Abstract
Listeria monocytogenes can respond rapidly to changing environmental conditions, as illustrated by its ability to transition from a saprophyte to an orally transmitted facultative intracellular pathogen. Differential associations between various alternative σ factors and a core RNA polymerase provide a transcriptional mechanism for regulating bacterial gene expression that is crucial for survival in rapidly changing conditions. Alternative σ factors are key components of complex L. monocytogenes regulatory networks that include multiple transcriptional regulators of stress-response and virulence genes, regulation of genes encoding other regulators, and regulation of small RNAs. In this article, the contributions of various σ factors to L. monocytogenes stress response and virulence are described.
Listeria monocytogenes – a facultative intracellular pathogen and an environmental saprophyte
The Gram-positive bacterium Listeria monocytogenes is described as having a Jekyll and Hyde lifestyle [1] because it can cause life-threatening invasive disease in humans and animals but can also effectively survive and multiply in many different non-host environments. In fact, the robust ability of L. monocytogenes to survive and multiply outside a host is crucial to its transmission, which typically occurs via the consumption of contaminated foods or animal feeds. L. monocytogenes has several regulatory mechanisms that contribute to its ability to respond to and survive under rapidly changing conditions. Regulatory mechanisms contributing to bacterial survival can function at various levels, including the transcriptional level, the post-transcriptional level and the post-translational level. This review focuses specifically on the contributions of alternative σ factors to transcriptional regulation in L. monocytogenes, including the contributions of these regulatory mechanisms to stress resistance, environmental survival and virulence.
The pathogenic lifestyle of L. monocytogenes
L. monocytogenes can cause disease (listeriosis) in several animal species. It predominantly occurs in mammals but is also seen in some poikilothermic animals and avian species [2]. Although L. monocytogenes can cause mild illness (e.g. diarrhea and flu-like symptoms) in humans, it also can cause severe invasive infections. Listeriosis manifestations in humans include meningitis, encephalitis and septicemia, and spontaneous abortions and stillbirths or infant septicemia and meningitis after the infection of pregnant women [3]. The majority (99%) of human listeriosis infections are foodborne [4]. Because initial contamination of food products usually occurs at low levels and the human infectious dose seems to be high [5], post-contamination multiplication of L. monocytogenes in food products usually needs to occur for the pathogen to reach high enough levels to cause human disease. Thus, the transmission of infectious doses of L. monocytogenes to human hosts is often dependent on the ability of this pathogen to survive and multiply in diverse environments, including in refrigerated foods.
After the consumption of contaminated foods, human listeriosis infections involve the following steps: (i) bacterial survival through the gastric passage; (ii) survival and colonization in the intestinal tract, with possible intestinal symptoms (diarrhea); (iii) invasion of intestinal epithelial cells and/or microfold (M) cells [6]; and (iv) systemic spread with intracellular infection of phagocytic and non-phagocytic cells. Cellular-infection stages also have been defined as follows: (i) internalization of L. monocytogenes within the host cell; (ii) bacterial escape from the host vacuole; (iii) bacterial multiplication within the host-cell cytoplasm and movement through the host cytoplasm by bacterially directed nucleation of host actin; (iv) bacterial movement to the host-cell surface and extrusion of bacterial cells in pseudopod-like structures; and (v) phagocytosis of these pseudopod-like structures by neighboring cells, followed by escape of the bacterium from the resulting double-membrane vacuole. The escape of the bacterium enables repetition of the cycle. Several L. monocytogenes virulence genes and their specific functions during these stages of intracellular infection have been identified and characterized, including prfA, which encodes positive regulatory factor A (PrfA) [5] (Box 1).
Box 1. Listeria monocytogenes stress-response and virulence genes and proteins, and abbreviations.
Transcription regulators
CodY: transcription repressor that responds to GTP and branched-chain amino acid levels in the cell.
CtsR: class III stress-response regulator; a transcription repressor.
Hfq: RNA-binding protein involved in stress resistance and virulence.
HrcA: heat regulation at controlling inverted-repeat of chaperone expression (CIRCE) elements; a transcription repressor.
PrfA: positive regulatory factor A; global regulator that controls transcription of many L. monocytogenes genes that are essential for virulence.
Genes involved in virulence
bilE (formerly lmo1421-lmo1422, or opuBAB): this two-gene operon (bilEAB) encodes a bile-exclusion system; responsible for bile resistance.
bsh: encodes bile salt hydrolase, which is important for bile tolerance.
clpC operon: comprises ctsR, mcsA, mcsB and clpC, encoding CtsR, McsA and McsB, which both regulate CtsR activity, and the ClpC endopeptidase and chaperone.
inlAB: this operon encodes internalin A and internalin B.
LIPI: Listeria pathogenicity island; comprises virulence genes prfA, plcA, hly, mpl, actA and plcB.
Genes involved in general-stress response
Acid stress
GAD: the glutamate decarboxylase system, which is encoded by the gad genes. gadA (formerly gadB, gadD, gadD3 or lmo2434) encodes glutamate decarboxylase. The gadCB (formerly gadD2T2 or lmo2363–lmo2362) operon encodes a glutamate decarboxylase and a glutamate-γ-aminobutyric acid (GABA) antiporter.
Osmotic stress
BetL: glycine betaine transporter encoded by betL.
Gbu: glycine betaine ABC transport system encoded by a three-gene operon (gbuABC).
OpuC: carnitine ATP-binding cassette (ABC) transport system encoded by a four-gene operon (opuCABCD).
Oxidative stress
lmo0669: encodes an oxidoreductase; transcribed in an operon with lmo0670, which encodes a hypothetical protein.
lmo1433: encodes glutathione reductase.
Energy stress and cellular metabolism
ldh: encodes L-lactate dehydrogenase.
lmo0398–lmo0400: operon encodes a fructose-specific phospho-transferase system (PTS).
lmo0784–lmo0781: operon encodes a mannose-specific PTS.
lmo2205: encodes phosphoglyceromutase.
lmo2460: encodes a transcriptional regulator similar to the central glycolytic gene regulator (CggR).
pdhB: encodes pyruvate dehydrogenase (E1 β subunit).
Cold stress
cspD: encodes cold-shock protein D.
ltrC: encodes low-temperature requirement C protein.
Other genes or proteins
clpB: encodes ClpB, similar to endopeptidase Clp ATP-binding chain B; transcribed in an operon with lmo2205.
clpP: encodes ClpP, an ATP-dependent Clp protease proteolytic subunit.
clpX: encodes ClpX, an ATP-dependent Clp protease ATP-binding subunit.
DnaKJ: heat-shock proteins encoded by the dnaK operon consisting of hrcA, grpE, dnaK, dnaJ, lmo1471 and lmo1470.
groESL: encode chaperone proteins GroES and GroEL, which regulate HrcA post-transcriptionally.
grpE: encodes heat-shock protein GrpE.
lmo1138: similar to ATP-dependent Clp protease proteolytic component.
lmo1601: encodes a general-stress protein; transcribed in an operon with lmo1602, which encodes an unknown protein.
lmo1602: encodes an unknown protein; transcribed in an operon with lmo1601.
lmo2230: encodes a protein similar to arsenate reductase.
lstR: encodes lineage-specific thermal regulator; transcribed in an operon with sigC.
rsbVWX: encode regulators of σB activity, RsbV, RsbW and RsbX.
Other abbreviations
CCCP: carbonyl cyanide 3-chloropenylhydrazone. A protonophore used to destroy the proton motive force across a plasma membrane to cause inhibition of ATP synthesis and depletion of cellular energy in a bacterial cell.
CHP: cumene hydroperoxide. A compound used to induce oxidative stress in the cell.
ppGpp: Guanosine tetraphosphate. An effector of the stringent response and a regulator of differential binding of alternative σ factors to RNA polymerase.
The nonpathogenic lifestyle of L. monocytogenes
L. monocytogenes is ubiquitously present in the environment. It has been isolated from several non-host-associated locations, including a variety of raw and ready-to-eat human foods, food-processing plants, retail food operations, urban environments, farm environments and natural environments that have had limited human impact [7]. Characterization of L. monocytogenes isolates from various hosts and environments by using a variety of different subtyping methods indicates that strains comprising the species L. monocytogenes represent at least three distinct genetic lineages [7]. Serotyping is a method that is commonly used to characterize L. monocytogenes strains. Thirteen L. monocytogenes serotypes have been differentiated; three serotypes (1/2a, 1/2b and 4b) represent the vast majority of human listeriosis isolates [3].
L. monocytogenes is capable of reproducing to high numbers in different natural environments. For example, naturally contaminated silage on farms has been reported to carry 1×108 colony-forming units (CFU) per g [8] and naturally contaminated human foods can contain >103 CFU per g [9], although the majority of food samples tested and reported in Ref. [9] contained <1 CFU per g. These findings clearly illustrate the ability of L. monocytogenes to compete and grow successfully in non-host environments.
Under laboratory conditions, L. monocytogenes can grow in a wide range of environmental conditions, including temperatures of +1 °C–45 °C, a pH range of 4.4–9.6 and salt concentrations up to 10% weight/volume NaCl [10]. L. monocytogenes can persist, although it might not grow, under an even wider range of conditions, including pH levels as low as 2.5 [11], bile and bile-acid concentrations higher than the 0.3% volume/volume and 5 mM, respectively, that are typically encountered in host animals [12], and high hydrostatic pressures [13]. Moreover, in addition to surviving under specific stress conditions, exposing L. monocytogenes to mild conditions of one type of stress can produce cross-protection against other stresses and result in robust and persistent strains in a given environment [14,15].
L. monocytogenes transcription regulation in external and intrahost environments
One important mechanism for regulating the transcription of appropriate genes under rapidly changing environmental conditions is the ability of different alternative σ factors to associate with a core RNA polymerase under specific environmental conditions. The σ factor is a dissociable subunit of the RNA polymerase holoenzyme that is responsible for enzyme recognition of a specific DNA sequence that represents a promoter region for a given gene and for transcription initiation at that promoter site. Therefore, differential associations between alternative σ factors and core polymerase, in essence, reprogram promoter-recognition specificities of the enzyme, thus enabling expression of new sets of target genes under different environmental conditions. Some alternative σ factors (e.g. σB) are maintained in inactive states through direct interactions between a given σ factor and its specific anti-σ-factor protein until the cell encounters environmental conditions requiring expression of genes recognized by that σ factor (as reviewed in Ref. [16]). Guanosine tetraphosphate (ppGpp) seems to have an important role in σ-factor competition for core RNA polymerase [17,18]). Alternative σ factors can produce dramatic changes in bacterial gene expression after exposure to environmental stress conditions, such as during the sporulation process of Bacillus subtilis, which is controlled by association of a specific cascade of alternative σ factors with core polymerase [19]. The L. monocytogenes genome encodes four alternative σ factors (σB, σC, σH and σL) [20]. By contrast, the genome of the human intestinal bacterium Bacteroides thetaiotaomicron encodes 50 alternative σ factors [21] and B. subtilis encodes 18 [22].
L. monocytogenes alternative σ factor σB
σB is encoded by sigB. Identification of the L. monocytogenes σB regulon, combined with proteomic [23] and phenotypic analyses of sigB-null mutants (ΔsigB), has provided considerable evidence that σB crucially contributes to the ability of L. monocytogenes to multiply and survive, both in mammalian hosts and under stress conditions encountered in non-host-associated environments [1]. σB was first identified and characterized in B. subtilis and subsequently has been identified in several other Gram-positive bacteria including L. monocytogenes, Staphylococcus aureus, Bacillus anthracis and Bacillus licheniformis [16]. Characterization of the L. monocytogenes σB regulon during stationary phase and under salt stress identified >170 positively regulated σB-dependent genes in L. monocytogenes, including 145 genes that are preceded by a putative σB-dependent promoter, indicating that they are directly regulated by σB [24,25]. The L. monocytogenes σB regulon seems to be similar in size to that of B. subtilis (125–150 genes [26,27]) and S. aureus (198 genes [28]).
Role of σB in L. monocytogenes’ general-stress response
Both gene-expression and phenotypic data support contributions of σB to L. monocytogenes response and survival under non-host-associated environmental stress conditions, including acid stress, osmotic stress, oxidative stress, cold stress and nutrient limitation or energy stress. Based solely on gene-expression profiles, σB also might be involved in arsenate resistance [24,25] (Table 1).
Table 1. Listeria monocytogenes.
σB contributions to stress resistance
| Stress condition | σB-dependent genes | Phenotypic data supporting σB contributions | Refs |
|---|---|---|---|
| Acid | gadA and gadCB operon | Survival of ΔsigB strain is five logs lower than wild type when logarithmic- phase cells are exposed to pH 2.5 for 1 h | [29] |
| Osmotic | betL, gbuA, bilEAB and opuCABCD | Reduced ability to use carnitine and betaine as osmoprotectants in ΔsigB strain | [35] |
| Oxidative | lmo0669a and lmo1433 | Survival of stationary-phase ΔsigB strain is one log lower than wild type when exposed to 13 mM CHP for 15 min | [30] |
| Energy | opuCA and carbon metabolism genes (e.g. ldh) | Viability of ΔsigB strain is impaired when glucose is limiting, upon entry into stationary phase and under conditions that deplete intracellular ATP | [30] |
| Cold or freezing | ltrC (low-temperature requirement C) | σB is not required during growth in rich media at 4 °C or for survival after freeze–thaw cycles, but contributes to growth in minimal media and in meats at low temperatures | [29,74–76] |
| High hydrostatic pressure | Not determined | Survival of ΔsigB strain is four logs lower than wild type at 350 MPa after 28 min | [29] |
| Arsenate | lmo2230 (encodes a protein similar to arsenate reductase) | Not determined | [24,25] |
lmo0669 might contribute to the acid-stress response at pH 4.5 [77].
σB clearly contributes to L. monocytogenes acid resistance. Specifically, phenotypic experiments have shown that an L. monocytogenes ΔsigB mutant has a severely reduced ability to survive acidic conditions (pH 2.5) compared with its parent strain [29,30]. σB regulates the transcription of genes contributing to acid resistance, including genes encoding components of the glutamate decarboxylase (GAD) system. In general, the GAD system consists of a glutamate decarboxylase enzyme (e.g. gadD2) and a glutamate-γ-aminobutyrate (GABA) antiporter (e.g. gadT2) that act in concert to reduce acidification within the cytoplasm of the cell. L. monocytogenes lmo2434 (also referred to as gadA [14,25,31], gadB [24], gadD [29] or gadD3 [32]) seems to be directly regulated by σB [24,25,29,31]. In addition, the higher transcript levels observed in ΔsigB relative to the parent strain for the lmo2363–lmo2362 operon (also called the gadCB [14] or the gadD2T2 operons [32]) indicate that these genes are indirectly regulated by σB [25].
σB contributes to the osmotic stress-resistance mechanisms of L. monocytogenes; σB activity increases with increased medium osmolarity [33,34]. The ability of L. monocytogenes to grow under conditions of high osmolarity is affected by its ability to accumulate compatible solutes by transport systems. Three σB-dependent systems (OpuC, Gbu and BetL) transport carnitine and glycine betaine across the bacterial membrane, from the environment into the cytoplasm, to combat osmotic stress [35,36]. An L. monocytogenes ΔsigB mutant (10403S, serotype 1/2a) is deficient in its ability to use carnitine and betaine as osmoprotectants compared with the corresponding wild-type strain [33]; however, this phenotype is not shared with a serotype 4c ΔsigB mutant [37]. Microarray analyses have revealed that several other transporters and transport systems are upregulated by σB in an L. monocytogenes 10403S parent strain relative to a ΔsigB mutant upon exposure of logarithmic-phase cells to 0.5 M KCl [24] or 0.3 M NaCl [25]. Furthermore, proteomic and reverse-transcription (RT)-PCR analyses showed σB-dependent expression of several proteins and genes in the presence of 0.5 M NaCl [23].
Transcriptional profiling of the σB regulon revealed several σB-dependent genes that encode proteins with putative roles in oxidative-stress resistance, including lmo0669 and lmo1433 [24,25,38]. Survival experiments with stationary-phase L. monocytogenes 10403S (serotype 1/2a) and its isogenic ΔsigB mutant during exposure to ~13 mM cumene hydroperoxide (CHP) showed considerably reduced survival of the ΔsigB strain relative to the parent strain and indicated a role for σB in oxidative-stress resistance in 10403S [30,39]. However, σB contributions to oxidative-stress survival seem to vary among L. monocytogenes strains of different serotypes. CHP experiments with a different serotype 1/2a parent strain (L. monocytogenes L61) and its isogenic ΔsigB mutant (ΔsigB61) showed no statistical difference in survival [37]. Interestingly, an L. monocytogenes serotype 4c strain (L99) was significantly less resistant than its corresponding ΔsigB mutant (ΔsigB99) upon exposure to CHP for 15 min [37]. These phenotypic experiments indicate considerable strain-to-strain variability in σB contributions to oxidative-stress resistance in L. monocytogenes.
Whole-genome transcriptional profiling of L. monocytogenes clearly indicated a major role for σB in regulating carbohydrate metabolism; identification of numerous σB-dependent genes that were upregulated during stationary phase indicates that σB is important for utilizing the limited nutrients present in the environment [25,38]. Several operons contributing to carbohydrate metabolism are σB-dependent, including lmo0784–lmo0781 and lmo0398–lmo0400, which encode mannose-specific and fructose-specific phosphotransferase systems, respectively [25]. In addition, the presence of a putative σB-dependent promoter upstream of lmo2460, which encodes a transcriptional regulator similar to the central glycolytic gene regulator (CggR) in B. subtilis, indicates that σB has a direct role in regulating genes involved in glycolysis and gluconeogenesis [25]. In phenotypic experiments, ΔsigB cell numbers declined rapidly compared to those of the L. monocytogenes 10403S parent strain during static incubation in brain heart infusion (BHI) broth at 37 °C for 36 h [30]. Enhanced survival of the 10403S parent relative to the ΔsigB strain under conditions of energy stress generated by limiting ATP synthesis through addition of the protonophore carbonyl cyanide 3-chloropenylhydrazone (CCCP) to the growth medium indicated that σB contributes to energy-stress survival [30]. Quantitative RT-PCR on RNA isolated from L. monocytogenes 10403S and otherwise isogenic ΔsigB cells exposed to CCCP for five minutes showed that RNA transcript levels for the σB-dependent opuCA were significantly higher in the parent strain than in the ΔsigB mutant, further indicating activation of σB in response to CCCP [40]. However, σB contributions to energy and carbon limitation seem to be strain dependent. Whereas the L. monocytogenes 10403S (serotype 1/2a) response to carbon limitation was σB-dependent [30,39], the long-term viability of L. monocytogenes L61 (serotype 1/2a) and L99 (serotype 4c) strains under carbon-starvation conditions seemed to be σB-independent [37].
Role of σB in L. monocytogenes virulence
Multiple lines of evidence indicate that σB contributes to L. monocytogenes virulence [16]. For example, σB directly regulates expression of some virulence genes and stress-response systems that are required for survival and multiplication under conditions that are encountered in a host, either during gastrointestinal passage (e.g. acidic pH in the gastric environment) or during intracellular survival and growth (e.g. oxidative stress in the host-cell phagosome).
Microarray analyses and single-gene transcriptional analyses have identified several virulence genes that are directly regulated by L. monocytogenes σB. Specifically, σB seems to contribute to the transcriptional regulation of genes important for bile resistance, including bsh, which encodes a bile-salt hydrolase [41], and the bilE (opuB) operon, which encodes a bile-exclusion system [42]. σB also regulates transcription of several genes that encode L. monocytogenes internalins, including inlAB that encode internalin A and B, which are crucial for L. monocytogenes attachment to and invasion of different host-cell types [24,43]. Importantly, σB also contributes to transcription of the gene encoding the global L. monocytogenes virulence gene regulator, PrfA [44,45] (Box 2).
Box 2. Regulation of PrfA expression by σB.
σB activates transcription at the prfAP2 promoter region, which includes overlapping σB- and σA-dependent promoters, and represents one of three promoter regions that contribute to prfA transcription. The other prfA promoters are the P1 promoter (which is located upstream of prfAP2) and the plcA promoter (which can generate a bicistronic transcript that includes plcA and prfA). Importantly, although translation of the prfAP1 transcript is temperature dependent (i.e. translated only at temperatures >30 °C), transcripts originating from the prfAP2 promoter do not show temperature-dependent translation [78]. Thus, transcription originating from prfAP2 provides a mechanism for σB-dependent production of PrfA, even at temperatures typically encountered outside a mammalian host. Inactivation of σB-dependent prfA transcription through deletion of the prfAP2 promoter does not affect L. monocytogenes virulence in intragastrically inoculated guinea pigs [50] or L. monocytogenes’ cell-to-cell spread in at least some mammalian cell lines [79]. The contribution of σB to prfA transcription thus seems to be important only at specific stages of infection and/or in specific hosts. Alternatively, σB transcription at prfAP2 might represent a redundant system that provides highly robust regulation of prfA expression. Interestingly, the role of σB in regulating a primary virulence gene regulator is not unique to L. monocytogenes. In S. aureus, σB contributes to regulation of virulence gene expression through transcriptional regulation of sar [28,80]. The sar locus partially controls expression of the agr locus; agr and sar are both global regulatory elements that control the synthesis of a variety of extracellular and cell-surface proteins involved in the pathogenesis of S. aureus [81].
In addition to regulation of virulence-gene expression (e.g. prfA, bsh and inlAB), L. monocytogenes σB also regulates transcription of stress-response genes that contribute to the ability of L. monocytogenes to infect mammalian hosts. For example, σB regulates the opuC operon, which encodes an ATP-binding cassette (ABC) transporter of osmoprotectant molecules (predominantly carnitine) that contribute to L. monocytogenes survival in the gastrointestinal tract of experimentally infected mice [46]. Another example of σB–dependent gene expression that contributes to bacterial survival is induction of glutamate decarboxylase (gad) genes when L. monocytogenes is exposed to mildly acidic conditions (pH 5.1) [32]. Furthermore, L. monocytogenes acid adaptation results in cross-protection against other cellular stresses [15], and acid-adapted L. monocytogenes also grows markedly better in interferon-γ (IFN-γ)-activated THP-1 macrophages than cultures not previously exposed to acid [14]. σB also seems to regulate expression of L. monocytogenes genes after entry into both phagocytic and non-phagocytic host cells [47,48]. Among the σB-dependent genes differentially transcribed in infected host cells, some genes seem to be predominantly regulated by σB (e.g. lmo1433, which encodes glutathione reductase) [25,47,48], whereas other genes seem to be co-regulated by σB and one or more transcriptional regulators, including PrfA [42,47].
Phenotypic evidence also supports important σB-mediated contributions to virulence. For example, σB clearly contributes to L. monocytogenes survival in artificial gastric fluid with a pH of 2.5 [39]. A ΔsigB strain was defective relative to the 10403S parent strain in invading Caco-2 and HepG-2 cells [49], and in causing infections in guinea pigs [50]. σB seems to contribute to virulence across multiple Gram-positive bacterial pathogens. For example, a B. anthracis ΔsigB strain was less virulent than its parent strain, indicating σB-mediated virulence contributions in this pathogen [51].
Gene regulation in L. monocytogenes by the alternative σ factor σH
L. monocytogenes σH, encoded by sigH, shows homology to the B. subtilis stationary-phase σ factor, σH, which regulates >400 genes (~240 genes positively and ~180 negatively) [52]. B. subtilis σH is involved in the transition from exponential phase to stationary phase, sporulation, nutrient transport, and the regulation of many other transcription factors and cell-wall-binding proteins. Although σH-dependent L. monocytogenes genes have not been identified, an L. monocytogenes sigH-null mutant (ΔsigH) showed reduced growth in a minimal medium and in alkaline conditions compared with wild type [53]. In addition, characterization of an L. monocytogenes ΔsigH mutant in an intraperitoneal inoculated mouse model also indicated a role for σH in infection [53]. In the closely related pathogen S. aureus, σH contributes to the expression of competence-related genes (e.g. comE and comG), indicating a possible indirect role for σH in S. aureus virulence, by enabling the acquisition of genetic material that is beneficial to bacterial survival and infection [54]. In B. anthracis, σH contributes to the expression of AtxA, a positive regulator of anthrax toxin components, which indicates a direct role for σH in B. anthracis virulence [55].
Gene regulation in L. monocytogenes by the alternative σ factor σL
L. monocytogenes σL (also known as σ54, or RpoN), which is closely related to the alternative σ factor σ54 in several other bacterial species [56], is encoded by sigL. sigL expression is sensitive to growth phase and temperature [57,58]. For example, sigL expression is induced in logarithmic-phase cells grown at 10 °C [57]; however, it is downregulated during stationary phase at 4 °C [58]. σL-dependent transcription requires energy to melt target promoter sequences; therefore, transcription initiation typically requires an additional activator (i.e. ManR) that is also an ATP–GTP-binding protein [59]. Global transcriptome analysis of an L. monocytogenes rpoN-null mutant (ΔsigL) identified 77 σL-dependent genes, including several genes that contribute to carbohydrate and amino acid metabolism [60]. Twenty-four σL-dependent genes are differentially expressed inside macrophage cells relative to their expression levels in BHI [47]. Genes encoding for pyruvate dehydrogenase, cell division and thioredoxin reductase are repressed, and those encoding for an oligo-peptide transporter and the ClpP serine protease are induced. These findings indicate roles for σL in intracellular replication and stress response. Phenotypic characterization of ΔsigL mutants has shown that σL contributes to L. monocytogenes osmotolerance [61] and influences susceptibility to the antibacterial peptide mesentericin Y105 [56].
Gene regulation in L. monocytogenes by the alternative σ factor σC
Extracytoplasmic function (ECF) σ factors comprise a distinct subgroup among σ factors. ECF σ factors typically regulate gene expression in response to envelope or extra-cytoplasmic stimuli, transport and secretion. The first recognized ECF σ factor (encoded by rpoE) was identified in Escherichia coli [62,63]. Seven ECF σ factors have been identified in B. subtilis [22]. Typically, although not in all cases, ECF σ factors are cotranscribed with corresponding anti-σ factors. L. monocytogenes encodes only a single ECF σ factor (σC). To date, sigC, which encodes σC, seems to be present only in strains classified into one L. monocytogenes evolutionary lineage (lineage II) [64]. L. monocytogenes σC seems to be activated upon heat stress, and a ΔsigC strain showed increased sensitivity to heat killing [64]. No virulence or virulence-associated phenotypes have been ascribed to L. monocytogenes σC. Expression of sigC and of lstR, which is immediately downstream of sigC, is repressed by the pleiotropic transcriptional regulator CodY during exponential growth, indicating interactions among these transcriptional regulators, and a possible role for CodY in mediating temperature stress (i.e. repressing thermal-resistance genes and cold-stress-response genes) [65].
L. monocytogenes regulatory networks and interactions involving alternative σ factors
L. monocytogenes alternative σ factors clearly have crucial roles in assuring appropriate gene expression during survival of non-host associated stress conditions and during infection. Emerging evidence indicates that alternative σ factors are crucial components of regulatory networks in L. monocytogenes and function to (i) co-regulate stress-response and virulence genes, (ii) regulate genes encoding other regulators and (iii) regulate small RNAs.
Co-regulation of stress-response and virulence genes by alternative σ factors
Transcriptional analyses have provided clear evidence that several virulence and stress-response genes are co-regulated by either multiple alternative σ factors, or alternative σ factors and other transcriptional regulators. One important theme that has emerged from various transcriptional studies is that, in addition to σB-dependent transcription of prfA itself, several other virulence genes are also co-regulated by PrfA and σB (e.g. inlA, inlB, bsh and the bilE operon) (Figure 1). Interestingly, some of these genes (e.g. inlA, bsh and bilEAB) encode proteins that seem to have specific roles in the gastrointestinal stage of the interactions between L. monocytogenes and its hosts, indicating the need for interactions between PrfA and σB during the transition of L. monocytogenes from the non-host environment to the systemic stages of infection.
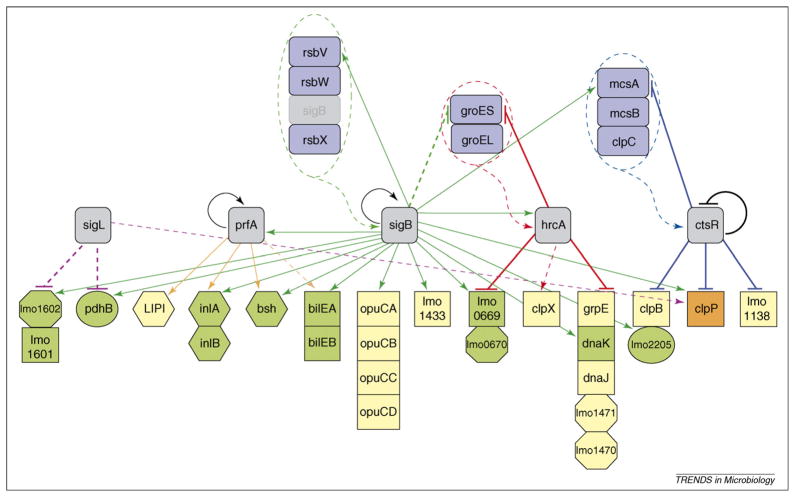
Figure 1.
Partial σL, PrfA, σB, HrcA and CtsR interaction network in L. monocytogenes. The network represented here is based on macroarray data for σL [60] and PrfA [82], and microarray data for σB [25], CtsR [66] and HrcA [67]; data for the bilE operon are based on Ref. [42]. The genes shown were selected specifically to highlight interactions between σL, PrfA, σB, HrcA and CtsR as identified by whole-genome transcriptome analyses and are only a subset of those regulated or co-regulated by the different regulator proteins. Different analytical approaches (e.g. through proteomic or metabolomic strategies) are likely to identify further additional interactions among these and other L. monocytogenes regulatory proteins. Color-coded arrows indicate genes regulated by σL (purple), PrfA (orange), σB (green), HrcA (red) or CtsR (blue). Solid lines indicate direct regulation of a gene by a given regulator as determined by the presence of a PrfA box, σB promoter, HrcA operator site or CtsR operator site; broken lines indicate indirect regulation. Target arrows (↓) indicate positive regulation by a given regulator; target stops (⊥) indicate negative regulation by a given regulator. Loops indicate autoregulation. Color-coded shapes identify (i) transcriptional regulators (gray rounded squares); (ii) genes that regulate a transcriptional regulator (blue rounded rectangles); and (iii) genes regulated by a single transcriptional regulator (yellow), two transcriptional regulators (green) or three transcriptional regulators (orange). Hexagons represent virulence genes (e.g. virulence genes directly regulated by PrfA [i.e. plcA, hly, mpl, actA and plcB] that comprise the Listeria pathogenicity island [LIPI]), squares represent stress-response genes (e.g. grpE encodes heat-shock protein GrpE), circles represent metabolism genes (e.g. lmo2205 and pdhB encode a phosphoglyceromutase and a subunit of pyruvate dehydrogenase, respectively) and octagons represent genes encoding proteins of unknown or other function (e.g. lmo1471 and lmo1470 encode an unknown protein and a ribosomal protein methyltransferase, respectively). Genes arranged in vertical columns represent operons (e.g. lmo0670, which encodes a hypothetical protein, is in an operon with lmo0669, which encodes an oxidoreductase). The green arrow targeting σB indicates post-translational regulation of σB by RsbV, RsbW and RsbX, which are encoded by rsb (regulator of sigma B) genes rsbV, rsbW and rsbX, respectively [30]. The red arrow targeting HrcA indicates post-transcriptional regulation of HrcA by GroES and GroEL, based on evidence reported for B. subtilis [83]. The blue arrow targeting CtsR indicates post-translational regulation of CtsR by McsA, McsB and ClpC, based on evidence reported for B. subtilis [68,69].
Genes regulated by more than one alternative σ factor (i.e. σB and σL) include those encoding cold-shock protein cspD and general-stress protein lmo1601. cspD is negatively regulated by σB under salt stress and by σL in late logarithmic-phase whereas lmo1601, which is positively regulated by σB under both salt stress and entry into stationary phase, is negatively regulated by σL [25,60]. σB and σL seem to modulate the expression of genes that are differentially expressed in intracellular bacteria, such as ldh and pdhB, which encode L-lactate dehydrogenase and a subunit of pyruvate dehydrogenase, respectively [25,47,60]. In addition to indicating interplay between σB and σL, these results also indicate that σB and σL control and fine-tune the expression of genes that is important for coping with physiological stresses and metabolic requirements in L. monocytogenes, including during host infection.
σB also co-regulates stress-response and virulence genes with negative regulators such as CtsR and HrcA, which both contribute to the regulation of expression of heat-shock genes and genes important for virulence [66,67] (Figure 1). Specific examples include: (i) the σB and CtsR co-regulation of genes encoding Clp proteins, which have endopeptidase and chaperone functions; and (ii) the σB and HrcA co-regulation of heat-shock proteins DnaKJ and GroESL, which act as chaperones. At least one gene (i.e. clpP) is co-regulated by σB, CtsR and σL. Furthermore, in addition to co-regulation by σB and σL, the expression of cspD is also positively regulated by HrcA. Networks between different transcriptional regulators, including alternative σ factors, thus seem to contribute to fine-tuning gene expression under various different stress conditions.
Regulation by alternative σ factors of genes encoding other regulators
Regulatory interactions among alternative σ factors and other regulators in L. monocytogenes are not limited to co-regulation of genes encoding effector proteins. Alternative σ factors also regulate the transcription of genes that encode proteins with regulatory functions, including other transcriptional regulators. One particularly intriguing and important interaction is the regulation of prfA transcription by σB (Box 2).
Emerging evidence indicates that σB contributes to the control of negative regulators CtsR and HrcA [66,67] and, hence, to the regulation of heat-shock proteins. σB seems to directly regulate HrcA through a σB promoter upstream of hrcA. Expression of hrcA from the σB-dependent promoter occurs in logarithmic-phase L. monocytogenes exposed to environmental stress (0.3 M NaCl), which indicates that the role of σB in hrcA expression might be growth-phase dependent [67]. σB seems to regulate CtsR indirectly. From a promoter upstream of mcsA, σB regulates transcription of the mcsA-mcsB-clpC operon, which encodes a set of modulators of CtsR activity [66]. In B. subtilis, McsA helps stabilize CtsR. Post-translational modification of CtsR by McsB results in release of CtsR from the operators of repressed genes. Consequently, released CtsR is targeted for degradation by the Clp proteins [68,69].
Contributions of alternatives σ factors to regulation of small RNAs
σ factors regulate small RNAs (sRNAs) directly by controlling their transcription. However, alternative σ factors also can regulate sRNAs indirectly by controlling the expression of binding proteins and chaperones such as Hfq. In E. coli and other bacteria, Hfq has two major roles: (i) pairing sRNA to targeted mRNA, resulting in mRNA inhibition, and (ii) binding directly to mRNA to stabilize the transcript. Hfq has been identified in many bacterial pathogens, including Vibrio cholerae and Salmonella Typhimurium, and contributes to virulence. In L. monocytogenes, Hfq contributes to stress response in the presence of salt and ethanol, in addition to contributing to virulence in mice, although no virulence role is apparent in tissue culture models [70]. In E. coli, Hfq regulates the translation of general-stress σ factor RpoS mRNA (reviewed in Ref. [71]). By contrast, hfq is regulated by the general-stress factor σB in L. monocytogenes [70] and by RpoS in Legionella pneumophilia, the causative agent of legionellosis [72]. Taken together, these findings indicate that, in addition to regulating gene expression at the level of transcription, alternative σ factors seem to exert post-transcriptional control indirectly through sRNAs and Hfq.
Concluding remarks
Developing an improved understanding of the roles of alternative σ factors within complex regulatory networks under the myriad environmental conditions that L. monocytogenes might encounter in its natural life represents a considerable challenge. Clearly, we are only starting to understand the roles that alternative σ factors have in enabling L. monocytogenes to thrive as a ‘Jekyll and Hyde’ in both host and non-host environments. Initial microarray studies indicate that the roles of stress-response regulators, and of σB in particular, differ across L. monocytogenes strains [73], indicating that the diversity of stress-response systems, even within a given species, is considerable. This is supported further by the observation that one alternative σ factor (σC) seems to be present only in a phylogenetically distinct lineage of L. monocytogenes. Taken together, the work in this field predicts that the roles of alternative σ factors in bacterial stress response and virulence will differ among strains within the same species. Therefore, considerable work remains to explore crucial regulatory networks present across the phylogenetic diversity of L. monocytogenes.
Acknowledgments
Listeria monocytogenes research in the authors’ laboratories is supported by the National Institutes of Health (AI052151 to K.J.B.) and by the Cooperative State Research, Education, and Extension Service, National Research Initiative Competitive Grants Program (NRI Proposal # 2005–35201–15330 to K.J.B.) of the United States Department of Agriculture. The contents of this manuscript are solely the responsibility of the authors and do not necessarily represent the official views of the NIH or the USDA.
References
- 1.Gray MJ, et al. How the bacterial pathogen Listeria monocytogenes mediates the switch from environmental Dr. Jekyll to pathogenic Mr Hyde. Infect Immun. 2006;74:2505–2512. doi: 10.1128/IAI.74.5.2505-2512.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lecuit M. Human listeriosis and animal models. Microbes Infect. 2007;9:1216–1225. doi: 10.1016/j.micinf.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 3.Farber JM, Losos JZ. Listeria monocytogenes: a foodborne pathogen. CMAJ. 1988;138:413–418. [PMC free article] [PubMed] [Google Scholar]
- 4.Mead PS, et al. Food-related illness and death in the United States. Emerg Infect Dis. 1999;5:607–625. doi: 10.3201/eid0505.990502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vazquez-Boland JA, et al. Listeria pathogenesis and molecular virulence determinants. Clin Micro Rev. 2001;14:584–640. doi: 10.1128/CMR.14.3.584-640.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jensen VB, et al. Interactions of the invasive pathogens Salmonella Typhimurium, Listeria monocytogenes, and Shigella flexneri with M cells and murine Peyer’s patches. Infect Immun. 1998;66:3758–3766. doi: 10.1128/iai.66.8.3758-3766.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sauders BD, Wiedmann M. Ecology of Listeria species and L. monocytogenes in the natural environment. In: Ryser ET, Marth EH, editors. Listeria, Listeriosis, and Food Safety. 3. CRC Press; 2007. pp. 21–53. [Google Scholar]
- 8.Wiedmann M, et al. Ribotype diversity of Listeria monocytogenes strains associated with outbreaks of listeriosis in ruminants. J Clin Microbiol. 1996;34:1086–1090. doi: 10.1128/jcm.34.5.1086-1090.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gombas DE, et al. Survey of Listeria monocytogenes in ready-to-eat foods. J Food Prot. 2003;66:559–569. doi: 10.4315/0362-028x-66.4.559. [DOI] [PubMed] [Google Scholar]
- 10.Rocourt J, Buchrieser C. The genus Listeria and Listeria monocytogenes: phylogenetic position, taxonomy, and identification. In: Ryser ET, Marth EH, editors. Listeria, Listeriosis, and Food Safety. 3. CRC Press; 2007. pp. 1–20. [Google Scholar]
- 11.Wiedmann M, et al. General stress transcription factor σB and its role in acid resistance and virulence of Listeria monocytogenes. J Bacteriol. 1998;180:3650–3656. doi: 10.1128/jb.180.14.3650-3656.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Begley M, et al. Bile stress response in Listeria monocytogenes LO28: adaptation, cross-protection, and identification of genetic loci involved in bile resistance. Appl Environ Microbiol. 2002;8:6005–6012. doi: 10.1128/AEM.68.12.6005-6012.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karatzas KA, Bennik MH. Characterization of a Listeria monocytogenes Scott A isolate with high tolerance towards high hydrostatic pressure. Appl Environ Microbiol. 2002;68:3183–3189. doi: 10.1128/AEM.68.7.3183-3189.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Conte MP, et al. Effect of acid adaptation on the fate of Listeria monocytogenes in THP-1 human macrophages activated by γ interferon. Infect Immun. 2002;70:4369–4378. doi: 10.1128/IAI.70.8.4369-4378.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferreira A, et al. Role of Listeria monocytogenes σB in survival of lethal acidic conditions and in the acquired acid tolerance response. Appl Environ Microbiol. 2003;69:2692–2698. doi: 10.1128/AEM.69.5.2692-2698.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kazmierczak MJ, et al. Alternative σ factors and their roles in bacterial virulence. Micro Mol Biol Rev. 2005;69:527–543. doi: 10.1128/MMBR.69.4.527-543.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Magnusson LU, et al. ppGpp: a global regulator in Escherichia coli. Trends Microbiol. 2005;13:236–242. doi: 10.1016/j.tim.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 18.Costanzo A, et al. ppGpp and DksA likely regulate the activity of the extracytoplasmic stress factor σE in Escherichia coli by both direct and indirect mechanisms. Mol Microbiol. 2008;67:619–632. doi: 10.1111/j.1365-2958.2007.06072.x. [DOI] [PubMed] [Google Scholar]
- 19.Stragier P, Losick R. Molecular genetics of sporulation in Bacillus subtilis. Annu Rev Gen. 1996;30:297–341. doi: 10.1146/annurev.genet.30.1.297. [DOI] [PubMed] [Google Scholar]
- 20.Glaser P, et al. Comparative genomics of Listeria species. Science. 2001;294:849–852. doi: 10.1126/science.1063447. [DOI] [PubMed] [Google Scholar]
- 21.Xu J, et al. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science. 2003;299:2074–2076. doi: 10.1126/science.1080029. [DOI] [PubMed] [Google Scholar]
- 22.Kunst F, et al. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 23.Abram F, et al. Proteomic analyses of a Listeria monocytogenes mutant lacking σB identify new components of the σB regulon and highlight a role for σB in the utilization of glycerol. Appl Environ Microbiol. 2008;74:594–604. doi: 10.1128/AEM.01921-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kazmierczak MJ, et al. Listeria monocytogenes σB regulates stress response and virulence functions. J Bacteriol. 2003;185:5722–5734. doi: 10.1128/JB.185.19.5722-5734.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raengpradub S, et al. Comparative analysis of the σB-dependent stress responses in Listeria monocytogenes and Listeria innocua strains exposed to selected stress conditions. Appl Environ Microbiol. 2008;74:158–171. doi: 10.1128/AEM.00951-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hecker M, et al. SigB-dependent general stress response in Bacillus subtilis and related gram-positive bacteria. Annu Rev Microbiol. 2007;61:215–236. doi: 10.1146/annurev.micro.61.080706.093445. [DOI] [PubMed] [Google Scholar]
- 27.Petersohn A, et al. Global analysis of the general stress response of Bacillus subtilis. J Bacteriol. 2001;183:5617–5631. doi: 10.1128/JB.183.19.5617-5631.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bischoff M, et al. Microarray-based analysis of the Staphylococcus aureus σB regulon. J Bacteriol. 2004;186:4085–4099. doi: 10.1128/JB.186.13.4085-4099.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wemekamp-Kamphuis HH, et al. Identification of sigma factor σB-controlled genes and their impact on acid stress, high hydrostatic pressure, and freeze survival in Listeria monocytogenes EGD-e. Appl Environ Microbiol. 2004;70:3457–3466. doi: 10.1128/AEM.70.6.3457-3466.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chaturongakul S, Boor KJ. RsbT and RsbV contribute to σB-dependent survival under environmental, energy and intracellular stress conditions in Listeria monocytogenes. Appl Environ Microbiol. 2004;70:5349–5356. doi: 10.1128/AEM.70.9.5349-5356.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kazmierczak MJ, et al. Contributions of Listeria monocytogenes σB and PrfA to expression of virulence and stress response genes during extra- and intracellular growth. Microbiology. 2006;152:1827–1838. doi: 10.1099/mic.0.28758-0. [DOI] [PubMed] [Google Scholar]
- 32.Cotter PD, et al. Presence of GadD1 glutamate decarboxylase in selected Listeria monocytogenes strains is associated with an ability to grow at low pH. Appl Environ Microbiol. 2005;71:2832–2839. doi: 10.1128/AEM.71.6.2832-2839.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Becker LA, et al. Identification of the gene encoding the alternative σ factor σB from Listeria monocytogenes and its role in osmotolerance. J Bacteriol. 1998;180:4547–4554. doi: 10.1128/jb.180.17.4547-4554.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sue D, et al. σB-dependent expression patterns of compatible solute transporter genes opuCA and lmo1421 and a conjugated bile salt hydrolase bsh in Listeria monocytogenes. Microbiology. 2003;149:3247–3256. doi: 10.1099/mic.0.26526-0. [DOI] [PubMed] [Google Scholar]
- 35.Fraser KR, et al. The role of σB in regulating compatible solute uptake systems of Listeria monocytogenes: osmotic induction of opuC is σB–dependent. Appl Environ Microbiol. 2003;69:2015–2022. doi: 10.1128/AEM.69.4.2015-2022.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cetin MS, et al. Regulation of transcription of compatible solute transporters by the general stress sigma factor, σB, in Listeria monocytogenes. J Bacteriol. 2004;186:794–802. doi: 10.1128/JB.186.3.794-802.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moorhead SM, Dykes GA. The role of the sigB gene in the general stress response of Listeria monocytogenes varies between a strain of serotype 1/2a and a strain of serotype 4c. Curr Microbiol. 2003;46:461–466. doi: 10.1007/s00284-002-3867-6. [DOI] [PubMed] [Google Scholar]
- 38.Hain T, et al. Temporal transcriptomic analysis of the Listeria monocytogenes EGD-e σB regulon. BMC Microbiol. 2008;8:20. doi: 10.1186/1471-2180-8-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ferreira A, et al. Role of σB in heat, ethanol, acid, and oxidative stress resistance and during carbon starvation in Listeria monocytogenes. Appl Environ Microbiol. 2001;67:4454–4457. doi: 10.1128/AEM.67.10.4454-4457.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chaturongakul S, Boor KJ. σB activation under environmental and energy stress conditions in Listeria monocytogenes. Appl Environ Microbiol. 2006;72:5197–5203. doi: 10.1128/AEM.03058-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dussurget O, et al. Listeria monocytogenes bile salt hydrolase is a PrfA-regulated virulence factor involved in the intestinal and hepatic phases of listeriosis. Mol Microbiol. 2002;45:1095–1106. doi: 10.1046/j.1365-2958.2002.03080.x. [DOI] [PubMed] [Google Scholar]
- 42.Sleator RD, et al. A PrfA-regulated bile exclusion system (BilE) is a novel virulence factor in Listeria monocytogenes. Mol Microbiol. 2005;55:1183–1195. doi: 10.1111/j.1365-2958.2004.04454.x. [DOI] [PubMed] [Google Scholar]
- 43.McGann P, et al. The alternative sigma factor σB and the virulence gene regulator PrfA both regulate transcription of Listeria monocytogenes internalins. Appl Environ Microbiol. 2007;73:2919–2930. doi: 10.1128/AEM.02664-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nadon CA, et al. σB contributes to PrfA-mediated virulence in Listeria monocytogenes. Infect Immun. 2002;70:3948–3952. doi: 10.1128/IAI.70.7.3948-3952.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schwab U, et al. The Listeria monocytogenes prfAP2 promoter is regulated by σB in a growth phase dependent manner. FEMS Microbiol Lett. 2005;245:329–336. doi: 10.1016/j.femsle.2005.03.025. [DOI] [PubMed] [Google Scholar]
- 46.Sleator RD, et al. Analysis of the role of OpuC, an osmolyte transport system, in salt tolerance and virulence potential of Listeria monocytogenes. Appl Environ Microbiol. 2001;67:2692–2698. doi: 10.1128/AEM.67.6.2692-2698.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chatterjee SS, et al. Intracellular gene expression profile of Listeria monocytogenes. Infect Immun. 2006;74:1323–1338. doi: 10.1128/IAI.74.2.1323-1338.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Joseph B, et al. Identification of Listeria monocytogenes genes contributing to intracellular replication by expression profiling and mutant screening. J Bacteriol. 2006;188:556–568. doi: 10.1128/JB.188.2.556-568.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim H, et al. σB contributes to Listeria monocytogenes invasion by controlling expression of inlA and inlB. Microbiology. 2005;151:3215–3222. doi: 10.1099/mic.0.28070-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Garner MR, et al. σB contributes to Listeria monocytogenes gastrointestinal infection but not to systemic spread in the guinea pig infection model. Infect Immun. 2006;74:876–886. doi: 10.1128/IAI.74.2.876-886.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fouet A, et al. Characterization of the operon encoding the alternative σB factor from Bacillus anthracis and its role in virulence. J Bacteriol. 2000;182:5036–5045. doi: 10.1128/jb.182.18.5036-5045.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Britton RA, et al. Genome-wide analysis of the stationary-phase sigma factor (Sigma-H) regulon of Bacillus subtilis. J Bacteriol. 2002;184:4881–4890. doi: 10.1128/JB.184.17.4881-4890.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rea RB, et al. Disruption of putative regulatory loci in Listeria monocytogenes demonstrates a significant role for Fur and PerR in virulence. Infect Immun. 2004;72:717–727. doi: 10.1128/IAI.72.2.717-727.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Morikawa K, et al. A new staphylococcal σ factor in the conserved gene cassette: functional significance and implication for the evolutionary processes. Genes Cells. 2003;8:699–712. doi: 10.1046/j.1365-2443.2003.00668.x. [DOI] [PubMed] [Google Scholar]
- 55.Hadjifrangiskou M, et al. The alternative sigma factor σH is required for toxin gene expression by Bacillus anthracis. J Bacteriol. 2007;189:1874–1883. doi: 10.1128/JB.01333-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Robichon D, et al. The rpoN (σ-54) gene from Listeria monocytogenes is involved in resistance to mesentericin Y105, an antibacterial peptide from Leuconostoc mesenteroides. J Bacteriol. 1997;179:7591–7594. doi: 10.1128/jb.179.23.7591-7594.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu S, et al. Identification of Listeria monocytogenes genes expressed in response to growth at low temperature. Appl Environ Microbiol. 2002;68:1697–1705. doi: 10.1128/AEM.68.4.1697-1705.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chan YC, et al. Microarray-based characterization of the Listeria monocytogenes cold regulon in log- and stationary-phase cells. Appl Environ Microbiol. 2007;73:6484–6498. doi: 10.1128/AEM.00897-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dalet K, et al. A σ54-dependent PTS permease of the mannose family is responsible for sensitivity of Listeria monocytogenes to mesentericin Y105. Microbiology. 2001;147:3263–3269. doi: 10.1099/00221287-147-12-3263. [DOI] [PubMed] [Google Scholar]
- 60.Arous S, et al. Global analysis of gene expression in an rpoN mutant of Listeria monocytogenes. Microbiology. 2004;150:1581–1590. doi: 10.1099/mic.0.26860-0. [DOI] [PubMed] [Google Scholar]
- 61.Okada Y, et al. The sigma factor RpoN (σ54) is involved in osmotolerance in Listeria monocytogenes. FEMS Microbiol Lett. 2006;263:54–60. doi: 10.1111/j.1574-6968.2006.00405.x. [DOI] [PubMed] [Google Scholar]
- 62.Erickson JW, Gross CA. Identification of the σE subunit of Escherichia coli RNA polymerase: a second alternative σ factor involved in high-temperature gene expression. Genes Dev. 1989;3:1462–1471. doi: 10.1101/gad.3.9.1462. [DOI] [PubMed] [Google Scholar]
- 63.Wang QP, Kaguni JM. A novel sigma factor is involved in expression of the rpoH gene of Escherichia coli. J Bacteriol. 1989;171:4248–4253. doi: 10.1128/jb.171.8.4248-4253.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang C, et al. Functional consequences of genome evolution in Listeria monocytogenes: the lmo0423 and lmo0422 genes encode σC and LstR, a lineage II-specific heat shock system. J Bacteriol. 2005;187:7243–7253. doi: 10.1128/JB.187.21.7243-7253.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bennett HJ, et al. Characterization of relA and codY mutants of Listeria monocytogenes: identification of the CodY regulon and its role in virulence. Mol Microbiol. 2007;63:1453–1467. doi: 10.1111/j.1365-2958.2007.05597.x. [DOI] [PubMed] [Google Scholar]
- 66.Hu Y, et al. Phenotypic and transcriptomic analyses demonstrate interactions between the transcriptional regulator CtsR and σB in Listeria monocytogenes. Appl Environ Microbiol. 2007;73:7967–7980. doi: 10.1128/AEM.01085-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hu Y, et al. Transcriptomic and phenotypic analyses suggest a network between the transcriptional regulators HrcA and σB in Listeria monocytogenes. Appl Environ Microbiol. 2007;73:7981–7991. doi: 10.1128/AEM.01281-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kirstein J, et al. A tyrosine kinase and its activator control the activity of the CtsR heat shock repressor in B. subtilis. EMBO J. 2005;24:3435–3445. doi: 10.1038/sj.emboj.7600780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kruger E, et al. Clp-mediated proteolysis in gram-positive bacteria is autoregulated by the stability of repressor. EMBO J. 2001;20:852–863. doi: 10.1093/emboj/20.4.852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Christiansen JK, et al. The RNA-binding protein Hfq of Listeria monocytogenes: role in stress tolerance and virulence. J Bacteriol. 2004;186:3355–3362. doi: 10.1128/JB.186.11.3355-3362.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gottesman S. The small RNA regulators of Escherichia coli: roles and mechanisms. Annu Rev Microbiol. 2004;58:303–328. doi: 10.1146/annurev.micro.58.030603.123841. [DOI] [PubMed] [Google Scholar]
- 72.McNealy TL, et al. The Hfq homolog in Legionella pneumophila demonstrates regulation by LetA and RpoS and interacts with the global regulator CsrA. J Bacteriol. 2005;187:1527–1532. doi: 10.1128/JB.187.4.1527-1532.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Severino P, et al. Comparative transcriptome analysis of Listeria monocytogenes strains of the two major lineages reveals differences in virulence, cell wall, and stress response. Appl Environ Microbiol. 2007;73:6078–6088. doi: 10.1128/AEM.02730-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Becker LA, et al. Role of σB in adaptation of Listeria monocytogenes to growth at low temperature. J Bacteriol. 2000;182:7083–7087. doi: 10.1128/jb.182.24.7083-7087.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Moorhead SM, Dykes GA. Influence of sigB gene on the cold stress survival and subsequent recovery of two Listeria monocytogenes serotypes. Int J Food Microbiol. 2004;91:63–72. doi: 10.1016/S0168-1605(03)00332-5. [DOI] [PubMed] [Google Scholar]
- 76.Chan YC, et al. σB-dependent and σB-independent mechanisms contribute to transcription of Listeria monocytogenes cold stress genes during cold shock and cold growth. Appl Environ Microbiol. 2007;73:6019–6029. doi: 10.1128/AEM.00714-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sue D, et al. σB-dependent gene expression and induction in Listeria monocytogenes during osmotic and acid stress conditions simulating the intestinal environment. Microbiology. 2004;150:3843–3855. doi: 10.1099/mic.0.27257-0. [DOI] [PubMed] [Google Scholar]
- 78.Johansson J, et al. An RNA thermosensor controls expression of virulence genes in Listeria monocytogenes. Cell. 2002;110:551–561. doi: 10.1016/s0092-8674(02)00905-4. [DOI] [PubMed] [Google Scholar]
- 79.Freitag NE, Portnoy DA. Dual promoters of the Listeria monocytogenes prfA transcriptional activator appear essential in vitro but are redundant in vivo. Mol Microbiol. 1994;12:845–853. doi: 10.1111/j.1365-2958.1994.tb01070.x. [DOI] [PubMed] [Google Scholar]
- 80.Bischoff M, et al. Influence of a functional sigB operon on the global regulators sar and agr in Staphylococcus aureus. J Bacteriol. 2001;183:5171–5179. doi: 10.1128/JB.183.17.5171-5179.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Arvidson S, Tegmark K. Regulation of virulence determinants in Staphylococcus aureus. Int J Med Microbiol. 2001;291:159–170. doi: 10.1078/1438-4221-00112. [DOI] [PubMed] [Google Scholar]
- 82.Milohanic E, et al. Transcriptome analysis of Listeria monocytogenes identifies three groups of genes differently regulated by PrfA. Mol Microbiol. 2003;47:1613–1625. doi: 10.1046/j.1365-2958.2003.03413.x. [DOI] [PubMed] [Google Scholar]
- 83.Mogk A, et al. The GroE chaperonin machine is a major modulator of the CIRCE heat shock regulon of Bacillus subtilis. EMBO J. 1997;16:4579–4590. doi: 10.1093/emboj/16.15.4579. [DOI] [PMC free article] [PubMed] [Google Scholar]