Abstract
Mammalian RNA complexity is regulated through interactions of RNA-binding proteins (RBPs) with their target transcripts. High-throughput sequencing together with UV-crosslinking and immunoprecipitation (HITS-CLIP) is able to globally map RBP-binding footprint regions at the resolution of ~30–60 nucleotides. Here we describe a systematic way to analyze HITS-CLIP data to identify exact crosslink sites, and thereby determine protein-RNA interactions at single-nucleotide resolution. We found that reverse transcriptase used in CLIP frequently skips the crosslinked amino-acid-RNA adduct, resulting in a nucleotide deletion. Genome-wide analysis of these cross-linking induced mutation sites (CIMS) in Nova and Argonaut (Ago) HITS-CLIP data demonstrated deletions in ~8–20% mRNA tags, which mapped to Nova and Ago binding sites on mRNA or miRNA in the mouse brain. CIMS analysis provides a general and more precise means of mapping protein-RNA interactions than currently available, as well as providing insight into the biochemical properties of such interactions in living tissues.
Introduction
The control of mammalian RNA processing is dictated by interactions of several hundred RNA-binding proteins (RBPs) with their target transcripts 1, 2. Precisely mapping direct protein-RNA interactions in living tissues is a key step towards understanding RNA-regulatory networks underlying normal physiological functions and disease 2, 3. Approaches to define RBP binding sites include mutation analysis 4, 5, in vitro RNA selection (SELEX) 6–14, RIP 15, and bioinformatic predictions 14, 16, which are typically limited in scale, signal-to-noise, resolution, or applicable experimental conditions. To overcome these limitations, we have developed cross-linking and immunoprecipitation (CLIP) to isolate transcript fragments directly interacting with a specific RBP in living organisms 17, 18, which are then amplified and can be analyzed by high-throughput sequencing (HITS-CLIP) 19–21. In the past few years, CLIP or HITS-CLIP has been used to generate genome-wide interaction maps for a diverse set of RBPs, including the mammalian Nova 17, 19, 22, 23, SRSF1 24, FOX-2 25, PTB 26, and TDP-43 27 proteins (reviewed in ref. 21). This technology has also been successful in isolating Argonaute (Ago)-miRNA-mRNA ternary complexes to map in vivo microRNA (miRNA) binding sites 20, 28, 29. In general, HITS-CLIP allows identification of footprint regions, i.e., ~30–60 nucleotide (nt) sequences around the peak of CLIP RNA tag clusters, which represent native RBP binding sites 20, 23.
Further increase in the resolution and efficiency in detecting protein-RNA interactions is of great interest. For example, significant progress has been made recently on anti-sense oligos as therapeutic tools, and their success is contingent on pinpointing protein-RNA interaction sites 30, 31. For this purpose, several modifications of CLIP have been proposed very recently 21. PAR-CLIP 32 introduces photoactive ribonucleoside analogs (e.g., 4-thiouridine) into RNA of cultured cells, to enhance the efficiency of cross-linking, and also to map the actual crosslink sites from thymidine-to-cytidine (T–C) transitions in the resulting cDNA. However, PAR-CLIP has several potential limitations, including its restriction to cell-based assays. iCLIP 33–35 is based on the observation that reverse transcriptase (RT) frequently stops at crosslink sites 36. Such RNAs with RT arrest will be missed by the standard CLIP method during PCR amplification, but are leveraged in iCLIP to determine the crosslink sites. iCLIP has been successfully applied to several RBPs 33–35, although the frequency and precision with which reverse transcription stalls versus bypasses crosslink sites have not been fully assessed, nor is it clear whether such stalling varies for different cross-linked amino acids of RBPs, RNA transcripts (e.g., different motifs, or snRNAs versus mRNAs), or experimental conditions (e.g., different temperatures or RTs).
The successful applications of the standard CLIP method to a number of RBPs with distinct binding specificity 19, 20, 24–28 strongly argue that RT efficiently bypasses and reads through the crosslink sites. Interestingly, anecdotal observations were made from a small set of Nova CLIP tags that CLIP tags frequently had mutations at the Nova-binding YCAY elements, likely introduced when RT bypassed the crosslink sites 18. This idea has been applied recently to determine the interaction sites of several snoRNAs or ribosomal RNAs (rRNAs) with RNP proteins 37–39. If such analyses could be generally applied to RBP-RNA interactions, identification of cross-linking induced mutation sites (CIMS) could provide a global and widely applicable means of mapping the exact sites of protein-RNA interactions. Here we explore this possibility by developing the systematic CIMS analysis method, applying it in two different HITS-CLIP experiments to precisely map Nova and Ago-RNA interaction sites at nucleotide resolution on a genome-wide scale.
Results
Analysis of HITS-CLIP data to identify CIMS
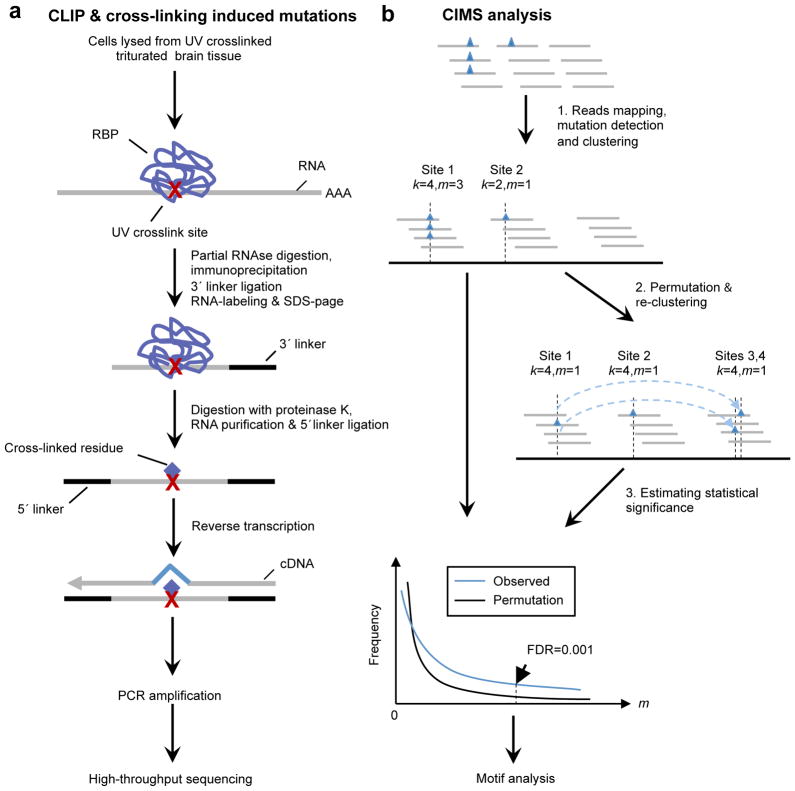
After RBP-RNA complexes are purified in the standard CLIP procedure (Fig. 1a), the bound RBPs are removed by proteinase digestion. However, due to irreversible UV-crosslinking, the residual amino-acid-RNA adducts impose a potential obstacle for RT to read through when RNA fragments are converted into cDNAs. At a certain frequency, RT stops at the crosslink site, resulting in a truncated cDNA. However, RT can also read through these sites 17, resulting in either a correct read or an error at the crosslink site, because of crosslink-induced interference with normal Watson-Crick base-pairing. These cross-linking induced mutation sites (CIMS) can be recovered by mapping sequenced reads to the reference genome that allow for deletions, insertions, or substitutions (collectively termed mutations) which were previously discarded in standard HITS-CLIP data analysis 21. Such mapping forms the basis of the CIMS analysis described here.
Figure 1. Overview of CIMS analysis.
a. Schematic representation of HITS-CLIP and cross-linking induced mutations. Protein-RNA complexes are purified by immunoprecipitation and stringent washing, followed by proteinase K treatment, a broad-specificity enzyme which cleaves peptide bonds. Remaining cross-linked amino acid(s) attached to RNA, as indicated by the red cross, impose an obstacle for RT, so that a mutation may be induced (~8–20% frequency; see below) during reverse transcription of RNA to cDNA. CLIP tags are then PCR amplified and read-out by high-throughput sequencing.
b. Schematic representation of the CIMS analysis method. Mutations detected during alignment, indicated by blue triangles, are clustered into discrete sites according to their genomic coordinates. Each site (cluster) is characterized by the total number of overlapping unique tags k and the number of tags with particular types of mutations m at the position. A permutation-based approach, which preserves the distribution of CLIP tags in the genome and also the positional bias of mutations relative to the 5′ end of reads, is used to evaluate the statistical significance of clustering for each given k and to estimate the FDR (see Online Methods for more details).
To distinguish cross-linking induced mutations from sequencing or alignment errors, we analyzed different types of CLIP tag mutations separately. We postulated that cross-linking induced mutations would occur at specific sites (CIMS) and would be reproducibly detected in multiple CLIP tags, while technical errors should map to random positions without reproducibility. Therefore, sites with clustered mutations were identified and the statistical significance (false discovery rate or FDR) of clustering was evaluated by permutation based on two parameters, k (the total number of overlapping unique tags at the nucleotide) and m (the number of unique tags with particular types of mutations at the nucleotide) (Fig. 1b and Online Methods). For substitutions, single nucleotide polymorphisms (SNPs) or RNA editing sites could also result in mutations clustered in the same positions; therefore, substitutions overlapping with known SNPs were excluded. We also predicted and excluded potential novel SNPs or RNA editing sites based on the identity of the non-reference allele (Online Methods).
Deletions, but not substitutions or insertions, represent bona fide CIMS that precisely mark Nova binding sites
Nova is a neuron-specific splicing factor important for synaptic functions 2, 23, 40, 41. It recognizes clusters of YCAY elements, initially characterized by in vitro RNA selection 10, 11 and confirmed with biochemical and protein-RNA crystallographic studies 4, 5, 17, 19, 42, 43. In-depth HITS-CLIP data comprising over 80 million raw RNA tags have mapped Nova-RNA interactions in mouse brain 23. Here we re-analyzed these reads, focusing on mutations in ~4 million unique CLIP RNA tags obtained after the raw reads were filtered, aligned to the mouse genome, and collapsed to remove PCR duplicates (Table 1; Online Methods). As a control for sequencing and alignment errors, we used a non-cross-linked mouse brain mRNA-Seq dataset composed of ~11 million unique reads obtained by the same sequencing and analysis pipeline.
Table 1.
Summary of mutations and clustered mutation sites. (a) The number of mutations shown are the total number for each indicated type; some tags have multiple mutations (for example, 42,811 deletions are present in 29,443 unique tags sequenced with the 454 platform). (b) Comparison of the frequency and significance of clustered deletion versus substitution mutation sites in Nova and Ago CLIP tags.
| a. Mutations in CLIP or mRNA-Seq reads | |||||
|---|---|---|---|---|---|
| Datasets | Unique tags | Mutations *
|
|||
| Substitutions | Deletions | Insertions | Total mutations | ||
| Nova CLIP | 3,966,800 | 1,752,573 (0.44) | 752,295 (0.19) | 32,358 (0.008) | 2,537,226 |
| Nova CLIP (454) | 141,706 | 22,790 (0.16) | 42,811 (0.30) | 3,848 (0.027) | 69,449 |
| Ago mRNA CLIP | 1,215,119 | 917,585 (0.76) | 136,100 (0.11) | 15,389 (0.013) | 1,069,074 |
| mRNA-Seq (control) | 10,974,156 | 3,357,690 (0.31) | 44,018 (0.004) | 87,238 (0.008) | 3,488,946 |
| b. Clustered mutation sites in CLIP tags | |||||
|---|---|---|---|---|---|
| Datasets | Total | m≥2 | FDR<0.001 | FDR<0.01 | FDR<0.1 |
| a) Deletions | |||||
| Nova CLIP | 474,895 | 87,423 | 24,482 | 35,548 | 72,668 |
| Ago mRNA CLIP | 127,002 | 5,841 | 886 | 1,201 | 5,267 |
| b) Substitutions | |||||
| Nova CLIP | 1,518,045 | 87,096 | 601 | 636 | 1,092 |
| Ago mRNA CLIP | 841,634 | 22,799 | 247 | 247 | 410 |
The average number of mutations per tag is shown in the parentheses.
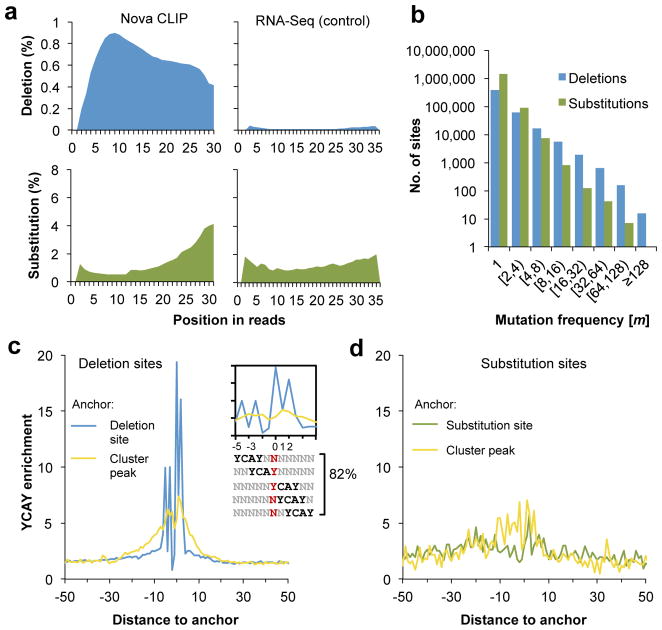
We first examined the frequency and distribution of different types of mutations in Illumina CLIP tags. Comparison of CLIP and mRNA-seq data revealed that substitutions were the most abundant (0.44 nt per tag vs. 0.31 nt per tag), while insertions were very rare (0.008 nt per tag vs. 0.008 nt per tag), at a comparable level in both datasets (Table 1). In contrast, deletions were specifically enriched in CLIP tags compared to mRNA-seq data (0.19 nt per tag vs 0.004 nt per tag), presumably due to cross-linking. In addition, substitutions in both datasets and deletions in the mRNA-seq data showed very similar U-shaped patterns (5′ and 3′ biased) in positional distributions relative to the 5′ end of sequence reads, which is characteristic of the sequencing error profile of the Illumina platform 44 (Fig. 2a). In contrast, deletions in the CLIP data showed a very different positional distribution, with the highest deletion rate observed between 5–10 nt from the 5′ end of the reads (Fig. 2a, top left panel). This is consistent with the possibility that a Nova binding footprint of ~5–10 nt was protected in the RBP-mRNA complex from RNase digestion, and hence mutations were preferentially preserved and detected in these positions.
Figure 2. Cross-linking induces deletions, but not substitutions that precisely map Nova-mRNA interactions.
a. The positional profiles of deletions (top panels, blue) and substitutions (bottom panels, green) relative to the 5′ end of reads are shown. Analysis of CLIP data is shown on the left whereas analysis of non-cross-linked mRNA-Seq data is shown on the right as a control.
b. Distribution of deletion sites or substitution sites is shown as a function of the number of tags supporting the mutation (m).
c. Enrichment of YCAY around clustered deletion sites (blue curve) is calculated from the number of YCAY starting at each position relative to the deletion sites, normalized by the frequency in flanking sequences. The same CLIP clusters are re-anchored at the CLIP tag cluster peak to calculate the enrichment of YCAY shown in the yellow curve.
d. Similar to (c) but the enrichment of YCAY around clustered substitution sites (green) or around CLIP cluster peaks of the corresponding CLIP clusters (yellow curve) is shown.
We then assessed the number of clustered mutation sites (as defined in Fig. 1b) in Nova CLIP tags. Far more clustered deletion sites were predicted than clustered substitution sites (24,482 vs. 601 sites, FDR≤0.001, or 72,684 vs. 1,092 sites, FDR ≤ 0.1), although substitutions were ~2 fold more prevalent than deletions, even after removal of substitutions sites overlapping with known or potential SNPs or RNA editing sites (Table 1 and Supplementary Table 1 online). This was also evident by examining the distribution of clustered mutation sites in terms of the number of tags showing deletions or substitutions (i.e., the parameter m), as a larger proportion of deletion sites had a larger m (Fig. 2b). Taken together, our data suggest that clustered deletions constitute a characteristic feature of bona fide Nova CIMS; in contrast, substitutions are in general dispersed, as expected from randomly distributed sequencing and alignment errors.
To evaluate how precisely CIMS can define RBP binding sites, we analyzed the high-confidence set of 24,482 clustered Nova CLIP deletion sites (FDR<0.001, Supplementary Table 2 online), and calculated the frequency of the high-affinity Nova-binding tetramer YCAY relative to the deletion sites. A remarkable enrichment of YCAYs was observed, and the majority of these elements aligned relative to the position of the deletion, especially at positions −5, −3, 0, 1 and 2. This alignment was such that crosslink sites were predominant in the first or last Y of YCAYs (Fig. 2c, blue curve). Overall, 82% of deletion sites harbored at least one YCAY element starting at one of these five positions, as compared to 6.9% expected by chance. In particular, 30% of all deletion sites had YCAY at position 0 (i.e., cross-linking at the first Y), representing a 17-fold enrichment compared to flanking background sequences. This is likely an underestimate because we frequently observed consecutive uridines near the deletion sites, preventing the unambiguous assignment of the deletion sites. As a baseline for comparison, we obtained the same set of CLIP tag clusters harboring deletion sites and re-anchored them at the CLIP tag peak position. Consistent with our previous analysis 23, we also found very significant enrichment of the YCAY elements at the CLIP cluster peak position (~6.5 fold), which was, however, substantially lower than that observed at CIMS and extended into a more dispersed region (+/−25 nt) (Fig. 2c, yellow curve). Similar results were obtained from even the least robust CIMS with deletion detected in only one tag (m=1) (Supplementary Fig. 1 online). These observations suggest that CIMS analysis greatly improved the resolution of mapping RBP binding sites to the single-nucleotide level.
As a comparison, we also examined 601 Nova CLIP substitution sites (FDR<0.001) for YCAY enrichment. In contrast to the deletion sites, these substitution sites showed only comparable or even lower motif enrichment than sequences around the CLIP tag cluster peak position (5.7 fold vs. 7 fold) (Fig. 2d), confirming that deletions, but not substitutions are due to cross-linking induced mutations. The significance of these substitution sites is unknown, and could reflect RNA editing sites, novel SNPs that pass our filtering procedure, or other mechanisms.
Estimating the frequency of cross-linking induced mutations
The 0.75 million deletion events were observed from 619,938 of 3.97 million Illumina tags, giving an estimate that 15.6% unique tags harbor ≥ 1 deletions; among these, 3.3% (132,357) unique tags have deletions in two consecutive nucleotides (Table 1). However, due to the small size of Illumina reads (32 nt for the Nova CLIP data) and the modal size of CLIP’ed RNA fragments around 50 nt, additional crosslink sites might extend beyond the read lengths (Fig. 2a), resulting in underestimation of the deletion frequency. To address this concern, we examined a set of Nova CLIP tags derived from 454 sequencing, which presumably represent full-length CLIP tags 19. Among the 141,706 unique tags derived from the dataset, 29,443 tags have deletions of one or more consecutive nucleotides (20.8%) (see also Table 1). Given that the frequency of deletions caused by sequencing or alignment errors is at least one magnitude lower (~0.4% in 36-nt tags), based on RNA-Seq data, we estimate that up to ~20% of Nova CLIP tags harbor cross-linking induced mutations.
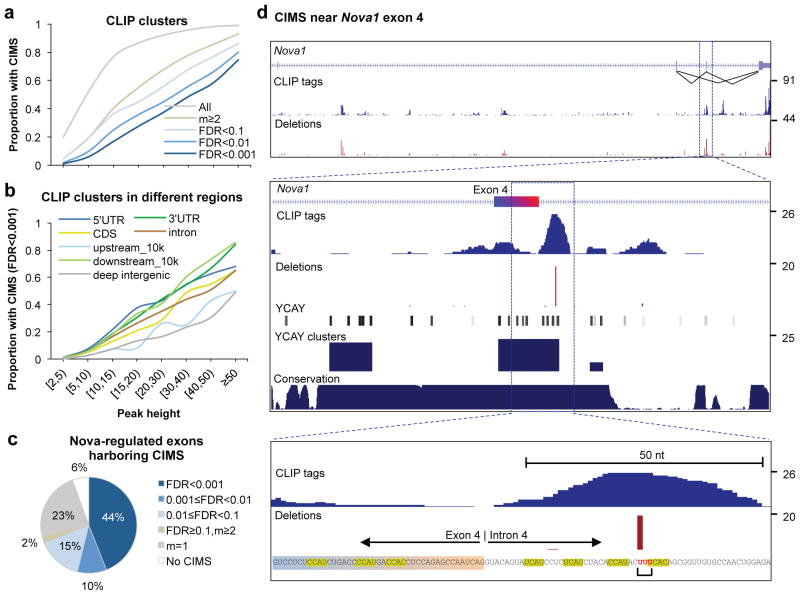
We examined the proportion of CLIP clusters that harbor CIMS. For the most robust CLIP clusters (i.e., clusters with peak height (PH) ≥ 50 tags), CIMS were detected in a majority of instances (74.6% at FDR < 0.001, 99% for all putative CIMS with m≥1) (Fig. 3a). For less stringent CLIP clusters with PH between 10–15 tags, a lower proportion harbored detected CIMS (16.8% at FDR < 0.001, 76.8% for all putative CIMS), suggesting that a higher sequencing coverage can further benefit the precise mapping of crosslink sites. We also compared CLIP clusters broken down into different genomic regions to see if CIMS differ according to cluster location. In general, clusters in different genomic regions had similar proportions of CIMS detected (Fig. 3b).
Figure 3. Frequency of CIMS in CLIP tag clusters and association of CIMS with Nova regulated alternative exons.
a. The proportion of CLIP tag clusters in genic or extended 3′ UTR regions (10k nt downstream of transcript termination) which harbor CIMS defined with varying stringency is shown as a function of CLIP tag cluster peak height (PH).
b. Similar to (a) but CIMS are defined with FDR<0.001 and CLIP tag clusters in different genomic regions are shown separately for comparison.
c. The breakdown of 325 non-redundant Nova-regulated cassette exons is shown, according to whether they have CIMS in the alternative exon, or upstream or downstream introns that are important for Nova-dependent alternative splicing regulation. CIMS are defined with varying stringency, similar to (a).
d. An example (Nova1 exon 4) of CIMS that precisely maps Nova-RNA interactions. Top panel: the Nova1 gene locus, with the number of CLIP tags and frequency of deletions shown in blue and red, respectively. Inclusion or exclusion of exon 4 is autoregulated by Nova 5. Middle panel: a zoom-in view of exon4 and flanking intronic sequences. In addition to CLIP tags and deletions, positions of YCAY elements, scores of bioinformatically predicted YCAY clusters 23, and cross-species sequence conservation in mammals are shown. Bottom panel: A further zoom-in view of sequences around the CIMS. YCAY elements and the nucleotides with deletions are highlighted. Note that although the alignment algorithm assigned the deletion to the first of the three uridines shown in red, the actual location could be any of the three nucleotides.
To link CIMS to functional outcomes of Nova-RNA interactions, we examined a set of non-redundant Nova regulated cassette exons that were either validated by RT-PCR or confidently predicted by a Bayesian network approach (Fig. 3c and Supplementary Table 3 online) 23. CIMS were robustly detected (FDR<0.01) in over half (54%) of Nova-regulated alternative exons, or in their upstream or downstream introns, regions important for Nova-dependent regulation of alternative splicing 19, 43. An additional 40% of targets had putative CIMS with lower stringency, and only 6% of exons had no CIMS detected. An illustrative example of CIMS near a well-studied Nova target exon is Nova1 exon 4 (Fig. 3d; see Supplementary Fig. 2 online for additional examples). This exon is auto-regulated by Nova through a YCAY cluster spanning exonic sequences and intronic sequences near the 5′ splice site 5, consistent with CLIP tags overlapping with the cluster. Interestingly, a CIMS supported by 20 tags (m=20) was identified in the intronic part of the cluster (CLIP cluster PH=26), resulting in deletions of one of the uridines in the sequence UUUCAC. The exonic part of the cluster, which is important for Nova-dependent splicing as well, does not have CIMS detected, presumably due to the relatively limited sequencing depth (PH=8). Taken together, these data demonstrate that CIMS analysis can precisely map Nova-RNA interactions in a substantial number of cases at the current sequencing depth.
CIMS analysis refines the Nova-binding motif
Single-nucleotide-resolution mapping of RBP binding sites derived from CIMS analysis has the potential to refine RBP motifs, especially since most RBPs recognize very short and degenerate sequences. Although Nova is known to bind clusters of YCAY elements separated by varying number of nucleotides 4, 5, 10, 11, 17, 19, 42, 43, the current model is qualitative regarding how a number of parameters affect Nova binding affinity in vivo.
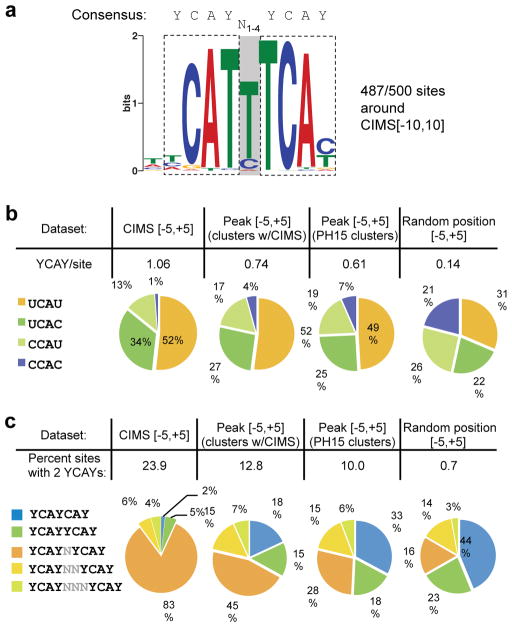
To address this question, we undertook a de novo motif analysis of sequences immediately around the 500 top CIMS (−10 to +10, 21 nt) using the GLAM2 program that allows gaps between aligned motif positions 45. This analysis revealed a dimeric YCAYN1–4YCAY motif pattern with several prominent features (Fig. 4a). The presence of U is much preferred over C in the first or last position of the YCAY element. There was also a strong preference for U in positions between the two ‘CA’ di-nucleotides. While the two YCAYs in the dimeric motif could be separated by a spacer of different sizes, a single-nucleotide spacer resulting in the YCAYNYCAY motif was by far predominant.
Figure 4. CIMS analysis refines the Nova binding motif.
a. A dimeric Nova binding motif with sites identified in 487 of 500 21-nt sequences (−10 to +10 nt) around the top CIMS by de novo motif analysis. The two YCAY elements (highlighted) are separated by a spacer region (shaded), which is variable in length, but predominantly 1 nt as depicted in the motif logo. The consensus of the motif is shown above the logo.
b. U is preferred over C in the first or last position of the Nova binding YCAY element around CIMS. The overall frequency (top) and composition (bottom) of the four tetramers conforming to the YCAY consensus are shown for 11-nt sequences around CIMS (−5 to +5 nt around deletion sites, FDR<0.001), 11-nt sequences around CLIP tag peaks of the same set of clusters with CIMS, 11-nt sequences around CLIP tag peaks of the most robust clusters independent of CIMS, or random positions in transcripts.
c. A single-nucleotide spacer between the two YCAY elements is preferred for dimeric Nova binding sites around CIMS. The overall frequency (top) and composition of dimeric motif sites with a spacer of different sizes (bottom) are shown for 11-nt sequences around CIMS or sequences around control groups, as in (b).
To characterize these features more quantitatively and further compare CIMS analysis with cluster peak analysis, we counted different tetramers conforming to the YCAY consensus in sequences around CIMS and different control groups. In the 11-nt region around CIMS (−5 to +5) with FDR < 0.001, 52% of YCAY elements are UCAU, followed by UCAC (34%) and CCAU (13%), with CCAC (1%) being the least frequent tetramer. This was in sharp contrast to the frequencies of the four tetramers observed in random transcript positions, which were approximately equal (Fig. 4b). Some previous studies have suggested that there may be a preference for UV to crosslink certain amino acids and nucleotides, particularly for thymidines in protein-DNA interactions studied with high intensity lasers 46, although this is not well established 47. To address whether the over-representation of U relative to C in YCAY might be due to preferential Nova binding or bias in cross-linking, we examined the 11-nt sequences around the peaks of the same clusters, or a set of the most robust clusters with PH≥15, independent of the presence of CIMS. In both cases, we observed a substantial over-representation of UCAU (~50%) and under-representation of CCAC (≤7%). Moreover, to further distinguish between preferential cross-linking and Nova binding, we examined sequences further away from CIMS (upstream sequences between positions −16 and −6, and downstream sequences between positions 6 and 16). These sites are very unlikely to have secondary Nova crosslinks in addition to the detected CIMS, due to the relatively low efficiency (~1–5%) of cross-linking 21. Nevertheless, these regions showed a greater frequency of YCAY than random (~0.28 versus 0.14 YCAYs per site), likely reflecting Nova’s tendency to bind YCAY multimers. In both these upstream and downstream sequences, we again observed that UCAU was the most abundant (45% and 40%, respectively), with CCAC being the least frequent YCAY sequence (11% and 14%, respectively) (Supplementary Fig. 3 online), suggesting that Nova has a bona fide preference for UCAU relative to CCAC. These observations together suggest that in vivo, UCAU and CCAC have the highest and lowest binding affinity to Nova, respectively, with UCAC and CCAU in between.
We next counted the number of dimeric YCAY motif sites with different spacers (overlapping YCAYs, or YCAYs separated by 0–3 nucleotides) in sequences around CIMS (−5 to +5, 11 nt) with FDR < 0.001 and different control groups. The overall frequency of dimeric motif sites was the highest in sequences around CIMS (23.9%), and much lower in all control groups, with the lowest in random positions (0.7%) (Fig. 4c). Importantly, the relative abundance of dimeric sites with different spacers differed dramatically among different groups. In sequences immediately around CIMS, 83% of dimeric sites had a single-nucleotide spacer (YCAYNYCAY), while overlapping YCAYs (YCAYCAY) were largely depleted (2%), in contrast to their frequency in random positions of the same transcripts (16% and 44%, respectively). The preference of YCAYNYCAY decreased substantially around peaks of the same clusters (45%), while the YCAYCAY motif became more frequent (18%). Of particular interest, when we ignored CIMS and focused on sequences around peaks of the most robust clusters, the preference of different spacers were more similar to the random positions (28% and 33% for YCAYNYCAY and YCAYCAY, respectively) than to sequences around crosslink sites. The preference of different spacers might be correlated with the arrangement of the three KH-type RNA binding domains in Nova, although further experiments are required to validate this hypothesis. Nevertheless, these observations together extend previous in vitro 11 and in vivo data 17, 43, and strongly suggest that Nova can quantitatively differentiate target sequences with subtle difference in base composition and motif arrangement, which is obvious only from CIMS analysis that produces nucleotide-resolution protein-RNA interaction map.
CIMS analysis precisely defines Ago-mRNA and Ago-miRNA interaction sites
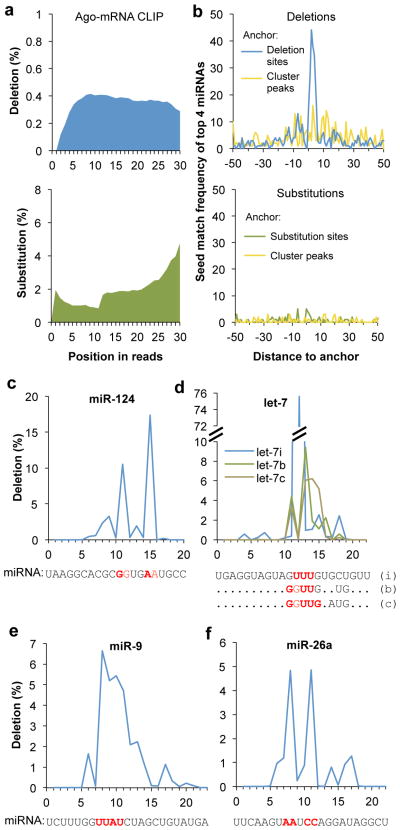
To assess whether the features of CIMS might extend more generally beyond the Nova CLIP data, we next performed CIMS analysis of the Ago CLIP data derived from analysis of mouse brain 20. Among the 1.2 million unique Ago CLIP tags, we detected 136,000 deletions in 101,092 tags (8.3%) and 917,585 substitutions in 827,127 tags (75.5%). Consistent with results obtained from Nova CLIP data, analysis of Ago-mRNA CLIP data demonstrated that cross-linking induced deletions, but not substitutions or insertions, as judged from the frequency (Table 1a) and positional profiles of mutations (Fig. 5a). We applied the same permutation-based method and defined 886 CIMS with the most reproducible deletions (FDR< 0.001) (Table 1b and Supplementary Table 4 online).
Figure 5. Cross-linking induces deletions, but not substitutions, that precisely map Ago-mRNA and Ago-miRNA interaction sites.
a. The positional profiles of deletions (top panel, blue) and substitutions (bottom panel, green) on Ago mRNA CLIP tags relative to 5′ end of reads.
b. Top panel: frequency of miRNA seed matches starting at each position around clustered deletion sites is shown for four top miRNAs (miR-124, miR-9, let-7, and miR-26) with the most seed enrichment and abundant in the brain (blue curve). The same CLIP clusters are re-anchored at the CLIP tag cluster peak to calculate the positional frequency of seed matches shown in the yellow curve. Bottom panel: similar to the top panel, but the frequency of miRNA seed matches around clustered substitution sites (green) or around CLIP tag cluster peak of the corresponding CLIP clusters (yellow curve) is shown.
c–f. Positional frequency of deletions for representative individual miRNAs abundant in brain: miR-124 (c), let-7i, b, and c (d), miR-9 (e), and miR-26a (f). For each miRNA, the sequence is shown at the bottom, with inferred crosslink sites highlighted in red.
We compared the ability to identify miRNA target sites in Ago-mRNA clusters using CIMS analysis, or methods currently available in which Ago clusters were aligned according to cluster peaks alone 20, 26. By unbiased de novo motif analysis, we discovered eight significant motifs in sequences around CIMS (−10 to +10, 21 nt) (Supplementary Fig. 4 online). Among these, five correspond to the seeds of known miRNAs, which rank in the top 20 in abundance according to the number of Ago miRNA CLIP tags, and therefore are expected to have a significant number of targets in cross-linked mRNAs 20. In contrast, when we used 21-nt sequences around peaks of the same clusters, we found only the top two motifs that correspond to miR-124 and miR-9.
To compare the signal-to-noise associated with CIMS and cluster peak analysis more quantitatively, we next focused on the four top miRNAs (miR-124, miR-9, let-7, and miR-26) with the most seed enrichment. The base composition of seed match sequences varies for these miRNAs (miR-124/UGCCUU, miR-9/CCAAAG, let-7/UACCUC, and miR-26/ACUUGA, corresponding to positions 2–7 of each miRNA), which avoids the potential complication of preferential cross-linking. When we examined the position of seed matches relative to the Ago-mRNA crosslink site, we found seed matches were sharply enriched in positions immediately around deletion sites, but not substitution sites, compared to sequences around CLIP cluster peaks, indicating that CIMS analysis greatly improved the signal-to-noise (Fig. 5b and Supplementary Table 5 online). For example, among the 886 robust deletion sites (FDR<0.001), 100 sites have the miR-124 seed matches located in the 21-nt sequences (−10 to +10) around CIMS, compared to 50 sequences if the same clusters were anchored at the peak position, and ~4 sites expected by chance (Supplementary Table 5 online, see the complete list in Supplementary Table 4 and examples in Supplementary Fig. 5 online). miRNA seed matches are particularly enriched immediately downstream of the Ago-mRNA crosslink site, although the position varies to a certain degree for individual transcripts (Fig. 5b and Supplementary Fig. 6a online). This profile is very similar to that derived from Ago PAR-CLIP data 32, although our data reflect native Ago-miRNA-mRNA ternary interactions in mouse brain. We also repeated the analysis with all 20 most abundant miRNAs that account for ~90% of Ago miRNA CLIP tags in the brain, and obtained very similar results (data not shown). These data together demonstrate that CIMS analysis of Ago-mRNA CLIP tag clusters provides a higher resolution for miRNA target detection, and also underscore the general applicability of CIMS analysis to different RBPs.
We also examined Ago-miRNA crosslink sites in the Ago-miRNA complex. Overall, the crosslink sites were the most frequent in the middle of miRNAs, between positions 9 and 15, but rare inside the seed region (Supplementary Fig. 6b online). For example, miR-124 was cross-linked most frequently at positions 11–12 (GG) and 15–16 (AA, Fig. 5c). This observation closely matched structural analysis of the Ago-miRNA-mRNA ternary complex, which suggested that this segment of miRNA forms hydrogen bonds with Ago, resulting in preferential cross-linking 48. In addition, a similar crosslink profile was also observed in PAR-CLIP data 32. Interestingly, individual miRNAs showed distinct sites preferable for cross-linking with Ago (Fig. 5c–f). Even paralogs of the same miRNA family showed differences in crosslink sites. For example, let-7i was most frequently cross-linked at positions 12–14 (UUU); for let-7b and let-7c, the CIMS sites extended into flanking nucleotides (GGUU or GGUUG) (Fig. 5d). Sequence divergence among the paralogs might account for subtle structural changes in the Ago-miRNA complex, which in turn determined the position of CIMS observed for each member.
Discussion
Here we present systematic analysis of CIMS to determine protein-RNA crosslink sites from standard HITS-CLIP data, as a means of mapping RBP binding sites in mRNAs and miRNAs at nucleotide resolution. CIMS analysis takes advantage of RT errors that are induced by the presence of cross-linked amino acids. Compared to several modifications of CLIP that have been used to determine crosslink sites, CIMS analysis does not rely on the assumption that RT always prematurely stops at the crosslink sites (iCLIP 33–35), or the introduction of artificial photoactive ribonucleoside analogs to induce preferential cross-linking, which entails potential adverse effects such as cytotoxicity 49 and changes in RNA structure, and currently limits the technique to a cell-based assay not readily applied to living tissues (PAR-CLIP 32).
We estimate the error rate of RT reading through crosslink sites to be approximately 8–20%, providing a balance between sufficient transcription efficiency and fidelity to yield many unique and accurate RNA tags in each HITS-CLIP experiments, as well as a wealth of information to determine the exact crosslink sites. The deletion rate appears to vary among different proteins, as demonstrated in Nova and Ago-mRNA CLIP data, as well as several other RBPs we analyzed (unpublished data, D. Licatalosi, J.C. Darnell and R.B. Darnell). This might reflect different amino acids and nucleotides at the protein-RNA interaction interface. In addition, this parameter might also relate to differences in the processivity or mutation rates with different enzymes, for example between AMV and MMLV RTs (the latter of which we have used exclusively), which do harbor intrinsically different thermodynamic characteristics 50. The cross-linking induced mutation frequency in standard CLIP is lower than that observed from PAR-CLIP, but more meaningful comparisons have to consider signal-to-noise, which is ~15–50 fold for CIMS analysis (~8–20% crosslinking mutation rate vs. ~0.4–0.5% background deletion rate due to sequencing or alignment errors), and 4–5 fold in PAR-CLIP (50–80% cross-linking induced T–C transition vs. 10–20% spontaneous transitions) 32.
The choice of well-studied proteins such as Nova and Ago allowed us to characterize and validate identified crosslink sites with independent data sources. In particular, the effectiveness of CIMS analysis is clearly reflected in the enrichment of Nova binding motif and miRNA seed matches at CIMS. The majority of Nova CIMS directly overlap with the high-affinity Nova-binding tetramer YCAY, and are predominantly located in the first and last positions of YCAY (Fig. 2c). These data are consistent with previous crystallography analysis, which demonstrated that the CA dinucleotide as well as the flanking nucleotides maintains tight contacts with the Nova KH domain 42. In addition, the improvement of resolution facilitated unbiased motif analysis to characterize RBP binding specificity more quantitatively. For example, previous in vitro RNA selection experiments revealed a strong preference for Nova binding to UCAU relative to either CCAU or UCAC, identified pyrimidines of variable length (up to 4 nt) as the preferred residues between YCAY elements, with an overall consensus sequence of UCAUY0 2UCAUY0 4NCAU 11. Results from CIMS analysis showed good agreement with these studies, extending them by highlighting the preference of Nova binding to a subset of dimeric YCAY motif separated by a single nucleotide in vivo. Together with the success of CIMS analysis to identify miRNA seed motifs de novo, our data suggests general applicability of CIMS analysis to other less characterized proteins, and its advantage compared to standard methods currently available.
UV cross-linking apparently induces only deletions, but not insertions or substitutions of natural ribonucleoside during reverse transcription. We do not know the physical basis for this bias, nor whether it applies equally to all RNA-protein crosslinks. Previous CLIP studies of several rRNPs and snoRNPs 37–39 found that cross-linking could induce both deletions and substitutions; interestingly, retrospective examination of this data showed that while four crosslink sites of three snoRNP proteins (including two sites cross-linked two different proteins) were identified by deletions, only one (1/6) was detected by substitutions 38. We expect studies of additional proteins with distinct specificity may provide a more comprehensive understanding of fundamental properties of protein-RNA cross-linking, which will potentially lead to further technological advances.
Online Methods
Data compilation
Nova CLIP data were generated previously using mouse brains by 454 19 and Illumina (32 nt reads) 23 sequencing. Nova splicing targets were defined previously 23. Ago CLIP data (32–36 nt reads) obtained from mouse brains were previously described in ref. 20. mRNA-Seq data were generated for a separate study by Illumina sequencing (36 nt reads), and a subset of 18 million reads sampled from 18 lanes (1 million per lane) were used here as a control.
Mapping reads
Raw Nova CLIP tags 23 and Ago-mRNA CLIP tags 20 resulted from Illumina sequencing (32–36 nt) were mapped back to the mouse genome (mm9) by the program novoalign (http://www.novocraft.com), using FASTA files as input. This program can perform exhaustive searches of hits tolerating substitutions, small insertions and deletions, collectively termed mutations here. It also allows iterative search for shorter matches by trimming nucleotides at the 3′ ends, when longer matches are not possible, presumably due to lower sequencing qualities or shorter CLIP tags. For this study, we required unambiguous mapping to the genome with≤2 substitutions, insertions or deletions in≥25 nt (parameters: -t 85 -l 25 -s 1). To remove potential duplicates resulted from PCR amplifications, we applied stringent filtering, and collapsed mappable reads with the same starting genomic positions. Representative unique reads were identified according to the following criteria in order:
The tag with the longest matches;
The tag with minimal mismatches;
Random pick among ties.
This provided a reasonable tradeoff between maximizing the chance of detecting mutations while minimizing the use of low-quality reads, although it might lose some bona fide mutations induced by cross-linking in discarded duplicate reads. From these unique reads, overlapping CLIP tags were then grouped to define CLIP clusters. The position and type of each mutation in unique reads were recorded to identify cross-linking induced mutations. Nova CLIP tags sequenced by the 454 platform 19 and control mRNA-Seq data were also mapped to the mouse genome by the same procedure.
Due to the small size of miRNAs (18–26 nt), Ago-miRNA tags 20 were mapped to miRNAs using a two-step strategy to improve sensitivity. In the first step, raw reads were mapped by blat 51 with modified parameters to identify candidate hits on miRNAs (parameters: -stepSize=2 -minScore=15 -tileSize=6). In the second step, candidate read-miRNA pairs identified by blat were re-aligned by the needle program 52 (parameters: -gapopen 20 -gapext 10) to refine the alignment. We required matches for whole-length miRNA, tolerating≤2 nt truncation at the ends and a maximum of 2 mutations (substitutions, insertions or deletions).
Modeling and identification of CIMS
For substitutions and deletions detected by read alignment, we used a statistical model to distinguish clustered mutation sites reproducibly detected in multiple CLIP tags, likely representing CIMS, and those randomly distributed in CLIP tags, likely representing sequencing or alignment errors. First, individual mutations on unique CLIP tags were clustered to identify mutation sites if they had the same genomic coordinates. Each site was then characterized by two parameters, the total number of overlapping unique CLIP tags k at that position, and the number of unique tags with mutations (of the particular type) m. We then assessed whether the observed mutation rate m/k for each site was significantly larger than one would expect from randomly distributed sequencing or alignment errors (Fig. 1b).
To be more specific, the statistical model is based on a permutation strategy to preserve the non-uniform distribution of CLIP tags in the genome, and also the non-uniform distribution of sequencing errors with respect to the distance to the 5′ end of reads. For each observed mutation in a read, we kept track of its offset from the 5′ end of the read. In the permutation procedure, each mutation was planted into a randomly picked CLIP tag in a position with the same offset observed from the original read. We then clustered the permuted mutations to identify permuted mutation sites, each characterized by the same two parameters, k and m. A null distribution of m, given k, were estimated empirically. To estimate the false discovery rate (FDR), we counted the cumulative number of clustered mutation sites with k tags at the position and ≥m tags with mutations c[m,k], and the corresponding cumulative number of permuted mutation sites c0[m,k]. By definition, the FDR of observing ≥m tags with mutations given a total of k tags is c0[m,k]/c[m,k]. To get a more precise estimate of the nulldistribution, the permutation was repeated (n=5 for this study) to increase the number of permuted mutation sites.
The model above distinguished clustered mutations versus randomly distributed mutations (due to sequencing or alignment errors), but did not remove substitutions due to known or novel SNPs, RNA editing sites or clustered mutation sites resulted from other mechanisms. To address this concern, for substitutions, we first removed those that overlap with known SNPs (v128, downloaded from UCSC genome browser 53). We then clustered the remaining substitution events to identify clustered substitution sites, also characterized by the two parameters k and m, as described above. We noted that for novel SNPs or RNA editing sites, the non-reference allele is a particular nucleotide, whereas cross-linking induced mutations will presumably generate different non-reference alleles randomly if a substitution should be introduced. Therefore, we recorded the number of reads supporting the major non-reference allele n and the ratio n/m. The ratio n/m was expected to ~1 for novel SNPs or RNA editing sites, and ~1/3 for CIMS. For this work, we removed all substitutions located in sites with n/m≥0.9 as putative novel SNPs or RNA editing sites. The remaining substitution events were then clustered again and tested using the permutation method described above.
Motif analysis
De novo motif analysis for Nova was performed using 21-nt sequences around the top 500 CIMS ranked by m and the GLAM2 program (parameters: -a 4 –b 10 –w 10, and the rest using defaults) 45. This program allows gaps of different sizes between aligned columns, and is therefore suitable to model YCAY clusters. De novo motif analysis for Ago mRNA data was performed using 21-nt sequences around all 886 CIMS with FDR<0.001, or around peaks of the corresponding clusters as a control, and the MEME program (parameters: -mod zoops -nmotifs 10 -minw 6 -maxw 8, and the rest using defaults) 54. For the cluster peak analysis, each cluster was included only once, even when it harbored multiple CIMS, to avoid repetitive counting.
To study the frequency of motif frequency relative to protein-mRNA crosslink sites, the starting positions of Nova-binding sites (YCAY) or miRNA seed matches (corresponding to positions of 2–7 of miRNAs) were recorded. Background motif frequency was estimated from control sequences of positions −500~−401 nt and 398–497 nt around deletion CIMS, to normalize the motif frequency. We also used the 11-nt sequences 500 nt away from CIMS (positions −505 to −495, and +495 to +505) for comparison of motif frequency with the 11-nt sequences around CIMS (Fig. 4b,c and Supplementary Fig. 3 online).
Supplementary Material
Acknowledgments
We thank J. Ule and all Darnell lab members for helpful discussion, and M.A. Frias, J. Luna, C.B. Marney and Y. Yuan for critical reading of the manuscript. This work was supported by grants from the NIH (NS34389 to R.B.D. and K99GM95713 to C.Z.), and the Rockefeller University Hospital CTSA (UL1 RR024143). R.B.D. is an HHMI Investigator.
Footnotes
Author contributions
C.Z. and R.B.D. conceived and designed the study; C.Z. performed the research; C.Z. and R.B.D. analyzed the data and wrote the paper.
References
- 1.Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463:457–463. doi: 10.1038/nature08909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Licatalosi DD, Darnell RB. RNA processing and its regulation: global insights into biological networks. Nat Rev Genet. 2010;11:75–87. doi: 10.1038/nrg2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cooper TA, Wan L, Dreyfuss G. RNA and disease. Cell. 2009;136:777–793. doi: 10.1016/j.cell.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dredge BK, Darnell RB. Nova regulates GABAA receptor γ2 alternative splicing via a distal downstream UCAU-rich intronic splicing enhancer. Mol Cell Biol. 2003;23:4687–4700. doi: 10.1128/MCB.23.13.4687-4700.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dredge BK, Stefani G, Engelhard CC, Darnell RB. Nova autoregulation reveals dual functions in neuronal splicing. EMBO J. 2005;24:1608–1620. doi: 10.1038/sj.emboj.7600630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilson DS, Szostak JW. In vitro selection of functional nucleic acids. Annu Rev Biochem. 2003;68:611–647. doi: 10.1146/annurev.biochem.68.1.611. [DOI] [PubMed] [Google Scholar]
- 7.Tacke R, Manley JL. The human splicing factors ASF/SF2 and SC35 possess distinct, functionally significant RNA binding specificities. EMBO J. 1995;14:3540–3551. doi: 10.1002/j.1460-2075.1995.tb07360.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perez I, Lin CH, McAfee JG, Patton JG. Mutation of PTB binding sites causes misregulation of alternative 3′ splice site selection in vivo. RNA. 1997;3:764–778. [PMC free article] [PubMed] [Google Scholar]
- 9.Burd CG, Dreyfuss G. RNA binding specificity of hnRNP A1: significance of hnRNP A1 high-affinity binding sites in pre-mRNA splicing. EMBO J. 1994;13:1197–1204. doi: 10.1002/j.1460-2075.1994.tb06369.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang YYL, Yin GL, Darnell RB. The neuronal RNA-binding protein Nova-2 is implicated as the autoantigen targeted in POMA patients with dementia. Proc Natl Acad Sci USA. 1998;95:13254–13259. doi: 10.1073/pnas.95.22.13254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buckanovich RJ, Darnell RB. The neuronal RNA binding protein Nova-1 recognizes specific RNA targets in vitro and in vivo. Mol Cell Biol. 1997;17:3194–3201. doi: 10.1128/mcb.17.6.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ponthier JL, et al. Fox-2 splicing factor binds to a conserved intron motif to promote inclusion of protein 4.1R alternative exon 16. J Biol Chem. 2006;281:12468–12474. doi: 10.1074/jbc.M511556200. [DOI] [PubMed] [Google Scholar]
- 13.Jin Y, et al. A vertebrate RNA-binding protein Fox-1 regulates tissue-specific splicing via the pentanucleotide GCAUG. EMBO J. 2003;22:905–912. doi: 10.1093/emboj/cdg089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galarneau A, Richard S. Target RNA motif and target mRNAs of the Quaking STAR protein. Nat Struct Mol Biol. 2005;12:691–698. doi: 10.1038/nsmb963. [DOI] [PubMed] [Google Scholar]
- 15.Keene JD, Komisarow JM, Friedersdorf MB. RIP-Chip: the isolation and identification of mRNAs, microRNAs and protein components of ribonucleoprotein complexes from cell extracts. Nat Protocols. 2006;1:302–307. doi: 10.1038/nprot.2006.47. [DOI] [PubMed] [Google Scholar]
- 16.Zhang C, et al. Defining the regulatory network of the tissue-specific splicing factors Fox-1 and Fox-2. Genes Dev. 2008;22:2550–2563. doi: 10.1101/gad.1703108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ule J, et al. CLIP identifies Nova-regulated RNA networks in the brain. Science. 2003;302:1212–1215. doi: 10.1126/science.1090095. [DOI] [PubMed] [Google Scholar]
- 18.Ule J, Jensen K, Mele A, Darnell RB. CLIP: A method for identifying protein-RNA interaction sites in living cells. Methods. 2005;37:376–386. doi: 10.1016/j.ymeth.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 19.Licatalosi DD, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Darnell RB. HITS-CLIP: panoramic views of protein-RNA regulation in living cells. Wiley Interdiscip Rev RNA. 2010;1:266–286. doi: 10.1002/wrna.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yano M, Hayakawa-Yano Y, Mele A, Darnell RB. Nova2 Regulates Neuronal Migration through an RNA Switch in Disabled-1 Signaling. Neuron. 2010;66:848–858. doi: 10.1016/j.neuron.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang C, et al. Integrative modeling defines the Nova splicing-regulatory network and its combinatorial controls. Science. 2010;329:439–443. doi: 10.1126/science.1191150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanford JR, et al. Splicing factor SFRS1 recognizes a functionally diverse landscape of RNA transcripts. Genome Res. 2009;19:381–394. doi: 10.1101/gr.082503.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yeo GW, et al. An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nat Struct Mol Biol. 2009;16:130–137. doi: 10.1038/nsmb.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xue Y, et al. Genome-wide analysis of PTB-RNA interactions reveals a strategy used by the general splicing repressor to modulate exon inclusion or skipping. Mol Cell. 2009;36:996–1006. doi: 10.1016/j.molcel.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Polymenidou M, et al. Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat Neurosci. 2011;14:459–468. doi: 10.1038/nn.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zisoulis DG, et al. Comprehensive discovery of endogenous Argonaute binding sites in Caenorhabditis elegans. Nat Struct Mol Biol. 2010;17:173–179. doi: 10.1038/nsmb.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leung AKL, et al. Genome-wide identification of Ago2 binding sites from mouse embryonic stem cells with and without mature microRNAs. Nat Struct Mol Biol. 2011;18:237–244. doi: 10.1038/nsmb.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hua Y, et al. Antisense correction of SMN2 splicing in the CNS rescues necrosis in a type III SMA mouse model. Genes Dev. 2010;24:1634–1644. doi: 10.1101/gad.1941310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coady TH, Lorson CL. Trans-splicing-mediated improvement in a severe mouse model of spinal muscular atrophy. J Neurosci. 30:126–130. doi: 10.1523/JNEUROSCI.4489-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hafner M, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Konig J, et al. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat Struct Mol Biol. 2010;17:909–915. doi: 10.1038/nsmb.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Z, et al. iCLIP predicts the dual splicing effects of TIA-RNA interactions. PLoS Biol. 2010;8:e1000530. doi: 10.1371/journal.pbio.1000530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tollervey JR, et al. Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat Neurosci. 2011;14:452–458. doi: 10.1038/nn.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Urlaub H, Hartmuth K, Lurmann R. A two-tracked approach to analyze RNA-protein crosslinking sites in native, nonlabeled small nuclear ribonucleoprotein particles. Methods. 2002;26:170–181. doi: 10.1016/S1046-2023(02)00020-8. [DOI] [PubMed] [Google Scholar]
- 37.Granneman S, Petfalski E, Swiatkowska A, Tollervey D. Cracking pre-40S ribosomal subunit structure by systematic analyses of RNA-protein cross-linking. EMBO J. 2010;29:2026–2036. doi: 10.1038/emboj.2010.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Granneman S, Kudla G, Petfalski E, Tollervey D. Identification of protein binding sites on U3 snoRNA and pre-rRNA by UV cross-linking and high-throughput analysis of cDNAs. Proc Natl Acad Sci U S A. 2009;106:9613–9618. doi: 10.1073/pnas.0901997106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bohnsack MT, et al. Prp43 bound at different sites on the pre-rRNA performs distinct functions in ribosome synthesis. Mol Cell. 2009;36:583–592. doi: 10.1016/j.molcel.2009.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Albert ML, Darnell RB. Paraneoplastic neurological degenerations: keys to tumour immunity. Nat Rev Cancer. 2004;4:36–44. doi: 10.1038/nrc1255. [DOI] [PubMed] [Google Scholar]
- 41.Ule J, Darnell RB. RNA binding proteins and the regulation of neuronal synaptic plasticity. CurrOpin Neurobiol. 2006;16:102–110. doi: 10.1016/j.conb.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 42.Lewis HA, et al. Sequence-specific RNA binding by a Nova KH domain: implications for paraneoplastic disease and the fragile X syndrome. Cell. 2000;100:323–332. doi: 10.1016/s0092-8674(00)80668-6. [DOI] [PubMed] [Google Scholar]
- 43.Ule J, et al. An RNA map predicting Nova-dependent splicing regulation. Nature. 2006;444:580–586. doi: 10.1038/nature05304. [DOI] [PubMed] [Google Scholar]
- 44.Wang ET, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Frith MC, Saunders NFW, Kobe B, Bailey TL. Discovering sequence motifs with arbitrary insertions and deletions. PLoS Comput Biol. 2008;4:e1000071. doi: 10.1371/journal.pcbi.1000071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hockensmith JW, Kubasek WL, Vorachek WR, von Hippel PH. Laser cross-linking of nucleic acids to proteins. Methodology and first applications to the phage T4 DNA replication system. J Biol Chem. 1986;261:3512–3518. [PubMed] [Google Scholar]
- 47.Fecko CJ, et al. Comparison of femtosecond laser and continuous wave UV sources for protein-nucleic acid crosslinking. Photochem Photobiol. 2007;83:1394–1404. doi: 10.1111/j.1751-1097.2007.00179.x. [DOI] [PubMed] [Google Scholar]
- 48.Wang Y, et al. Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes. Nature. 2009;461:754–761. doi: 10.1038/nature08434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lozzio CB, Wigler PW. Cytotoxic effects of thiopyrimidines. J Cell Physiol. 1971;78:25–31. doi: 10.1002/jcp.1040780105. [DOI] [PubMed] [Google Scholar]
- 50.Yasukawa K, Nemoto D, Inouye K. Comparison of the thermal stabilities of reverse transcriptases from avian myeloblastosis virus and Moloney murine leukaemia virus. J Biochem. 2008;143:261–268. doi: 10.1093/jb/mvm217. [DOI] [PubMed] [Google Scholar]
- 51.Kent WJ. BLAT---The BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rice P, Longden I, Bleasby A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 53.Rhead B, et al. The UCSC Genome Browser database: update. Nucl Acids Res. 2010;38:D613–619. doi: 10.1093/nar/gkp939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bailey T, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 1994:28–36. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.