alphapapillomavirus | Alpha-7 | Human Papillomavirus 18 | DQ080080 | Chen et al., 2007a |
| HPV 98 | Betapapillomavirus | Beta-1 | Human Papillomavirus 5 | FM955837 | de Villiers & Gunst, 2009 |
| HPV 99 | Betapapillomavirus | Beta-1 | Human Papillomavirus 5 | FM955838 | de Villiers & Gunst, 2009 |
| HPV 100 | Betapapillomavirus | Beta-2 | Human Papillomavirus 9 | FM955839 | de Villiers & Gunst, 2009 |
| HPV 101 | Gammapapillomavirus | Gamma-6 | Human Papillomavirus 101 | NC_008189 | Chen et al., 2007c |
| HPV 102 | Alphapapillomavirus | Alpha-3 | Human Papillomavirus 61 | DQ080083 | Chen et al., 2007b |
| HPV 103 | Gammapapillomavirus | Gamma-6 | Human Papillomavirus 101 | NC_008188 | Chen et al., 2007c |
| HPV 104 | Betapapillomavirus | Beta-2 | Human Papillomavirus 9 | FM955840 | de Villiers & Gunst, 2009 |
| HPV 105 | Betapapillomavirus | Beta-1 | Human Papillomavirus 5 | FM955841 | de Villiers & Gunst, 2009 |
| HPV 106 | Alphapapillomavirus | Alpha-14 | Human Papillomavirus 90 | DQ080082 | Chen et al., 2007b |
| HPV 107 | Betapapillomavirus | Beta-2 | Human Papillomavirus 9 | EF422221 | Vasiljevic et al., 2008 |
| HPV 108 | Gammapapillomavirus | Gamma-6 | Human Papillomavirus 101 | NC_012213 | Nobre et al., 2009 |
| HPV 109 | Gammapapillomavirus | Gamma-7 | Human Papillomavirus 109 | NC_012485 | Ekstrom et al., in press |
| HPV 110 | Betapapillomavirus | Beta-2 | Human Papillomavirus 9 | EU410348 | Vasiljevic et al., 2008 |
| HPV 111 | Betapapillomavirus | Beta-2 | Human Papillomavirus 9 | EU410349 | Vasiljevic et al., 2008 |
| HPV 112 | Gammapapillomavirus | Gamma-8 | Human Papillomavirus 112 | EU541442 | Ekstrom et al., in press |
| HPV 113 | Betapapillomavirus | Beta-2 | Human Papillomavirus 9 | FM955842 | de Villiers & Gunst, 2009 |
| HPV 114 | Alphapapillomavirus | Alpha-3 | Human Papillomavirus 61 | GQ244463 | Ekstrom et al., in press |
| HPV 115 | Betapapillomavirus | Beta-3 | Human Papillomavirus 49 | FJ947080 | Chouhy et al., in press |
| HPV 116 | Gammapapillomavirus | Gamma-9 | Human Papillomavirus 116 | FJ804072 | Li et al., 2009 |
| HPV 117 | Alphapapillomavirus | Alpha-2 | Human Papillomavirus 10 | GQ246950 | Köhler et al., in press. |
| HPV 118 | Betapapillomavirus | Beta-1 | Human Papillomavirus 5 | GQ246951 | Nindl et al., unpubl. |
| HPV 119 | Gammapapillomavirus | Gamma-8 | Human Papillomavirus 112 | GQ845441 | Chen et al., unpubl. |
| HPV 120 | Betapapillomavirus | Beta-2 | Human Papillomavirus 9 | GQ845442 | Chen et al., unpubl. |
| HPV 121 | Gammapapillomavirus | Gamma-10 | Human Papillomavirus 121 | GQ845443 | Chen et al., unpubl. |
| HPV 122 | Betapapillomavirus | Beta-2 | Human Papillomavirus 9 | GQ845444 | Chen et al., unpubl. |
| HPV 123 | Gammapapillomavirus | Gamma-7 | Human Papillomavirus 109 | GQ845445 | Chen et al., unpubl. |
| HPV 124 | Betapapillomavirus | Beta-1 | Human Papillomavirus 5 | GQ845446 | Chen et al., unpubl. |
Subtypes and variants of PV types
The definition and properties of PV subtypes and variants as DNA isolates with less than 10% sequence diversity in the L1 gene have been discussed (de Villiers et al., 2004; Calleja-Macias et al., 2005; Bernard et al., 2006). These issues are not further addressed here, as the ICTV does not implement taxonomic systems below the species level. A nomenclature of variants of Alpha-PV types associated with cervix cancer, which will be based on complete genome sequences and extending beyond the historic classification of HPV16 and 18 variant lineages that were based on geographic association (Bernard et al., 2006), is being developed by some of us (Burk et al., in prep.).
Reference Centers for Human and Animal Papillomaviruses
The Reference Center for Human Papillomaviruses at the German Cancer Research Center in Heidelberg under leadership of E.M. de Villiers has for the past 25 years been instrumental in confirming the nucleotide sequence of novel HPV types, assigning the appropriate HPV numbers, depositing and maintaining reference samples, and, if permitted by proprietary rules, distributing DNA samples. This process has been essential to assure an orderly expansion of HPV types and avoid misinterpretation of incomplete or heterologous DNA clones and isolates not meeting the established criteria. The continued maintenance of this reference center and service to the community is of great importance.
In order to avoid misclassifications and maintain a unique nomenclature, we have made efforts to support the establishment of a Repository for Animal Papillomaviruses since no similar reference center for animal PVs has yet been established. One of us (R.D.B.) in collaboration with Dr. Rob DeSalle of the Sackler Institute for Comparative Genomics at the American Museum for Natural History will facilitate the creation of this service. The function of such a center for animal PVs will be to (i) streamline the curation of non-human PVs, (ii) establish a repository for all characterized non-human PVs, and (iii) establish a reference center for all non-human PVs by obtaining the cloned genomes and re-sequencing the provided clones to confirm the existence of each novel non-human PV. As curation of novel HPVs has been necessary for an orderly HPV nomenclature, this repository will implement a similar system for animal PVs, where, prior to publication of a novel animal PV, an “official name” has been designated and the cloned genome been publicly deposited.
Phylogenetic and biological considerations regarding the nature of “PV species” and “PV types”
Defining PV taxa below the level of genera resulted in some inconsistencies between the official ICTV PV nomenclature (Fauquet et al., 2005) and that used by the scientific community (de Villiers et al., 2004). A goal of the current Study Group of Papillomaviruses is to harmonize the official ICTV designations with the known genetics and biology of PVs. The ICTV only names species after a specific virus, such as HPV16, and related types including the “type species” are designated as strains within the species (see Table 1) (Fauquet et al., 2005). For example, the commonly used term “PV species Alpha-9” (de Villiers et al., 2004) is a synonym for what the ICTV called the “HPV16 species”, which contains the HPV types (strains) 16, 31, 35, 33, 52, 58 and 67. Although it can be argued (see below) that a PV “type” has many characteristics of a “species”, we recommend maintaining the current allocation of the taxonomic levels of genus and species. Nevertheless, since the species designations of de Villiers et al. (2004) have been widely accepted and useful to the scientific community, we support their continued use.
The ICTV proposed guidelines defining that a species should be “a polythetic class of viruses that constitute a replicating lineage and occupies a particular ecological niche” (van Regenmortel et al., 1991). The term “polythetic” means that several or all possible criteria or attributes be used to determine a viral species. This definition suggests that a single property, even genomic sequences, might be insufficient to define a viral species. This definition has been challenged (van Regenmortel et al., 2000; Drebot et al., 2002; Gibbs and Gibbs, 2006). Concepts of defining viral species have always been complicated and are not yet mature. Among several complimentary approaches, nucleotide sequence-based comparisons developed in the last few decades have impacted taxonomic research throughout biology, led to completely new insights into the species concept, and have become widely accepted as a solid taxonomic criterion. Papillomaviruses were the first viruses to be significantly classified by comparison of viral genomes, in part, because of the lack of a culture or serologic system.
The age of PV types is a major component of equating PV types with PV species (Rector et al., 2007). Even closely related PV types have evolved over time scales equivalent to those that gave rise to their host species, i.e. over millions of years. This is fundamentally different from the emergence of quasi-species of RNA viruses over very short periods of time, such as the human immunodeficiency viruses (Rambaut et al., 2004), or from the evolution of all human rhinoviruses in the relatively short time span since the origin of humans (Palmenberg et al., 2009). In addition, the distribution of PV type identities within a species (intra-species identity) as shown in Fig. 1 and 2 has a normal distribution suggestive of a natural taxonomic order. Moreover, reports of phenotypic idiosyncrasies of closely related PV types, classified within a single species, continue to accumulate. Some notable examples are found amongst the members of the Alpha-PV 10 species (species HPV6 and related “strains” in ICTV nomenclature). For example, HPV6 and 11 show significantly different tissue tropism, HPV6 being more common in genital warts, HPV11 more common in laryngeal papillomas. The closely related type, HPV13, causes focal epithelial hyperplasia of the oral cavity and has neither been found in genital warts nor laryngeal papillomas (de Villiers, 1994; de Villiers, 2001). HPV16, 31 and 35 are grouped in the species Alpha-PV 9 (species HPV16 and related “strains” in ICTV nomenclature). HPV16 is significantly stronger associated with cancer than either HPV31 or HPV35 (Munoz et al., 2003). HPV16 is uniquely associated with tumors of the oropharyngeal region.
In summary, we presented arguments how a phylogenetic species concept could be applied to PV taxonomy. This concept has increasing impact throughout biology, but is presently not implemented in virology, and we therefore do not yet recommend implementation at this point. Once widely accepted, a phylogenetic species concept would lead to a promotion of PV types (strains) to species, and of the present species to sub-genera, while genera would remain unchanged.
Online Papillomavirus Database
PV nucleotide sequences are deposited in the GenBank and EMBL nucleotide sequence databases. A compilation of all sequences available in the mid-1990s, was compiled and reviewed in the “Papillomavirus Database” sponsored by NIAID and published online and as hardcopy by the Los Alamos National Laboratory (Myers et al., 1994). A new interactive database “Papillomavirus Episteme” (http://pave.niaid.nih.gov/#home) is currently under development.
Materials and Methods
Origin of sequences
This paper is based on published PV sequences, which can be accessed either through the listed references, or the GenBank accession numbers in the tables 2 and 4.
Supplementary Material
Supplemental Table. Animal papillomavirus ICTV nomenclature.
Acknowledgments
This paper is written by four authors that form the PV Study Group of the ICTV appointed in 2009 (H.U.B., R.D.B., H.z.H., E.M.deV.) and two colleagues (Z.C., K.v.D). We thank Ignacio G. Bravo and Benjamin Smith for stimulating discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahola H, Bergman P, Ström AC, Moreno-Lopéz J, Pettersson U. Organization and expression of the transforming region from the European elk papillomavirus (EEPV) Gene. 1986;50:195–205. doi: 10.1016/0378-1119(86)90324-0. [DOI] [PubMed] [Google Scholar]
- Bennett MD, Woolford L, Stevens H, Van Ranst M, Oldfield T, Slaven M, O’Hara AJ, Warren KS, Nicholls PK. Genomic characterization of a novel virus found in papillomatous lesions from a southern brown bandicoot (Isoodon obesulus) in Western Australia. Virology. 2008;376:173–182. doi: 10.1016/j.virol.2008.03.014. [DOI] [PubMed] [Google Scholar]
- Bernard HU, Calleja-Macias IE, Dunn ST. Genome variation of human papillomavirus types: Phylogenetic and medical implications. Internat J Cancer. 2006;118:1071–1076. doi: 10.1002/ijc.21655. [DOI] [PubMed] [Google Scholar]
- Calleja-Macias IE, Kalantari M, Allan B, Williamson AL, Chung LP, Collins RJ, Zuna RE, Dunn ST, Ortiz-Lopez R, Barrera-Saldaña HA, Cubie HA, Villa LL, Bernard HU. Papillomavirus subtypes are natural and old taxa: Phylogeny of the human papillomavirus (HPV) types 44/55 and 68a/b. J Virol. 2005;79:6565–6569. doi: 10.1128/JVI.79.10.6565-6569.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SY, Bernard HU, Ong CK, Chan SP, Hofmann B, Delius H. Phylogenetic analysis of 48 papillomavirus types and 28 subtypes and variants: A showcase for the molecular evolution of DNA viruses. J Virol. 1992;66:5714–5725. doi: 10.1128/jvi.66.10.5714-5725.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SY, Delius H, Halpern AL, Bernard HU. Analysis of genomic sequences of 95 papillomavirus types: uniting typing, phylogeny, and taxonomy. J Virol. 1995;69:3074–3083. doi: 10.1128/jvi.69.5.3074-3083.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen EY, Howley PM, Levinson AD, Seeburg PH. The primary structure and genetic organization of the bovine papillomavirus type 1 genome. Nature. 1982;299:529–534. doi: 10.1038/299529a0. [DOI] [PubMed] [Google Scholar]
- Chen Z, Fu L, Herrero R, Schiffman M, Burk RD. Identification of a novel human papillomavirus (HPV97) related to HPV18 and HPV45. Int J Cancer. 2007a;121:193–198. doi: 10.1002/ijc.22632. [DOI] [PubMed] [Google Scholar]
- Chen Z, Schiffman M, Herrero R, Burk RD. Identification and characterization of two novel human papillomaviruses (HPVs) by overlapping PCR: HPV102 and HPV106. J Gen Virol. 2007b;88:2952–2955. doi: 10.1099/vir.0.83178-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Schiffman M, Herrero R, Desalle R, Burk RD. Human papillomavirus (HPV) types 101 and 103 isolated from cervicovaginal cells lack an E6 open reading frame (ORF) and are related to gamma-papillomaviruses. Virology. 2007c;360:447–453. doi: 10.1016/j.virol.2006.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, van Doorslaer K, Desalle R, Wood CE, Kaplan JR, Wagner JD, Burk RD. Genomic diversity and interspecies host infection of alpha12 Macaca fascicularis papillomaviruses (MfPVs) Virology. 2009 doi: 10.1016/j.virol.2009.07.012. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen ND, Cladel NM, Reed CA, Han R. Rabbit oral papillomavirus complete genome sequence and immunity following genital infection. Virology. 2000;269:451–461. doi: 10.1006/viro.2000.0237. [DOI] [PubMed] [Google Scholar]
- Clertant P, Seif I. A common function for polyoma virus large-T and papillomavirus E1 proteins? Nature. 1984;311:276–279. doi: 10.1038/311276a0. [DOI] [PubMed] [Google Scholar]
- Delius H, Van Ranst MA, Jenson AB, zur Hausen H, Sundberg JP. Canine oral papillomavirus genomic sequence: a unique 1.5-kb intervening sequence between the E2 and L2 open reading frames. Virology. 1994;204:447–452. doi: 10.1006/viro.1994.1552. [DOI] [PubMed] [Google Scholar]
- de Villiers EM. Human pathogenic papillomavirus types, an update. Curr Top Microbiol Immunol. 1994;186:1–12. doi: 10.1007/978-3-642-78487-3_1. [DOI] [PubMed] [Google Scholar]
- de Villiers EM. Taxonomic Classification of Papillomaviruses. Papillomavirus Report. 2001;12:57–63. [Google Scholar]
- de Villiers EM, Fauquet C, Broker TR, Bernard HU, zur Hausen H. Classification of papillomaviruses. Virology. 2004;324:17–27. doi: 10.1016/j.virol.2004.03.033. [DOI] [PubMed] [Google Scholar]
- de Villiers EM, Gunst K. Characterization of seven novel human papillomavirus types isolated from cutaneous tissue, but also present in mucosal lesions. J Gen Virol. 2009;90:1999–2004. doi: 10.1099/vir.0.011478-0. [DOI] [PubMed] [Google Scholar]
- Drebot MA, Henchal E, Hjelle B, LeDuc JW, Repik PM, Roehrig JT, Schmaljohn CS, Shope RE, Tesh RB, Weaver SC, Calisher CH. Improved clarity of meaning from the use of both formal species names and common (vernacular) virus names in virological literature. Arch Virol. 2002;147:2464–2472. doi: 10.1007/s00705-002-0938-8. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdelyi K, Balint A, Dencso L, Dan A, Ursu K. Characterisation of the first complete genome sequence of the roe deer (Capreolus capreolus) papillomavirus. Virus Res. 2008;135:307–311. doi: 10.1016/j.virusres.2008.03.002. [DOI] [PubMed] [Google Scholar]
- Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA. Family Papillomaviridae. Elsevier; 2005. Virus taxonomy. The Eighth Report of the International Committee on Taxonomy of Viruses; pp. 239–255. [Google Scholar]
- Galtier N, Gouy M, Gautier C. SEAVIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Comput Appl Biosci. 1996;12:543–548. doi: 10.1093/bioinformatics/12.6.543. [DOI] [PubMed] [Google Scholar]
- Gibbs AJ, Gibbs MJ. A broader definition of ‘the virus species’. Arch Virol. 2006;151:1419–1422. doi: 10.1007/s00705-006-0775-2. [DOI] [PubMed] [Google Scholar]
- Ghim SJ, Rector A, Delius H, Sundberg JP, Jenson AB, Van Ranst M. Equine papillomavirus type 1: complete nucleotide sequence and characterization of recombinant virus-like particles composed of the EcPV-1 L1 major capsid protein. Biochem Biophys Res Commun. 2004;324:1108–1115. doi: 10.1016/j.bbrc.2004.09.154. [DOI] [PubMed] [Google Scholar]
- Giri I, Danos O, Yaniv M. Genomic structure of the cottontail rabbit (Shope) papillomavirus. Proc Natl Acad Sci USA. 1985;82:1580–1584. doi: 10.1073/pnas.82.6.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groff DE, Lancaster WD. Molecular cloning and nucleotide sequence of deer papillomavirus. J Virol. 1985;56:85–91. doi: 10.1128/jvi.56.1.85-91.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatama S, Nobumoto K, Kanno T. Genomic and phylogenetic analysis of two novel bovine papillomaviruses, BPV-9 and BPV-10. J Gen Virol. 2008;89:158–163. doi: 10.1099/vir.0.83334-0. [DOI] [PubMed] [Google Scholar]
- Herbst LH, Lenz J, Van Doorslaer K, Chen Z, Stacy BA, Wellehan JF, Jr, Manire CA, Burk RD. Genomic characterization of two novel reptilian papillomaviruses, Chelonia mydas papillomavirus 1 and Caretta caretta papillomavirus 1. Virology. 2009;383:131–135. doi: 10.1016/j.virol.2008.09.022. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Jarrett WF, Campo MS, O’Neil BW, Laird HM, Coggins LW. A novel bovine papillomavirus (BPV-6) causing true epithelial papillomas of the mammary gland skin: a member of a proposed new BPV subgroup. Virology. 1984;136:255–264. doi: 10.1016/0042-6822(84)90162-4. [DOI] [PubMed] [Google Scholar]
- Joh J, Hopper K, Van Doorslaer K, Sundberg JP, Jenson AB, Ghim SJ. Macaca fascicularis papillomavirus type 1: a non-human primate betapapillomavirus causing rapidly progressive hand and foot papillomatosis. J Gen Virol. 90:987–994. doi: 10.1099/vir.0.006544-0. [DOI] [PubMed] [Google Scholar]
- Kloster BE, Manias DA, Ostrow RS, Shaver MK, McPherson SW, Rangen SR, Uno H, Faras AJ. Molecular cloning and characterization of the DNA of two papillomaviruses from monkeys. Virology. 1988;166:30–40. doi: 10.1016/0042-6822(88)90143-2. [DOI] [PubMed] [Google Scholar]
- Köhler A, Gottschling M, Förster J, Röwert-Huber J, Stockfleth E, Nindl I. Genomic characterization of a novel human papillomavirus (HPV-117) with a high viral load in a persisting wart. Virology. 2010;2010 doi: 10.1016/j.virol.2009.12.023. in press. [DOI] [PubMed] [Google Scholar]
- Lange CE, Tobler K, Favrot C, Muller M, Nothling JO, Ackermann M. Detection of antibodies against epidermodysplasia verruciformis-associated canine papillomavirus 3 in sera of dogs from Europe and Africa by enzyme-linked immunosorbent assay. Clin Vaccine Immunol. 2009a;16:66–72. doi: 10.1128/CVI.00346-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange CE, Tobler K, Markau T, Alhaidari Z, Bornand V, Stockli R, Trussel M, Ackermann M, Favrot C. Sequence and classification of FdPV2, a papillomavirus isolated from feline Bowenoid in situ carcinomas. Vet Microbiol. 2009b;137:60–65. doi: 10.1016/j.vetmic.2009.01.002. [DOI] [PubMed] [Google Scholar]
- Li L, Barry P, Yeh E, Glaser C, Schnurr D, Delwart E. Identification of a novel human gammapapillomavirus species. J Gen Virol. 2009;90:2413–2417. doi: 10.1099/vir.0.012344-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz N, Bosch FX, de Sanjosé S, Herrero R, Castellsagué X, Shah KV, Snijders PJF, Meijer CJLM. Epidemiological classification of human papillomavirus types associated with cervical cancer. New Engl J Med. 2003;348:518–527. doi: 10.1056/NEJMoa021641. [DOI] [PubMed] [Google Scholar]
- Myers G, Bernard HU, Delius H, Favre M, Iconogle J, van Ranst M, Wheeler C, editors. Human papillomaviruses 1994 Compendium. Los Alamos National Laboratory; Los Alamos, New Mexico, USA: 1994. ( http://hpv-web.lanl.gov/) [Google Scholar]
- Nafz J, Kohler A, Ohnesorge M, Nindl I, Stockfleth E, Rosl F. Persistence of Mastomys natalensis papillomavirus in multiple organs identifies novel targets for infection. J Gen Virol. 2007;88:2670–2678. doi: 10.1099/vir.0.82955-0. [DOI] [PubMed] [Google Scholar]
- Nobre RJ, Herráez-Hernández E, Fei JW, Langbein L, Kaden S, Gröne HJ, de Villiers EM. E7 oncoprotein of novel human papillomavirus type 108 lacking the E6 gene induces dysplasia in organotypic keratinocyte cultures. J Virol. 2009;83:2907–2916. doi: 10.1128/JVI.02490-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrow RS, LaBresh KV, Faras AJ. Characterization of the complete RhPV 1 genomic sequence and an integration locus from a metastatic tumor. Virology. 1991;181:424–429. doi: 10.1016/0042-6822(91)90519-h. [DOI] [PubMed] [Google Scholar]
- Palmenberg AC, Spiro D, Kuzmickas R, Wang S, Djikeng A, Rathe JA, Fraser-Liggett CM, Liggett SB. Sequencing and analyses of all known human rhinovirus genomes reveal structure and evolution. Science. 2009;324:55–59. doi: 10.1126/science.1165557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel KR, Smith KT, Campo MS. The nucleotide sequence and genome organization of bovine papillomavirus type 4. J Gen Virol. 1987;68:2117–2128. doi: 10.1099/0022-1317-68-8-2117. [DOI] [PubMed] [Google Scholar]
- Posada D, Crandall KA. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Rambaut A, Posada D, Crandall KA, Holmes EC. The causes and consequences of HIV evolution. Nat Rev Genet. 2004;5:52–61. doi: 10.1038/nrg1246. [DOI] [PubMed] [Google Scholar]
- Rebrikov DV, Bogdanova EA, Bulina ME, Lukyanov SA. A new planarian extrachromosomal virus-like element revealed by subtractive hybridization. Mol Biol. 2002;36:813–820. [PubMed] [Google Scholar]
- Rector A, Bossart GD, Ghim SJ, Sundberg JP, Jenson AB, Van Ranst M. Characterization of a novel close-to-root papillomavirus from a Florida manatee by using multiply primed rolling-circle amplification: Trichechus manatus latirostris papillomavirus type 1. J Virol. 2004;78:12698–12702. doi: 10.1128/JVI.78.22.12698-12702.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rector A, Lemey P, Tachezy R, Mostmans S, Ghim SJ, Van Doorslaer K, Roelke M, Bush M, Montali RJ, Joslin J, Burk RD, Jenson AB, Sundberg JP, Shapiro B, Van Ranst M. Ancient papillomavirus-host co-speciation in Felidae. Genome Biol. 2007;8:R57. doi: 10.1186/gb-2007-8-4-r57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rector A, Mostmans S, Van Doorslaer K, McKnight CA, Maes RK, Wise AG, Kiupel M, Van Ranst M. Genetic characterization of the first chiropteran papillomavirus, isolated from a basosquamous carcinoma in an Egyptian fruit bat: the Rousettus aegyptiacus papillomavirus type 1. Vet Microbiol. 2006;117:267–275. doi: 10.1016/j.vetmic.2006.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rector A, Stevens H, Lacave G, Lemey P, Mostmans S, Salbany A, Vos M, Van Doorslaer K, Ghim SJ, Rehtanz M, Bossart GD, Jenson AB, Van Ranst M. Genomic characterization of novel dolphin papillomaviruses provides indications for recombination within the Papillomaviridae. Virology. 2008;378:151–161. doi: 10.1016/j.virol.2008.05.020. [DOI] [PubMed] [Google Scholar]
- Rector A, Tachezy R, Van Doorslaer K, MacNamara T, Burk RD, Sundberg JP, Van Ranst M. Isolation and cloning of a papillomavirus from a North American porcupine by using multiply primed rolling-circle amplification: the Erethizon dorsatum papillomavirus type 1. Virology. 2005a;331:449–456. doi: 10.1016/j.virol.2004.10.033. [DOI] [PubMed] [Google Scholar]
- Rector A, Van DK, Bertelsen M, Barker IK, Olberg RA, Lemey P, Sundberg JP, Van Ranst M. Isolation and cloning of the raccoon (Procyon lotor) papillomavirus type 1 by using degenerate papillomavirus-specific primers. J Gen Virol. 2005b;86:2029–2033. doi: 10.1099/vir.0.80874-0. [DOI] [PubMed] [Google Scholar]
- Rehtanz M, Ghim SJ, Rector A, Van Ranst M, Fair PA, Bossart GD, Jenson AB. Isolation and characterization of the first American bottlenose dolphin papillomavirus: Tursiops truncatus papillomavirus type 2. J Gen Virol. 2006;87:3559–3565. doi: 10.1099/vir.0.82388-0. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Schulz E, Gottschling M, Bravo IG, Wittstatt U, Stockfleth E, Nindl I. Genomic characterization of the first insectivoran papillomavirus reveals an unusually long, second non-coding region and indicates a close relationship to betapapillomavirus. J Gen Virol. 2009a;90:626–633. doi: 10.1099/vir.0.008011-0. [DOI] [PubMed] [Google Scholar]
- Schulz E, Gottschling M, Wibbelt G, Stockfleth E, Nindl I. Isolation and genomic characterization of the first Norway rat (Rattus norvegicus) papillomavirus and its phylogenetic position within Pipapillomavirus primarily infecting rodents. J Gen Virol. 2009b doi: 10.1099/vir.0.012583-0. in press. [DOI] [PubMed] [Google Scholar]
- Stevens H, Rector A, Bertelsen MF, Leifsson PS, Van Ranst M. Novel papillomavirus isolated from the oral mucosa of a polar bear does not cluster with other papillomaviruses of carnivores. Vet Microbiol. 2008a;129:108–116. doi: 10.1016/j.vetmic.2007.11.037. [DOI] [PubMed] [Google Scholar]
- Stevens H, Rector A, Van Der Kroght K, Van Ranst M. Isolation and cloning of two variant papillomaviruses from domestic pigs: Sus scrofa papillomaviruses type 1 variants a and b. J Gen Virol. 2008b;89:2475–2481. doi: 10.1099/vir.0.2008/003186-0. [DOI] [PubMed] [Google Scholar]
- Tachezy R, Duson G, Rector A, Jenson AB, Sundberg JP, Van Ranst M. Cloning and genomic characterization of Felis domesticus papillomavirus type 1. Virology. 2002a;301:313–321. doi: 10.1006/viro.2002.1566. [DOI] [PubMed] [Google Scholar]
- Tachezy R, Rector A, Havelkova M, Wollants E, Fiten P, Opdenakker G, Jenson B, Sundberg J, Van Ranst M. Avian papillomaviruses: the parrot Psittacus erithacus papillomavirus (PePV) genome has a unique organization of the early protein region and is phylogenetically related to the chaffinch papillomavirus. BMC Microbiol. 2002b;2:19–27. doi: 10.1186/1471-2180-2-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan CH, Tachezy R, Van Ranst M, Chan SY, Bernard HU, Burk RD. The Mastomys natalensis papillomavirus: nucleotide sequence, genome organization, and phylogenetic relationship of a rodent papillomavirus involved in tumorigenesis of cutaneous epithelia. Virology. 1994;198:534–541. doi: 10.1006/viro.1994.1064. [DOI] [PubMed] [Google Scholar]
- Terai M, DeSalle R, Burk RD. Lack of canonical E6 and E7 open reading frames in bird papillomaviruses: Fringilla coelebs papillomavirus and Psittacus erithacus timneh papillomavirus. J Virol. 2002;76:10020–10023. doi: 10.1128/JVI.76.19.10020-10023.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobler K, Favrot C, Nespeca G, Ackermann M. Detection of the prototype of a potential novel genus in the family Papillomaviridae in association with canine epidermodysplasia verruciformis. J Gen Virol. 2006;87:3551–3557. doi: 10.1099/vir.0.82305-0. [DOI] [PubMed] [Google Scholar]
- Tomita Y, Literak I, Ogawa T, Jin Z, Shirasawa H. Complete genomes and phylogenetic positions of bovine papillomavirus type 8 and a variant type from a European bison. Virus Genes. 2007;3:243–249. doi: 10.1007/s11262-006-0055-y. [DOI] [PubMed] [Google Scholar]
- Van Bressem MF, Cassonnet P, Rector A, Desaintes C, Van Waerebeek K, Alfaro-Shigueto J, Van Ranst M, Orth G. Genital warts in Burmeister’s porpoises: characterization of Phocoena spinipinnis papillomavirus type 1 (PsPV-1) and evidence for a second, distantly related PsPV. J Gen Virol. 2007;88:1928–1933. doi: 10.1099/vir.0.82694-0. [DOI] [PubMed] [Google Scholar]
- Van Doorslaer K, Rector A, Jenson AB, Sundberg JP, Van Ranst M, Ghim SJ. Complete genomic characterization of a murine papillomavirus isolated from papillomatous lesions of a European harvest mouse (Micromys minutus) J Gen Virol. 2007;88:1484–1488. doi: 10.1099/vir.0.82615-0. [DOI] [PubMed] [Google Scholar]
- Van Doorslaer K, Rector A, Vos P, Van Ranst M. Genetic characterization of the Capra hircus papillomavirus: a novel close-to-root artiodactyl papillomavirus. Virus Res. 2006;118:164–169. doi: 10.1016/j.virusres.2005.12.007. [DOI] [PubMed] [Google Scholar]
- Van Doorslaer K, Sidi AO, Zanier K, Rybin V, Deryckere F, Rector A, Burk RD, Lienau EK, van Ranst M, Trave G. Identification of unusual E6 and E7 proteins within avian papillomaviruses: cellular localization, biophysical characterization, and phylogenetic analysis. J Virol. 2009;83:8759–8770. doi: 10.1128/JVI.01777-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Ranst M, Fuse A, Fiten P, Beuken E, Pfister H, Burk RD, Opdenakker G. Human papillomavirus type 13 and pygmy chimpanzee papillomavirus type 1: comparison of the genome organizations. Virology. 1992a;190:587–596. doi: 10.1016/0042-6822(92)90896-w. [DOI] [PubMed] [Google Scholar]
- Van Ranst M, Kaplan JB, Burk RD. Phylogenetic classification of human papillomaviruses: correlation with clinical manifestations. J Gen Virol. 1992b;73:2653–2660. doi: 10.1099/0022-1317-73-10-2653. [DOI] [PubMed] [Google Scholar]
- Van Ranst M, Tachezy R, Delius H, Burk RD. Taxonomy of the human papillomaviruses. Papillomavirus Rep. 1993;4:61–65. [Google Scholar]
- Van Regenmortel MH, Maniloff J, Calisher CH. The concept of virus species. Arch Virol. 1991;120:313–314. doi: 10.1007/BF01310487. [DOI] [PubMed] [Google Scholar]
- Van Regenmortel MH, Mayo MA, Fauquet CM, Maniloff J. Virus nomenclature: consensus versus chaos. Arch Virol. 2000;145:2227–2232. doi: 10.1007/s007050070053. [DOI] [PubMed] [Google Scholar]
- Van Regenmortel MHV, Fauquet CM, Bishop DHL, Calisher CH, Carsten EB, Estes MK, Lemon SM, Maniloff J, Mayo MA, McGeoch DJ, Pringle CR, Wickner RB. Virus Taxonomy. Seventh Report of the International Committee for the Taxonomy of Viruses”; Academic Press, New-York, San Diego. 2002. [Google Scholar]
- Vasiljevic N, Hazard K, Dillner J, Forslund O. Four novel human betapapillomaviruses of species 2 preferentially found in actinic keratosis. J Gen Virol. 2008;89:2467–2474. doi: 10.1099/vir.0.2008/001925-0. [DOI] [PubMed] [Google Scholar]
- Wettstein FO, Stevens JG. Distribution and state of viral nucleic acid in tumors induced by Shope papilloma virus. Cold Spring Harbor Conf Cell Proliferation. 1980;7:301–307. [Google Scholar]
- Woolford L, Rector A, Van Ranst M, Ducki A, Bennett MD, Nicholls PK, Warren KS, Swan RA, Wilcox GE, O’Hara AJ. A novel virus detected in papillomas and carcinomas of the endangered western barred bandicoot (Perameles bougainville) exhibits genomic features of both the Papillomaviridae and Polyomaviridae. J Virol. 2007;81:13280–13290. doi: 10.1128/JVI.01662-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H, Ghim S, Newsome J, Apolinario T, Olcese V, Martin M, Delius H, Felsburg P, Jenson B, Schlegel R. An epidermotropic canine papillomavirus with malignant potential contains an E5 gene and establishes a unique genus. Virology. 2007;359:28–36. doi: 10.1016/j.virol.2006.08.029. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table. Animal papillomavirus ICTV nomenclature.


