Abstract
The transcriptional coactivator complex Mediator facilitates transcription of nuclear hormone receptors and other transcription factors. We have previously isolated the Mediator complex from primary keratinocytes as the vitamin D receptor interacting protein complex. We identified a role for Mediator in keratinocyte proliferation and differentiation in cultured keratinocytes. Here, we investigated the in vivo role of Mediator by generating conditional null mice, where a critical subunit of the Mediator complex, MED1, is deleted from their keratinocytes. The MED1 ablation resulted in aberrant hair differentiation and cycling leading to hair loss. During the first hair follicle cycle, MED1 deletion resulted in a rapid regression of the hair follicles. Hair differentiation was reduced, and β-catenin/vitamin D receptor (VDR) regulated gene expression was dramatically decreased. In the subsequent adult hair cycle, MED1 ablation activated the initiation of hair follicle cycling. Shh signaling was increased, but terminal differentiation was not sufficient. Deletion of MED1 also caused hyper-proliferation of interfollicular epidermal keratinocytes, and increased the expression of epidermal differentiation markers. These results indicate that MED1 plays a critical role in regulating hair/epidermal proliferation and differentiation.
Introduction
The transcription coactivator Mediator is a multi-protein complex that activates transcription of a number of nuclear hormone receptors and transcription factors (Kornberg, 2005; Malik and Roeder, 2005) (Blazek et al., 2005; Bourbon et al., 2004; Kornberg, 2005). We have previously identified Mediator as a binding protein complex for the vitamin D receptor (VDR), initially referred to as DRIP (VDR interacting protein) (Oda et al., 2003; Rachez et al., 1999). This complex was isolated from primary keratinocytes using GST-VDR affinity beads, and its multiple subunits were identified by mass spectrometry (Oda et al., 2010; Oda et al., 2003). The complex contained a critical subunit MED1 (also named DRIP205) (Oda et al., 2007; Rachez et al., 1999; Yuan et al., 1998; Zhu et al., 1997))(Viswakarma et al., 2010) that directly binds to VDR to anchor the rest of the complex to facilitate transcriptional activation. Mediator also activates other nuclear hormone receptors or transcription factors such as peroxisome proliferator activated receptor (PPAR) (Viswakarma et al., 2010), thyroid hormone receptor (TR) (Ito and Roeder, 2001; Ito et al., 2000). Mediator also activates other transcription factors including the GATA family (Crawford et al., 2002; Stumpf et al., 2006) and C/EBPβ.
Mediator has an important role in specific biological processes. We have previously shown that MED1 regulates keratinocyte proliferation and differentiation using cultured keratinocytes (Hawker et al., 2007; Oda et al., 2010). Silencing of MED1 resulted in hyper-proliferation and decreased calcium induced keratinocyte differentiation (Oda et al., 2010). In contrast, steroid receptor coactivator 3 (SRC3), a member of the p160/SRC family does not affect keratinocyte proliferation but participated in the terminal differentiation process (Oda et al., 2009). Based on these observations, we hypothesized that Mediator has a critical role in epidermal homeostasis through temporal and spatial regulation to control keratinocyte proliferation and differentiation.
Here, we investigated the in vivo role of the Mediator in epidermal homeostasis. We generated a mouse model in which a critical subunit of the Mediator complex, MED1, is deleted from keratinocytes. Previously, MED1 conditional null mice revealed a role for MED1 in erythroid differentiation (Stumpf et al., 2010), liver degeneration (Matsumoto et al., 2007), adipogenesis (Ge et al., 2008; Ge et al., 2002), mammary grand development (Jia et al., 2005), liver tumorigenesis (Matsumoto et al.), and glucose and energy metabolism (Chen et al.). However, the role of MED1 in skin has not previously been investigated. The skin contains different populations of keratinocytes in 1) interfollicular epidermis (IFE) and 2) hair follicle (HF). Morphogenesis and maintenance of IFE and HF are differentially regulated. The epidermis is maintained by proliferation of basal cells and their differentiation into suprabasal cells. The HF undergoes a perpetual cycle of growth and regression. In the mouse, all the HFs synchronously enter a cycle of growth (anagen) after birth, and go through a regression phase (catagen) that leads to the quiescent stage (telogen). In mice with melanin in the hair shaft, melanogenesis is coupled to anagen (Slominski and Paus, 1993; Slominski et al., 2005). The initial cycle extends from the late stage embryo through P21 (morphogenic hair cycle) (Paus and Foitzik, 2004). Adult hair cycles involve longer telogens, resulting in less synchronous cycling (post-morphogenic cycle) (Paus and Foitzik, 2004).
In vivo the role of MED1 was investigated by using a mouse model, in which the expression of the MED1 is deleted from their keratinocytes. MED1 deletion resulted in abnormalities in hair differentiation and cycling leading to hair loss. The mRNA expression of various components of signaling pathways involved in hair progression and differentiation was also evaluated. Wnt/β-catenin signaling has a major role in HF morphogenesis and regeneration (Blanpain and Fuchs, 2006; Huelsken et al., 2001). VDR induces transcription of hair differentiation genes through the Wnt/β-catenin signaling pathway (Palmer et al., 2008). Several genes including peptidyl arginine deiminase 1 (PADI1) and 3 (PADI3), tubulin Tubb3, calcium binding protein S100A3, homeo box gene Dlx3 were identified as such targets (Palmer et al., 2008). The hedgehog (Hh) signaling pathway regulates keratinocyte proliferation and promotes HF progression during the adult hair cycle as well as morphogenic cycle (Blanpain and Fuchs, 2006). Hh pathway components Patch 1 and Patch 2 and Hh targets of oncogene glioma-associated oncogene homolog 1 (Gli 1) are regulated by VDR (Oda et al., 2010; Palmer et al., 2008; Teichert et al., 2010). Bone morphogenetic protein (BMP) signaling through BMP2, BMP4 and the BMP receptor 1a (BMPr1a) also has a critical role in hair differentiation (Botchkarev and Sharov, 2004; Sharov et al., 2006). We explored several of these signaling pathways as to their regulation by MED1 in the current study.
Ablation of MED1 results in disrupted or aberrant hair differentiation and cycling leading to hair loss. On the other hand, markers of interfollicular epidermal differentiation increased. Our results indicate that the transcriptional coactivator MED1 has a unique function to maintain epidermal homeostasis in vivo.
Results
Keratinocyte specific MED1 knockout mice
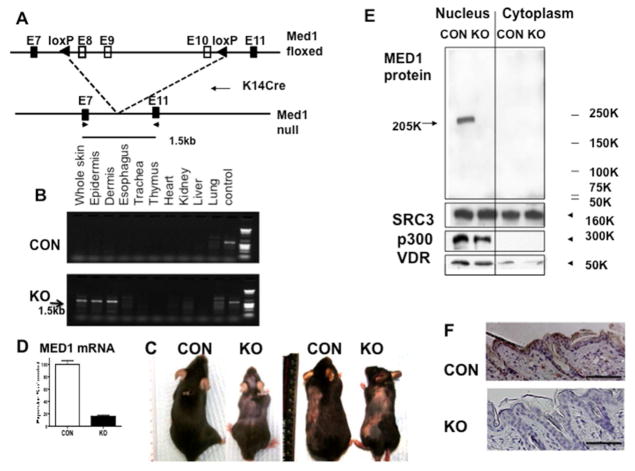
We generated conditional MED1 null mice using a Cre-lox system to specifically delete MED1 expression in keratinocytes. Floxed MED1 mice, in which two lox P sites were inserted into the introns upstream and downstream of exons 8, 9 and 10 of the MED1 gene (C57/BL6 background) (Jia et al., 2004) were bred with transgenic mice expressing Cre recombinase under the control of the Keratin 14 (K14) promoter (C57/BL6 background, Jackson Lab) (Fig. 1A). Homozygous floxed mice with the Cre transgene (KO) were compared to control littermates that had floxed MED1 alleles but no Cre (CON). The recombination was detected in the whole skin as well as in epidermis and dermis containing the hair follicles (Fig. 1B). However, excision was not detected in other tissues except small amounts in lung probably in bronchial epithelia expressing K14 (Fig. 1B), confirming tissue specific excision. A marked reduction in MED1 mRNA was seen in the epidermis of null mice (Fig. 1D). Deletion of MED1 protein was shown by western analysis. A 200 kDa MED1 protein band was absent in keratinocytes separated from the null skin (Fig. 1E KO). As MED1 is a nuclear protein, it was detected in the nuclear fraction but not in the cytoplasm (Fig. 1E). A different coactivator, SRC3, also involved in transcriptional regulation, was equally detected in both control and null keratinocytes. SRC3 is localized in both the nucleus and cytoplasm. The coactivator p300, VDR and SRC3 were also equally detected (Fig. 1E). These results were confirmed by immuno-histochemistry. As shown in Fig. 1F, MED1 was absent in keratinocytes in the null skin. In contrast, it was detected in keratinocytes of both IFE and HF the control skin at P17. These results demonstrate that both transcript and protein of MED1 were deleted from keratinocytes.
Fig. 1. Generation of conditional MED1 null mice.
(A) The gene-targeting strategy to delete the MED1 from keratinocytes by using Cre-loxP system is illustrated. (B) Tissue specific excision of exons (8–10) was detected by PCR amplifying ~1.5kb cDNA fragments (arrow) from genomic DNA isolated from various tissues of null mice (KO) and control littermate mice (CON). (C) The MED1 null mice showed hair loss at 6 months. Right two panels show mice in which the left side of the back is shaved to grossly assess anagen in the same mouse in which the quality of the hair coat on the right (unshaved) side is shown. (D) The mRNA expression of MED1 was reduced in the null skin. (E) A 205 kDa MED1 protein was absent from the nuclear extracts of null keratinocytes compared to control keratinocytes (taken from 3 mice each) (F) Decreased expression of the MED1 in the skin is shown by immuno-histochemistry at P17 (n=3, representative image). Bar=200 μm
Disruption of the MED1 gene resulted in abnormalities in hair differentiation and cycling
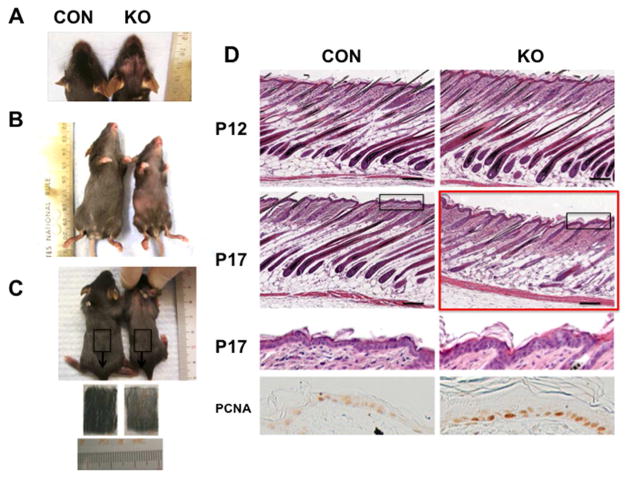
The MED1 floxed mice with or without Cre transgene were born in the expected Mendelian ratios. The growth of the null mice was slightly retarded (10–30% smaller than control). The MED1 null mice did not show abnormalities in the appearance of the skin and hair at least through P12 (anagen). However, changes in their hair coats were first recognized at P17, at which point hair loss was visually observed in the skin of head and neck (Fig. 2A). The hair density decreased in ventral skin (Fig. 2B), and in dorsal skin as shown by hairs remaining on the depilation tapes after hairs were plucked (Fig. 2C). Hair loss was relatively mild, but changes in hair coats were sufficient to distinguish them before PCR genotyping. Hair loss increased gradually with age (Fig. 1C). As these animals were generated in the C57/BL6 background, skin pigmentation and epidermal thickness were correlated to hair follicle cycling and used as a tool to stage their hair cycle (Slominski and Paus, 1993). Histological examination showed that null skin was normal in neonates (P3) (data not shown) to first anagen (P12) (Fig. 2D P12). The control skin underwent catagen beginning at the head and moving caudally. Even by P17, the lower back showed late anagen morphology (Fig. 2D P17 CON). In contrast, most of the null skin was pale pink macroscopically, and the hair follicles were in an advanced stage of the regression (Fig. 2D P17 KO). The IFE was hyperplastic (Fig. 2D P17 KO). Telogen at P21 was similar in KO and controls (data not shown).
Fig. 2. Deletion of MED1 resulted in hair loss.
(A) Hair loss in the head and neck of MED1 null mice (KO) and controls (CON) at P17 is depicted. (B) Hair profiles of KO and CON in ventral skins at 4 wk are shown. (C) Decreased hair density in dorsal skins is shown. (D) Histological assessment of the null skin of CON and KO at P12 and P17 (HE staining) is depicted. Mildly thickened epidermis (enlarged images of boxed area of P17) and hyper-proliferation of IEF at P17 (PCNA staining, bottom panels) are shown. Greater regression of HF in KO at P17 is shown (red box). Representative images from 3 KO and 3 CON are shown. Reproducibility was confirmed in two litters of null mice. Bar=200 μm
Deletion of MED1 caused rapid regression of HFs in the morphogenic hair cycle
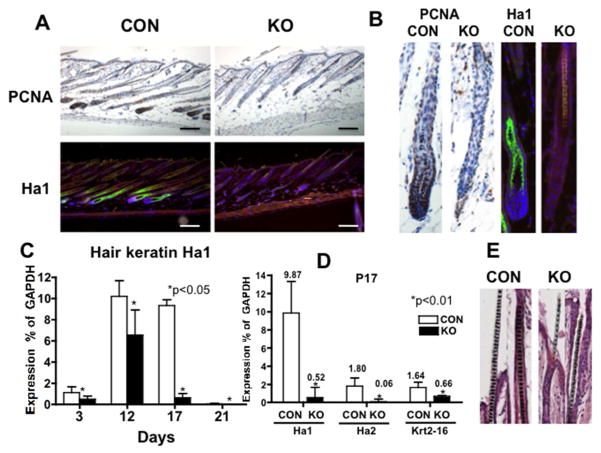
Proliferation and hair follicle differentiation were assessed at P17 (Fig. 3). The control skin was in late anagen at P17 but HFs in KO were regressing (Fig. 3A, enlarged images are shown in B). Hair bulbs containing PCNA positive cells were already lost (Fig. 3A, B PCNA staining in brown), and hair shafts were thinner (Fig. 3E) in the KO. Hair differentiation was assessed by hair keratin Ha1/KRT31 (Ha1). The Ha1 was highly expressed in the inner root sheath (IRS) in the control skin (Fig. 3A, B Ha1staining in green), but absent in the null skin (Fig. 3A, B). Hair differentiation was also evaluated by measuring the mRNA levels of hair keratins during synchronous hair follicle cycling (P3 to P21) by isolating RNA from whole skin. Ha1 expression increased from P3 to P12 and remained high to P17, then sharply decreased by P21 (Fig. 3C open bar) paralleling the activation and regression during hair cycling in the controls. However, MED1 null skin showed lower Ha1 levels during all the stages, most significantly at P17 (Fig. 3C closed bar). Expression of other hair keratins, Ha2 and Krt2-16, were also decreased significantly at P17 (Fig. 3D).
Fig. 3. Ablation of MED1 decreased hair differentiation at P17.
(A) PCNA staining of the lower dorsal skin from null mice (KO) and littermate controls (CON) at P17 is shown: brown PCNA signals with blue counter staining. Hair differentiation is illustrated by Ha1 (green) with DAPI (blue) to mark the nuclei. Bar = 200 μm (B) Representative images from 3 mice are shown. (C) The mRNA levels of Ha1 in P3 to P21 are plotted as % expression of GAPDH of KO (closed bar) and CON (open bar). (D) The mRNA expression of other hair keratins at P17 is shown. Statistical significance evaluated by t-test (*p values <0.01 or <0.05) is shown with asterisks (n=3). The experiments were repeated to confirm reproducibility with two litters.
Deletion of MED1 increased epidermal differentiation markers
The expression of the early epidermal differentiation marker keratin 1 (K1), and late differentiation marker filaggrin (FLG) was evaluated at P17. The protein levels of both K1 (Fig. 4A upper panels) and FLG (lower panels) increased in the null skin. The mRNA expression of K1 and FLG was highest at P3 and markedly decreased at P17 in the control skin (Fig. 4B open bar). In contrast, their expressions were higher in KO compared to control at P3, P17 (Fig. 4B) and 10 weeks (Fig. 4C).
Fig. 4. Epidermal differentiation and proliferation increased upon the deletion of MED1.
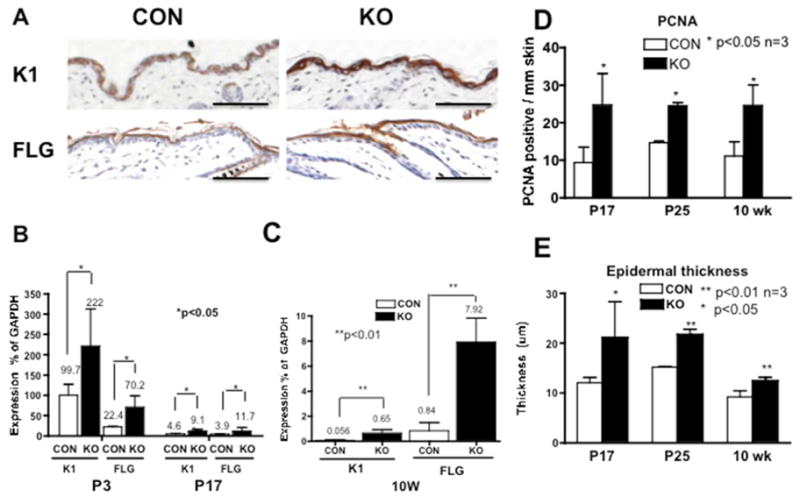
(A) Expression of K1 (upper panels) and FLG (lower panels) in the null skin (KO) and littermate control (CON) at P17 is shown. Bar=200 μm. (B, C) The relative expression of K1 and FLG (% expression of GAPDH) was evaluated in the null skin (closed bar) and the controls (open bar) at P3, P17 (B) and at 10 weeks (C). Proliferation of IFE was quantitatively evaluated by counting PCNA positive cells per mm skin (D) and by epidermal thickness (E) at P17 (Fig. 2D), P25 (Fig. 5B), and at 10 weeks (Fig. 5H). Details of the image analysis are shown in Supplement Table SIII and Fig. S1. Statistical significance was evaluated by t-test (*p values <0.01 or <0.05, n=3).
Ablation of MED1 increased keratinocyte proliferation
Keratinocyte proliferation in IFE was evaluated by PCNA staining and epidermal thickness. There is not a significant difference between null and control mice in the skin of neonates to P12 (data not shown). However, the null skin showed hyperplasia of IFE at P17 (Fig. 2D), P25 (Fig. 5B, C) and at 10 weeks (Fig. 5G, H). These differences were quantified and shown to be significant at P17, P25 and 10 weeks (Fig 4D, E, Supplemental Table SIII, Fig. S1).
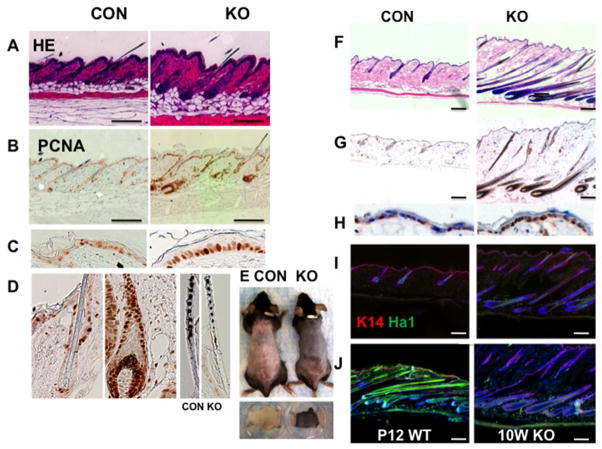
Fig. 5. MED1 ablation resulted in abnormal HF anagen activation in the adult hair follicle cycle.
(A) Histological analyses at P25 are shown. (B–D) Brown PCNA staining with no counter staining is shown. Enlarged images of IFE (C) and HF (D) are shown. (E) Pigmentation of the null skin at 9 weeks in mice (E, upper panels, after hair was shaved) and in excised skins (E, lower panels) is shown. (F–J) Histological analyses were performed at 10 weeks: (F) HE staining (G, H), brown PCNA with blue counter staining, and enlarged images of IFE (H), (I, J) Ha1 staining (green) with counter-staining of K14 (red) and DAPI (blue), (J) Ha1 staining of KO at 10 weeks (J, right 10W KO) and CON at P12 (J, left P12 WT). These figures are representative images of multiple analyses (n=2–3). Bar=200 μm
The deletion of MED1 activated HF progression during the adult hair cycle
At P25, the control skin was mostly in telogen (Fig. 5D CON). In contrast, large areas of the null skin were now in anagen characterized by thicker epidermis (Fig. 5 A KO) and numerous PCNA positive cells in enlarged hair bulbs and the outer root sheath (ORS) (5D KO). However, hair shafts were thinner in KO (Fig. 5D left two panels). At 6 weeks, control skin remained in prolonged telogen, and by 9–10 weeks, the entire back of control mice was pale pink (when shaved) indicating telogen (Fig. 5E). In contrast, large areas of null skin were pigmented with epidermis thickening, indicating anagen stage VI (Fig. 5E, F). At 6 month, the null skin was still in anagen even though hair loss progressed (Fig. 1C left two panels) shown by pigmented anagen skin after hairs were shaved (1C right two panels). These changes were consistent within a litter (at least 3 mice per group) and between litters (2 or more at each time point).
The MED1 ablation resulted in abnormal hair follicle differentiation
Hair differentiation was assessed by Ha1 expression. The control skin did not express Ha1 in the telogen HF at 10 week (Fig. 5 I CON). In contrast, Ha1 was expressed in the IRS of the null skin (Fig. 5 I KO). However, Ha1 levels at 10 weeks (10W KO) when compared to those in normal anagen (P12 CON) indicated a reduction in Ha1 expression in the null HF at 10 weeks (Fig. 5J). These results suggest that MED1 ablation activates HF progression, but terminal differentiation is defective.
MED1 deletion did not affect calcium metabolism
We analyzed serum levels of calcium because of concern for hypocalcemia seen in VDR null mice, which may secondarily affect skin physiology. Serum levels of calcium were normal in MED1 null mice fed standard mouse chow (Supplemental Table SI).
Differential regulation of signaling pathways involved in HF progression and differentiation
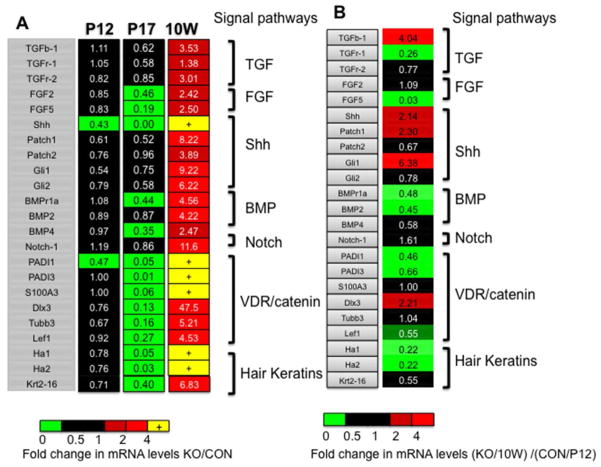
RNA was isolated from whole skin of null and littermate control mice. The mRNA levels of genes known to be involved in HF progression and differentiation that we examined are listed in Fig. 6. Their expression was measured at i) P12 when the null skin was in normal anagen; ii) P17 when the null HF was rapidly regressing; iii) 10 weeks when the HF was abnormally activated. Fold changes of null skin (KO) compared to control skin (CON) were calculated (Fig. 6A). At P12, the expression of most genes was not significantly changed (Fig. 6A P12 shown as black). At P17, the expression of hair differentiation genes such as PADI1, PADI3, S100A3, Tubb3, Dlx3 and hair keratins was dramatically decreased (Fig. 6A P17 green). In contrast, the exression of all the genes examined increased at 10 weeks. Most significant increases were in Hh signaling (Fig. 6A 10W, light red). At 10 weeks, Shh, PADI1, PADI3, S100A3, and hair keratins were undetectable in CON, but increased in the KO (Fig 6A 10W, yellow (+) indicating that the mRNA levels were undetectable in CON but increased in KO preventing a calculation of the ratio). The mRNA levels of these genes in KO at 10 week (10W KO) were then compared to those in normal anagen (P12 CON) (Fig. 6B). The expression of Shh, Patch1, and Gli1 was higher in the 10 week anagen of the KO compared to the P12 anagen of CON (Fig. 6B). However, hair keratins were lower in the 10wk KO anagen than in the P12 CON anagen suggesting incomplete hair differentiation (Fig. 6B). The BMP components, Lef1, PADI1, PADI3 were also lower. These results indicate that deletion of MED1 accelerates HF regression at P17, with a reduction in the expression of hair keratins and VDR/β-catenin target genes, whereas at 10 weeks, HF progression is activated and Hh pathway components are elevated but hair differentiation is incomplete.
Fig. 6. Signaling pathways impacted by the deletion of MED1.
(A) Expression profiling of signaling pathways at different stages (P12, P17 and 10 week (10W)) was performed. The mRNA levels of the genes listed relative to control GAPDH were measured by RT-QPCR. Fold changes of MED1 null skin compared to littermate control skin are shown (n=3–6). Statistically significant increases (red and yellow) and decreases (green), or no difference (black) are shown. (B) Gene expression comparing anagen in the KO at 10 weeks to normal anagen at P12 are shown. Fold changes of the KO (10 weeks) compared to the CON at P12 are shown.
Discussion
MED1 maintains hair differentiation through VDR/β-catenin during the morphogenic hair cycle
We suggest that MED1 may be involved in VDR regulated β-catenin signaling in hair follicle differentiation. Hair follicle genes such as PADI and S100A3, which decreased dramatically in MED1 null skin at P17, are regulated by VDR through vitamin D response elements (VDRE) adjacent or apart from Lef1/TCF sites in their promoters (Palmer et al., 2008). Chromatin immuno-precipitation showed that β-catenin is specifically recruited to the VDRE sites in the promoters of these genes, where VDR and MED1 are expected to mediate transcription (Palmer et al., 2008). These observations suggest that MED1 may cooperatively stimulate VDR transcription with β-catenin. VDR mutational analysis showed that VDR binds to both MED1 and β-catenin through activation domain 2 (AF-2), although different sites or structural requirements may be involved (Shah et al., 2006). Therefore, we postulate that MED1 may regulate hair follicle gene transcription through VDR and β-catenin. However, the precise mechanism(s) by which MED1 regulates hair follicle genes remains to be elucidated. The mode of action of MED1 need not be limited to its coactivator role for VDR. Although the timing at which HF cycling is initially disrupted in VDR null mice is comparable (P17) to that of MED1 null mice, the MED1 null mice do not generate abnormal hair follicle structures such as FLG expressing uticles and oil retaining dermal cysts seen in the VDR null mice (Bikle et al., 2006). Thus, Mediator may partially work through VDR, but it is still possible that other nuclear receptors such as TR, PPAR, RAR and other transcription factors may be involved in MED1 regulation.
MED1 restrains the onset of anagen but promotes terminal differentiation during adult hair follicle cycling
Deletion of MED1 resulted in accelerated and sustained activation of anagen in the post-morphogenic cycle. This differs from its action in the morphogenic cycle. Several mechanisms may be involved here. MED1 may suppress Hh signaling to maintain keratinocytes in quiescence and keep HFs in telogen. Our previous in vitro studies indicated that MED1 suppressed Gli1 expression, and silencing of MED1 caused hyper-proliferation of keratinocytes (Oda et al., 2010). These results are consistent with our in vivo results. Despite the accelerated cycling of HF into anagen, HF differentiation was incomplete in the null skin. In addition this failure may be due to a role of MED1 in promoting terminal differentiation through BMP signaling. BMP signaling is critical for hair shaft differentiation, and disturbed BMP signaling also causes hair loss with premature anagen similar to our MED1 null model (Kobielak et al., 2003; Segrelles et al., 2008). These potential mechanisms remain under investigation.
MED1 suppresses interfollicular epidermal differentiation and proliferation
Deletion of MED1 resulted in increased expression of epidermal differentiation markers of K1 and FLG. These results are different from our previous in vitro data that showed that MED1 mediates calcium induced keratinocyte differentiation (Oda et al., 2010). However, the in vivo situation is clearly different. The skin possesses stem cells that can differentiate in three directions: HF, IFE, and sebaceous gland. Ablation of MED1 appears to shift differentiation away from the HF and toward the IFE lineage. A similar shift is observed in VDR null mice (Bikle et al., 2006). The epidermal marker FLG is over-expressed in hair follicles (utricles) as is caspase 14 in the IFE in VDR null skin (Bikle et al., 2006). On the other hand, deletion of MED1 caused hyper-proliferation of keratinocytes in the epidermis, consistent to our in vitro results (Oda et al., 2010) and similar to that seen in the VDR null mouse skin. Increased proliferation of keratinocytes in IFE in the MED1 null skin may predispose to epidermal tumorigenesis as seen in the VDR null mouse model (Ellison et al., 2008; Teichert et al., 2011; Zinser et al., 2002).
MED1 is involved in cell type or stage specific regulation in keratinocyte proliferation and differentiation
Our studies reveal the temporal and spatial specific role of MED1, supporting our previous hypothesis that MED1 is a key molecule mediating cell or stage specific transcriptional regulation and control of proliferation and differentiation (Oda et al., 2003). Since then, other studies also supported our concept in various differentiation systems. The MED1 regulates myotube differentiation through specific gene regulation at specific cells (Deato et al., 2008). It controls erythroid differentiation but not lymphoid differentiation (Stumpf et al., 2010). Transgenic mutant target-in MED1 (LxxLL) mice showed its role in luminal cell differentiation but not in other mammary epithelial cells (Jiang et al., 2010). These phenotypes were not necessarily parallel to those of partner molecules, indicating that Mediator has unique multiple functions. Further research will likely identify other genes and signaling pathways that it regulates.
Here, we demonstrated a critical role for MED1 in hair/epidermal proliferation, differentiation and cycling using an in vivo mouse model. These studies further increase our understanding of the function of MED1 and provide a model to identify potential therapeutics to treat diseases such as alopecia where hair development is disturbed, or hyper-proliferative skin diseases where proliferation is perturbed.
Materials and Methods
Generation and genotyping of conditional MED1 null nice
To delete the expression of MED1 from keratinocytes, floxed MED1 null mice (originally named as floxed PBP) were mated with transgenic mice expressing Cre recombinase under the control of the K14 promoter (The Jackson Laboratory, C57/BL6 background).
Histological analysis and K1/FLG immunostaining
The lower portion of the dorsal skin was excised from the MED1 null and their littermate control mice. K1 and FLG were detected by immuno-staining using primary antibodies against K1 (1:1000) and FLG (1:500), and signals were detected using ImmunoCruz rabbit™ LSAB Staining System (sc-2051, Santa Cruz Biotechnology).
Evaluation of keratinocyte proliferation
Proliferative keratinocytes were quantitatively evaluated by both PCNA staining and epidermal thickness using the image software Bioquant. The dorsal skin sections of MED1 null and littermate control mice (n=3) were stained using the PCNA staining kit (93–1143, Invitrogen, Carlsbad, CA) at various stages.
Hair keratin Immunofluorescence
Paraffin embedded skin sections were incubated with primary antibodies against Ha1 (Progen, made in guinea pig 1:1000) and K14 (Covance, made in rabbit 1:4000), and subsequently incubated with secondary antibodies, Alexa 594 (red) conjugated anti rabbit IgG and Alexa 488 (green) conjugated anti guinea pig IgG (10 mg/ml), and counterstained with DAPI.
Measurement of mRNA expression by RT-QPCR
Total RNA was isolated from whole skin excised from the lower dorsal part of the null mice skin and from equivalent regions of skin from their littermate controls. cDNA synthesis and real time PCR were performed as described in the supplement. Primer sequences for QPCR are shown in supplement Table SII.
Details of these methods are provided in the supplemental information.
Supplementary Material
Acknowledgments
We thank Sandra Huling at Imaging Core Facility of Northern California Institute of Research Education for histological analyses, and Chak K. Fong for image analysis and technical assistance. We are also grateful to Chia-Ling Tu, Zhongjian Xie, Yoshikazu Uchida, and Peter Elias for their support and discussion. This work was supported by NIH grants R01 AR050023 (D. D. B) and DK083163 (J.K.R.).
Abbreviations
- MED
Mediator
- DRIP
Vitamin D receptor interacting protein
- VDR
vitamin D receptor
- SRC
steroid receptor coactivator
- K1
keratin 1
- K10
keratin 10
- K14
keratin 14
- FLG
filaggrin
- Ha1
hair keratin Ha1/KRT31
- PADI
peptidyl arginine deiminase
- Hh
hedgehog
- Gli 1
glioma-associated oncogene homolog
- HF
hair follicles
- IFE
interfollicular epidermis
- IRS
inner root sheath
- ORS
outer root sheath
- BMP
bone morphogenetic protein
- TGF
transforming growth factor
- FGF
fibroblast growth factor
- P
postnatal
- PPAR
peroxisome proliferator activated receptor
- TR
thyroid receptor
References
- Bikle DD, Elalieh H, Chang S, Xie Z, Sundberg JP. Development and progression of alopecia in the vitamin D receptor null mouse. J Cell Physiol. 2006;207:340–353. doi: 10.1002/jcp.20578. [DOI] [PubMed] [Google Scholar]
- Blanpain C, Fuchs E. Epidermal stem cells of the skin. Annu Rev Cell Dev Biol. 2006;22:339–373. doi: 10.1146/annurev.cellbio.22.010305.104357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazek E, Mittler G, Meisterernst M. The mediator of RNA polymerase II. Chromosoma. 2005;113:399–408. doi: 10.1007/s00412-005-0329-5. [DOI] [PubMed] [Google Scholar]
- Botchkarev VA, Sharov AA. BMP signaling in the control of skin development and hair follicle growth. Differentiation. 2004;72:512–526. doi: 10.1111/j.1432-0436.2004.07209005.x. [DOI] [PubMed] [Google Scholar]
- Bourbon HM, Aguilera A, Ansari AZ, Asturias FJ, Berk AJ, Bjorklund S, et al. A unified nomenclature for protein subunits of mediator complexes linking transcriptional regulators to RNA polymerase II. Molecular cell. 2004;14:553–557. doi: 10.1016/j.molcel.2004.05.011. [DOI] [PubMed] [Google Scholar]
- Chen W, Zhang X, Birsoy K, Roeder RG. A muscle-specific knockout implicates nuclear receptor coactivator MED1 in the regulation of glucose and energy metabolism. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:10196–10201. doi: 10.1073/pnas.1005626107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford SE, Qi C, Misra P, Stellmach V, Rao MS, Engel JD, et al. Defects of the heart, eye, and megakaryocytes in peroxisome proliferator activator receptor-binding protein (PBP) null embryos implicate GATA family of transcription factors. The Journal of biological chemistry. 2002;277:3585–3592. doi: 10.1074/jbc.M107995200. [DOI] [PubMed] [Google Scholar]
- Deato MD, Marr MT, Sottero T, Inouye C, Hu P, Tjian R. MyoD targets TAF3/TRF3 to activate myogenin transcription. Molecular cell. 2008;32:96–105. doi: 10.1016/j.molcel.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison TI, Smith MK, Gilliam AC, MacDonald PN. Inactivation of the vitamin D receptor enhances susceptibility of murine skin to UV-induced tumorigenesis. The Journal of investigative dermatology. 2008;128:2508–2517. doi: 10.1038/jid.2008.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge K, Cho YW, Guo H, Hong TB, Guermah M, Ito M, et al. Alternative mechanisms by which mediator subunit MED1/TRAP220 regulates peroxisome proliferator-activated receptor gamma-stimulated adipogenesis and target gene expression. Molecular and cellular biology. 2008;28:1081–1091. doi: 10.1128/MCB.00967-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge K, Guermah M, Yuan CX, Ito M, Wallberg AE, Spiegelman BM, et al. Transcription coactivator TRAP220 is required for PPAR gamma 2-stimulated adipogenesis. Nature. 2002;417:563–567. doi: 10.1038/417563a. [DOI] [PubMed] [Google Scholar]
- Hawker NP, Pennypacker SD, Chang SM, Bikle DD. Regulation of human epidermal keratinocyte differentiation by the vitamin D receptor and its coactivators DRIP205, SRC2, and SRC3. The Journal of investigative dermatology. 2007;127:874–880. doi: 10.1038/sj.jid.5700624. [DOI] [PubMed] [Google Scholar]
- Huelsken J, Vogel R, Erdmann B, Cotsarelis G, Birchmeier W. beta-Catenin controls hair follicle morphogenesis and stem cell differentiation in the skin. Cell. 2001;105:533–545. doi: 10.1016/s0092-8674(01)00336-1. [DOI] [PubMed] [Google Scholar]
- Ito M, Roeder RG. The TRAP/SMCC/Mediator complex and thyroid hormone receptor function. Trends in Endocrinology & Metabolism. 2001;12:127–134. doi: 10.1016/s1043-2760(00)00355-6. [DOI] [PubMed] [Google Scholar]
- Ito M, Yuan CX, Okano HJ, Darnell RB, Roeder RG. Involvement of the TRAP220 component of the TRAP/SMCC coactivator complex in embryonic development and thyroid hormone action. Molecular cell. 2000;5:683–693. doi: 10.1016/s1097-2765(00)80247-6. [DOI] [PubMed] [Google Scholar]
- Jia Y, Qi C, Kashireddi P, Surapureddi S, Zhu YJ, Rao MS, et al. Transcription coactivator PBP, the peroxisome proliferator-activated receptor (PPAR)-binding protein, is required for PPARalpha-regulated gene expression in liver. The Journal of biological chemistry. 2004;279:24427–24434. doi: 10.1074/jbc.M402391200. [DOI] [PubMed] [Google Scholar]
- Jia Y, Qi C, Zhang Z, Zhu YT, Rao SM, Zhu YJ. Peroxisome proliferator-activated receptor-binding protein null mutation results in defective mammary gland development. The Journal of biological chemistry. 2005;280:10766–10773. doi: 10.1074/jbc.M413331200. [DOI] [PubMed] [Google Scholar]
- Jiang P, Hu Q, Ito M, Meyer S, Waltz S, Khan S, et al. Key roles for MED1 LxxLL motifs in pubertal mammary gland development and luminal-cell differentiation. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:6765–6770. doi: 10.1073/pnas.1001814107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobielak K, Pasolli HA, Alonso L, Polak L, Fuchs E. Defining BMP functions in the hair follicle by conditional ablation of BMP receptor IA. The Journal of cell biology. 2003;163:609–623. doi: 10.1083/jcb.200309042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg RD. Mediator and the mechanism of transcriptional activation. Trends Biochem Sci. 2005;30:235–239. doi: 10.1016/j.tibs.2005.03.011. [DOI] [PubMed] [Google Scholar]
- Malik S, Roeder RG. Dynamic regulation of pol II transcription by the mammalian Mediator complex. Trends Biochem Sci. 2005;30:256–263. doi: 10.1016/j.tibs.2005.03.009. [DOI] [PubMed] [Google Scholar]
- Matsumoto K, Huang J, Viswakarma N, Bai L, Jia Y, Zhu YT, et al. Transcription coactivator PBP/MED1-deficient hepatocytes are not susceptible to diethylnitrosamine-induced hepatocarcinogenesis in the mouse. Carcinogenesis. 2010;31:318–325. doi: 10.1093/carcin/bgp306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto K, Yu S, Jia Y, Ahmed MR, Viswakarma N, Sarkar J, et al. Critical role for transcription coactivator peroxisome proliferator-activated receptor (PPAR)-binding protein/TRAP220 in liver regeneration and PPARalpha ligand-induced liver tumor development. The Journal of biological chemistry. 2007;282:17053–17060. doi: 10.1074/jbc.M701956200. [DOI] [PubMed] [Google Scholar]
- Oda Y, Chalkley RJ, Burlingame AL, Bikle DD. The transcriptional coactivator DRIP/mediator complex is involved in vitamin D receptor function and regulates keratinocyte proliferation and differentiation. The Journal of investigative dermatology. 2010;130:2377–2388. doi: 10.1038/jid.2010.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda Y, Ishikawa MH, Hawker NP, Yun QC, Bikle DD. Differential role of two VDR coactivators, DRIP205 and SRC-3, in keratinocyte proliferation and differentiation. J Steroid Biochem Mol Biol. 2007;103:776–780. doi: 10.1016/j.jsbmb.2006.12.069. [DOI] [PubMed] [Google Scholar]
- Oda Y, Sihlbom C, Chalkley RJ, Huang L, Rachez C, Chang CP, et al. Two distinct coactivators, DRIP/mediator and SRC/p160, are differentially involved in vitamin D receptor transactivation during keratinocyte differentiation. Molecular endocrinology (Baltimore, Md. 2003;17:2329–2339. doi: 10.1210/me.2003-0063. [DOI] [PubMed] [Google Scholar]
- Oda Y, Uchida Y, Moradian S, Crumrine D, Elias PM, Bikle DD. Vitamin D receptor and coactivators SRC2 and 3 regulate epidermis-specific sphingolipid production and permeability barrier formation. The Journal of investigative dermatology. 2009;129:1367–1378. doi: 10.1038/jid.2008.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer HG, Anjos-Afonso F, Carmeliet G, Takeda H, Watt FM. The vitamin D receptor is a Wnt effector that controls hair follicle differentiation and specifies tumor type in adult epidermis. PLoS ONE. 2008;3:e1483. doi: 10.1371/journal.pone.0001483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paus R, Foitzik K. In search of the “hair cycle clock”: a guided tour. Differentiation. 2004;72:489–511. doi: 10.1111/j.1432-0436.2004.07209004.x. [DOI] [PubMed] [Google Scholar]
- Rachez C, Lemon BD, Suldan Z, Bromleigh V, Gamble M, Näär AM, et al. Ligand-dependent transcription activation by nuclear receptors requires the DRIP complex. Nature. 1999;398:824–828. doi: 10.1038/19783. [DOI] [PubMed] [Google Scholar]
- Segrelles C, Moral M, Lorz C, Santos M, Lu J, Cascallana JL, et al. Constitutively active Akt induces ectodermal defects and impaired bone morphogenetic protein signaling. Mol Biol Cell. 2008;19:137–149. doi: 10.1091/mbc.E07-08-0764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah S, Islam MN, Dakshanamurthy S, Rizvi I, Rao M, Herrell R, et al. The molecular basis of vitamin D receptor and beta-catenin crossregulation. Molecular cell. 2006;21:799–809. doi: 10.1016/j.molcel.2006.01.037. [DOI] [PubMed] [Google Scholar]
- Sharov AA, Sharova TY, Mardaryev AN, Tommasi di Vignano A, Atoyan R, Weiner L, et al. Bone morphogenetic protein signaling regulates the size of hair follicles and modulates the expression of cell cycle-associated genes. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:18166–18171. doi: 10.1073/pnas.0608899103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slominski A, Paus R. Melanogenesis is coupled to murine anagen: toward new concepts for the role of melanocytes and the regulation of melanogenesis in hair growth. The Journal of investigative dermatology. 1993;101:90S–97S. doi: 10.1111/1523-1747.ep12362991. [DOI] [PubMed] [Google Scholar]
- Slominski A, Wortsman J, Plonka PM, Schallreuter KU, Paus R, Tobin DJ. Hair follicle pigmentation. The Journal of investigative dermatology. 2005;124:13–21. doi: 10.1111/j.0022-202X.2004.23528.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpf M, Waskow C, Krotschel M, van Essen D, Rodriguez P, Zhang X, et al. The mediator complex functions as a coactivator for GATA-1 in erythropoiesis via subunit Med1/TRAP220. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:18504–18509. doi: 10.1073/pnas.0604494103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpf M, Yue X, Schmitz S, Luche H, Reddy JK, Borggrefe T. Specific erythroid-lineage defect in mice conditionally deficient for Mediator subunit Med1. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:21541–21546. doi: 10.1073/pnas.1005794107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teichert A, Elalieh H, Bikle D. Disruption of the hedgehog signaling pathway contributes to the hair follicle cycling deficiency in Vdr knockout mice. J Cell Physiol. 2010;225:482–489. doi: 10.1002/jcp.22227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teichert AE, Elalieh H, Elias PM, Welsh J, Bikle DD. Overexpression of Hedgehog Signaling Is Associated with Epidermal Tumor Formation in Vitamin D Receptor-Null Mice. The Journal of investigative dermatology. 2011 doi: 10.1038/jid.2011.196. on line. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswakarma N, Jia Y, Bai L, Vluggens A, Borensztajn J, Xu J, et al. Coactivators in PPAR-Regulated Gene Expression. PPAR Res. 2010 doi: 10.1155/2010/250126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan CX, Ito M, Fondell JD, Fu ZY, Roeder RG. The TRAP220 component of a thyroid hormone receptor-associated protein (TRAP) coactivator complex interacts directly with nuclear receptors in a ligand-dependent fashion. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:7939–7944. doi: 10.1073/pnas.95.14.7939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Qi C, Jain S, Rao MS, Reddy JK. Isolation and characterization of PBP, a protein that interacts with peroxisome proliferator-activated receptor. The Journal of biological chemistry. 1997;272:25500–25506. doi: 10.1074/jbc.272.41.25500. [DOI] [PubMed] [Google Scholar]
- Zinser GM, Sundberg JP, Welsh J. Vitamin D(3) receptor ablation sensitizes skin to chemically induced tumorigenesis. Carcinogenesis. 2002;23:2103–2109. doi: 10.1093/carcin/23.12.2103. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.