Abstract
Buildings are complex ecosystems that house trillions of microorganisms interacting with each other, with humans and with their environment. Understanding the ecological and evolutionary processes that determine the diversity and composition of the built environment microbiome—the community of microorganisms that live indoors—is important for understanding the relationship between building design, biodiversity and human health. In this study, we used high-throughput sequencing of the bacterial 16S rRNA gene to quantify relationships between building attributes and airborne bacterial communities at a health-care facility. We quantified airborne bacterial community structure and environmental conditions in patient rooms exposed to mechanical or window ventilation and in outdoor air. The phylogenetic diversity of airborne bacterial communities was lower indoors than outdoors, and mechanically ventilated rooms contained less diverse microbial communities than did window-ventilated rooms. Bacterial communities in indoor environments contained many taxa that are absent or rare outdoors, including taxa closely related to potential human pathogens. Building attributes, specifically the source of ventilation air, airflow rates, relative humidity and temperature, were correlated with the diversity and composition of indoor bacterial communities. The relative abundance of bacteria closely related to human pathogens was higher indoors than outdoors, and higher in rooms with lower airflow rates and lower relative humidity. The observed relationship between building design and airborne bacterial diversity suggests that we can manage indoor environments, altering through building design and operation the community of microbial species that potentially colonize the human microbiome during our time indoors.
Keywords: aeromicrobiology, bacteria, built environment microbiome, community ecology, dispersal, environmental filtering
Introduction
Humans spend up to 90% of their lives indoors (Klepeis et al., 2001). Consequently, the way we design and operate the indoor environment has a profound impact on our health (Guenther and Vittori, 2008). One step toward better understanding of how building design impacts human health is to study buildings as ecosystems. Built environments are complex ecosystems that contain numerous organisms including trillions of microorganisms (Rintala et al., 2008; Tringe et al., 2008; Amend et al., 2010). The collection of microbial life that exists indoors—the built environment microbiome—includes human pathogens and commensals interacting with each other and with their environment (Eames et al., 2009). There have been few attempts to comprehensively survey the built environment microbiome (Rintala et al., 2008; Tringe et al., 2008; Amend et al., 2010), with most studies focused on measures of total bioaerosol concentrations or the abundance of culturable or pathogenic strains (Berglund et al., 1992; Toivola et al., 2002; Mentese et al., 2009), rather than a more comprehensive measure of microbial diversity in indoor spaces. For this reason, the factors that determine the diversity and composition of the built environment microbiome are poorly understood. However, the situation is changing. The development of culture-independent, high-throughput molecular sequencing approaches has transformed the study of microbial diversity in a variety of environments, as demonstrated by the recent explosion of research on the microbial ecology of aquatic and terrestrial ecosystems (Nemergut et al., 2011) and the human microbiome (Turnbaugh et al., 2007; Badger et al., 2011; Pflughoeft and Versalovic, 2011). In this study, we apply these approaches to study the ecology and diversity of the built environment microbiome.
The health benefits of well-planned architecture have been recognized for many years (Sternberg, 2009). A prominent example is the work of Florence Nightingale, who over 150 years ago wrote that open windows were the hallmark of a healthy hospital ward (Nightingale, 1859). Today, ventilation remains a key design strategy to mitigate the spread of infectious disease indoors (Arundel et al., 1986; Li et al., 2007; Guenther and Vittori, 2008). Despite the growing body of data linking architecture and human health, we continue to live in an era where many buildings are associated with significant health risks. These risks include, but are not limited to, sick building syndrome, other health risks resulting from exposure to indoor pollutants (Institute of Medicine, 2011) and hospital-acquired infections, which remain among the leading causes of death in developed countries (Institute of Medicine, 2001, 2004). Scientific studies and data are increasingly focused on understanding how improved design can make buildings less risky for their occupants (Ulrich et al., 2008).
As for any other biome, the composition of the built environment microbiome is determined by some combination of two simultaneous ecological processes: the dispersal of microbes from a pool of available species and selection of certain microbial types by the environment (Martiny et al., 2006). The microbial species available for dispersal into most built environments are likely to come primarily from outside air (introduced through ventilation), indoor surfaces and the bodies of humans and other micro- and macroorganisms residing and moving through indoor spaces (Pakarinen et al., 2008; Rintala et al., 2008; Grice and Segre, 2011). It is unclear which of these sources is the most important, or what factors might determine their relative importance within and among buildings (Rintala et al., 2008). It has been hypothesized that filtration by mechanical ventilation is a form of dispersal limitation, resulting in indoor microbial communities that represent a subset of outdoor microbes (Lee et al., 2006). However, indoor environments have been found to harbor microbial taxa not commonly found outdoors (Tringe et al., 2008; Amend et al., 2010). Selection of specific microbial taxa by environmental conditions has been suggested to occur in built environments, but this has been demonstrated for only a few taxa (Shaman and Kohn, 2009). For example, it has been reported that air temperature and relative humidity (Arundel et al., 1986; Tang, 2009), as well as the source of ventilation air and occupant density (Qian et al., 2010), can influence the abundance and transmission of some pathogenic microbes indoors.
In this study, we used high-throughput, culture-independent approaches to survey the built environment microbiome of a health-care facility. We chose a health-care facility because it allowed us to sample across a range of design and environmental factors (including ventilation source, temperature and humidity) and because there is keen interest in the role that the microbiome of health-care facilities has in human health (Guenther and Vittori, 2008). We focused our survey on bacteria, the most common cause of hospital-associated infections (Edmond et al., 1999). Our study addresses three general questions: First, what is the composition of airborne microbial communities indoors? Second, how does building design, in particular ventilation source, influence the diversity and structure of the built environment microbiome? Third, are ventilation sources or environmental conditions correlated with the abundance of human-associated microbes in the built environment?
Materials and methods
Setting and study design
Airborne microbial communities and environmental conditions were sampled six times at Providence Milwaukie Hospital, Milwaukie, OR, USA, on 27–28 February 2010 (Supplementary Table S1). At each sampling time, a sample was collected from outdoor air, indoor air from a mechanically ventilated room and indoor air from a ‘naturally' ventilated (that is, primarily window ventilated) room simultaneously. Outdoor samples were collected from the roof of the hospital immediately adjacent to the air intake for the building's heating, ventilating and air conditioning (HVAC) system. Indoor samples were collected in researcher-occupied patient rooms. Mechanically ventilated rooms had ventilation air supplied by the HVAC system through a supply duct and removed through a return duct and bathroom exhaust. Window-ventilated rooms had ventilation air supplied directly from the outside through a window and removed through a return duct, bathroom exhaust and the window. A detailed description of the architectural attributes of the building is provided in the Supplementary Information.
Environmental measurements
During each sampling period, environmental conditions, including air temperature, relative humidity, absolute humidity and air flow rate, were measured using TSI Inc. (Shoreview, MN, USA) VelociCalc multi-function ventilation meters (Series 9555 with probe 964) placed at the patient bed and in air supply in patient rooms, and Davis Instruments (Hayward, CA, USA) Vantage Pro2 meters placed adjacent to BioSamplers (SKC Inc., Eight Four, PA, USA) outdoors. Sampling occurred every second, and 1-min averages were stored for indoor samples. For outdoor readings, the variables were sampled and stored every 15 min. Environmental conditions for each sample represent the average of all measurements for the entire sampling period. Air changes per hour were calculated for patient rooms taking into account room volume, air speed and volume flowing into the room through the window (window-ventilated rooms) or diffuser (mechanically ventilated rooms).
Microbial community sampling
Each microbial sample was collected by drawing air through two liquid impingers (BioSamplers) filled with sterile molecular-grade water for 1 h at a rate of 12.5 l min−1, resulting in a total sampled air volume of 1500 l per sample. Impingers were refilled with sterile water midway through each sampling to maintain a constant liquid volume and collection efficiency. The impingers and all tubing used to connect the impingers to vacuum pumps were autoclaved and maintained in a sterile condition before sampling. Outdoor samples were collected with impingers placed at the roof surface immediately adjacent to the hospital's HVAC air intake vents. Indoor samples were collected from impingers placed at ∼30 cm above the middle of the bed in patient rooms occupied by two researchers for the duration of each sampling. Window-ventilated rooms were ventilated exclusively by window for at least 1 h before sampling.
Bacterial cell density estimation, DNA extraction and bacterial 16S gene amplification procedures are described in detail in the Supplementary Information. Total bacterial cell density counts were performed using epifluorescence microscopy of 4',6-diamidino-2-phenylindole-stained samples. We extracted DNA from impinger liquid using standard methods, and gene fragments from the V2–V3 region of the bacterial 16S SSU-rRNA gene (Pace, 1997; Hamady et al., 2008; Huse et al., 2008) were amplified using universal bacterial 16S primers 27F and 338R modified for use with the GS FLX Titanium platform (454 Life Sciences, Branford, CT, USA).
Sequence processing
Pyrosequencing of the DNA library resulted in 179 146 sequences. Raw sequences from 454 pyrosequencing were processed with the QIIME pipeline (Caporaso et al., 2010) using standard quality control guidelines to eliminate low-quality sequences and assign sequences to different samples based on their barcodes. Criteria for sequence inclusion in subsequent analyses were based on QIIME version 1.1 defaults, with the exception of a minimum sequence quality score of 20 and up to three ambiguous bases and three primer mismatches permitted per sequence. After quality control, 107 820 sequences remained and were included in subsequent analyses. Quality-controlled sequences were denoised using the QIIME denoiser using default settings and binned into operational taxonomic units (OTUs) at a 97% sequence similarity cutoff using uclust (Edgar, 2010), resulting in 10 585 OTUs. The 97% sequence similarity cutoff is the highest similarity cutoff that can be used to accurately bin sequences into OTUs due to the sequencing error rates inherent in the 454 sequencing technology (Kunin et al., 2010). The longest, highest-quality sequence from each OTU was chosen as a representative sequence for that OTU in subsequent analyses. After quality control and OTU binning, the median sequence length was 329 nucleotides (mean length (±s.d.)=310±49 nucleotides). A check for chimeric sequences with the ChimeraSlayer algorithm identified <1% of OTUs and sequences as potentially chimeric; these sequences were excluded from subsequent analyses.
Because our primary interest concerned the structure of airborne bacterial communities, we excluded chloroplast and all other nonbacterial 16S sequences from subsequent analyses. To ensure adequate sampling depth for statistical comparison among samples, we eliminated samples containing fewer than 700 sequences after quality control. Additionally, a single outdoor air sample was eliminated from subsequent analyses because it contained >95% identical sequences that likely represented contamination during sample processing. The median number of sequences in each of the 13 remaining samples (5 from mechanically ventilated patient rooms, 4 from window-ventilated patient rooms and 4 from outdoors) was 2756 (range: 702–6830 sequences per sample).
Representative sequences for each OTU were identified taxonomically using the Ribosomal Database Project Bayesian classifier algorithm (Cole et al., 2009), with a 50% support cutoff. Phylogenetic relationships among OTUs were inferred based on the representative sequence for each OTU. Representative sequences for each OTU were aligned using the Infernal aligner (Nawrocki et al., 2009), with default settings provided by the Ribosomal Database Project (RDP) pyro pipeline (Cole et al., 2009). Representative sequence alignments were masked with the RDP hard mask (Cole et al., 2009), sites with <50% coverage were removed and sequences with <50 nucleotides remaining after masking were removed. We inferred a phylogeny among the 10 486 remaining representative OTU sequences using FastTree version 2.0.1 (Price et al., 2010) with a GTR+CAT model of evolution and pseudocount distances.
Microbial community analyses
After sequence processing, all data were imported into the R version 2.11. statistical computing environment (R Development Core Team, 2010), and all subsequent analyses and visualizations were performed in R using functions from the picante (Kembel et al., 2010) version 1.2, vegan (Oksanen et al., 2007) version 1.17, and ggplot2 (Wickham, 2009) version 0.8.9 packages. Bacterial community dissimilarity was quantified using the normalized weighted UniFrac distance metric (Lozupone et al., 2006; Hamady et al., 2009), which measures the phylogenetic distinctness of organisms in different communities, based on abundances and phylogenetic relationships of the representative OTU sequences. The compositional similarity of all samples was visualized using a nonmetric multidimensional scaling (NMDS) ordination of weighted UniFrac dissimilarities. The relative strength of relationships between airborne bacterial community structure and environmental variables were quantified using an analysis of molecular variance analysis (Excoffier et al., 1992) of the weighted UniFrac dissimilarities, which quantifies the variance in community dissimilarity explained by different explanatory variables. For each environmental variable, we measured the variance in community dissimilarity explained by that variable. Because environmental conditions differed among rooms exposed to different ventilation sources, we also measured the variance in community dissimilarity explained by each environmental variable after accounting for the variance explained by ventilation source. We repeated these analyses including indoor and outdoor samples, as well as for indoor samples only.
We tested for a relationship between ventilation source and diversity using mixed models, with rarefied diversity as the response variable, ventilation source as a fixed effect and time of sample collection as a random effect. Phylogenetic diversity (PD) was calculated as Faith's PD (Faith, 1992), the total phylogenetic branch length separating OTUs in each rarefied sample. To allow robust comparisons among samples containing different numbers of sequences, sample diversity was calculated based on samples rarefied to contain 700 sequences, as the sample with the fewest sequences contained 702 sequences. Pairwise differences in diversity and abundance of different taxa among environments and ventilation sources were tested using Tukey's honestly significant difference (HSD) tests. Repeated analyses of rarefied data yielded nearly identical results; hence results of a single representative rarefaction of the data are presented. The taxonomic composition of bacterial communities in each sample was quantified by calculating the relative abundance of sequences assigned to different taxa by the RDP naive Bayesian taxonomic classifier algorithm at a 50% cutoff (Cole et al., 2009).
We estimated the relative abundance of potentially pathogenic bacteria in each sample by identifying sequences that were related to bacterial strains that are known human pathogens, based on published lists of human pathogens including the UK select agents list (UK Health and Safety Executive Advisory Committee on Dangerous Pathogens, 2004), the Microbial Rosetta Stone Database of global and emerging infectious microorganisms and bioterrorist threat agents (Ecker et al., 2005), and several studies that provide lists of human pathogens associated with health-care facilities and other buildings (Rinttilä et al., 2004; Brodie et al., 2007; Luna et al., 2007). We conducted a Basic Local Alignment Search Tool (BLAST) sequence similarity search (Altschul et al., 1990) comparing each OTU with a database of reference sequences (Pruitt et al., 2005). We classified an OTU as a potential human pathogen if it shared 97% or greater sequence identity with a strain in the reference database that has been classified as a potential human pathogen (UK Health and Safety Executive Advisory Committee on Dangerous Pathogens, 2004; Rinttilä et al., 2004; Ecker et al., 2005; Brodie et al., 2007; Luna et al., 2007). We repeated this analysis using a variety of different taxonomic classification similarity (that is, confamilials or congeners of known pathogens) and sequence similarity cutoffs (95 and 97% sequence similarity), and the results were nearly identical; we present only the results based on the 97% sequence similarity cutoff.
We identified OTUs that were characteristic of mechanically ventilated indoor, window-ventilated indoor and outdoor environments using indicator species analysis (Dufrêne and Legendre, 1997). Indicator species analysis uses a randomization test approach to identify OTUs that have higher fidelity (relative abundance and occurrence frequency) in an environment than expected (P<0.05) based on 1000 random assignments of samples to different environments.
Results
Does architectural design influence the built environment microbiome?
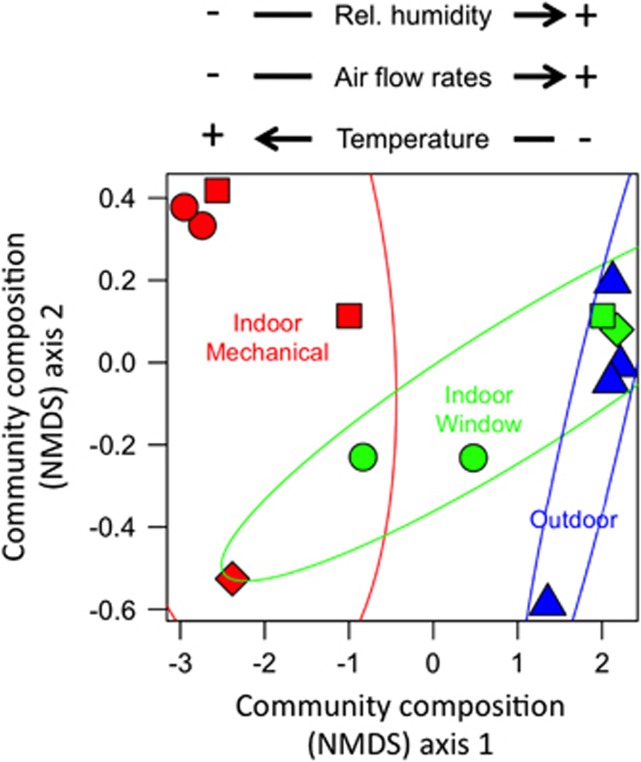
The composition of airborne bacterial communities differed among outdoor air, mechanically ventilated rooms and window-ventilated rooms (analysis of molecular variance; weighted UniFrac phylogenetic community dissimilarity; R2=0.57, P<0.01). Microbial community composition in mechanically ventilated patient rooms was distinct from the composition of communities in outdoor air, as shown by the lack of overlap among samples from these environments in terms of their phylogenetic similarity (first axis of NMDS ordination; Figure 1). Community composition in window-ventilated patient rooms was intermediate between mechanically ventilated patient rooms and outdoor air. Window-ventilated rooms with higher air temperature, lower relative humidity and lower rates of air flow contained bacterial communities more similar to mechanically ventilated rooms than to outdoor air (correlations between first axis of NMDS ordination and environmental conditions; Figure 1). Thus, there exists a gradient in the composition of airborne microbial communities with mechanically ventilated patient rooms with relatively warm and dry air at one extreme, relatively cool and moist outdoor air communities at the other extreme, and window-ventilated rooms having greater compositional similarity to mechanically ventilated rooms or outdoor air depending on the environmental conditions in the room (Figure 1).
Figure 1.
Ordination diagram (axis 1 and 2 from a NMDS ordination) summarizing similarity of airborne bacterial community composition (weighted UniFrac community phylogenetic dissimilarity) in samples from outdoors (blue), indoor mechanically ventilated patient rooms (red) and indoor window-ventilated patient rooms (green) at a health-care facility. Distances among communities indicate the phylogenetic similarity of bacteria in those communities. Symbols indicate sample location (○=room 229, □=room 231, ◊=room 235 and △=roof; Supplementary Table S1). Ellipses are 95% confidence intervals around samples from each environment. Arrows indicate direction of correlation between axis 1 scores from the NMDS ordination versus relative humidity (% r=0.67, P=0.01), temperature (°C; r=−0.68, P=0.01) and air flow velocity (m s−1; r=0.50, P=0.07).
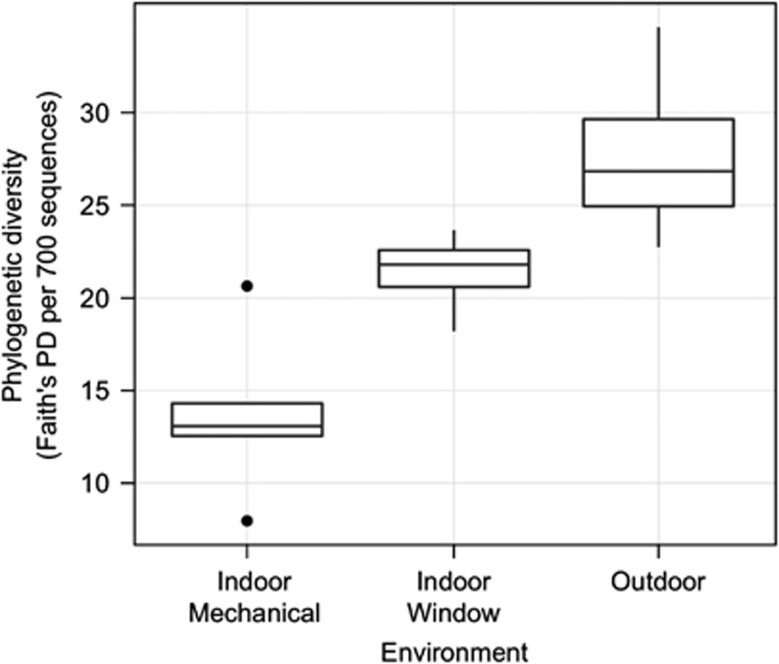
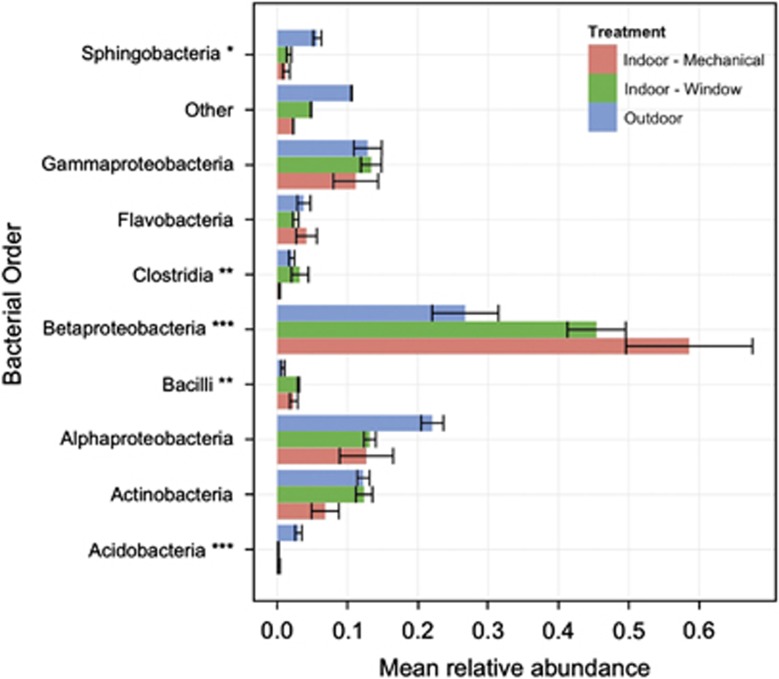
Airborne bacterial cell density in samples varied from 502 000 to 2 580 000 cells m−3 (Supplementary Table S1), but cell density did not vary significantly among environments (analysis of variance (ANOVA); log10(airborne bacterial cell density) versus environment; R2=0.02, F2,10=1.1, P=0.36). The PD of airborne bacterial communities differed significantly among environments (ANOVA; F2,6=15.5, P=0.005), with highest diversity in outdoor air and lowest in indoor air from rooms that were mechanically ventilated (Figure 2). The taxonomic composition of airborne bacterial communities also varied with ventilation source (Figure 3). Betaproteobacteria were the dominant taxa in all the airborne communities, but were more abundant indoors than outdoors. In outdoor air, Actinobacteria, Gammaproteobacteria and Alphaproteobacteria were the dominant taxa, and Acidobacteria and Sphingobacteria were more abundant in outdoor air than indoors.
Figure 2.
PD (total phylogenetic branch length; Faith's PD per 700 sequences) in different environments at a health-care facility: outdoors and indoors in patient rooms exposed to different ventilation sources (mechanical or window ventilation). PD (based on samples rarefied to 700 sequences per sample) was significantly different among all environments (Tukey's HSD; mixed model with fixed effect of environment, random effect of measurement time, overall model significant (P<0.05), pairwise differences significant (P<0.05)).
Figure 3.
Taxonomic composition of airborne bacterial communities in different environments at a health-care facility: outdoors (blue) and indoors in patient rooms exposed to different ventilation sources (mechanical (red) or window (green) ventilation). Composition estimates (mean±s.d.) are based on relative abundances of bacterial 16S sequences assigned to different phyla. Asterisk symbols indicate taxonomic groups whose relative abundance differed significantly among ventilation treatments (ANOVA; *P<0.1, **P<0.05 and ***P<0.01).
We observed a significant relationship between indoor ventilation source and environmental conditions versus airborne bacterial community structure. Ventilation source (mechanical versus window) explained the majority of variation in bacterial community structure among rooms in the hospital we studied (analysis of molecular variance; weighted UniFrac phylogenetic community dissimilarity versus ventilation source; R2=0.66, P<0.01; Table 1). However, after accounting for the effect of ventilation source, bacterial community composition indoors was related to the environmental conditions in a room (Table 1), especially relative humidity (R2=0.11, P=0.05) and air changes per hour (R2=0.11, P=0.1).
Table 1. Variance in phylogenetic similarity of airborne bacterial communities (weighted UniFrac distance (35)) explained by different environmental factors for samples from different environments at a health-care facility ((A) indoors and outdoors; (B) indoors in window-ventilated rooms and mechanically ventilated rooms).
|
Variance explained (total) |
Variance explained (after accounting for environment) |
|||
|---|---|---|---|---|
| R2 | P-value | R2 | P-value | |
| (A) All samples (indoor and outdoor) | ||||
| Environment | 0.57 | <0.01 | ||
| Relative humidity | 0.31 | <0.01 | 0.14 | 0.08 |
| Humidity ratio | 0.22 | 0.01 | 0.07 | 0.59 |
| Temperature | 0.31 | <0.01 | 0.14 | 0.12 |
| Air flow velocity | 0.17 | 0.03 | 0.13 | 0.20 |
| Time | 0.09 | 0.07 | 0.09 | 0.32 |
|
Variance explained (total) |
Variance explained (after accounting for ventilation method) |
|||
|---|---|---|---|---|
| R2 | P-value | R2 | P-value | |
| (B) Indoor samples only (mechanical and window-ventilated rooms) | ||||
| Ventilation method | 0.66 | <0.01 | ||
| Relative humidity | 0.38 | 0.01 | 0.11 | 0.05 |
| Humidity ratio | 0.07 | 0.31 | 0.02 | 0.77 |
| Temperature | 0.14 | 0.08 | 0.10 | 0.15 |
| Air changes per hour | 0.38 | 0.02 | 0.11 | 0.10 |
| Air flow velocity at bed | 0.10 | 0.13 | 0.07 | 0.22 |
| Air flow velocity at supply | 0.37 | 0.03 | 0.03 | 0.60 |
| Time of sampling | 0.06 | 0.22 | 0.12 | 0.11 |
| Room | 0.12 | 0.27 | 0.09 | 0.34 |
Abbreviation: AMOVA, analysis of molecular variance.
Variance explained (total) indicates variance explained by that variable alone, variance explained (after accounting for environment) represents variance explained after accounting for environment effects; for AMOVA, analyses (34) of variance in weighted UniFrac distance among samples explained by different variables.
Correlates of human-associated bacterial abundance in the built environment
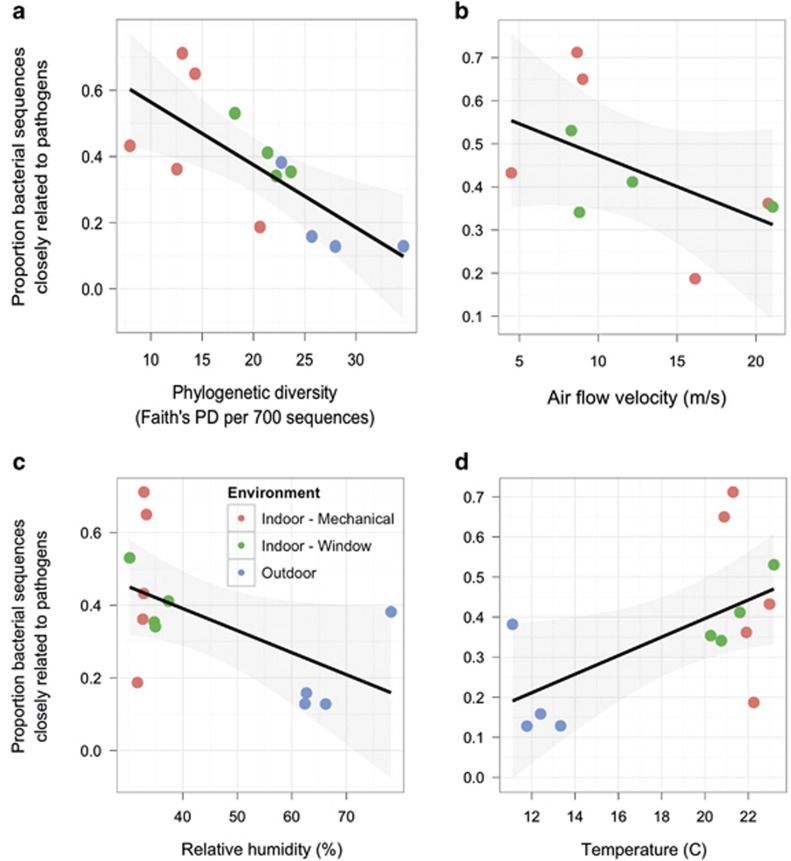
Air in mechanically ventilated rooms contained significantly less chloroplast DNA compared with window-ventilated rooms and outdoor air (ANOVA and Tukey's HSD test; P<0.01). The relative abundance of potentially pathogenic bacteria (bacterial OTUs with a 97% or greater sequence similarity to known human pathogens; UK Health and Safety Executive Advisory Committee on Dangerous Pathogens, 2004; Rinttilä et al., 2004; Ecker et al., 2005; Brodie et al., 2007; Luna et al., 2007) was higher in indoor air than in outdoor air (ANOVA; P<0.01). Indoor air contained communities that were dominated by a few closely related bacteria that were related to known human pathogens and human-associated bacteria (Figure 4a). The relative abundance of potentially pathogenic bacteria in patient rooms was independent of ventilation source (ANOVA; P=0.2), but decreased with increasing air flow rates (Figure 4b; P=0.04), air changes per hour (P=0.004) and relative humidity (Figure 4c; P=0.02). Because airborne bacterial cell density did not differ among rooms with different ventilation sources, this suggests that the absolute abundance of potential pathogens was lower with higher rates of airflow.
Figure 4.
Relative abundance of sequences from bacterial sequences closely related to human pathogens (95% or greater sequence similarity) in airborne microbial samples versus PD and environmental conditions at a health-care facility. Solid line is best fit (with shaded 95% confidence interval) from a linear model of relative abundance of potentially pathogenic sequences versus (a) PD (total phylogenetic branch length; Faith's PD (23) per 700 sequences; % R2=0.53, P=0.005), (b) air flow velocity measured at the patient bed (m s−1; R2=0.27 P=0.04), (c) relative humidity (% R2=0.29, P=0.02) and (d) temperature (°C; R2=0.33, P=0.01).
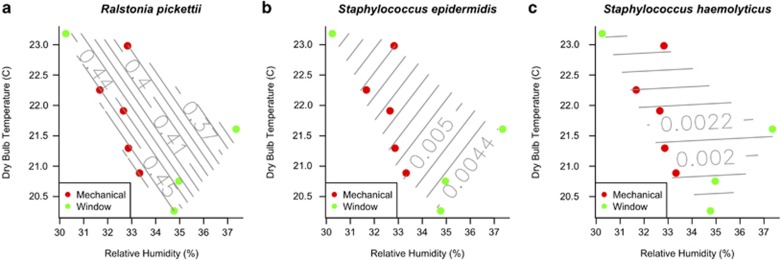
The abundance of individual bacterial taxa also responded to indoor environmental conditions and ventilation source. Indicator species analyses indicated that several bacterial taxa that are commonly found in the human microbiome (Turnbaugh et al., 2007; Costello et al., 2009; Grice and Segre, 2011), including members of the families Burkholderiaceae, Pseudomonadaceae, Staphylococcaceae and genera Micrococcus, Pseudomonas, Ralstonia and Staphylococcus, were common and abundant in indoor air, especially in air from mechanically ventilated rooms, but nearly absent from outdoor air (Figure 5, Table 2). Several indicator OTUs found in mechanically ventilated patient rooms were closely related (>97% sequence similarity) to the facultative human pathogens Staphylococcus epidermidis, S. haemolyticus and Ralstonia pickettii. Bacteria commonly found in soil and water, including members of the Acidobacteria, Planctomycetales, Gemmatimonadales, Methylococcales and Sphingobacteriales, were abundant and common in outdoor air, but relatively rare or absent indoors.
Figure 5.
Temperature and relative humidity of air in hospital patient rooms exposed to mechanical and window ventilation. Contours indicate relative abundance of sequences closely related (97% or greater sequence similarity) to the potential human pathogens (a) R. pickettii, (b) S. epidermidis and (c) S. haemolyticus, based on a polynomial spline surface fit to sample environmental coordinates.
Table 2. Taxonomic identity of indicator OTUs in different environments at a hospital.
| Environment | Indicator OTU class | Indicator OTU family | Indicator OTU closest BLAST hit | Indicator OTU % ID |
|---|---|---|---|---|
| Indoor | Actinobacteria | Actinomycetales | Kytococcus sedentarius | 98.8 |
| Mechanical | Bacteroidetes | Flavobacteriaceae | ||
| Firmicutes | Staphylococcaceae | Staphylococcus epidermidis* | 99.1 | |
| S. haemolyticus* | 98.5 | |||
| Proteobacteria | Burkholderiaceae | Ralstonia pickettii* | 98.2 | |
| Caulobacteraceae | ||||
| Enterobacteriaceae | Enterobacter spp. | 98.5 | ||
| Indoor | Actinobacteria | Actinomycetales | Kocuria rhizophila | 96.1 |
| Window | Micrococcus luteus | 99.7 | ||
| Cyanobacteria | Bacillariophyta | |||
| Proteobacteria | Acetobacteraceae | |||
| Moraxellaceae | ||||
| Rhodobacteraceae | ||||
| Outdoor | Acidobacteria | |||
| Actinobacteria | Acidimicrobiales | |||
| Actinomycetales | ||||
| Solirubrobacterales | ||||
| Bacteroidetes | Chitinophagaceae | |||
| Cytophagaceae | ||||
| Chloroflexi | Chloroflexaceae | |||
| Cyanobacteria | Bangiophyceae | Microcystis aeruginosa | 95.1 | |
| Streptophyta | Prochlorococcus marinus | 95 | ||
| Deinococcus-Thermus | Deinococcaceae | |||
| Gemmatimonadetes | Gemmatimonadaceae | |||
| Proteobacteria | Acetobacteraceae | |||
| Beijerinckiaceae | Methylocella silvestris | 95.2 | ||
| Bradyrhizobiaceae | ||||
| Caulobacteraceae | ||||
| Erythrobacteraceae | ||||
| Halothiobacillaceae | ||||
| Methylobacteriaceae | Methylobacterium extorquens | 96.7 | ||
| M. radiotolerans | 99.0 | |||
| Methylococcaceae | ||||
| Methylocystaceae | ||||
| Oxalobacteraceae | ||||
| Pasteurellaceae | ||||
| Rhodospirillaceae | ||||
| Sinobacteraceae | ||||
| Sphingomonadaceae | ||||
| Verrucomicrobia |
Abbreviations: BLAST, Basic Local Alignment Search Tool; OUT, operational taxonomic unit; RDP, Ribosomal Database Project.
Indicator OTUs are OTUs with greater abundance and occurrence frequency in an environment than expected by chance. Taxonomic identity based on assignment to class/family by the RDP taxonomic classifier. The closest BLAST hit in the reference database is shown for indicator OTUs with 95% or greater sequence similarity to reference taxa. Asterisk symbols indicate potentially pathogenic indicator taxa with 97% or greater similarity to known human pathogens.
Discussion
Our results indicate that architectural design, in particular the source of ventilation air, does influence the diversity and composition of the built environment microbiome. Most of the observed variation in airborne microbial community structure in patient rooms at the sampled health-care facility was explained by ventilation source. Mechanically ventilated patient rooms contained an ecologically distinctive set of microbial taxa from those found in outdoor air, and window-ventilated rooms contained airborne bacterial communities intermediate in structure between mechanically ventilated patient rooms and outdoor air. Although indoor air had a lower PD of microbes relative to outdoor air, this is not readily explained by the filtering of bacterial taxa dispersed from outdoors. The outdoor air communities were dominated by bacterial taxa common in aquatic and soil habitats (Fierer et al., 2008; Womack et al., 2010). In contrast, the indoor air communities were dominated by a small number of bacterial taxa from clades that are commonly associated with humans as commensals or pathogens, and as a result they were characterized by low PD. These findings suggest that humans can be important dispersal vectors for microbes that colonize the built environment (Klevens et al., 2007).
Architects and engineers design buildings for human comfort by controlling factors such as humidity, temperature and airflow (Olgyay, 1973), but we understand little about how these factors influence the diversity and distribution of microorganisms indoors. We observed a significant relationship between indoor environmental conditions—including relative humidity and temperature—and airborne bacterial community structure. This relationship could be due to a direct link between the growth or survival of certain taxa and environmental conditions in patient rooms, or an increase in the dispersal of microbes from humans or material surfaces to the built environment under these conditions. It has been suggested that the indoor climate can influence human health through direct effects on microbial populations (Arundel et al., 1986) and communities, and our data are consistent with this hypothesis. In general, our findings are consistent with a role for both species-neutral processes such as dispersal as well as niche-based processes such as environmental filtering (Martiny et al., 2006), in the assembly of the built environment microbiome.
Ventilation method and airflow have long been known to impact allergen, pollutant and pathogen load in the built environment (Nightingale, 1863; Berglund et al., 1992; Sundell, 2004; Li et al., 2007). Mechanical ventilation greatly reduced the relative abundance of chloroplast DNA in the hospital air, and this was likely due to the filtration of pollen by the mechanical air ventilation system. Ventilation method, however, did not significantly impact the potential pathogen load indoors. In both mechanically and window-ventilated rooms, the abundance of potentially pathogenic airborne bacteria was negatively correlated with airflow rates. Our finding that increased airflow rates decreased the potential pathogen load is consistent with the hypothesis that ventilation is beneficial to human health, as modeled by the classic Wells–Riley equation (Wells, 1955; Riley et al., 1978). Given the observed similarity between indoor- and human-associated bacterial communities, it is parsimonious to assume that many of the potentially pathogenic bacteria detected indoors were emitted from humans or material surfaces indoors, and that increased airflow diluted the concentration of these bacteria relative to the non-human-associated bacteria that are more common in outdoor air.
There has been significant interest in alternatives to mechanical ventilation for infection control in health-care settings, including natural ventilation and displacement ventilation (Escombe et al., 2007; Atkinson et al., 2009), due to the lower costs of construction and operation of natural ventilation systems (Omer, 2008). Our study did not directly examine natural ventilation, but we were able to assess the effect of ventilating rooms with outdoor air entering directly through open windows. Our findings suggest that it is worthwhile to explore the effects of natural ventilation on microbial communities more rigorously using modern molecular tools, as we found that the abundance of potentially pathogenic bacteria was not higher in window-ventilated patient rooms than in mechanically ventilated rooms. The use of high-throughput molecular sequencing methods can reveal indoor microbial biodiversity that was previously difficult or impossible to observe, and to fully understand the microbiology of the built environment, additional studies in different geographic regions, seasons and architectural settings will be required.
In conclusion, we note that architects use a ‘comfort zone' model to design indoor spaces within an envelope of temperature, humidity, airflow and light availability that is physically comfortable for humans (Olgyay, 1973). An improved understanding of the ecology of the built environment microbiome could allow this model to be expanded to design indoor spaces that maximize human health and well-being by linking architectural and environmental conditions to the ecology of indoor microbes. Our data suggest that reducing direct contact with the outdoor environment may not always be an optimal design strategy for bacterial pathogen management. Just as we currently manage natural ecosystems to promote the growth of certain species and inhibit the growth of others, an evidence-based understanding of the ecology of the built environment microbiome opens the possibility that we can similarly manage indoor environments, altering through building design and operation the pool of species that potentially colonize the human microbiome during our time indoors.
Acknowledgments
This study was funded by a grant from the Alfred P. Sloan foundation to the Biology and the Built Environment Center, University of Oregon. We thank Richard Beam, Garth Didlick, William Heston, Dee Putzier, Doug Spencer and Rodney Waage for logistical assistance at the hospital. We thank Colin Bohannan, Adam Burns, Ed Clark, Mitzi Liu, Gwynhwyfer Mhuireach and Tim O'Connor for assistance with sample collection and processing.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Amend AS, Seifert KA, Samson R, Bruns TD. Indoor fungal composition is geographically patterned and more diverse in temperate zones than in the tropics. Proc Natl Acad Sci USA. 2010;107:13748–13753. doi: 10.1073/pnas.1000454107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arundel AV, Sterling EM, Biggin JH, Sterling TD. Indirect health effects of relative humidity in indoor environments. Environ Health Perspect. 1986;65:351–361. doi: 10.1289/ehp.8665351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson J, Chartier Y, Pessoa-Silva CL, Jensen P, Li Y, Seto W-H.eds). (2009Natural Ventilation for Infection Control in Health-Care Settings World Health Organization: Geneva, Switzerland; [PubMed] [Google Scholar]
- Badger JH, Ng PC, Venter JC.2011The human genome, microbiomes, and diseaseIn Nelson KE, (ed).Metagenomics of the Human Body Springer New York: New York, NY; 1–14. [Google Scholar]
- Berglund B, Brunekreef B, Knöppe H, Lindvall T, Maroni M, Mølhave L, et al. Effects of indoor air pollution on human health. Indoor Air. 1992;2:2–25. [Google Scholar]
- Brodie EL, DeSantis TZ, Parker JPM, Zubietta IX, Piceno YM, Andersen GL. Urban aerosols harbor diverse and dynamic bacterial populations. Proc Natl Acad Sci USA. 2007;104:299–304. doi: 10.1073/pnas.0608255104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, et al. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37:D141–D145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. Bacterial community variation in human body habitats across space and time. Science. 2009;326:1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufrêne M, Legendre P. Species assemblages and indicator species: the need for a flexible asymmetrical approach. Ecol Monogr. 1997;67:345–366. [Google Scholar]
- Eames I, Tang JW, Li Y, Wilson P. Airborne transmission of disease in hospitals. J R Soc Interface. 2009;6 (Suppl 6:S697–S702. doi: 10.1098/rsif.2009.0407.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecker DJ, Sampath R, Willett P, Wyatt JR, Samant V, Massire C, et al. The Microbial Rosetta Stone Database: a compilation of global and emerging infectious microorganisms and bioterrorist threat agents. BMC Microbiol. 2005;5:19. doi: 10.1186/1471-2180-5-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- Edmond MB, Wallace SE, McClish DK, Pfaller MA, Jones RN, Wenzel RP. Nosocomial bloodstream infections in United States hospitals: a three-year analysis. Clin Infect Dis. 1999;29:239–244. doi: 10.1086/520192. [DOI] [PubMed] [Google Scholar]
- Escombe AR, Oeser CC, Gilman RH, Navincopa M, Ticona E, Pan W, et al. Natural ventilation for the prevention of airborne contagion. PLoS Med. 2007;4:e68. doi: 10.1371/journal.pmed.0040068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Smouse PE, Quattro JM. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faith DP. Conservation evaluation and phylogenetic diversity. Biol Conserv. 1992;61:1–10. [Google Scholar]
- Fierer N, Liu Z, Rodriguez-Hernandez M, Knight R, Henn M, Hernandez MT. Short-term temporal variability in airborne bacterial and fungal populations. Appl Environ Microbiol. 2008;74:200–207. doi: 10.1128/AEM.01467-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grice EA, Segre JA. The skin microbiome. Nat Rev Micro. 2011;9:244–253. doi: 10.1038/nrmicro2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther R, Vittori G. Sustainable Healthcare Architecture. John Wiley and Sons: New Jersey; 2008. [Google Scholar]
- Hamady M, Lozupone C, Knight R. Fast UniFrac: facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J. 2009;4:17–27. doi: 10.1038/ismej.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamady M, Walker JJ, Harris JK, Gold NJ, Knight R. Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat Methods. 2008;5:235–237. doi: 10.1038/nmeth.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huse SM, Dethlefsen L, Huber JA, Welch DM, Relman DA, Sogin ML. Exploring microbial diversity and taxonomy using SSU rRNA hypervariable tag sequencing. PLoS Genet. 2008;4:e1000255. doi: 10.1371/journal.pgen.1000255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Institute of Medicine . Crossing the Quality Chasm: A New Health System for the 21st Century. National Academy Press: Washington, DC; 2001. [PubMed] [Google Scholar]
- Institute of Medicine . Keeping Patients Safe: Transforming the Work Environment of Nurses. National Academy Press: Washington, DC; 2004. [Google Scholar]
- Institute of Medicine . Climate Change, the Indoor Environment, and Health. National Academy Press: Washington, DC; 2011. [Google Scholar]
- Kembel SW, Cowan PD, Helmus MR, Cornwell WK, Morlon H, Ackerly DD, et al. Picante: R tools for integrating phylogenies and ecology. Bioinformatics. 2010;26:1463–1464. doi: 10.1093/bioinformatics/btq166. [DOI] [PubMed] [Google Scholar]
- Klepeis NE, Nelson WC, Ott WR, Robinson JP, Tsang AM, Switzer P, et al. The National Human Activity Pattern Survey (NHAPS): a resource for assessing exposure to environmental pollutants. J Expo Anal Environ Epidemiol. 2001;11:231–252. doi: 10.1038/sj.jea.7500165. [DOI] [PubMed] [Google Scholar]
- Klevens RM, Edwards JR, Richards CL, Horan TC, Gaynes RP, Pollock DA, et al. Estimating health care-associated infections and deaths in US hospitals, 2002. Public Health Rep. 2007;122:160–166. doi: 10.1177/003335490712200205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunin V, Engelbrektson A, Ochman H, Hugenholtz P. Wrinkles in the rare biosphere: pyrosequencing errors can lead to artificial inflation of diversity estimates. Environ Microbiol. 2010;12:118–123. doi: 10.1111/j.1462-2920.2009.02051.x. [DOI] [PubMed] [Google Scholar]
- Lee T, Grinshpun SA, Martuzevicius D, Adhikari A, Crawford CM, Luo J, et al. Relationship between indoor and outdoor bio-aerosols collected with a button inhalable aerosol sampler in urban homes. Indoor Air. 2006;16:37–47. doi: 10.1111/j.1600-0668.2005.00396.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Leung GM, Tang JW, Yang X, Chao CYH, Lin JZ, et al. Role of ventilation in airborne transmission of infectious agents in the built environment—a multidisciplinary systematic review. Indoor Air. 2007;17:2–18. doi: 10.1111/j.1600-0668.2006.00445.x. [DOI] [PubMed] [Google Scholar]
- Lozupone C, Hamady M, Knight R. UniFrac—an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics. 2006;7:371. doi: 10.1186/1471-2105-7-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna RA, Fasciano LR, Jones SC, Boyanton BL, Ton TT, Versalovic J. DNA pyrosequencing-based bacterial pathogen identification in a pediatric hospital setting. J Clin Microbiol. 2007;45:2985–2992. doi: 10.1128/JCM.00630-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martiny JBH, Bohannan BJM, Brown JH, Colwell RK, Fuhrman JA, Green JL, et al. Microbial biogeography: putting microorganisms on the map. Nat Rev Micro. 2006;4:102–112. doi: 10.1038/nrmicro1341. [DOI] [PubMed] [Google Scholar]
- Mentese S, Arisoy M, Rad AY, Güllü G. Bacteria and fungi levels in various indoor and outdoor environments in Ankara, Turkey. Clean Soil Air Water. 2009;37:487–493. [Google Scholar]
- Nawrocki EP, Kolbe DL, Eddy SR. Infernal 1.0: inference of RNA alignments. Bioinformatics. 2009;25:1335–1337. doi: 10.1093/bioinformatics/btp157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemergut DR, Costello EK, Hamady M, Lozupone C, Jiang L, Schmidt SK, et al. Global patterns in the biogeography of bacterial taxa. Environ Microbiol. 2011;13:135–144. doi: 10.1111/j.1462-2920.2010.02315.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nightingale F. Notes on Hospitals. Longman, Green, Longman, Roberts, and Green: London; 1863. [Google Scholar]
- Nightingale F. Notes on Nursing. D. Appleton: London; 1859. [Google Scholar]
- Oksanen J, Kindt R, Legendre P, O'Hara B, Simpson GL, Solymos P, et al. Vegan: community ecology package. R Package Version 1.17. 2007.
- Olgyay V. Design with Climate: Bioclimatic Approach to Architectural Regionalism. Princeton University Press: Princeton, NJ; 1973. [Google Scholar]
- Omer AM. Energy, environment and sustainable development. Renew Sustain Energy Rev. 2008;12:2265–2300. [Google Scholar]
- Pace NR. A molecular view of microbial diversity and the biosphere. Science. 1997;276:734–740. doi: 10.1126/science.276.5313.734. [DOI] [PubMed] [Google Scholar]
- Pakarinen J, Hyvärinen A, Salkinoja-Salonen M, Laitinen S, Nevalainen A, Mäkelä MJ, et al. Predominance of gram-positive bacteria in house dust in the low-allergy risk Russian Karelia. Environ Microbiol. 2008;10:3317–3325. doi: 10.1111/j.1462-2920.2008.01723.x. [DOI] [PubMed] [Google Scholar]
- Pflughoeft KJ, Versalovic J. Human microbiome in health and disease. Annu Rev Pathol. 2011;7:99–122. doi: 10.1146/annurev-pathol-011811-132421. [DOI] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. FastTree 2: approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt KD, Tatusova T, Maglott DR. NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2005;33:D501–D504. doi: 10.1093/nar/gki025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian H, Li Y, Seto WH, Ching P, Ching WH, Sun HQ. Natural ventilation for reducing airborne infection in hospitals. Build Environ. 2010;45:559–565. doi: 10.1016/j.buildenv.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team 2010R: A Language and Environment for Statistical Computing Vienna, Austria; , http://www.R-project.org . [Google Scholar]
- Riley EC, Murphy G, Riley RL. Airborne spread of measles in a suburban elementary school. Am J Epidemiol. 1978;107:421–432. doi: 10.1093/oxfordjournals.aje.a112560. [DOI] [PubMed] [Google Scholar]
- Rintala H, Pitkaranta M, Toivola M, Paulin L, Nevalainen A. Diversity and seasonal dynamics of bacterial community in indoor environment. BMC Microbiol. 2008;8:56. doi: 10.1186/1471-2180-8-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinttilä T, Kassinen A, Malinen E, Krogius L, Palva A. Development of an extensive set of 16S rDNA-targeted primers for quantification of pathogenic and indigenous bacteria in faecal samples by real-time PCR. J Appl Microbiol. 2004;97:1166–1177. doi: 10.1111/j.1365-2672.2004.02409.x. [DOI] [PubMed] [Google Scholar]
- Shaman J, Kohn M. Absolute humidity modulates influenza survival, transmission, and seasonality. Proc Natl Acad Sci USA. 2009;106:3243–3248. doi: 10.1073/pnas.0806852106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg EM. Healing Spaces: the Science of Place and Well-Being. Belknap Press: Cambridge; 2009. [Google Scholar]
- Sundell J. On the history of indoor air quality and health. Indoor Air. 2004;14:51–58. doi: 10.1111/j.1600-0668.2004.00273.x. [DOI] [PubMed] [Google Scholar]
- Tang JW. The effect of environmental parameters on the survival of airborne infectious agents. J R Soc Interface. 2009;6:S737–S746. doi: 10.1098/rsif.2009.0227.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toivola M, Alm S, Reponen T, Kolari S, Nevalainen A. Personal exposures and microenvironmental concentrations of particles and bioaerosols. J Environ Monit. 2002;4:166–174. doi: 10.1039/b108682k. [DOI] [PubMed] [Google Scholar]
- Tringe SG, Zhang T, Liu X, Yu Y, Lee WH, Yap J, et al. The airborne metagenome in an indoor urban environment. PLoS One. 2008;3:e1862. doi: 10.1371/journal.pone.0001862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature. 2007;449:804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UK Health and Safety Executive Advisory Committee on Dangerous Pathogens 2004The Approved List of Biological AgentsUK Health and Safety Executive. . http://www.hse.gov.uk/pubns/misc208.pdf .
- Ulrich R, Zimring C, Zhu X, DuBose J, Seo H, Choi Y, et al. A review of the research literature on evidence-based healthcare design. HERD. 2008;1:61–125. doi: 10.1177/193758670800100306. [DOI] [PubMed] [Google Scholar]
- Wells WF. Airborne Contagion and Air Hygiene: An Ecological Study of Droplet Infections. Harvard University Press: Cambridge, MA; 1955. [Google Scholar]
- Wickham H. ggplot2: Elegant Graphics for Data Analysis. Springer: New York; 2009. [Google Scholar]
- Womack AM, Bohannan BJM, Green JL. Biodiversity and biogeography of the atmosphere. Philos Trans R Soc Lond B Biol Sci. 2010;365:3645–3653. doi: 10.1098/rstb.2010.0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.