Abstract
The release of energy from particulate substrates such as dietary fiber and resistant starch (RS) in the human colon may depend on the presence of specialist primary degraders (or ‘keystone species') within the microbial community. We have explored the roles of four dominant amylolytic bacteria found in the human colon in the degradation and utilization of resistant starches. Eubacterium rectale and Bacteroides thetaiotaomicron showed limited ability to utilize RS2- and RS3-resistant starches by comparison with Bifidobacterium adolescentis and Ruminococcus bromii. In co-culture, however, R. bromii proved unique in stimulating RS2 and RS3 utilization by the other three bacterial species, even in a medium that does not permit growth of R. bromii itself. Having previously demonstrated low RS3 fermentation in vivo in two individuals with undetectable populations of R. bromii-related bacteria, we show here that supplementation of mixed fecal bacteria from one of these volunteers with R. bromii, but not with the other three species, greatly enhanced the extent of RS3 fermentation in vitro. This argues strongly that R. bromii has a pivotal role in fermentation of RS3 in the human large intestine, and that variation in the occurrence of this species and its close relatives may be a primary cause of variable energy recovery from this important component of the diet. This work also indicates that R. bromii possesses an exceptional ability to colonize and degrade starch particles when compared with previously studied amylolytic bacteria from the human colon.
Keywords: energy harvest, resistant starch, human colon, anaerobes, gut microbiota, primary degrader
Introduction
Non-digestible carbohydrates generally provide the main sources of energy for the microbial communities that inhabit the human large intestine. These include plant cell wall polysaccharides such as cellulose, xylan and pectin that comprise plant fiber, but on most diets the largest contribution is thought to come from the fraction of dietary starch that survives digestion in the upper gastrointestinal tract (resistant starch, RS) (Macfarlane and Englyst, 1986). The reasons for starch being resistant include physical inaccessibility (RS1), native granular structure (RS2) and retrogradation resulting from heating and cooling (RS3), and chemical modification (RS4) (Englyst et al., 1992). There is evidence that RS fermentation can confer health benefits, helping to reduce insulin resistance (Robertson et al., 2005), reverse infectious diarrhea (Ramakrishna et al., 2000; Niderman-Meyer et al., 2010), and prevent colorectal cancer (Young et al., 2005; Le Leu et al., 2009). Little is known about the factors that influence the fermentation of starches that reach the large intestine, although factors that result in resistance may also tend to reduce fermentability.
Although many amylolytic bacteria have been described from the human GI tract, our understanding of the microbial ecology of starch fermentation in the human large intestine is currently limited. Previous work has emphasized the numerical abundance of amylolytic Bacteroides spp. (Salyers et al., 1977) and the ability of certain Bifidobacterium spp. to degrade resistant starches (Macfarlane and Englyst, 1986; Ryan et al., 2006), and has produced information on the amylases of B. thetaiotaomicron (D'Elia and Salyers, 1996) and of Bifidobacterium spp. (Motherway et al., 2008). More recent work with mixed human fecal communities in vitro, however, revealed that 80% of 16S rRNA sequences recovered from resistant starch (RS) particles belonged to the Firmicutes species Ruminococcus bromii, and Eubacterium rectale, along with Bifidobacterium spp. (Leitch et al., 2007), and the same groups of bacteria were found to utilize 13C-labelled starch (Kovatcheva-Datchary et al., 2009). E. rectale and related Roseburia spp. are reported to possess amylolytic activity (Aminov et al., 2006; Ramsay et al., 2006), while R. bromii, although originally identified as a starch-degrading species (Moore et al., 1972; Herbeck and Bryant, 1974), has received no subsequent study. Recent human dietary intervention studies have reported increased populations of R. bromii and other Gram-positive bacteria in fecal samples during consumption of diets high in RS (Abell et al., 2008; Martínez et al., 2010; Walker et al., 2011). One study detected a mean increase of 10-fold in the proportion of fecal R. bromii in obese subjects consuming RS3, but also noted marked inter-individual variation; remarkably 2 out of the 14 individuals showed no detectable ruminococci in fecal samples, and less than 40% fermentation of RS compared with more than 95% in the other subjects (Walker et al., 2011). These findings suggest that R. bromii might have a key role in RS breakdown.
In the present study, we examine the abilities of four amylolytic bacteria that represent highly abundant groups within the human colon to degrade and utilize different starches. These include starches that differ in the content of amylose (straight chain α(1,4)-linked glucose) and amylopectin (branched α(1,4)-glucose chains connected via α(1,6)-linkages), in resistance to small intestinal degradation, and in heat pretreatment. Our investigations reveal that R. bromii possesses a superior degradative ability with respect to RS when compared with the other three species. They further suggest that R. bromii is a ‘keystone' species whose activities are required to release products from RS that can be utilized by other gut bacteria.
Materials and methods
Bacterial strains
All strains were of human fecal origin. Isolation of Eubacterium rectale A1-86T (DSM 17629) was reported previously (Barcenilla et al., 2000). Ruminococcus bromii L2-63 and Bifidobacterium adolescentis L2-32 were non-butyrate-producing isolates from the same study, from stool samples of a healthy male child. B. adolescentis L2-32 was found to be the most active of 10 starch-utilizing Bifidobacterium strains tested (Belenguer et al., 2006). Bacteroides thetaiotaomicron 5482 (ATCC 29148) was a gift from A Salyers, whereas R. bromii 27255 (Moore et al., 1972) and B. breve 20213 were from ATCC. All strains were routinely maintained in M2GSC medium containing 0.75% agar (Miyazaki et al., 1997). M2GSC medium contains 30% clarified bovine rumen fluid (Stewart et al., 1997).
Starch substrates
Hi-maize 240 and Hi-maize 958 are high-amylose RS2 starches made using a hydrothermal methodology. Hi-maize 240 is prepared from Hylon VII (Le Leu et al., 2009), whereas Hi-maize 958 is a natural, unmodified, food-grade RS2 containing 22% dietary fiber (analyzed by AOAC) and 61.8% RS (Ferguson et al., 2000; Le Leu et al., 2005). Novelose 330 is a retrograded RS3 generated from the hydrolyzed products of corn starch (Jacobasch et al., 2006; Le Leu et al., 2009). Sigma 4180 is a high amylose corn starch with only 30% of amylopectin, whereas S9679 is an unmodified waxy corn starch comprising almost 100% amylopectin. S5127 is an unmodified wheat starch that contains 75% amylopectin and S7260 a rice starch with 75–87% amylopectin. Raw starch solutions (2% wt per vol) were prepared using autoclaved dH20, some of which was then boiled for 10 min (boiled starch). Autoclaved starch refers to starch added to the growth medium to a final concentration of 0.2% before autoclaving at 120 °C for 15 min.
Growth media
Modified YCFA medium consists of (per 100 ml): casitone (0.25 g), yeast extract (0.1 g), NaHCO3 (0.4 g), cysteine (0.1 g), K2HPO4 (0.045 g), KH2PO4 (0.045 g) NaCl, (0.09 g) (NH4)2SO4 (0.09 g), MgSO4.7H2O (0.009 g), CaCl2 (0.009 g), resazurin (0.1 mg), haemin (1 mg), biotin (1 μg), cobalamin (1 μg), p-aminobenzoic acid (3 μg), folic acid (5 μg), pyridoxamine (15 μg). In addition, the following SCFA are included (final concentrations): acetate (33 mM), propionate (9 mM), iso-butyrate, iso-valerate and valerate (each 1 mM). Cysteine is added to the medium following boiling and dispensed into Hungate tubes, while the tubes are flushed with CO2. Filter sterilized solutions of thiamine and riboflavin were added to give final concentrations of 0.05 μg ml−1 of each after autoclaving the tubes. M2 (Miyazaki et al., 1997) medium was used to obtain optimal growth of R. bromii L2-63, which in common with R. bromii ATCC 27255 was unable to grow on YCFAS medium. Finally, 1 ml of 2% carbohydrate solutions was added to 9 ml of basal medium to give a final concentration of 0.2%. Cultures providing the inocula were pregrown overnight in M2GSC medium; all growth experiments were performed in triplicate.
Co-culture experiments were performed by inoculating pairwise combinations of the four strains, and single strains as controls, into YCFA medium that was supplemented with boiled RS2 (HM-958) or RS3 (Novelose 330). Co-culture of R. bromii and E. rectale was also performed in M2S medium.
Batch culture incubations involving fecal inocula
Fecal samples were collected from three healthy controls (v2, v6 and v7; one male and two female, consuming western diets) and one low starch-utilizing (v25, male) volunteer. 20% (wt per vol) of fecal slurries were prepared in sterile 1 × PBS solution within an hour of collection and 1 ml used to inoculate 8 ml fermentor medium (Macfarlane et al., 1989) (modified to contain 0.2% peptides) plus 1 ml of 2% boiled RS3 solution. Individual bacterial cultures (pregrown overnight in M2GSC medium) were adjusted to OD650=0.7 before adding 1 ml to the above mixture (adjusted to contain 7 ml fermentor medium). Samples were taken at intervals up to 48 h for total sugar and real-time PCR analysis. All experiments were performed in triplicate. Total sugars present in cultures were determined by the phenol sulphuric assay (Dubois et al., 1956) and reducing sugars by the method of Lever (1977).
Effects of fecal water upon bacterial growth
Fecal waters were separated from solid matter by centrifugation (50 000 g, 2 h, 10 °C) using Beckman Coulter Optima L-100XP ultracentrifuge. The cleared supernatant was sterilized by filtration through a 0.2-μm filter and included in bacterial growth media before autoclaving, as described in the text and in Supplementary Figure S5. Oxytetracycline concentrations were determined by Liquid Chromatography Mass Spectrometry (LCMS) on 100 μl fecal water mixed with 900 μl acetonitrile (containing 500 pg μl−1 of the internal standard demeclocycline hydrochloride).
Quantitative real-time PCR
The relative abundance of different bacterial groups was determined by quantitative real-time PCR. DNA extraction used a FastDNA SPIN for Soil Kit (MP Biomedical, Illkrich, France) following the manufacturer's instructions. Real-time PCR was performed on triplicate samples using iQ SYBR Green Supermix (Bio-Rad, Hercules, CA, USA) using the primers and conditions described by Walker et al., 2011. Results were analyzed using iQ5 Optical System Software version 2.0 (Bio-Rad).
Statistical analysis
Comparison of the ability of four bacterial strains to degrade different starches was tested by univariate analysis of variance, regarding degradation as a dependent variable and bacterial strain, starch treatment as fixed factors. Post hoc multiple comparisons were performed when the effect of fixed factors were significant. Tests of between- subjects effects were performed to find out factor interactions. One-way ANOVA was used to compare means within one factor when the other two factors were fixed. All statistical analyses were performed by using IBM SPSS Statistics 19. Significance was set at P-value of <0.05.
Results
Degradation of RS by human colonic bacteria in pure culture
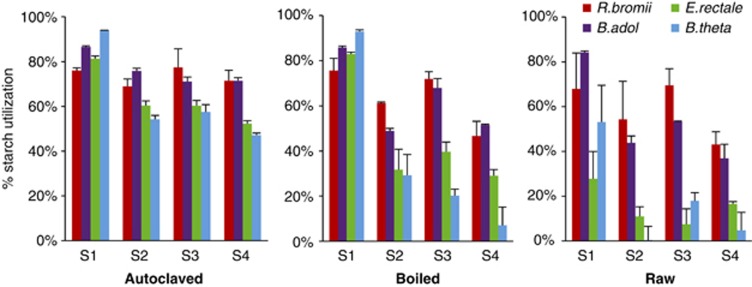
Four of the most abundant bacterial species found in the human large intestine (based on cultural (Moore and Moore, 1995) and molecular (Tap et al., 2009; Walker et al., 2011) analyses) were chosen as representatives of the major groups of amylolytic bacteria. These were Eubacterium rectale and Ruminococcus bromii (Firmicutes), Bacteroides thetaiotaomicron (Bacteroidetes) and Bifidobacterium adolescentis (Actinobacteria). Starch utilization was followed by measuring the total sugar that remained in bacterial cultures. Strains were inoculated into anaerobic YCFAS medium, except for R. bromii that grew optimally in M2S medium (containing 30% rumen fluid) (see Materials and methods). The four species showed varying abilities to utilize seven different starches, including two RS2 and one RS3 (Supplementary Table S1, Figure 1). In all cases, the highest utilization (47–94%) was observed for starches added to the growth medium before autoclaving. Autoclaving is a more extreme treatment than normal cooking, however, and therefore, we also examined starch that was pretreated by boiling for 10 min or left untreated (raw). Control tubes did not reveal any growth in the absence of a bacterial inoculum.
Figure 1.
Utilization of different starches by four species of amylolytic bacteria that are abundant in the human colon. The % decrease in total sugar in cultures is shown after 72 h incubation with Ruminococcus bromii L2-63 (R.bromii), Bifidobacterium adolescentis L2-32 (B.adol), Eubacterium rectale A1-86 (E.rectale) or Bacteroides thetaiotaomicron 5482 (B.theta). Strains were inoculated into YCFAS medium or M2S medium in the case of R.bromii that contained 0.2% starch. Starches shown are: high amylopectin corn starch (S9679 - S1); high amylose corn starch (S4180 - S2); and RS2- (HM-958 - S3) and RS3 (NL-330 - S4) -resistant starches. Results are shown for three different pretreatment regimes (autoclaved, boiled and raw). Data are given for three further starches in Supplementary Table S1.
For the three most degradable starches (S9629, S5127, S7260) B. adolescentis L2-32 and R. bromii L2-63 showed similar utilization regardless of the pretreatment. In contrast, E. rectale A1-86 and B. thetaiotaomicron 5482 showed much lower utilization of raw starches. For the 70% amylose corn starch (S4180) and the three RS, R. bromii L2-63 and B adolescentis L2-32 utilized 44–72% of sugar from boiled starch and 26–70% from raw starch. In contrast, utilization by E. rectale A1-86 and B. thetaiotaomicron 5482 for these four starches was only 7–40% for boiled and 0.25–18% for raw starch. B. breve 20213 was found to have similar activities to B. adolescentis L2-32 (Supplementary Table S1). Subsequent experiments employed boiled RS3 and RS2 (HM958) as the test substrates.
Utilization of starch-derived sugars
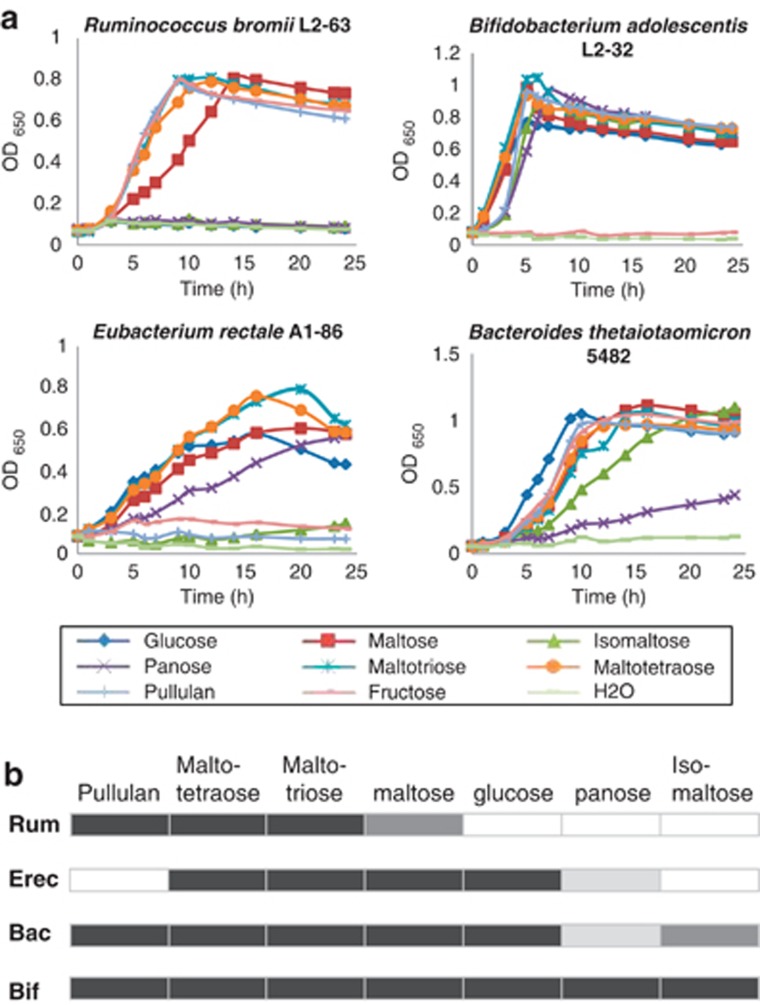
The four species were also compared for their ability to utilize potential breakdown products of starch. R. bromii L2-63 was unable to grow with 0.2% glucose as energy source, although able to utilize fructose as reported previously for other R. bromii strains (Herbeck and Bryant, 1974). Growth of R. bromii L2-63 in M2 medium with maltotriose or maltotetraose occurred at a rate similar to that on fructose, but growth on maltose was slower (Figure 2). R. bromii L2-63 also grew rapidly with added pullulan (DT=1.55 h), which is a repeating polymer consisting of two α(1,4)-linked and one α(1,6)-linked glucose residues, but did not grow with either isomaltose (two α(1,6)-linked glucose residues) or panose (a trisaccharide consisting of α(1,4)- and α(1,6)-linked glucose residues) as the sole added sugar. A second R. bromii strain, ATCC27255, showed a pattern of growth very similar to that of R. bromii L2-63, although growing slightly faster on pullulan (Supplementary Figure S1); this strain also gave a similar extent of total sugar utilization from boiled RS3 (50.2% for ATCC27255 compared with 48.5% for L2-63, by 96 h). By comparison, B. adolescentis L2-32 and B. thetaiotaomicron 5482 utilized pullulan and isomaltose, and B. adolescentis L2-32 grew well with panose, whereas E. rectale A1-86 did not utilize pullulan or isomaltose (Figure 2).
Figure 2.
Growth of human colonic amylolytic bacteria on starch-derived sugars. (a) Growth (monitored by OD650) is shown for R. bromii in M2 medium containing 0.2% of the carbohydrate indicated. Fructose was included as a reference substrate as R. bromii was unable to utilize glucose. Growth of B. adolescentis L2-32, E. rectale A1-86 and B. thetaiotaomicron 5482 was on YCFA medium containing 0.2% carbohydrates. (b) Summary of the abilities of B. adolescentis (Bif), B. thetaiotaomicron (Bac), E. rectale (Erec) and R. bromii (Rum) to utilize different sugars. The darkest shading indicates the substrate supporting the most rapid growth rate for each strain, whereas lighter shadings indicate less rapid growth (see also Supplementary Figure S1). Blanks indicate no growth.
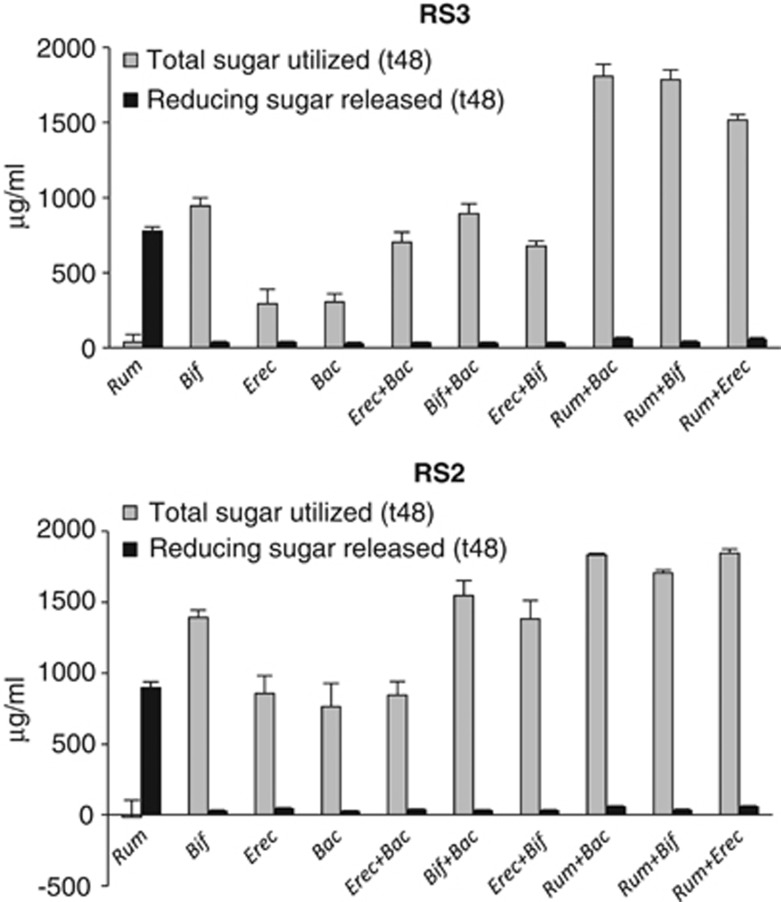
Interactions in co-culture
Interactions between the four amylolytic strains R. bromii L2-63, E. rectale A1-86, B. adolescentis L2-32 and B. thetaioaomicron 5482 were investigated by setting up pairwise combinations in YCFAS medium containing boiled RS2 or RS3 (Figure 3). Co-incubation of R. bromii with B. thetaiotaomicron, B. adolescentis or E. rectale resulted in a significant increase in utilization of RS2, and especially RS3, compared with monocultures of each strain. The extent of starch utilization was lower in co-cultures between B. adolescentis, E. rectale and B. thetaiotomicron. This result is remarkable as R. bromii was unable to grow on YCFAS medium and showed no utilization of starch in monoculture. On the other hand, reducing sugar accumulated during incubation, indicating extensive cleavage of the starch by R. bromii amylases (Figure 3, Supplementary Figure S2). Little reducing sugar was detected in co-cultures, where the other species were evidently able to grow on the products released by the R. bromii enzymes. TLC analysis showed that glucose and maltose were the major products of RS2 and RS3 breakdown by R. bromii incubated in YCFAS medium (Supplementary Figure S3).
Figure 3.
Stimulation of starch utilization in co-cultures containing R. bromii. Total sugar utilized and the concentration of free soluble reducing sugar are shown after 48 h incubation in YCFA medium containing RS2 (High-maize 958) or RS3 starch. Monocultures and co-cultures of the four bacterial strains are indicated as follows: Rum, R. bromii; Bif, B. adolescentis; Erec, E. rectale and Bac, B. thetaiotaomicron. Initial (t0) total sugar concentrations were 2039±39 and 2043±81 μg ml−1 and reducing sugar concentrations were 91±39 and 63±25 μg ml−1 for RS3 and RS2, respectively.
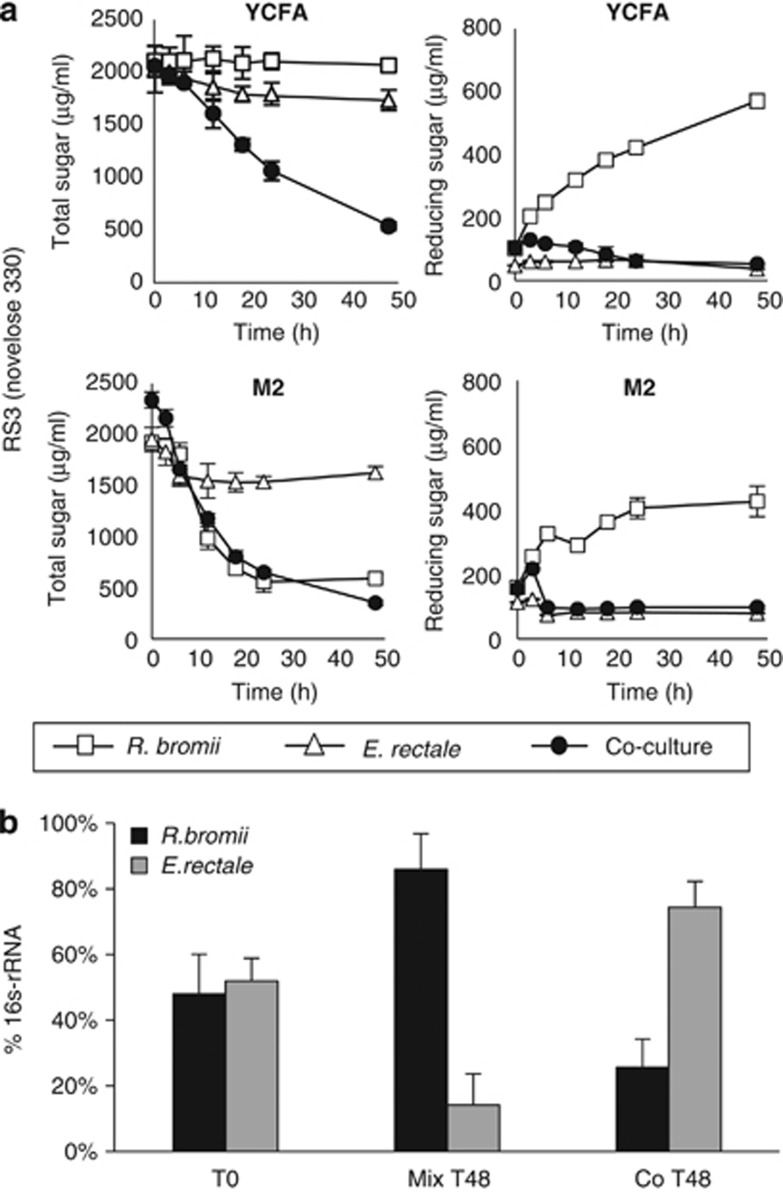
Time-courses were followed for R. bromii L2-63 and E. rectale A1-86 inoculated together or separately into YCFAS and M2S medium containing boiled RS3 (Figure 4a) or RS2 (Supplementary Figure S2). Although R. bromii growth occurred in the M2S medium, utilization of released reducing sugar was again incomplete in the monoculture in part due to the inability to utilize glucose. The relative abundance of R. bromii and E. rectale in monocultures and co-culture in M2S medium was estimated by 16S rRNA-based qPCR (Figure 4b). This confirmed that the presence of R. bromii stimulated the growth of E. rectale on this medium.
Figure 4.
Interaction between R. bromii and E. rectale in co-culture. (a) Time course of total sugar utilization and reducing sugar release on YCFAS and M2S medium containing 0.2% boiled RS3 by monocultures and co-cultures of R. bromii L2-63 and E. rectale A1-86. (b) Comparison by qPCR, using primer pairs specific to each bacterium, of the relative growth of the two bacteria for the experiment shown in (a) in M2S medium. The signal obtained with each specific primer pair is expressed relative to the signal obtained with the ‘universal' primer pair. Equal volumes of the two monocultures were mixed immediately after inoculation (t0) or after 48 h growth (t48) confirming the greater growth of R. bromii relative to E. rectale on this substrate. The same analysis on the co-culture, however, showed that E. rectale had outgrown R. bromii after 48 h.
Degradation of RS by mixed fecal bacteria
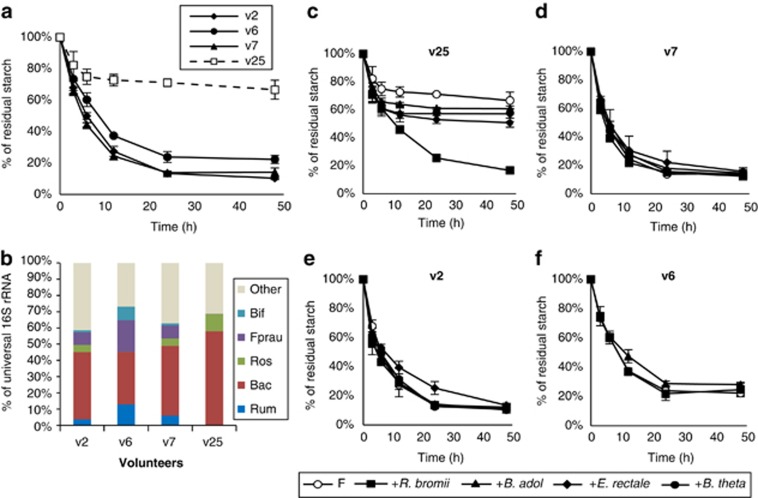
The rate of breakdown of boiled RS3 was similar in in vitro incubations performed with fecal samples provided by three healthy donors (Figure 5a). A previous human intervention study reported that fermentation of RS in vivo was incomplete for 2 out of 14 obese men examined (<40% fermentation compared with >96% for the remaining 12 people) (Walker et al., 2011). Fecal samples from these two volunteers showed undetectable levels of bacteria related to R. bromii (Walker et al., 2011). A fecal sample provided by one of these two volunteers (v25) 36 months after the original study gave a considerably lower extent of RS fermentation in vitro by comparison with the three controls (Figure 5a). R. bromii relatives, as well as bifidobacteria, were again undetectable by qPCR in the v25 fecal inoculum but were present in the three control samples (Figure 5b). In the same experiment, replicate incubations were performed after the addition of each of the four amylolytic strains considered above. Remarkably, only R. bromii addition restored RS fermentation in incubations with the v25 fecal sample to the extent seen in the controls, whereas the other three bacteria had limited impact on the extent of RS utilization (Figure 5c). Addition of any one of the four strains had no effect on fermentation in fecal samples from the normal controls (Figures 5d–f).
Figure 5.
Fermentation of RS3 in vitro by mixed human fecal bacteria. (a) Utilization of RS3 is shown for fecal samples from three healthy control volunteers (v2, v6 and v7) and from one volunteer (v25) previously found to show low starch fermentation in vivo. (b) Representation of different bacterial groups estimated by 16S-rRNA-targeted qPCR in the fecal samples employed in (a). (Bif, bifidobacteria, Fprau, F. prausnitzii; Rum, cluster IV ruminococci; Bac, Bacteroidetes; Ros, relatives of E. rectale/Roseburia). Abundance for each group is expressed as a percentage of the 16S-rRNA signal obtained using universal primers. (c–f) Effect on RS3 utilization of adding each of the four isolated amylolytic bacteria to fecal inocula (F) from v25, v7, v2 and v6.
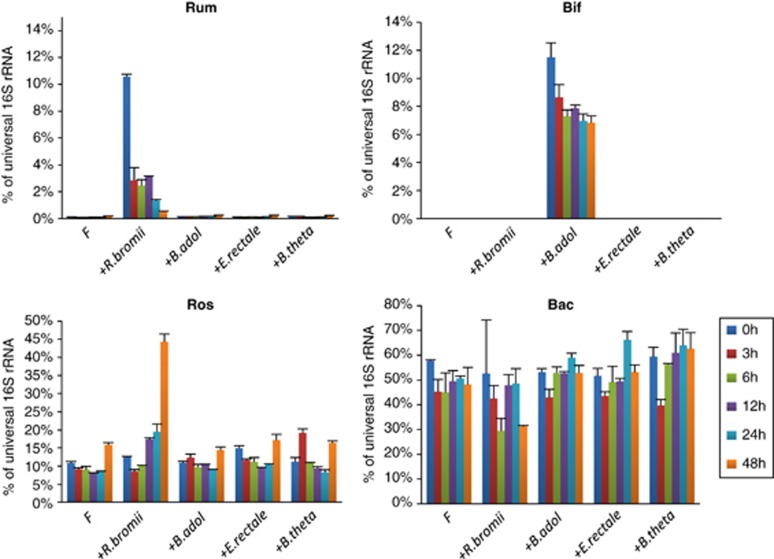
Quantitative PCR revealed that the inoculation of v25 fecal sample incubations with R. bromii boosted the initial proportion of this species among total bacteria (Figure 6). This proportion subsequently declined, although other groups, such as E. rectale, increased with time. Thus, the stimulation of RS fermentation in the mixed fecal community was apparently not dependent on growth of the R. bromii strain. This is consistent with the findings for defined co-cultures (Figure 3), again suggesting that R. bromii possesses particularly potent enzyme systems that can make the substrate available to other groups of bacteria. Results for a control sample (v7) are shown in Supplementary Figure S4.
Figure 6.
Effect of bacterial inoculation on relative abundance in incubations with fecal samples from v25. Changes in four groups of bacteria (Rum, cluster IV ruminococci, Bif, bifidobacteria, Ros, E. rectale and Roseburia spp., Bac, Bacteroidetes) were monitored by 16S rRNA-targeted qPCR for the experiment shown in Figure 5c. Abundance for each group is expressed as a percentage of the 16S rRNA signal obtained using universal primers. The impact of inoculation is easily seen for B. adolescentis and for R. bromii, as these groups were almost undetectable in the initial fecal samples, but is less evident for relatives of E. rectale and B. thetaiotaomicron because of the large populations in the fecal sample. Interestingly, a major increase in E. rectale-related bacteria was evident following inoculation with R. bromii. Data for one control sample (v7) are shown for comparison in Supplementary Figure S5.
Substitution of rumen fluid by fecal water from control volunteer samples (v2, v6 and v7) in the M2 growth medium (before autoclaving) resulted in growth of R. bromii L2-63 at variable rates dependent on the sample (Supplementary Figure S5). Fecal water from v25, however, resulted in no detectable growth. This volunteer reported taking oxytetracycline (three doses of 250 mg per day) during the period of sample donation, and oxytetracycline was detected at concentrations above 20 μg ml−1 in fecal samples. Growth of R. bromii was completely inhibited by oxytetracycline concentrations as low as 0.2 μg ml−1 (prepared by filter sterilizing), but the addition of oxytetracycline to a concentration of 20 μg ml−1 before autoclaving was also strongly inhibitory, giving an OD650<0.1 after 24 h growth of R. bromii in 0.2% fructose (M2F medium). This is consistent with recent evidence that oxytetracycline is partially heat stable (Hsieh et al., 2011). Although all four of the amylolytic strains used in the experiment shown in Figure 5 were tetracycline sensitive, the bacterial community present in fecal samples from v25 must be assumed to be resistant to tetracycline (Kazimierczak et al., 2008).
Discussion
Bacteria that belong to clostridial cluster IV (Ruminococccaeae) are numerically abundant in the human large intestine, typically accounting for 10–40% of total bacterial 16S rRNA sequences, but are under-represented by cultured isolates (Lay et al., 2005). There are strong indications, however, that this family has a primary role in the degradation of particulate substrates, as it includes cellulolytic species, such as R. flavefaciens and R. albus, described from the rumen (Stewart et al., 1997; Flint et al., 2008), and R. champanellensis isolated from the human large intestine (Chassard et al., 2011). The breakdown of plant fiber in the rumen is dependent on such cellulolytic microorganisms, with many other species utilizing the solubilized products (Dehority, 1991). In the human colon, cluster IV ruminococci were the only group of 16S rRNA sequences found to be significantly enriched on fiber particles, accounting for 12.2% of fiber-attached sequences, but only 3% of sequences in the liquid fraction of fecal samples (Walker et al., 2008). The evidence presented here establishes a primary role for this group in the degradation of dietary-RS, which generally provides the largest source of energy for microbial growth in the human colon.
The relative activities of the four amylolytic species studied (B. thetaiotaomicron, B. adolescentis, R. bromii and E. rectale) varied with the type of starch and its pretreatment, with R. bromii and B. adolescentis the most active on raw or boiled resistant starches. Inoculation of R. bromii L2-63 into unmodified YCFAS medium did not allow growth but resulted in extensive release of reducing sugar from boiled RS3 and RS2. Furthermore, R. bromii greatly enhanced the overall utilization of RS in the presence of the three other amylolytic species on YCFAS medium, whereas other combinations of these species without R. bromii gave lower utilization. These experiments show that R. bromii had the greatest capacity to degrade resistant starches, and especially RS3, among the four species examined, and was able to release products that could be readily utilized by the other three species. Our results show that R. bromii has a preference for α(1-4)-linked oligosaccharides larger than maltose, as it grew faster in rumen fluid medium on maltotriose or maltotetraose than on maltose, and was unable to utilize glucose. Although it grew well on pullulan, it was unable to utilize panose and isomaltose (oligosaccharides that, like pullulan, contain α(1-6) linkages). These characteristics create potential for nutritional cross feeding as many other species (including B. adolescentis and B. thetaiotaomicron) are able to utilize glucose, maltose, panose and isomaltose for growth. In the case of resistant starches, however, the main basis for the observed synergy is likely to be the ability of R. bromii to initiate degradation of insoluble RS particles.
The starch-utilization system of B. thetaiotaomicron has been studied extensively and is thought to represent a sequestration system that is particularly suited to the utilization of soluble starch molecules (Flint et al., 2008). In isolation, B. thetaiotaomicron gave the lowest extent of utilization of the resistant starches studied here. Among Gram-positive bacteria, B. breve and relatives of E. rectale have been shown to produce large extracellular amylases that include multiple catalytic and substrate-binding domains (Ramsay et al., 2006; Motherway et al., 2008). No research has been carried out so far on the amylases of R. bromii, but it seems clear that this system must display key differences when compared with those of the other three amylolytic species examined here. The genome of R. bromii L2-63 encodes at least 15 GH13 amylases, compared with 13, 7 and 13 in E. rectale, B. thetaiotaomicron and B. adolescentis, respectively (CAZY website: http://www.cazy.org/fam/acc_GH.html). The key differences may, however, lie in the organization of these enzymes and in the ability of this species to adhere closely to the insoluble substrate (Flint et al., 2008), both of which are under investigation.
Although we do not exclude the possibility that non-cultured organisms might have important roles in RS degradation, the groups studied here accounted for most of the 16S rRNA sequences associated with starch utilization in recent in vitro studies on mixed human fecal bacteria (Leitch et al., 2007; Kovatcheva-Datchary et al., 2009). Furthermore, we were able to use the microbiota from a low-RS3 fermenting volunteer with low numbers of cluster IV ruminococci (Walker et al., 2011) as a test system. While growth of the four added amylolytic bacteria is likely to have been curtailed by residual oxytetracycline present in autoclaved v25 fecal fluid, this experiment nevertheless demonstrated that addition of R. bromii alone restored the extent of RS3 fermentation to that seen for ‘normal' control microbiota. No tetracyclines were detected in fecal samples from a second volunteer (v14) who also showed low ruminococcal populations together with reduced starch fermentation in vivo (Walker et al., 2011). The possible causes of inter-individual variation in ruminococcal populations, which include both immediate and long-term effects of antibiotic therapy, however, remain to be established by future studies. Given that bacteria related to R. bromii appear to have a key role in the degradation of dietary RS, it seems likely that low numbers of cluster IV ruminococci will prove to be linked more generally to low rates of fermentation of RS in the human colon. More extensive investigations with larger cohorts of human volunteers will be required for a critical examination of this hypothesis.
Acknowledgments
Xiaolei Ze received support from the Scottish Overseas research scholarship, University of Aberdeen. We acknowledge the Scottish Government RESAS for their support and wish to thank Richard Le Leu for kindly providing Hi-maize 240 and 958 and Grietje Holtrop for statistical advice.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Abell GCJ, Cooke CM, Bennett CN, Conlon MA, McOrist AL. Phylotypes related to Ruminococcus bromii are abundant in the large bowel of humans and increase in response to a diet high in resistant starch. FEMS Microbiol Ecol. 2008;66:505–515. doi: 10.1111/j.1574-6941.2008.00527.x. [DOI] [PubMed] [Google Scholar]
- Aminov RI, Walker AW, Duncan SH, Harmsen HJM, Welling GW, Flint HJ. Molecular diversity, cultivation, and improved detection by fluorescent in situ hybridization of a dominant group of human gut bacteria related to Roseburia spp. or Eubacterium rectale. Appl Environ Microbiol. 2006;72:6371–6376. doi: 10.1128/AEM.00701-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barcenilla A, Pryde SE, Martin J, Duncan SH, Stewart CS, Henderson C, et al. Phylogenetic relationships of butyrate-producing bacteria from the human gut. Appl Environ Microbiol. 2000;66:1654–1661. doi: 10.1128/aem.66.4.1654-1661.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belenguer A, Duncan SH, Calder G, Holtrop G, Louis P, Lobley GE, et al. Two routes of metabolic cross-feeding between Bifidobacterium adolescentis and butyrate-producing anaerobes from the human gut. Appl Environ Microbiol. 2006;72:3593–3599. doi: 10.1128/AEM.72.5.3593-3599.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chassard C, Delmas E, Robert C, Lawson PA, Bernalier-Donadille A. Ruminococcus champanellensis sp.nov., a cellulose-degrading bacterium from the human gut microbiota. Int J Syst Evol Microbiol. 2011;62 (Pt 1:138–143. doi: 10.1099/ijs.0.027375-0. [DOI] [PubMed] [Google Scholar]
- Dehority BA. Effects of microbial synergism on fibre digestion in the rumen. Proc Nutr Soc. 1991;50:149–159. doi: 10.1079/pns19910026. [DOI] [PubMed] [Google Scholar]
- D'Elia JN, Salyers AA. Contribution of a neopullulanase, a pullulanase, and an α-glucosidase to growth of Bacteroides thetaiotaomicron on starch. J Bacteriol. 1996;178:7173–7179. doi: 10.1128/jb.178.24.7173-7179.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois M, Gilles KA, Hamilton JK, Rebers PA, Smith F. Colorimetric method for determination of sugars and related substances. Anal Chem. 1956;28:350–356. [Google Scholar]
- Englyst HN, Kingman SM, Cummings JH. Classification and measurement of nutritionally important starch fractions. Eur J Clin Nutr. 1992;46:S33–S50. [PubMed] [Google Scholar]
- Ferguson LR, Tasman-Jones C, Englyst H, Harris PJ. Comparative effects of three resistant starch preparations on transit time and short-chain fatty acid production in rats. Nutr Cancer. 2000;36:230–237. doi: 10.1207/S15327914NC3602_13. [DOI] [PubMed] [Google Scholar]
- Flint HJ, Bayer EA, Rincon MT, Lamed R, White BA. Polysaccharide utilization by gut bacteria: potential for new insights from genomic analysis. Nature Rev Microbiol. 2008;6:121–131. doi: 10.1038/nrmicro1817. [DOI] [PubMed] [Google Scholar]
- Herbeck JL, Bryant MP. Nutritional features of the intestinal anaerobe Ruminococcus bromii. J Appl Microbiol. 1974;28:1018–1022. doi: 10.1128/am.28.6.1018-1022.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh MK, Shyu CL, Liao JW, Franje CA, Huang YJ, Chang SK, et al. Correlation analysis of heat stability of veterinary antibiotics by structural degradation, changes in antimicrobial activity and genotoxicity. Veterinarni Medicina. 2011;56:274–285. [Google Scholar]
- Jacobasch G, Dongowski G, Schmiedl D, Müller-Schmehl K. Hydrothermal treatment of novelose 330 results in high yield of resistant starch type 3 with beneficial prebiotic properties and decreased secondary bile acid formation in rats. Br J Nutr. 2006;95:1063–1074. doi: 10.1079/bjn20061713. [DOI] [PubMed] [Google Scholar]
- Kazimierczak KA, Rincon MT, Patterson AJ, Martin JC, Young P, Flint HJ, et al. A new tetracycline efflux gene, tet(40), is located in tandem with tet(O/32/O) in a human gut Firmicute bacterium and in metagenomic clone libraries. Antimicrob Agents Chemother. 2008;52:4001–4009. doi: 10.1128/AAC.00308-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovatcheva-Datchary P, Egert M, Maathuis A, Rajilic-Stojanovic M, de Graaf AA, Smidt H, et al. Linking phylogenetic identities of bacteria to starch fermentation in an in vitro model of the large intestine by RNA-based stable isotope probing. Environ Microbiol. 2009;11:914–926. doi: 10.1111/j.1462-2920.2008.01815.x. [DOI] [PubMed] [Google Scholar]
- Lay C, Sutren M, Violaine Rochet V, Saunier K, Doré J, Rigottier-Gois L, et al. Design and validation of 16S rRNA probes to enumerate members of the Clostridium leptum subgroup in human faecal microbiota. Environ Microbiol. 2005;7:933–946. doi: 10.1111/j.1462-2920.2005.00763.x. [DOI] [PubMed] [Google Scholar]
- Leitch ECM, Walker AW, Duncan SH, Holtrop G, Flint HJ. Selective colonization of insoluble substrates by human faecal bacteria. Environ Microbiol. 2007;9:667–679. doi: 10.1111/j.1462-2920.2006.01186.x. [DOI] [PubMed] [Google Scholar]
- Le Leu RK, Brown IL, Hu Y, Bird AR, Jackson M, Esterman A, et al. A synbiotic combination of resistant starch and Bifidobacterium lactis facilitates apoptotic deletion of carcinogen-damaged dells in rat colon. J Nutr. 2005;135:996–1001. doi: 10.1093/jn/135.5.996. [DOI] [PubMed] [Google Scholar]
- Le Leu RK, Hu Y, Brown IL, Young GP.2009Effect of high amylose maize starches on colonic fermentation and apoptotic response to DNA-damage in the colon of rats Nutr Metab 611doi: 10.1186/1743-7075-6-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lever M. Carbohydrate determination with 4 hydroxybenzoic acid hydrazide (PAHBAH): effect of bismuth on the reaction. Anal Biochem. 1977;81:21–27. doi: 10.1016/0003-2697(77)90594-2. [DOI] [PubMed] [Google Scholar]
- Macfarlane GT, Englyst HN. Starch utilization by the human large intestinal microflora. J Appl Bacteriol. 1986;60:195–201. doi: 10.1111/j.1365-2672.1986.tb01073.x. [DOI] [PubMed] [Google Scholar]
- MacFarlane GT, Hay S, Gibson GR. Influence of mucin on glycosidase, protease and arylamidase activities of human gut bacteria grown in a 3-stage continuous culture system. J Appl Bacteriol. 1989;66:407–417. doi: 10.1111/j.1365-2672.1989.tb05110.x. [DOI] [PubMed] [Google Scholar]
- Martínez I, Kim J, Duffy PR, Schlegel VL, Walter J. Resistant starches types 2 and 4 have differential effects on the composition of the fecal microbiota in human subjects. PLoS ONE. 2010;5:e15046. doi: 10.1371/journal.pone.0015046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki K, Martin JC, Marinsek-Logar R, Flint HJ. Degradation and utilization of xylans by the rumen anaerobe Prevotella bryantii (formerly P. ruminicola subsp. brevis) B14. Anaerobe. 1997;3:373–381. doi: 10.1006/anae.1997.0125. [DOI] [PubMed] [Google Scholar]
- Moore WEC, Cato EP, Holdeman LV. Ruminococcus bromii sp. n and emendation of the description of Ruminococcus Sijpestein. Int J Syst Bacteriol. 1972;22:78–80. [Google Scholar]
- Moore WEC, Moore LH. Intestinal floras of populations that have a high risk of colon cancer. Appl Environ Microbiol. 1995;61:3202–3207. doi: 10.1128/aem.61.9.3202-3207.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motherway MO, Fitzgerald GF, Neirynck S, Ryan S, Steidler L, van Sinderen D, et al. Characterization of ApuB, an extracellular type II amylopullulanase from Bifidobacterium breve UCC2003. Appl Environ Microbiol. 2008;74:6271–6279. doi: 10.1128/AEM.01169-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niderman-Meyer O, Zeidman T, Shimoni E, Kashi Y. Mechanisms involved in governing adherence of Vibrio cholerae to granular starch. Appl Environ Microbiol. 2010;76:1034–1043. doi: 10.1128/AEM.01533-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishna BS, Venkataraman S, Srinivasan P, Dash P, Young GP, Binder HJ, et al. Amylase-resistant starch plus oral rehydration solution for cholera. N Engl J Med. 2000;342:308–313. doi: 10.1056/NEJM200002033420502. [DOI] [PubMed] [Google Scholar]
- Ramsay AG, Scott KP, Martin JC, Rincon MT, Flint HJ. Cell-associated α-amylases of butyrate-producing firmicute bacteria from the human colon. Microbiology. 2006;152:3281–3290. doi: 10.1099/mic.0.29233-0. [DOI] [PubMed] [Google Scholar]
- Robertson MD, Bickerton AS, Dennis AL, Vidal H, Frayn KN. Insulin-sensitizing effects of dietary resistant starch and effects on skeletal muscle and adipose tissue metabolism. Am J Clin Nutr. 2005;82:559–567. doi: 10.1093/ajcn.82.3.559. [DOI] [PubMed] [Google Scholar]
- Ryan SM, Fitzgerald GF, Van Sinderen D. Screening for and identification of starch-, amylopectin-, and pullulan-degrading activities in Bifidobacterial strains. Appl Environ Microbiol. 2006;72:5289–5296. doi: 10.1128/AEM.00257-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salyers AA, West SEH, Vercellotti JR, Wilkins TD. Fermentation of mucins and plant polysaccharides by anaerobic bacteria from the human colon. Appl Environ Microbiol. 1977;34:529–533. doi: 10.1128/aem.34.5.529-533.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart CS, Flint HJ, Bryant MP.1997The rumen bacteriaIn: Hobson PN, Stewart CS (eds).The Rumen Microbial Ecosystem2nd edn.Blackie Academic & Professional: London, UK; 10–72. [Google Scholar]
- Tap J, Mondot S, Levenez F, Pelletier E, Caron C, Furet JP, et al. Towards the human intestinal microbiota phylogenetic core. Environ Microbiol. 2009;11:2574–2584. doi: 10.1111/j.1462-2920.2009.01982.x. [DOI] [PubMed] [Google Scholar]
- Walker AW, Duncan SH, Harmsen HJM, Holtrop G, Welling GW, Flint HJ. The species composition of the human intestinal microbiota differs between particle-associated and liquid phase communities. Environ Microbiol. 2008;10:3275–3283. doi: 10.1111/j.1462-2920.2008.01717.x. [DOI] [PubMed] [Google Scholar]
- Walker AW, Ince J, Duncan SH, Webster LM, Holtrop G, Ze X, et al. Dominant and diet-responsive groups of bacteria within the human colonic microbiota. ISMEJ. 2011;5:220–230. doi: 10.1038/ismej.2010.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young GP, Hu Y, Le Leu RK, Nyskohus L. Dietary fibre and colorectal cancer: a model for environment - gene interactions. Mol Nutr Food Res. 2005;49:571–584. doi: 10.1002/mnfr.200500026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.