Abstract
Recent successes in deriving human-induced pluripotent stem cells (hiPSCs) allow for the possibility of studying human neurons derived from patients with neurological diseases. Concomitant inhibition of the BMP and TGF-β1 branches of the TGF-β signaling pathways by the endogenous antagonist, Noggin, and the small molecule SB431542, respectively, induces efficient neuralization of hiPSCs, a method known as dual-SMAD inhibition. The use of small molecule inhibitors instead of their endogenous counterparts has several advantages including lower cost, consistent activity, and the maintenance of xeno-free culture conditions. We tested the efficacy of DMH1, a highly selective small molecule BMP-inhibitor for its potential to replace Noggin in the neuralization of hiPSCs. We compare Noggin and DMH1-induced neuralization of hiPSCs by measuring protein and mRNA levels of pluripotency and neural precursor markers over a period of seven days. The regulation of five of the six markers assessed was indistinguishable in the presence of concentrations of Noggin or DMH1 that have been shown to effectively inhibit BMP signaling in other systems. We observed that by varying the DMH1 or Noggin concentration, we could selectively modulate the number of SOX1 expressing cells, whereas PAX6, another neural precursor marker, remained the same. The level and timing of SOX1 expression have been shown to affect neural induction as well as neural lineage. Our observations, therefore, suggest that BMP-inhibitor concentrations need to be carefully monitored to ensure appropriate expression levels of all transcription factors necessary for the induction of a particular neuronal lineage. We further demonstrate that DMH1-induced neural progenitors can be differentiated into β3-tubulin expressing neurons, a subset of which also express tyrosine hydroxylase. Thus, the combined use of DMH1, a highly specific BMP-pathway inhibitor, and SB431542, a TGF-β1-pathway specific inhibitor, provides us with the tools to independently regulate these two pathways through the exclusive use of small molecule inhibitors.
Keywords: Induced pluripotent stem cells, iPS, neural induction, neural differentiation, TGF-β, transcription factors
Recently developed methods to derive patient-specific stem cells (human-induced pluripotent stem cells, hiPSCs) provide the scientific and medical community the tantalizing possibility of developing experimental model systems using fully differentiated human cells.1,2 hiPSCs are derived from somatic cells such as fibroblasts or keratinocytes and, by definition, can be differentiated into cell types representing all three germ layers. Thus, cells such as myocardiocytes, red blood cells, and several types of neurons, including dopaminergic and GABAergic neurons, motor neurons, and neural crest cells have been derived by manipulation of signaling pathways known to play a role in the in vivo differentiation of these cell types.3−11
Signal transduction by the transforming growth factor-β (TGF-β) superfamily members plays many diverse roles in the maintenance as well as differentiation of hiPSCs. There are over 30 different vertebrate TGF-β superfamily ligands that can be divided into two main groups, the TGF-β1 group (TGF-βs, activins, and nodals) and bone-morphogenic protein (BMP)-like ligands (BMPs, GDFs, and MIS).12,13 These ligands induce the association of ligand-specific type I and type II cell surface receptors upon binding, an interaction that activates type II receptor to phosphorylate type I receptor, which then propagates the signal by phosphorylating receptor-activated SMAD (R-SMADS) proteins. The TGF-β1- and BMP-like ligands bind to different receptor complexes resulting in the phosphorylation of SMAD 2/3 and SMAD1/5/8 proteins, respectively.12,14 The maintenance of stem cell pluripotency requires signaling through the TGF-β1-group pathway but repression of the BMP pathway.15−18 Differentiation of stem cells into various cell lineages can be achieved by differential inhibition and/or activation of these two pathways.19−22 Recent studies have shown that the combined inhibition of the BMP and TGF-β1 pathways, a process termed dual-SMAD inhibition, results in highly efficient conversion of hES and hiPSCs into neural precursor cells.5,23
BMP and TGF-β1 ligands are opposed in vivo by endogenous extracellular ligand trap proteins (antagonists) such as Noggin, Follistatin, and Chordin.13,14 Recombinant forms of these antagonists have been successfully used for the differentiation of hiPSCs; however, the use of small molecules in place of these recombinant proteins has several advantages. For example, small chemical compounds can be produced relatively cheaply, in large quantities and of high purity. In addition, since they diffuse more readily into tissues and are more stable, their effectiveness is more consistent than the variable activity often observed with different lots of recombinant proteins.
Here, we compare the formation of neural precursor cells derived from hiPSCs with dual SMAD-inhibition using the highly specific TGF-β1-inhibitor SB43154224 in combination with either recombinant Noggin or DMH1, a recently developed highly selective small molecule BMP-inhibitor.25 We assessed DMH1 and Noggin-induced mRNA and protein expression levels of pluripotency and neuronal precursor markers of 9 different hiPSCs lines, which were derived from a control subject, two patients with compound heterozygous PARK2 mutations, and a patient with tuberous sclerosis complex (TSC) due to a TSC1 mutation. The expression of all markers was similarly regulated by Noggin and DMH1. SOX1 expression, however, was dependent on the concentration of the BMP-inhibitors, DMH1 or Noggin. We demonstrate that DMH-1-induced neural precursor cells are competent to further differentiate into β3-tubulin and tyrosine-hydroxylase expressing neurons. The combined use of SB431542 and DMH-1 enables us to regulate the two TGF-β signaling pathways independently in hiPSCs with the exclusive use of small molecules.
Results and Discussion
To ensure the successful use of hiPSC-derived neural precursors or differentiated neurons for cell-based therapy or drug discovery, standardized, chemically defined, and economically affordable methods to derive these cells need to be developed. Small molecule agonists/antagonists have the advantages that they can be produced cheaply and in large quantities, are stable, show increased tissue penetration, and allow for xeno-free culture conditions. Here, we tested the efficacy of DMH1, a highly selective, small molecule analogue of dorsomorphin, to replace Noggin, the endogenous BMP-antagonists for the neuralization of hiPSCs.
Generation and Validation of hiPSCs
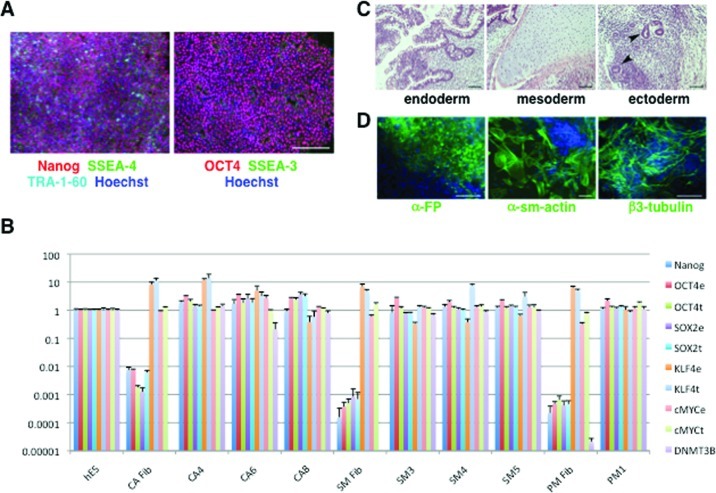
The efficiency with which neural precursors can be derived varies substantially between hiPSC lines,26,27 and it is therefore necessary to assess and validate methods of hiPSC differentiation in sufficiently large sets of cell lines derived from different subjects. Thus, in order to compare Noggin and DMH1-induced neuralization, we tested nine different hiPSC-lines derived from four subjects (Supporting Information, Table S1), three lines (CA4, CA6, and CA8) from a control subject (CA), four lines (SM3, SM4, SM5, and PM1) from two brothers, SM and PM, who share a heterozygous compound PARK2 mutation, and two lines (TSC-12B2 and TSC-12B4) from a patient (TSC-12) with tuberous sclerosis complex (TSC). All lines were validated using an array of assays. Immunofluorescence staining confirmed the expression of the pluripotency markers OCT4, NANOG, TRA-1-60, SSEA-3, and SSEA-4 in all cell lines used in this study (Figure 1A and data not shown). mRNA levels of the pluripotency markers OCT4, SOX2, KLF4, C-MYC, and DNMT3B were determined by qRT-PCR and compared to those of the human embryonic stem cell line HES-2 (Figure 1B and data not shown). The pluripotency of the derived lines was further examined by assessing their potential to differentiate into cell lineages of all three germ layers in teratomas (CA6) (Figure 1C) or in embryoid body-mediated differentiation (CA6, TSC-12B2, and TSC-12B4) (Figure 1D). All hiPSC lines were successfully differentiated into neural precursors. All hiPSC lines showed normal karyotype except for CA8, which was trisomic for chromosomes 12 and 21, and CA4, which was mosaic with 13.3% of the cells having trisomy for the long (20q) and monosomy for the short arm (20p) of chromosome 20, and 5% of the cells being trisomic for chromosome 8 (Supporting Information, Figure S1 and data not shown). We determined hiPSC population doubling times during the exponential growth phase by two independent methods in each experiment in parallel. In one assay, cell numbers were determined by counting cells with a cell counter. The other assay was performed by measuring the fluorescence intensity of lysed cells stained with PicoGreen, a dye that fluoresces when bound to DNA. The doubling times determined by these two methods were comparable and similar to values published for other hiPSCs and hES cells (Supporting Information, Figure S2).28,29
Figure 1.
Pluripotency of hiPSC lines. (A) The expression of 5 pluripotency marker proteins (Nanog, SSEA-3, SSEA-4, TRA-1-60, and OCT4) was assessed in all hiPSC lines derived from patients CA, SM, PM, and TSC-12 by immunocytochemistry. All lines showed expression of all five markers. Shown here is the SM4 line derived from a patient SM. Scale bar = 200 μm. (B) Expression levels of six pluripotency markers were assessed in all hiPSC lines by real time quantitative RT-PCR (qRT-PCR). mRNA levels are expressed relative to the levels measured for the human embryonic stem cell line HES-2. The marker DNMT3B was not detectable in fibroblasts, except for the PM-fibroblast in which we measured very low levels. e = endogenous; t = total (endogenous and viral expression) (shown are mean ± STD of 4 technical replicates). (C) Teratomas generated from hiPSC contain cell lineages from all three germ layers with gastrointestinal structures (endoderm, scale bar = 100 μm), cartilage (mesoderm, scale bar = 200 μm), and neural rosettes (ectoderm, arrow heads, scale bar = 200 μm). (D) Embryoid bodies (CA6) were stained with antibodies against α-fetoprotein (α-FP, endoderm), α-smooth muscle actin (αsm-actin, mesoderm), and β3-tubulin (ectoderm). The cultures were counter stained with Hoechst dye (blue). Scale bars = 100 μm.
Comparison of Noggin and DMH1-Induced Neural Precursor Formation
Assessment of Pluripotency and Neural Precursor Marker Protein Expression
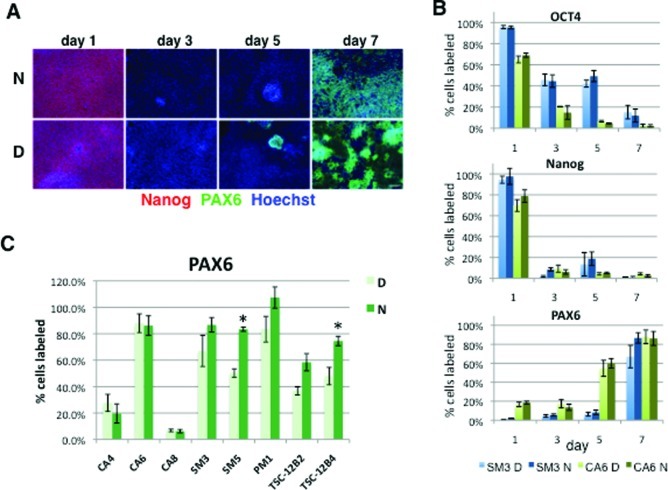
We next compared the efficacy of DMH1, a highly selective, small molecule analogue of dorsomorphin, and Noggin, the endogenous BMP-antagonists, to induce neuralization of hiPSC via dual-SMAD inhibition. We used the highly specific TGF-β1 inhibitor SB43154230 in combination with either Noggin (500 ng/mL) or DMH1 (0.5 μM), concentrations that have been shown to efficiently block BMP signaling.25,31,32 The percentage of cells expressing the pluripotency marker proteins OCT4 and Nanog was significantly reduced by day 3 in both SM3 and CA6 cells (p < 0.05), while PAX6 expression was significantly up-regulated by day 5 and day 7 in CA6 and SM3 cells, respectively (p < 0.05); these differences were independent of the treatment (p > 0.05 for Noggin versus DMH1: OCT4 F3,24 = 0.4085, Nanog F3,24 = 0.114, and PAX6 F3,24 = 1.775) (Figure 2A,B). In six of the eight cell lines examined, neuralization with DMH1 and Noggin resulted in a comparable percentage of PAX6 positive cells at day 7 regardless of the efficiency of PAX6 expression, which ranged from ∼10–20% in the two cell lines with abnormal karyotypes (CA4 and CA8) to 100% in the PM1 line (Figure 2C). In two cell lines, SM5 and TSC-12B4, the number of PAX6 positive cells was higher in cultures neuralized with Noggin (Figure 2C).
Figure 2.
Noggin and DMH1-induced regulation of OCT4, Nanog, and PAX6 protein expression. Expression of OCT4, Nanog, and PAX6 proteins was assessed on days 1, 3, 5, and 7 of neuralization induced with SB431542 and either Noggin or DMH1. (A) Cells (SM3) were stained with antibodies against OCT4, Nanog, and PAX6 and counterstained with Hoechst dye. Scale bar in = 100 μm. (B) The percentage of OCT4-, Nanog-, and PAX6-positive cells was determined for days 1, 3, 5, and 7. (C) Noggin- and DMH1-induced PAX6 expression at day 7 is shown for eight cell lines. PAX6 data for day 7 in B (SM3 and CA6) are the same as in C. Weak signal for some of the Hoechst stained cells resulted in a slight underestimation of nuclei by automated cell counting, explaining the slightly greater than 100% PAX6 positive cells calculated for PM1. Shown are the mean ± SEM, *p < 0.05.
Assessment of Pluripotency and Neural Precursor Marker mRNA Levels
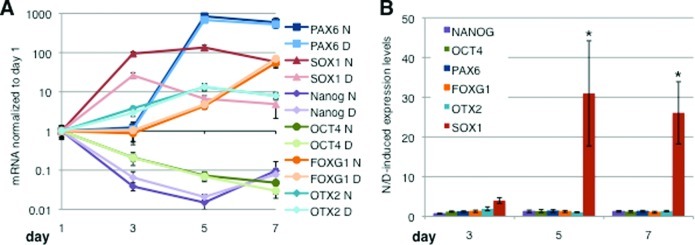
In order to determine overall expression levels of the above markers as well as additional neuralization markers, we quantified mRNA levels of two pluripotency markers OCT4 and NANOG and four neural precursor markers (PAX6, FOXG1, OTX2, and SOX1) by qRT-PCR over the first 7 days of neural induction for all nine cell lines. The degree and time frame of the down-regulation of the hiPSC markers OCT4 and NANOG were similar in the presence of Noggin and DMH1 in all nine cell lines examined (Figure 3A and data not shown). Overall, we observed a 10–100-fold down-regulation of both OCT4 and NANOG by day 7 of neuralization with DMH1 or Noggin in all the cell lines, except CA4, where OCT4 levels decreased to a lesser degree in the presence of both, DMH1 and Noggin (data not shown). Assessment of OCT4 and NANOG levels at the cell line level at day 7 revealed less than 2.5-fold difference between cells neuralized with DMH1 and Noggin (Figure 3 and Supporting Information, Figure S3). Thus, the Noggin- and DMH1-induced down-regulation of the pluripotency marker mRNAs was comparable for all the cell lines we assessed.
Figure 3.
Noggin and DMH1-induced regulation of pluripotency and neural precursor marker mRNAs. (A) Expression of two hiPSC markers (OCT4, Nanog) and four neural precursor markers (PAX6, SOX1, FOXG1, and OTX2) was assessed during neuralization using dual-SMAD inhibition with SB431542 and either Noggin or DMH1 by qRT PCR over a period of 7 days. Shown are the mean values and 99.5% confidence intervals for the cell line SM5 (n = 6), D = DMH1, and N = Noggin. (B) The averaged ratios of Noggin-induced/DMH1-induced marker mRNA levels of all 9 cell lines are plotted for day 3, 5, and 7 of neuralization. Values >1 indicate higher levels in the presence of Noggin, while values <1 indicate higher expression in the presence of DMH1. Shown are the means ± SEM. Marker ratios were independent of time (day) but dependent on the marker protein (*p < 0.001, n = 9).
The levels of PAX6 mRNA, an early neuroectoderm and neuronal precursor marker, increased dramatically between days 3 and 5 and then either increased at a slower rate or remained constant from days 5 and day 7 in all nine cell lines examined (Figure 3A and data not shown). The relative difference of Noggin and DMH1-induced PAX6 levels was small (<4-fold) for all cell lines and at all time points assessed (Figure 3 and Supporting Information, Figure S3). Similarly, mRNA levels of FOXG1 (also known as BF1), which, like PAX6, regulates proliferation of telencephalic progenitors in the early neural plate and tube,33,34 were elevated by day 5 in the presence of Noggin or DMH1. The differences between Noggin and DMH1-induced FOXG1 mRNA levels were small (<3-fold) for all cell lines (Figure 3 and Supporting Information, Figure S3). Levels of OTX2, a marker of fore-and midbrain neural progenitors,35,36 were modestly increased by day 3, continued to increase until day 5, and then leveled off in most cell lines (Figure 3A and data not shown). Noggin and DMH1-induced OTX2 expression levels were comparable and differed by <5-fold for all cell lines and at all time points assessed (Figure 3A and Supporting Information, Figure S3). The degree and timing of the Noggin-induced marker expression in our study replicated observations made by other authors using Noggin-induced neuralizations of hES or hiPS cells cultured and neuralized under very similar conditions.5
Levels of SOX1, a marker of early neuroectoderm, rose before PAX6 and were increased 10–100-fold by day 3 in all cell lines except in CA4 in which SOX1 levels increased minimally (Figure 3A and data not shown). While both Noggin and DMH1 cause an increase of SOX1 expression by day 3, this increase was smaller in the presence of DMH1 in all cell lines (Figure 3A,B, Supporting Information, Figure S3, and data not shown). In the presence of DMH1, SOX1 levels were down-regulated again by day 5 and 7, whereas in the presence of Noggin, SOX1 levels remained elevated, a difference that is manifested by the significant differences in Noggin and DMH1-induced SOX1 expression at days 5 and 7 (Figure 3A,B and Supporting Information, Figure S3). Indeed, in all cell lines, except CA4, Noggin-induced SOX1 levels were 10- to 100-fold higher than DMH1-induced levels at day 7 (Figure 3B and Supporting Information, Figure S3). Examination of all six markers was replicated in independent experiments for lines CA6 and SM3 (data not shown) confirming our observations for all nine lines that SOX1 is the only marker that was differentially regulated by Noggin and DMH1. The Noggin/DMH1-induced expression level ratios of all cell lines were averaged and plotted as a function of time (Figure 3B). Except for SOX1, the ratios for all markers (transcription factors) on all days were close to 1 (0.7 ≤ ratio ≤1.9) indicating that these transcription factors are similarly regulated by Noggin and DMH1. Two-way ANOVA shows a significant interaction between the expression level of these markers and the day of neuralization (F10,144 = 2.607). Bonferroni posthoc tests indicate that the ratio of Noggin/DMH1-induced SOX1 expression levels is significantly different from all the other markers at days 5 and 7 (*p < 0.001), while the ratios for all the other markers were comparable at all time points (Figure 3B).
The differential regulation of SOX1 mRNA in the presence of Noggin and DMH1 led us to examine this transcription factor at the protein level in more detail. Similar to our observations of SOX1 mRNA levels, the number of cells expressing SOX1 protein at day 7 was dramatically higher in cultures neuralized with Noggin than those with DMH1 (at 0.5 μM) (Figure 4A). In the majority of the cells, SOX1 expression colocalized with PAX6 expression. Flow cytometric analyses confirmed our qPCR and immunocytochemical observations that SOX1-protein expression is higher in the presence of Noggin, while PAX6-protein expression is comparable in the presence of Noggin and DMH1 at 0.5 μM (Figure 4B).
Figure 4.
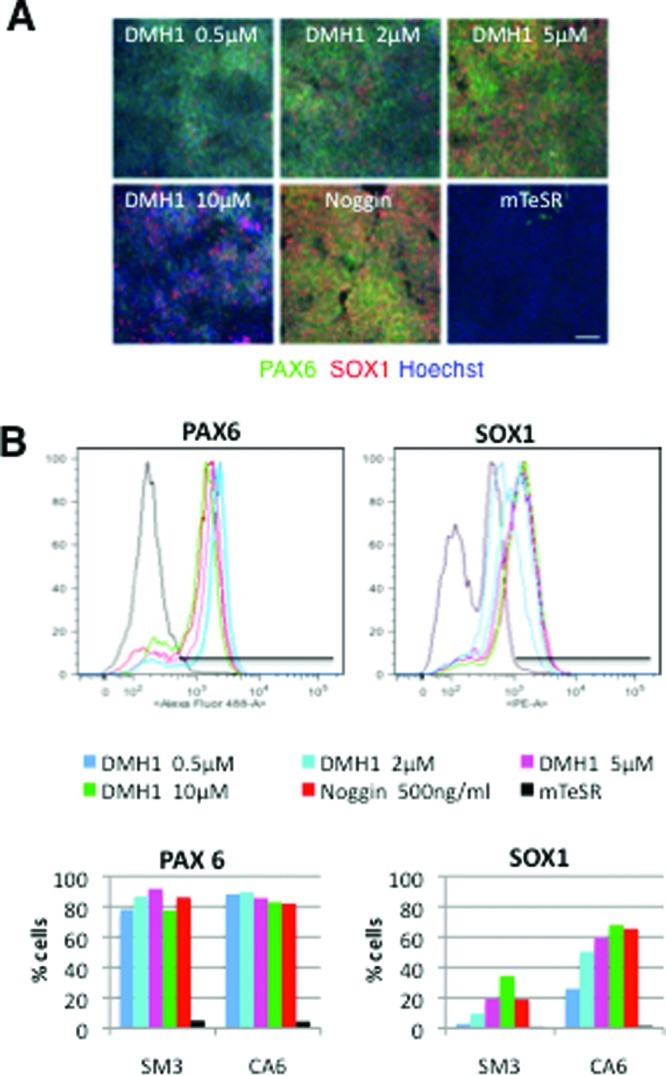
DMH1 concentration dependent SOX1 expression. hiPSCs were neuralized with DMH1 at 0.5 μM, 2 μM, 5 μM, 10 μM, or Noggin (500 ng/mL) for 7 days. Control cells were cultured in mTeSR medium for the same length of time. (A) The micrographs show SM3 cells that were stained with antibodies against PAX6 (green) and SOX1 (red) and counterstained with Hoechst dye (blue) at day 7 of neuralization. Scale bar = 100 μm. (B) PAX6- and SOX1-positive CA6 and SM3 cells were quantified by flow cytometry (traces shown are for CA6 cells). Relative frequencies of cells (y-axis) against fluorescence intensity (x-axis) and the gating used for quantification are plotted for each antibody. The percentage of PAX6 and SOX1 labeled cells for each treatment group and cell line are plotted in bar graphs.
The DMH1 concentration (0.5 μM) used has been shown to effectively inhibit signaling of the BMP type-1 receptors ALK2 and ALK3 in other systems25,31 and, as described above, with the exception of SOX1, regulates the expression of the pluripotency and neuralization markers in a manner comparable to that of Noggin. In another set of experiments, we tested if increasing the DMH1 concentration would alter SOX1 expression levels. Immunocytochemical (Figure 4A) as well as flow cytometric analysis (Figure 4B) revealed a DMH1 concentration-dependent increase of SOX1 expression with a 10-fold increase of DMH1 (5 μM) resulting in a percentage of SOX1 expressing cells comparable to the number observed in cultures neuralized with Noggin (500 ng/mL), while PAX6 expression was not affected by DMH1 concentration (Figure 4). Increasing the DMH1 concentration to 10 μM further increased the percentage of cells expressing SOX1 (Figure 4) but caused significant cellular toxicity manifested in a much lower cell density after 7 days of neural induction (Figure 4A). These observations suggest that the expression of SOX1 can be regulated independently of PAX6 by manipulating the DMH1 concentration during the neural induction of hiPSCs.
Next, we tested if SOX1 expression is similarly dependent on the Noggin concentration. We exposed four of our cell lines to DMH1 at 0.5 μM, Noggin at 500 ng/mL (the concentration used in the above-described experiments), to a 10-fold lower Noggin concentration (50 ng/mL), or a combination of DMH1 (0.5 μM) and 10-fold lower levels of Noggin (50 ng/mL). Similarly to DMH1, reducing the Noggin concentration by a factor of 10 resulted in a decrease in the number of cells expressing SOX1 (Figure 5A,B). DMH1 (0.5 μM) combined with a low concentration of Noggin (50 ng/mL) induced a higher percentage of SOX1-positive cells than either DMH1 (0.5 μM) or low Noggin alone but did not reach the levels observed with Noggin at 500 ng/mL (Figure 5). PAX6 expression was similar under all neuralization conditions, except in the case of the cell line CA4, which showed reduced PAX6 expression in the presence of low Noggin (Figure 5). Together, these data suggest that SOX1, but not PAX6 expression, levels depend on the concentration of the BMP-antagonist used for neural induction.
Figure 5.
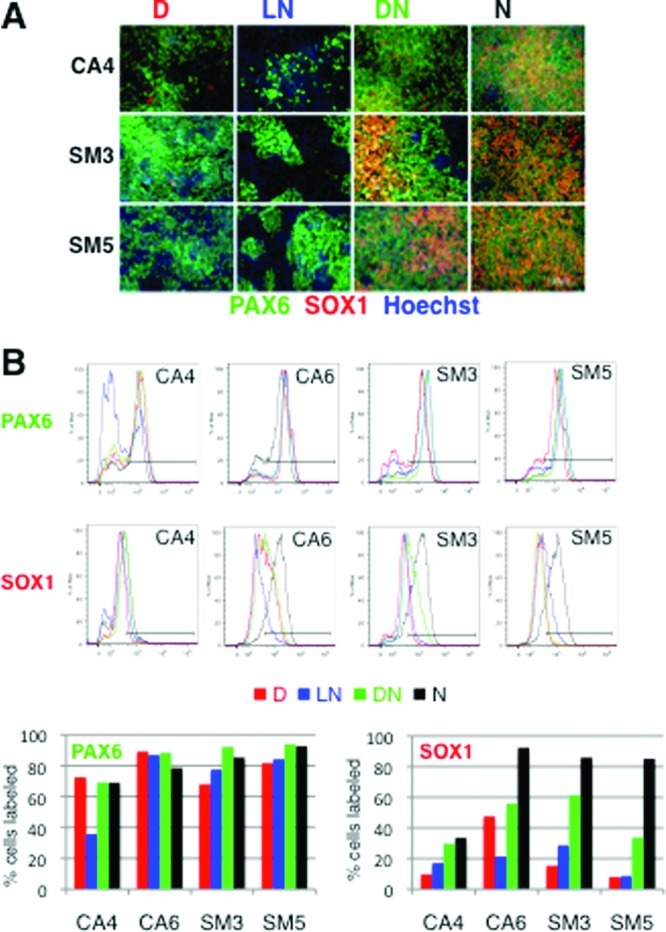
Lowering the Noggin concentration results in a decrease of SOX1 protein expression. Four different cell lines (CA4, CA6, SM3, and SM5) were neuralized with DMH1 (0.5 μM, D), a low concentration of Noggin (LN, 50 ng/mL), a combination of DMH1 and a low concentration of Noggin (50 ng/mL; DN), or a high concentration of Noggin (500 ng/mL; N). (A) The cells were stained with antibodies against PAX6 (green) and SOX1 (red), counterstained with Hoechst dye (blue), and expression assessed by fluorescence microscopy. Scale bar = 100 μm. (B) Cells stained for PAX6 and SOX1 were quantified by flow cytometry. Relative frequencies of cells (y-axis) against fluorescence intensity (x-axis) and the gating used for quantification are plotted for each antibody and cell line. The percentage of PAX6 and SOX1 labeled cells for each treatment group and cell line are plotted in bar graphs.
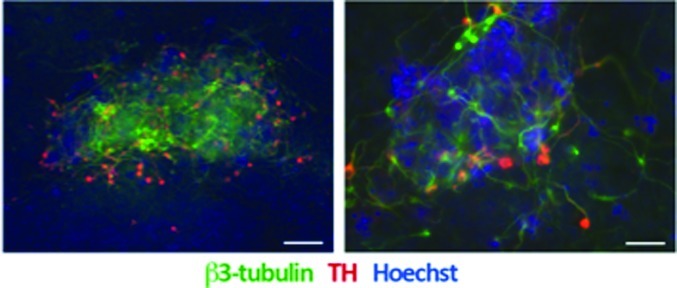
In a final set of experiments, we assessed the differentiation potential of DMH1-induced neural progenitor cells and demonstrate that they can be further differentiated into β3-tubulin neurons, some of which coexpress tyrosine-hydroxylase indicative of a dopaminergic fate (Figure 6).
Figure 6.
DMH1-induced neural precursors can be differentiated into β3-tubulin, tyrosine-hydroxylase positive neurons. We further differentiated DMH1-induced neural precursor cells into β3-tubulin (green), some of which expressed tyrosine-hydroxylase (red) positive neurons. Neurons tended to occur in clusters (left panel). Higher magnification images (right panel) show coexpression of β3-tubulin in tyrosine-hydroxylase positive neurons. Cultures were counterstained with Hoechst dye (blue). Scale bar in A = 100 μm and in B = 50 μm.
Conclusions
Efficient and reproducible methods to differentiate hiPSCs into specific cell types, such as neurons, are critical to ensure the successful use of these cells for the study of human developmental processes, cellular mechanisms underlying human disease, or the development of drug screening platforms. Here, we tested the efficacy of DMH1, a highly selective, small molecule BMP-inhibitor to induce neuralization of hiPSCs. We report that Noggin and DMH1-induced hiPSC neuralization occurs within the same time frame and that the derived neural precursor cells display a very similar transcription factor expression profile. DMH1 is a highly specific BMP-antagonist that inhibits signaling through ALK1, ALK2, and ALK3 receptors, with negligible effect on the ALK6 receptor and no other off-target effects.25,31 Dorsomorphin and LDN-193189, two other small molecule BMP-inhibitors, have been used to neuralize hiPSCs;23,27,37−39 however, both of these compounds have been shown to inhibit the BMP- as well as the TGF-β1 branches of the TGF-β pathways23,31 and have many additional targets including AMP-activated kinase (AMPK),40,41 receptor tyrosine kinases for PDGF and VEGF, and many other kinases.23,25,42,43 Thus, using DMH1 in combination with SB431542 allows us for the first time to inhibit the BMP- and TGF-β1 pathways specifically and independently through the exclusive use of small molecules.
We observed that the number of neural precursor cells expressing SOX1 increased with increasing concentrations of DMH1 or Noggin, while PAX6 expression was independent of the BMP-inhibitor concentrations examined. Levels and timing of SOX1 expression have been reported to affect neural development and neural lineage specification.44−48 Thus, our observations highlight the importance of determining the optimal small molecule concentration that results in appropriate expression levels and timing of all transcription factors relevant for the differentiation of a specific neuronal lineage.
Methods
hiPSC-Line Generation
Skin biopsies (3 mm) were obtained from four different subjects (patient information is provided in Supporting Information, Table S1) for the purpose of generating hiPSCs via informed consent under a protocol approved by the Vanderbilt Institutional Review Board. The tissue was cultured in complete DMEM medium (Invitrogen, Inc., Carlsbad, CA) containing 10% FBS (Sigma, St. Louis, MO), penicillin/streptomycin, (Mediatech, Inc., Manassas, VA), and nonessential amino acids (Sigma). After 14 days, the residual skin tissue was removed and the emerging dermal fibroblasts propagated in the same medium. hiPSCs from patients coded as CA, SM, PM, and TSC-12 were generated by adapting published methods.49,50 In brief, fibroblasts were first transduced with a lentivirus for the mouse receptor Slc7a1. This virus was produced in 293FT cells cotransfected with the pLenti6/UbC/mSlc7a1 plasmid DNA (Addgene, Cambridge, MA) and ViraPower (Invitrogen) using Lipofectamine 2000 (Invitrogen). The fibroblasts were subsequently transduced with equal volumes of supernatants from PLAT-E cells that had been transfected via Fugene 6 (Roche, Indianapolis, IN) with one of the following plasmids: pMXs-hc-MYC, pMXs-hKLF4, pMXs-hOCT3/4, or pMXs-hSOX2 (Addgene). Fibroblasts from subject CA were transduced with 4 factors (OCT4, SOX2, KLF4, and cMYC) and the fibroblasts from patients PM and SM with 3 factors (OCT4, SOX2, and KLF4). Nine days after transduction, CA cells were seeded onto SNL feeder cells (MMRC, UC Davis, CA) and cultured in hES medium (DMEM/F12 (Invitrogen), 20% Knockout serum replacement (Invitrogen), 2 mM Glutamax (Invitrogen), nonessential amino acids (Sigma), penicillin/streptomycin (Mediatech), 55 μM β-mercaptoethanol (Sigma), and recombinant human FGF-2 (Promega, Madison, WI)). Six days after transduction, PM and SM cells were plated onto Matrigel (BD Biosciences, San Jose, CA) coated plates and cultured in hES medium containing LIF (10 ng/mL, Millipore, Billerica, MA), PD0325901 (0.5 μM, Cayman, Ann Arbor, MI), SB431542 (2 μM, Tocris, Ellisville, MO), and thiazovivin (0.5 μM, Stemgent, Cambridge, MA).51 Fibroblasts from patient TSC-12 were transduced on two consecutive days with lentiviral particles expressing KLF4, OCT4, and SOX2 (Stemgent).52 Seven days after transduction, the fibroblasts were seeded onto SNL feeder cells. After about 3–6 weeks, emerging hiPSC colonies were picked, plated on Matrigel (BD Biosciences, Bedford, MA), and propagated in mTeSR1 medium (StemCell Technologies, Vancouver, Canada).50
hiPSC-Line Validation
The expression of the pluripotency markers OCT4, NANOG, SOX2, KLF4, DNMT3B, SSEA-3, SSEA-4, and TRA-1-60 was confirmed by quantitative real time PCR and/or immunofluorescence (see below). Total RNA from the hES-line HES-253 was used for comparison (a gift from Dr. Mark Magnuson, Vanderbilt University). Genetics Associates, Nashville, TN performed the karyotype analyses using standard protocols with at least 20 metaphase spreads per cell line being screened for chromosomal abnormalities. Pluripotency was assessed in vivo by injection of hiPSC into immune-compromised mice using established techniques. Briefly, approximately 1 × 106 CA6 hiPSCs were injected below the left kidney of each mouse following a retroperitoneal incision from the back. Teratoma development was monitored by ultrasound at one and two months postinjection. All mice were euthanized 2 months postinjection and the left kidney and associated teratomas dissected. The tissue was fixed overnight in 4% paraformaldehyde and then embedded in paraffin. Embryoid bodies were formed in AggreWell 800 plates (StemCell Technologies) according to the manufacturer’s instructions. Briefly, hiPSC cultures were washed once with DMEM/F12 medium (Invitrogen), dissociated with accutase (Innovative Cell Technologies, San Diego, CA) for 7 min at 37 °C min, sedimented, and the cell pellet resuspended in hES medium containing Rho Associated Kinase (ROCK) inhibitor, Y-27632 (10 μM, Tocris). Two milliliters of hES medium containing 3 × 106 cells were transferred into each well of an AggreWell 800 plate and the plate centrifuged at 100g for 3 min at room temperature. After 24 h, the embryoid bodies were dislodged by gentle pipetting and separated from single cells using a reversible restrainer. Embryoid bodies formed in a single AggreWell plate were resuspended in 12 mL of hES medium and evenly distributed into a 24 well plate coated with 0.1% gelatin. After 6 days, embryoid bodies were fixed and prepared for immunofluorescence staining. For the determination of the doubling times, hiPSCs were plated at low density (6000 cells per well of a 96 well plate) and cell numbers determined every 24 h in four biological replicates for four days by two independent methods: by automated cell counting with a Cellometer AutoT4 cell counter (Nexcelom Biosciences LLC., Lawrence, MA) and by measuring fluorescence intensity of cells that had been labeled with PicoGreen, a dye that binds to double stranded DNA. Fluorescence intensity was assessed in three technical replicates as described.54 A minimum of two independent experiments was performed for each cell line, and doubling times (Td) were determined while cells were in the exponential growth phase.
Neural Induction
hiPSC cultures were dissociated with accutase (Innovative Cell Technologies) for 8 min, washed twice with mTeSR1 medium (StemCell Technologies) containing ROCK inhibitor, Y-27632 (10 μM, Tocris), and plated at 6 × 103 cells per well of Matrigel-coated 96 well plates in the same medium. When the cells reached confluence (4–6 days) neuralization was initiated in neuralization medium (Knockout DMEM/F12, 15% KSR, glutamax (all from Invitrogen), penicillin/streptomycin (Mediatech), nonessential amino acids, and 55 μM β-mercaptoethanol (both from Sigma)) containing 10 μM of SB431542 (Tocris) and either human recombinant Noggin (R&D systems, Minneapolis, MN) or DMH1 that was synthesized as described.25 On days 5–7, the cells were cultured in neuralization medium containing 25% N2-medium (DMEM/F12, N2 supplement, and high d-glucose at a final concentration of 4.5 g/L, all from Invitrogen) as described.5 Neural precursor marker expression was assessed on days 1, 3, 5, and 7 by quantitative real time PCR (qRT-PCR) and immunocytochemistry as described below.
Neuronal Differentiation
Dopaminergic neurons were derived as described previously with minor modifications.5,55 At day 5 of neural induction SB431542 was omitted, while 0.5 μM DMH1 was continued until day 11, and increasing amounts of N2-medium were combined with neuralization medium such that day 5/6, 7/8, and 9/10 media contained 25%, 50%, and 75% N2-medium, respectively. As of day 11, the cells were kept in 100% N2-medium. From days 5–11, sonic hedgehog (200 ng/mL, SHH-C24II) was included in the medium, and day 9–11 medium also contained recombinant human BDNF (20 ng/mL), recombinant mouse FGF-8b (100 ng/mL), and ascorbate (200 μM). From day 12 on, the N2-medium was supplemented with BDNF (20 ng/mL), recombinant human GDNF (20 ng/mL), recombinant human TGFβ-3 (1 ng/mL) (all growth factors were obtained from R&D Systems), ascorbate (200 μM), and dcAMP (0.5 mM, both from Sigma). Differentiation was terminated and neurons fixed at days 21–28.
Quantitative Real Time PCR (qRT-PCR)
For validation of hiPSC-lines, total RNA from hiPSC-lines was prepared using the RNeasy kit (Qiagen, Valencia, CA) according to the manufacturer’s instructions. To determine mRNA levels of pluripotency and neural precursor markers, mRNA was prepared using the mRNA Catcher Plus kit (Invitrogen) according to the manufacturer's descriptions. Isolated mRNA was then reverse transcribed into cDNA on a Mycycler Thermal Cycler (Bio-Rad, Hercules, CA) using SuperScript III First-Strand Synthesis System (Invitrogen) according to the protocol provided by manufacturer except that the concentration of the superscript III enzyme was reduced 5-fold. qRT-PCR was performed with Power SYBR Green Master Mix (Applied Biosystems, Carlsbad, CA) on an ABI 7900HT fast real-time PCR detection system (Applied Biosystems, Carlsbad, CA). Primer sequences used are provided in Supporting Information, Table S2. The expression of the housekeeping genes GAPDH, PGK1, UBC, and ACTIN were assessed and ACTIN and UBC expression found to be the most consistent; measured mRNA levels were all normalized to the expression of ACTIN mRNA levels.
Immunofluorescence
The cells were fixed in PBS containing 4% paraformaldehyde for 30 min at room temperature. After permeabilization with 0.2% Triton-X100 for 20 min at room temperature, cells were incubated in PBS containing 5% donkey serum and 0.05% Triton-X100 for 2 h at room temperature or overnight at 4 °C. The following primary antibodies were used: Nanog (1:20, R&D Systems), Oct4 (1:200, Millipore,), Pax6 (1:200, Covance, Emeryville, CA), Sox1 (1:50, R&D Systems), SSEA-3 (1:500, Millipore), SSEA-4 (1:500, BD Biosciences), Tra-1-60 (1:200, Millipore), Tu-20 (1:500, Millipore), tyrosine-hydroxylase (1:500, Pel-Freez, Rogers, AR), α-fetoprotein (1:100, R&D Systems), desmin (1:100), α-smooth muscle actin (1:80), GATA4 (1:200), and Foxa2 (1:500) (all from Abcam, Temecula, CA). Secondary antibodies conjugated to DyLight 488 (1:200), DyLight 549 (1:400), or DyLight 649 (1:200) (Jackson ImmunoResearch, West Grove, PA) were used. Images were Obtained with a Zeiss ObserverZ1 microscope and AxioVs40 software (version 4.7.2). To quantify neural precursor marker expression, four images of each of 4 replicate cultures per condition were acquired with a 10× objective (EC Plan-NeoFluar 10×/0,3) and cell numbers determined using ImageJ plugin ITCN (Image Based Tool for Counting Nuclei). A total of 40,000 cells for each experimental condition were counted.
Flow Cytometry
Neural precursor cells were removed from the culture surface by treatment with accutase (Innovative Cell Technologies) for 10 min and were then triturated with a 1 mL glass pipet to obtain a single cell suspension. The cells were fixed in 2% paraformaldehyde in PBS for 12 min, permeabilized in 0.2% Triton-X100 in PBS for 30 min on ice, and then incubated with Alexa Fluor 488-conjugated mouse antihuman PAX6- and PE-conjugated mouse anti-Human SOX1 antibodies (BD Biosciences) for 30 min on ice. The samples were analyzed on a Custom Becton Dickson five-laser Fortessa analytical cytometer using BD FACSDiva acquisition (BD Biosciences) and FlowJo analysis software (Tree Star, Inc., Ashland, OR). A total of 10,000 events were acquired and analysis restricted to live single cell events determined by light scatter properties.
Statistical Analysis
Comparisons of the relative changes in marker mRNA levels were performed by testing for nonoverlap of the 99.5% confidence intervals to account for multiple comparisons using Microsoft Excel 2008. All other comparisons of marker expression levels were assessed by ANOVA using GraphPad Prism software (version 5.0b, La Jolla, CA 92037). Pair-wise posthoc analyses were done by t test with a Bonferroni multitesting correction. The alpha level for all analyses was set at p < 0.05.
Acknowledgments
We thank Dr. V. G. Dev of Genetics Associates, Inc (Nashville, TN) for karyotyping analyses, Dr. Mark A. Magnuson for generously providing total RNA from the HES-2 cell line, Dr. Peter Hedera for identification and recruitment of subjects PM and SM, Dr. Harold Lovvorn for injecting hiPSCs into mice, and Dr. Stuart Chambers and Dr. Lorenz Studer (Sloan-Kettering, NY) for helpful discussions. We thank Bingying Han, Brandon Goodman and Kevin Kumar for superb technical assistance and discussions of experimental protocols.
Supporting Information Available
Patient information, sequences of primers used for qRT-PCR experiments, karyotyping of hiPSC lines, and determination of hiPSC growth rates, ratios of Noggin-/DMH1-induced expression of transcription factors during neuralization of hiPSCs. This material is available free of charge via the Internet at http://pubs.acs.org.
This work was supported by the following grants: Doris Duke Charitable Foundation (KCE), Peterson Foundation for Parkinsons (ABB), Hazinski-Turner Award (KCE), Veterans Affairs CDTA and Merit Award 101BX000771 (CCH), and NIH grants 5U01HL100398 and 1R01HL104010 (CCH). VICTR Grant (ABB, KCE), Subaward RR166-737/4787736 (ABB and KCE) under NIH/NIGMS 5PO1 GM08535403, and Vanderbilt Center for Molecular Toxicology Pilot Project (ABB, MDN) under NIH/NIEHS 5P30 ES000267, NIH/NIEHS ES016931 (ABB), and ES016931-02S1 (ABB). Flow cytometry experiments were performed in the Vanderbilt Medical Center Flow Cytometry Shared Resource, which is supported by the Vanderbilt Ingram Cancer Center (P30 CA68485) and the Vanderbilt Digestive Disease Research Center (DK058404). Additional support was provided by the Vanderbilt Kennedy Center, NIH/NICHD P30HD15052.
The authors declare the following competing financial interest(s):Author Disclosure Statement: M. Diana Neely, Aaron B. Bowman, Kevin C. Ess and Charles C. Hong indicate a potential conflict of interest for intellectual property rights of DMH1.
Author Contributions
M.D.N.: Conception and design, collection and assembly of data, data analysis and interpretation, manuscript writing, and final approval of manuscript. M.J.L.: Collection and assembly of data, data analysis and interpretation. A.A.A.: Collection and assembly of data, data analysis, and interpretation. A.M.T.: Collection and assembly of data, data analysis, and interpretation. G.G.L.: Collection and assembly of data, data analysis, and interpretation. C.R.H.: Provision of study material. R.C.: Collection and assembly of data, data analysis, and interpretation. C.C.H.: Conception and design, provision of study material, data analysis and interpretation, manuscript writing. K.C.E.: Conception and design, financial support, provision of study material, collection and assembly of data, data analysis and interpretation, manuscript writing. A.B.B.: Conception and design, financial support, administrative support, data analysis and interpretation, manuscript writing, and final approval of manuscript.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Takahashi K.; Tanabe K.; Ohnuki M.; Narita M.; Ichisaka T.; Tomoda K.; Yamanaka S. (2007) Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861–872. [DOI] [PubMed] [Google Scholar]
- Yu J.; Vodyanik M. A.; Smuga-Otto K.; Antosiewicz-Bourget J.; Frane J. L.; Tian S.; Nie J.; Jonsdottir G. A.; Ruotti V.; Stewart R.; Slukvin I. I.; Thomson J. A. (2007) Induced pluripotent stem cell lines derived from human somatic cells. Science 318, 1917–1920. [DOI] [PubMed] [Google Scholar]
- Aubry L.; Bugi A.; Lefort N.; Rousseau F.; Peschanski M.; Perrier A. L. (2008) Striatal progenitors derived from human ES cells mature into DARPP32 neurons in vitro and in quinolinic acid-lesioned rats. Proc. Natl. Acad. Sci. U.S.A. 105, 16707–16712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burridge P. W.; Thompson S.; Millrod M. A.; Weinberg S.; Yuan X.; Peters A.; Mahairaki V.; Koliatsos V. E.; Tung L.; Zambidis E. T. (2011) A universal system for highly efficient cardiac differentiation of human induced pluripotent stem cells that eliminates interline variability. PLoS One 6, e18293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers S. M.; Fasano C. A.; Papapetrou E. P.; Tomishima M.; Sadelain M.; Studer L. (2009) Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol. 27, 275–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper O.; Hargus G.; Deleidi M.; Blak A.; Osborn T.; Marlow E.; Lee K.; Levy A.; Perez-Torres E.; Yow A.; Isacson O. (2010) Differentiation of human ES and Parkinson’s disease iPS cells into ventral midbrain dopaminergic neurons requires a high activity form of SHH, FGF8a and specific regionalization by retinoic acid. Mol. Cell Neurosci. 45, 258–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias J.; Gumenyuk M.; Kang H.; Vodyanik M.; Yu J.; Thomson J.; Slukvin I. (2011) Generation of red blood cells from human induced pluripotent stem cells. Stem Cells Dev. 20, 1639–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert A. D.; Yu J.; Rose F. F. Jr.; Mattis V. B.; Lorson C. L.; Thomson J. A.; Svendsen C. N. (2009) Induced pluripotent stem cells from a spinal muscular atrophy patient. Nature 457, 277–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G.; Chambers S. M.; Tomishima M. J.; Studer L. (2010) Derivation of neural crest cells from human pluripotent stem cells. Nat. Protoc. 5, 688–701. [DOI] [PubMed] [Google Scholar]
- Vandekerckhove B.; Vanhee S.; Van Coppernolle S.; Snauwaert S.; Velghe I.; Taghon T.; Leclercq G.; Kerre T.; Plum J. (2011) In vitro generation of immune cells from pluripotent stem cells. Front. Biosci. 16, 1488–1504. [DOI] [PubMed] [Google Scholar]
- Zeng H.; Guo M.; Martins-Taylor K.; Wang X.; Zhang Z.; Park J. W.; Zhan S.; Kronenberg M. S.; Lichtler A.; Liu H. X.; Chen F. P.; Yue L.; Li X. J.; Xu R. H. (2010) Specification of region-specific neurons including forebrain glutamatergic neurons from human induced pluripotent stem cells. PLoS One 5, e11853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attisano L.; Wrana J. L. (2002) Signal transduction by the TGF-beta superfamily. Science 296, 1646–1647. [DOI] [PubMed] [Google Scholar]
- Massague J. (2000) How cells read TGF-beta signals. Nat. Rev. Mol. Cell. Biol. 1, 169–178. [DOI] [PubMed] [Google Scholar]
- Shi Y.; Massague J. (2003) Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell 113, 685–700. [DOI] [PubMed] [Google Scholar]
- James D.; Levine A. J.; Besser D.; Hemmati-Brivanlou A. (2005) TGFbeta/activin/nodal signaling is necessary for the maintenance of pluripotency in human embryonic stem cells. Development 132, 1273–1282. [DOI] [PubMed] [Google Scholar]
- Vallier L.; Alexander M.; Pedersen R. A. (2005) Activin/Nodal and FGF pathways cooperate to maintain pluripotency of human embryonic stem cells. J. Cell Sci. 118, 4495–4509. [DOI] [PubMed] [Google Scholar]
- Vallier L.; Touboul T.; Brown S.; Cho C.; Bilican B.; Alexander M.; Cedervall J.; Chandran S.; Ahrlund-Richter L.; Weber A.; Pedersen R. A. (2009) Signaling pathways controlling pluripotency and early cell fate decisions of human induced pluripotent stem cells. Stem Cells 27, 2655–2666. [DOI] [PubMed] [Google Scholar]
- Xu R. H.; Sampsell-Barron T. L.; Gu F.; Root S.; Peck R. M.; Pan G.; Yu J.; Antosiewicz-Bourget J.; Tian S.; Stewart R.; Thomson J. A. (2008) NANOG is a direct target of TGFbeta/activin-mediated SMAD signaling in human ESCs. Cell Stem Cell. 3, 196–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadwick K.; Wang L.; Li L.; Menendez P.; Murdoch B.; Rouleau A.; Bhatia M. (2003) Cytokines and BMP-4 promote hematopoietic differentiation of human embryonic stem cells. Blood 102, 906–915. [DOI] [PubMed] [Google Scholar]
- Xu R. H.; Chen X.; Li D. S.; Li R.; Addicks G. C.; Glennon C.; Zwaka T. P.; Thomson J. A. (2002) BMP4 initiates human embryonic stem cell differentiation to trophoblast. Nat. Biotechnol. 20, 1261–1264. [DOI] [PubMed] [Google Scholar]
- Zhang P.; Li J.; Tan Z.; Wang C.; Liu T.; Chen L.; Yong J.; Jiang W.; Sun X.; Du L.; Ding M.; Deng H. (2008) Short-term BMP-4 treatment initiates mesoderm induction in human embryonic stem cells. Blood 111, 1933–1941. [DOI] [PubMed] [Google Scholar]
- D’Amour K. A.; Agulnick A. D.; Eliazer S.; Kelly O. G.; Kroon E.; Baetge E. E. (2005) Efficient differentiation of human embryonic stem cells to definitive endoderm. Nat. Biotechnol. 23, 1534–1541. [DOI] [PubMed] [Google Scholar]
- Zhou J.; Su P.; Li D.; Tsang S.; Duan E.; Wang F. (2010) High-efficiency induction of neural conversion in human ESCs and human induced pluripotent stem cells with a single chemical inhibitor of transforming growth factor beta superfamily receptors. Stem Cells 28, 1741–1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inman G. J.; Nicolás F. J.; Callahan J. F.; Harling J. D.; Gaster L. M.; Reith A. D.; Laping N. J.; Hill C. S. (2002) SB-431542 is a potent and specific inhibitor of transforming growth factor-Œ≤ superfamily type I activin receptor-like kinase (ALK) receptors ALK4, ALK5, and ALK7. Mol. Pharmacol. 62, 65–74. [DOI] [PubMed] [Google Scholar]
- Hao J.; Ho J. N.; Lewis J. A.; Karim K. A.; Daniels R. N.; Gentry P. R.; Hopkins C. R.; Lindsley C. W.; Hong C. C. (2010) In vivo structure-activity relationship study of dorsomorphin analogues identifies selective VEGF and BMP inhibitors. ACS Chem. Biol. 5, 245–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B. Y.; Weick J. P.; Yu J.; Ma L. X.; Zhang X. Q.; Thomson J. A.; Zhang S. C. (2010) Neural differentiation of human induced pluripotent stem cells follows developmental principles but with variable potency. Proc. Natl. Acad. Sci. U.S.A. 107, 4335–4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D. S.; Lee J. S.; Leem J. W.; Huh Y. J.; Kim J. Y.; Kim H. S.; Park I. H.; Daley G. Q.; Hwang D. Y.; Kim D. W. (2010) Robust enhancement of neural differentiation from human ES and iPS cells regardless of their innate difference in differentiation propensity. Stem Cell Rev. 6, 270–281. [DOI] [PubMed] [Google Scholar]
- Nagaoka M.; Si-Tayeb K.; Akaike T.; Duncan S. A. (2010) Culture of human pluripotent stem cells using completely defined conditions on a recombinant E-cadherin substratum. BMC Dev. Biol. 10, 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan C. A.; Klimanskaya I.; McMahon J.; Atienza J.; Witmyer J.; Zucker J. P.; Wang S.; Morton C. C.; McMahon A. P.; Powers D.; Melton D. A. (2004) Derivation of embryonic stem-cell lines from human blastocysts. N. Engl. J. Med. 350, 1353–1356. [DOI] [PubMed] [Google Scholar]
- Inman G. J.; Nicolas F. J.; Callahan J. F.; Harling J. D.; Gaster L. M.; Reith A. D.; Laping N. J.; Hill C. S. (2002) SB-431542 is a potent and specific inhibitor of transforming growth factor-beta superfamily type I activin receptor-like kinase (ALK) receptors ALK4, ALK5, and ALK7. Mol. Pharmacol. 62, 65–74. [DOI] [PubMed] [Google Scholar]
- Cross E. E.; Thomason R. T.; Martinez M.; Hopkins C. R.; Hong C. C.; Bader D. M. (2011) Application of small organic molecules reveals cooperative TGFbeta and BMP regulation of mesothelial cell behaviors. ACS Chem. Biol. 6, 952–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman L. B.; De Jesus-Escobar J. M.; Harland R. M. (1996) The Spemann organizer signal noggin binds and inactivates bone morphogenetic protein 4. Cell 86, 599–606. [DOI] [PubMed] [Google Scholar]
- Dou C. L.; Li S.; Lai E. (1999) Dual role of brain factor-1 in regulating growth and patterning of the cerebral hemispheres. Cereb. Cortex 9, 543–550. [DOI] [PubMed] [Google Scholar]
- Manuel M. N.; Martynoga B.; Molinek M. D.; Quinn J. C.; Kroemmer C.; Mason J. O.; Price D. J. (2011) The transcription factor Foxg1 regulates telencephalic progenitor proliferation cell autonomously, in part by controlling Pax6 expression levels. Neural Dev. 6, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broccoli V.; Boncinelli E.; Wurst W. (1999) The caudal limit of Otx2 expression positions the isthmic organizer. Nature 401, 164–168. [DOI] [PubMed] [Google Scholar]
- Ono Y.; Nakatani T.; Sakamoto Y.; Mizuhara E.; Minaki Y.; Kumai M.; Hamaguchi A.; Nishimura M.; Inoue Y.; Hayashi H.; Takahashi J.; Imai T. (2007) Differences in neurogenic potential in floor plate cells along an anteroposterior location: midbrain dopaminergic neurons originate from mesencephalic floor plate cells. Development 134, 3213–3225. [DOI] [PubMed] [Google Scholar]
- Morizane A.; Doi D.; Kikuchi T.; Nishimura K.; Takahashi J. (2011) Small-molecule inhibitors of bone morphogenic protein and activin/nodal signals promote highly efficient neural induction from human pluripotent stem cells. J. Neurosci. Res. 89, 117–126. [DOI] [PubMed] [Google Scholar]
- Wada T.; Honda M.; Minami I.; Tooi N.; Amagai Y.; Nakatsuji N.; Aiba K. (2009) Highly efficient differentiation and enrichment of spinal motor neurons derived from human and monkey embryonic stem cells. PLoS One 4, e6722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriks S.; Shim J. W.; Piao J.; Ganat Y. M.; Wakeman D. R.; Xie Z.; Carrillo-Reid L.; Auyeung G.; Antonacci C.; Buch A.; Yang L.; Beal M. F.; Surmeier D. J.; Kordower J. H.; Tabar V.; Studer L. (2011) Dopamine neurons derived from human ES cells efficiently engraft in animal models of Parkinson’s disease. Nature 480, 547–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou G.; Myers R.; Li Y.; Chen Y.; Shen X.; Fenyk-Melody J.; Wu M.; Ventre J.; Doebber T.; Fujii N.; Musi N.; Hirshman M. F.; Goodyear L. J.; Moller D. E. (2001) Role of AMP-activated protein kinase in mechanism of metformin action. J. Clin. Invest. 108, 1167–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E. K.; Miller I.; Aja S.; Landree L. E.; Pinn M.; McFadden J.; Kuhajda F. P.; Moran T. H.; Ronnett G. V. (2004) C75, a fatty acid synthase inhibitor, reduces food intake via hypothalamic AMP-activated protein kinase. J. Biol. Chem. 279, 19970–19976. [DOI] [PubMed] [Google Scholar]
- Yu P. B.; Deng D. Y.; Lai C. S.; Hong C. C.; Cuny G. D.; Bouxsein M. L.; Hong D. W.; McManus P. M.; Katagiri T.; Sachidanandan C.; Kamiya N.; Fukuda T.; Mishina Y.; Peterson R. T.; Bloch K. D. (2008) BMP type I receptor inhibition reduces heterotopic [corrected] ossification. Nat. Med. 14, 1363–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt J.; Traynor R.; Sapkota G. P. (2011) The specificities of small molecule inhibitors of the TGFss and BMP pathways. Cell Signal. 23, 1831–1842. [DOI] [PubMed] [Google Scholar]
- Pevny L. H.; Sockanathan S.; Placzek M.; Lovell-Badge R. (1998) A role for SOX1 in neural determination. Development 125, 1967–1978. [DOI] [PubMed] [Google Scholar]
- Chung S.; Shin B. S.; Hedlund E.; Pruszak J.; Ferree A.; Kang U. J.; Isacson O.; Kim K. S. (2006) Genetic selection of sox1GFP-expressing neural precursors removes residual tumorigenic pluripotent stem cells and attenuates tumor formation after transplantation. J. Neurochem. 97, 1467–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang R. J.; Haynes J. M.; Kelly J.; Johnson J.; Greenhalgh J.; O’Brien C.; Mulholland E. M.; Baker L.; Munsie M.; Pouton C. W. (2004) Electrical and neurotransmitter activity of mature neurons derived from mouse embryonic stem cells by Sox-1 lineage selection and directed differentiation. Eur. J. Neurosci. 20, 3209–3221. [DOI] [PubMed] [Google Scholar]
- Fukuda H.; Takahashi J.; Watanabe K.; Hayashi H.; Morizane A.; Koyanagi M.; Sasai Y.; Hashimoto N. (2006) Fluorescence-activated cell sorting-based purification of embryonic stem cell-derived neural precursors averts tumor formation after transplantation. Stem Cells 24, 763–771. [DOI] [PubMed] [Google Scholar]
- Fasano C. A.; Chambers S. M.; Lee G.; Tomishima M. J.; Studer L. (2010) Efficient derivation of functional floor plate tissue from human embryonic stem cells. Cell Stem Cell. 6, 336–347. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Ohnuki M., Takahashi K., and Yamanaka S. (2009) Generation and characterization of human induced pluripotent stem cells. Curr. Protoc. Stem Cell. Biol. Chapter 4, Unit 4A 2. [DOI] [PubMed] [Google Scholar]
- Neely M. D., Tidball A. M., Aboud A. A., Ess K. C., and Bowman A. B. (2011) Induced Pluripotent Stem Cells (iPSCs): An Emerging Model System for the Study of Human Neurotoxicology, in Cell Culture Techniques (Aschner M., Sunol C., and Bal-Price A., Eds.) pp 27–62, Springer, New York. [Google Scholar]
- Lin T.; Ambasudhan R.; Yuan X.; Li W.; Hilcove S.; Abujarour R.; Lin X.; Hahm H. S.; Hao E.; Hayek A.; Ding S. (2009) A chemical platform for improved induction of human iPSCs. Nat. Methods 6, 805–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paya M.; Segovia J. C.; Santiago B.; Galindo M.; del Rio P.; Pablos J. L.; Ramirez J. C. (2006) Optimising stable retroviral transduction of primary human synovial fibroblasts. J. Virol. Methods 137, 95–102. [DOI] [PubMed] [Google Scholar]
- Reubinoff B. E.; Itsykson P.; Turetsky T.; Pera M. F.; Reinhartz E.; Itzik A.; Ben-Hur T. (2001) Neural progenitors from human embryonic stem cells. Nat. Biotechnol. 19, 1134–1140. [DOI] [PubMed] [Google Scholar]
- Kwakye G. F., Li D., Kabobel O. A., and Bowman A. B. (2011) Cellular fura-2 manganese extraction assay (CFMEA). Curr. Protoc. Toxicol., Chapter 12, Unit12 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrier A. L.; Tabar V.; Barberi T.; Rubio M. E.; Bruses J.; Topf N.; Harrison N. L.; Studer L. (2004) Derivation of midbrain dopamine neurons from human embryonic stem cells. Proc. Natl. Acad. Sci. U.S.A. 101, 12543–12548. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.