Abstract
RNA splicing is a major regulatory mechanism for controlling eukaryotic gene expression. By generating various splice isoforms from a single pre–mRNA, alternative splicing plays a key role in promoting the evolving complexity of metazoans. Numerous splicing factors have been identified. However, the in vivo functions of many splicing factors remain to be understood. In vivo studies are essential for understanding the molecular mechanisms of RNA splicing and the biology of numerous RNA splicing-related diseases. We previously isolated a Caenorhabditis elegans mutant defective in an essential gene from a genetic screen for suppressors of the rubberband Unc phenotype of unc-93(e1500) animals. This mutant contains missense mutations in two adjacent codons of the C. elegans microfibrillar-associated protein 1 gene mfap-1. mfap-1(n4564 n5214) suppresses the Unc phenotypes of different rubberband Unc mutants in a pattern similar to that of mutations in the splicing factor genes uaf-1 (the C. elegans U2AF large subunit gene) and sfa-1 (the C. elegans SF1/BBP gene). We used the endogenous gene tos-1 as a reporter for splicing and detected increased intron 1 retention and exon 3 skipping of tos-1 transcripts in mfap-1(n4564 n5214) animals. Using a yeast two-hybrid screen, we isolated splicing factors as potential MFAP-1 interactors. Our studies indicate that C. elegans mfap-1 encodes a splicing factor that can affect alternative splicing.
Author Summary
RNA splicing removes intervening intronic sequences from pre–mRNA transcripts and joins adjacent exonic sequences to generate functional messenger RNAs. The in vivo functions of numerous factors that regulate splicing remain to be understood. From a genetic screen for suppressors of the rubberband Unc phenotype caused by the Caenorhabditis elegans unc-93(e1500) mutation, we isolated a mutation that affects a highly conserved essential gene, mfap-1. MFAP-1 is a nuclear protein that is broadly expressed. MFAP-1 can affect the alternative splicing of tos-1, an endogenous reporter gene for splicing, and is required for the altered splicing at a cryptic 3′ splice site of tos-1. mfap-1 enhances the effects of the gene uaf-1 (splicing factor U2AF large subunit) in suppressing the rubberband Unc phenotype of unc-93(e1500) animals. Our studies provide in vivo evidence that MFAP-1 functions as a splicing factor.
Introduction
RNA splicing removes non-coding introns and joins adjacent coding exons from pre-mRNAs to generate functional coding mRNAs. Alternative splicing can generate numerous splice isoforms from the same pre-mRNA [1]. The complex proteome encoded by mRNA splice isoforms is believed to be a major driving force for the evolving complexity of metazoans [1], [2]. Numerous proteins and non-coding RNAs regulate RNA splicing [3]. The U1 snRNP complex and the SF1/U2AF65/U2AF35 protein complex recognize the 5′ and 3′ splice sites of an intron, respectively [4]–[11], and the U2 and U4/U5/U6 snRNP complexes assemble in a step-wise manner and undergo compositional and conformational rearrangements to drive the two steps of the trans-esterification reaction in RNA splicing [7], [12]. Mutations in trans-splicing factors or cis-regulatory splicing elements cause numerous diseases [13], [14].
Over 200 protein factors have been shown to regulate splicing or associate with the splicing machinery or other splicing factors [3]. Splicing factors have been identified mostly using biochemical approaches, and the in vivo functions of many splicing factors remain largely unknown. In vivo analysis of these factors remains a challenge, since many are essential for viability and the analyses of their in vivo functions can be limited by the lethality caused by mutations in these factors.
Recent studies indicate that conclusions concerning splicing factors derived from in vitro analyses should be complemented by in vivo analyses. For example, in vivo studies suggest that the splicing factor SF1/BBP, once thought to be ubiquitously required for splicing, might be required for the splicing of only a subset of genes [15]–[17]. Some Saccharomyces cerevisiae core splicing factors, once thought to be essential for the splicing of all introns, were found to affect splicing of only a subgroup of introns when the effects of loss-of-function mutations in these factors were examined [18]. Also, the functions of some splicing factors extend beyond regulating RNA splicing. For example, the U2AF large subunit is required for the efficient export of intronless mRNAs in Drosophila in addition to its essential role in regulating 3′ splice site recognition [19]. Thus, in vivo functional analyses are essential for an accurate understanding of the biological functions of splicing factors.
In Caenorhabditis elegans, the genes unc-93, sup-9 and sup-10 encode components of a presumptive two-pore domain K+ channel complex that affects muscle activity [20]–[23]. Animals carrying rare gain-of-function (gf) mutations in any of these three genes are sluggish, defective in egg laying and exhibit a rubberband phenotype: when touched on the head, the animal contracts and relaxes along its entire body without moving backwards. Complete loss-of-function (lf) mutations of unc-93, sup-9 or sup-10 do not cause obvious abnormalities [21], [22]. The SUP-9 protein is similar to the mammalian Two-pore Acid Sensitive K+ channels TASK-1 and TASK-3 [20]. sup-10 encodes a novel single-transmembrane domain protein without identified mammalian homologs [20], and unc-93 encodes a multiple transmembrane-domain protein that defines a novel family of proteins conserved from C. elegans to mammals [20], [23]. A mammalian UNC-93 homolog, UNC-93b, interacts with Toll-like receptors and regulates innate immune responses [24]–[28].
We previously screened for new suppressors of the “rubberband” Unc phenotype of unc-93(e1500) animals and isolated mutations affecting the splicing factors U2AF large subunit (UAF-1) and SF1/BBP (SFA-1) [29]. Our analysis suggested that mutations in uaf-1 and sfa-1 result in the suppression of the rubberband Unc phenotype of unc-93(e1500) animals by altering the splicing of the pre-mRNA of an unknown gene [29]. We identified the pre-mRNA of the gene tos-1 as abnormally spliced in uaf-1 and sfa-1 mutants and determined that tos-1 is a sensitive endogenous reporter for analyzing in vivo functions of splicing factors [30]. In this study, we describe a third isolate, n4564 n5214, from the genetic screen in which we identified the uaf-1 and sfa-1 mutations [29]. We found that the gene affected by n4564 n5214 encodes a novel splicing factor that affects alternative splicing in C. elegans.
Results
n4564 n5214 suppresses the rubberband Unc phenotype of unc-93(e1500) animals
Previously we performed a genetic screen for mutations that caused sterility and/or lethality and concurrently suppressed the rubberband Unc phenotype caused by the unc-93(e1500) mutation [29]. Besides uaf-1(n4588) and sfa-1(n4562), we also isolated the mutation n4564 [29], which we renamed n4564 n5214 in this study (see below). Although the precise mechanisms underlying the suppression of the rubberband Unc phenotypes by the uaf-1, sfa-1 [29] and n4564 n5214 (see below) mutations remain to be determined, the uaf-1 and sfa-1 mutations we isolated and the splicing of the uaf-1 target tos-1 can provide tools for studying in vivo functions of splicing factor genes [29], [30], which we now report include the gene affected by n4564 n5214.
n4564 n5214 mutants exhibited temperature-sensitive lethality: at 15°C, n4564 n5214 homozygous animals grew and behaved similarly to the wild type; at 20°C, mutant animals grew more slowly, had few progeny and were hyperactive (Table 1); at 25°C, the mutant strain was embryonically lethal. n4564 n5214 weakly suppressed the Unc phenotype of unc-93(e1500) animals at 15°C (L. Ma and H. R. Horvitz, unpublished observations) and was a stronger suppressor at 20°C (Table 1). n4564 n5214/+; unc-93(e1500) animals were as Unc as unc-93(e1500) animals (L. Ma and H. R. Horvitz, unpublished observations), indicating that n4564 n5214 is a recessive suppressor of the Unc phenotype of unc-93(e1500) animals and suggesting that n4564 n5214 causes a reduction or loss of mfap-1 function. We tested whether n4564 n5214 also suppressed the Unc phenotype caused by the rubberband mutants unc-93(n200), sup-9(n1550) and sup-10(n983). n200 is a weakly semi-dominant unc-93 allele that causes a weak rubberband Unc phenotype [22]. The sup-9(n1550) and sup-10(n983) gain-of-function mutations cause strong and moderate rubberband Unc phenotypes, respectively [20], [21], [23], [31]. n4564 n5214 strongly suppressed sup-10(n983) but did not suppress unc-93(n200) or sup-9(n1550) (Table 1). Thus, n4564 n5214 suppressed the same rubberband Unc mutants as do the uaf-1(n4588) and sfa-1(n4562) mutations, both of which suppress the rubberband Unc phenotypes of unc-93(e1500) and sup-10(n983) animals but not the rubberband Unc phenotypes of unc-93(n200) or sup-9(n1550) animals [29].
Table 1. Suppression of the rubberband Unc phenotype by mfap-1 mutations.
| Genotype | Bodybends per 30 sec (± SD) | n |
| wild-type | 21.8±3.4 | 20 |
| wild-type (25°C) | 28.3±4.4 | 20 |
| unc-93(e1500) | 0.9±1.3 | 20 |
| unc-93(e1500) (25°C) | 0.8±0.8 | 20 |
| unc-93(n200) [29] | 15.6±3.5 | 20 |
| sup-10(n983) [29] | 4.3±1.9 | 20 |
| sup-9(n1550); sup-18(n1014) [29] | 0.1±0.4 | 20 |
| mfap-1(n4564 n5214) | 27.1±2.2 | 20 |
| mfap-1(n5214) | 28.4±3.3 | 20 |
| mfap-1(n5214) (25°C) | 28.3±4.0 | 20 |
| mfap-1(n4564 n5214); unc-93(e1500) | 21.8±4.5 | 20 |
| mfap-1(n4564 n5214); unc-93(n200) | 13.9±3.0 | 20 |
| mfap-1(n4564 n5214); sup-10(n983) | 18.0±4.1 | 20 |
| mfap-1(n4564 n5214); sup-9(n1550); sup-18(n1014) | 0.3±0.5 | 20 |
| mfap-1(n5214); unc-93(e1500) | 1.8±1.3 | 20 |
| mfap-1(n5214); unc-93(e1500) (25°C) | 8.0±3.6 | 20 |
n4564 n5214 contains two missense mutations in the gene mfap-1, which encodes a highly conserved nuclear protein
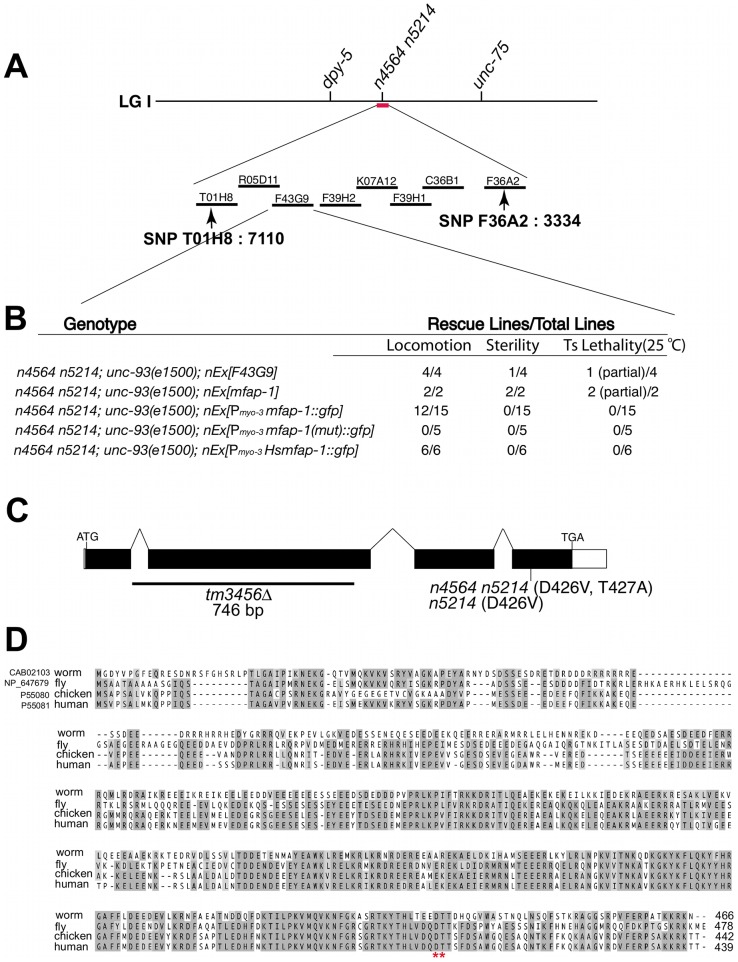
We cloned the gene affected by n4564 n5214 by genetic mapping and transgene rescue experiments (Figure 1A, 1B; see Materials and Methods). The n4564 n5214 strain contains two missense mutations in the gene F43G9.10, changing the adjacent conserved amino acids Asp 426 and Thr 427 to Val (n5214) and Ala (n4564) in the predicted protein, respectively (Figure 1C). F43G9.10 encodes the C. elegans ortholog of a highly conserved mammalian protein that has been called “microfibrillar-associated protein 1” (41% identity between the C. elegans and human orthologs) (Figure 1D). We named F43G9.10 mfap-1 (microfibrillar-associated protein 1). The chicken ortholog of MFAP-1 was suggested to be an extracellular matrix protein [32]. However, the Drosophila ortholog of MFAP-1 (dMFAP1) interacts with the splicing factor dPrp38, is required for normal expression of γ-tubulin and stg/cdc25 mRNAs and has been proposed to act as a splicing factor [33]. We found that the expression of a human MFAP1::GFP fusion protein (Hsmfap-1) in C. elegans body-wall muscles rescued the suppression of unc-93(e1500) by mfap-1(n4564 n5214) (Figure 1B), indicating that the function of MFAP-1 is conserved from nematodes to humans.
Figure 1. Genetic mapping, cloning, and identification of mfap-1.
(A) Genomic location of the n4564 n5214 mutation based on genetic mapping using visible markers and SNPs (e.g., SNP T01H8: 7110 on the left and SNP F36A2: 3334 on the right). Cosmids tested in rescue experiments are labeled. Only cosmid F43G9 rescued the three phenotypic characteristics of mfap-(n4564 n5214) individuals: suppression of the Unc phenotype of unc-93(e1500) animals; partial sterility at 20°C; and temperature-sensitive lethality at 25°C. (B) Transgene rescue experiments. Transgenes (nEx) were injected into the gonads of mfap-1(n4564 n5214); unc-93(e1500) animals, and lines stably transmitting the transgenes were established. Transgenic lines were analyzed for the rescue of each of three abnormalities: suppression of the Unc phenotype of unc-93(e1500) animals; partial sterility at 20°C; and temperature-sensitive lethality at 25°C. mut: the n4564 n5214 mutation. (C) Gene structure of mfap-1. Black boxes: coding exons. Open boxes: 5′ and 3′ UTRs. Positions of start (ATG) and stop codons (TGA) are indicated. The sites of the n4564 n5214 mutation, the n5214 mutation and the tm3456Δ deletion are labeled. (D) Sequence alignment of predicted MFAP-1 proteins from C. elegans, Drosophila, chicken and human. *: amino acids mutated in mfap-1(n4564 n5214) mutants. Amino acids conserved in at least three orthologs are darkly shaded, while amino acids with similar physical properties or conserved in two orthologs are lightly shaded.
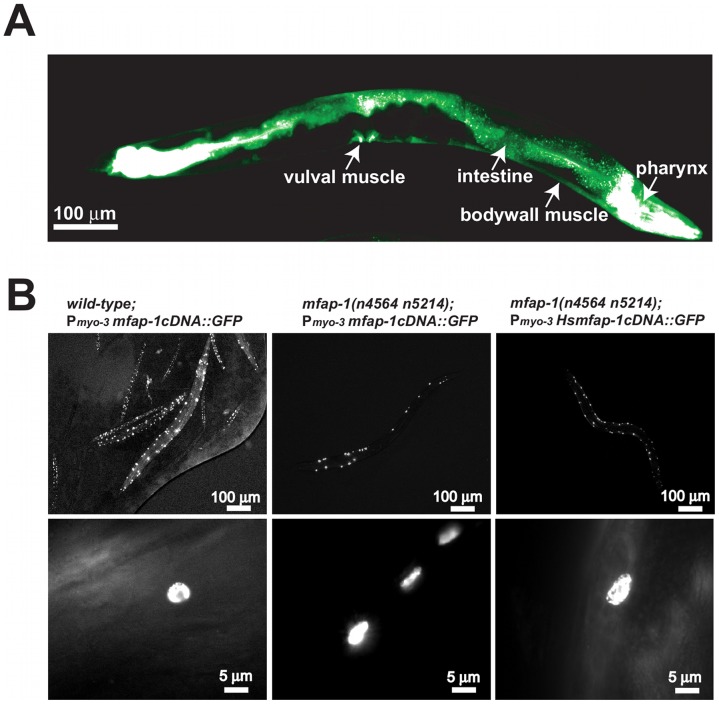
We examined the expression pattern of mfap-1 in transgenic animals using a transcriptional fusion reporter construct that drives GFP expression under the control of an approximately 2.5 kb mfap-1 promoter (Figure 2A). We observed strong GFP expression in the intestine, pharynx and vulval muscles (Figure 2A). We also observed GFP expression in the body-wall muscles (Figure 2A), consistent with the finding that body-wall muscle-specific expression of mfap-1 rescued the suppression of the unc-93(e1500) Unc phenotype by mfap-1(n4564 n5214) (Figure 1). Both C. elegans MFAP-1::GFP and human MFAP-1::GFP, when expressed in body-wall muscles, were exclusively localized in nuclei (Figure 2B and unpublished observations), indicating that MFAP-1 is a nuclear protein.
Figure 2. MFAP-1 is a nuclear protein expressed in multiple tissues.
(A) A 2.5 kb 5′ promoter region upstream of the start codon of mfap-1 was used to drive the expression of a transcriptional fusion GFP reporter in transgenic animals. The tissues in which the GFP reporter was expressed are labeled. This figure shows a montage of two photographs, which were pseudo-colored from a gray-scale image to a green-scale image using Adobe Photoshop to reflect the level of GFP expression. (B) Transgene experiments were performed as described in Materials and Methods. The C. elegans MFAP-1::GFP fusion protein, when expressed in body-wall muscles using a myo-3 promoter in either wild-type or mfap-1(n4564 n5214) animals of mixed developmental stages, was exclusively localized in nuclei (left and middle panels). Similarly, human MFAP-1::GFP fusion protein, when expressed in the body-wall muscles of mfap-1(n4564 n5214) animals, was also exclusively localized in nuclei (right panels). Lower panels show typical nuclear GFP signals in adult transgenic animals. Genetic backgrounds and transgenes are indicated.
We obtained a 746 bp mfap-1 deletion allele, tm3456Δ (kindly provided by S. Mitani, personal communication), which removes the entire first intron and most of the second exon of mfap-1 (Figure 1C). tm3456Δ is predicted to encode a truncated protein with a frameshift after amino acid 53, suggesting that tm3456Δ is likely a null allele of mfap-1. mfap-1(tm3456Δ)/+animals grew and behaved like the wild type, and mfap-1(tm3456Δ) homozygous animals arrested developmentally at the L1 or L2 larval stages. tm3456Δ/n4564 n5214 animals similarly arrested at the L1/L2 larval stages, suggesting that the lethal phenotype of tm3456Δ homozygotes is caused by the mfap-1(tm3456Δ) mutation and consistent with the hypothesis that mfap-1(n4564 n5214) causes a reduction or loss of mfap-1 function.
The D426V(n5214) and T427A(n4564) mutations act together to suppress unc-93(e1500)
The temperature-sensitive lethal phenotype of mfap-1(n4564 n5214) at 25°C provided an approach to the identification of genes that interact with mfap-1 by seeking suppressors of the temperature-sensitive lethality. We screened about 50,000 haploid genomes and identified two independent suppressors, which we later found to cause the same intragenic change in mfap-1 (see Materials and Methods). Specifically, both suppressors caused a reversion of the T427A (n4564) mutation (see Materials and Methods) to the wild-type codon (ACA) and amino acid (A427T) and retained the D426V (n5214) mutation. mfap-1(n5214) did not obviously suppress the rubberband Unc phenotype of unc-93(e1500) animals at 20°C but did do so at 25°C (Table 1), indicating that mfap-1(n5214) is a temperature-sensitive allele.
We examined how the two mfap-1 mutations, n4564 and n5214, contribute to the activity of mfap-1(n4564 n5214) as a suppressor of the Unc phenotype of unc-93(e1500) animals. We expressed MFAP-1::GFP fusion transgenes containing the n4564 (T427A) mutation, the n5214 (D426V) mutation or both mutations in the body-wall muscles of mfap-1(n4564 n5214); unc-93(e1500) animals and assayed the locomotion of transgenic animals (Table 2). Both the mfap-1(n4564)::GFP and the mfap-1(n5214)::GFP transgenes greatly rescued the suppression of unc-93(e1500) by mfap-1(n4564 n5214), while the mfap-1(n4564 n5214)::GFP transgene only partially rescued the suppression (Table 2). These results suggest that individually the mutations n4564 or n5214 only slightly if at all affect the function of mfap-1 and that the strong suppression of unc-93(e1500) by mfap-1(n4564 n5214) probably results from an additive or synergistic effect of these two mutations, likely in reducing mfap-1 function.
Table 2. mfap-1(n4564) and mfap-1(n5214) are weak mutations that act together to suppress the rubberband Unc phenotype of unc-93(e1500) animals.
| Genotype | Bodybends per 30 sec (±SD) | n |
| nEx[Pmyo-3 mfap-1 cDNA (wt)::GFP] line 1 | 1.3±1.4 | 20 |
| nEx[Pmyo-3 mfap-1 cDNA (wt)::GFP] line 2 | 2.1±2.1 | 20 |
| nEx[Pmyo-3 mfap-1 cDNA (wt)::GFP] line 3 | 1.7±2.0 | 20 |
| nEx[Pmyo-3 mfap-1 cDNA (n4564)::GFP] line 1 | 1.3±1.1 | 20 |
| nEx[Pmyo-3 mfap-1 cDNA (n4564)::GFP] line 2 | 1.6±1.7 | 20 |
| nEx[Pmyo-3 mfap-1 cDNA (n4564)::GFP] line 3 | 3.8±1.4 | 20 |
| nEx[Pmyo-3 mfap-1 cDNA (n5214)::GFP] line 1 | 3.4±1.6 | 20 |
| nEx[Pmyo-3 mfap-1 cDNA (n5214)::GFP] line 2 | 2.2±2.5 | 9 |
| nEx[Pmyo-3 mfap-1 cDNA (n5214)::GFP] line 3 | 3.1±1.6 | 20 |
| nEx[Pmyo-3 mfap-1 cDNA (n4564 n5214)::GFP] line 1 | 10.1±4.7 | 20 |
| nEx[Pmyo-3 mfap-1 cDNA (n4564 n5214)::GFP] line 2 | 14.3±6.4 | 20 |
All transgenes were expressed in mfap-1(n4564 n5214); unc-93(e1500) animals. Two lines were obtained and analyzed for transgene Pmyo-3 mfap-1 cDNA (n4564 n5214)::GFP.
mfap-1 and uaf-1 interact to affect the locomotion of unc-93(e1500) animals
To test for an interaction between mfap-1 and uaf-1, we generated mfap-1(n4564 n5214); uaf-1 mutant animals carrying the unc-93(e1500) mutation. Like mfap-1(n4564 n5214) animals, mfap-1(n4564 n5214); uaf-1(n5123) animals were viable at 20°C. mfap-1(n4564 n5214); uaf-1(n4588 n5125) mutants and mfap-1(n4564 n5214); uaf-1(n4588 n5127) mutants were viable at 15°C and became sterile at 20°C, and mfap-1(n4564 n5214); uaf-1(n4588) mutants were likely inviable at both 15°C and 20°C, since we failed to obtain progeny with this genotype from mfap-1(n4564 n5214)/+; uaf-1(n4588)/+ hermaphrodites (Table 3). All viable mfap-1; uaf-1 multiple mutant animals exhibited strong suppression of the unc-93(e1500) Unc phenotype, similar to that of the mfap-1(n4564 n5214) mutant animals (Table 3). That mfap-1; uaf-1 multiple mutant animals, except mfap-1(n4564 n5214); uaf-1(n5123), exhibited a stronger temperature-sensitive sterile or lethal phenotype than either mfap-1 or uaf-1 mutant animals suggests that the mfap-1(n4564 n5214) and uaf-1 mutations might have additive effects on animal survival.
Table 3. Mutations in mfap-1 and uaf-1 interact to affect the locomotion of unc-93(e1500) animals.
| Genotype | Body-bends per 30 sec ± SD | n |
| wild-type [29] | 20.4±3.7 | 20 |
| unc-93(e1500) [29] | 0.9±1.2 | 20 |
| mfap-1(n5214); unc-93(e1500) | 1.8±1.3 | 20 |
| mfap-1(n4564 n5214); unc-93(e1500) | 21.8±4.5 | 20 |
| uaf-1(n5123) unc-93(e1500) [29] | 0.6±0.8 | 20 |
| mfap-1(n5214); uaf-1(n5123) unc-93(e1500) | 2.2±1.9 | 20 |
| mfap-1(n4564 n5214); uaf-1(n5123) unc-93(e1500) | 20.9±2.9 | 20 |
| uaf-1(n4588 n5125) unc-93(e1500) [29] | 5.4±2.1 | 20 |
| mfap-1(n5214); uaf-1(n4588 n5125) unc-93(e1500) | 10.2±5.7 | 20 |
| mfap-1(n4564 n5214); uaf-1(n4588 n5125) unc-93(e1500) * | 20.5±4.5 | 20 |
| uaf-1(n4588 n5127) unc-93(e1500) [29] | 21.5±3.6 | 20 |
| mfap-1(n5214); uaf-1(n4588 n5127) unc-93(e1500) | 21.2±4.4 | 20 |
| mfap-1(n4564 n5214); uaf-1(n4588 n5127) unc-93(e1500) * | 20.5±4.7 | 20 |
| uaf-1(n4588) unc-93(e1500) [29] | 21.0±5.5 | 20 |
| mfap-1(n5214); uaf-1(n4588) unc-93(e1500) * | 17.7±4.0 | 20 |
| mfap-1(n4564 n5214); uaf-1(n4588) unc-93(e1500) ** | Inviable | 20 |
| mfap-1(n5214) | 28.4±3.3 | 20 |
| mfap-1(n4564 n5214) | 27.1±2.2 | 20 |
| uaf-1(n5123) [29] | 19.1±3.1 | 20 |
| uaf-1(n4588 n5125) [29] | 19.5±3.3 | 20 |
| uaf-1(n4588 n5127) [29] | 22.1±4.0 | 20 |
| uaf-1(n4588) [29] | 23.1±4.1 | 20 |
*: Sterile at 20°C.
**: Inviable.
We next examined whether mfap-1(n5214) could enhance the suppressor activities of uaf-1 mutations for the Unc phenotype of unc-93(e1500) animals by generating mfap-1(n5214); uaf-1 double mutant animals carrying the unc-93(e1500) mutation (Table 3). mfap-1(n5214); uaf-1(n5123) double mutants were similar to mfap-1(n5214) or uaf-1(n5123) single mutants and were not substantially suppressed for the Unc phenotype of unc-93(e1500) animals (Table 3). mfap-1(n5214) enhanced the suppressor activity of the uaf-1(n4588 n5125) mutation. As expected, both mfap-1(n5214); uaf-1(n4588 n5127) and mfap-1(n5214); uaf-1(n4588) strongly suppressed unc-93(e1500), since both uaf-1(n4588 n5127) and uaf-1(n4588) are strong suppressors for unc-93(e1500) [29]. Although mfap-1(n5214) or uaf-1(n4588 n5127) mutants were apparently healthy at 20°C, mfap-1(n5214); uaf-1(n4588 n5127) mutant animals became partially sterile at 20°C (L. Ma and H. R. Horvitz, unpublished observations). Furthermore, mfap-1(n5214); uaf-1(n4588) double mutant animals were viable at 15°C but became sterile when grown at 20°C, which is a more severe temperature-sensitive phenotype than that of uaf-1(n4588) single mutants. That mfap-1 mutations enhance the temperature-sensitive phenotypes of uaf-1 mutants suggests that mfap-1 and uaf-1 might act in parallel in affecting unc-93 function and animal survival.
We tested if mfap-1(n4564 n5214), like the uaf-1(n4588) mutation [29], would cause the recognition of a cryptic 3′ splice site in exon 9 of unc-93(e1500). We did not observe any obviously altered recognition of this cryptic 3′ splice site (1.3% for wild type vs. 2% for mfap-1(n4564 n5214) animals) (Figure S1). Therefore, the suppression of the Unc phenotype of unc-93(e1500) by mfap-1(n4564 n5214) might not be caused by altered splicing of the unc-93(e1500) transcript.
mfap-1(n4564 n5214) alters the splicing of tos-1 by increasing intron 1 retention and exon 3 skipping
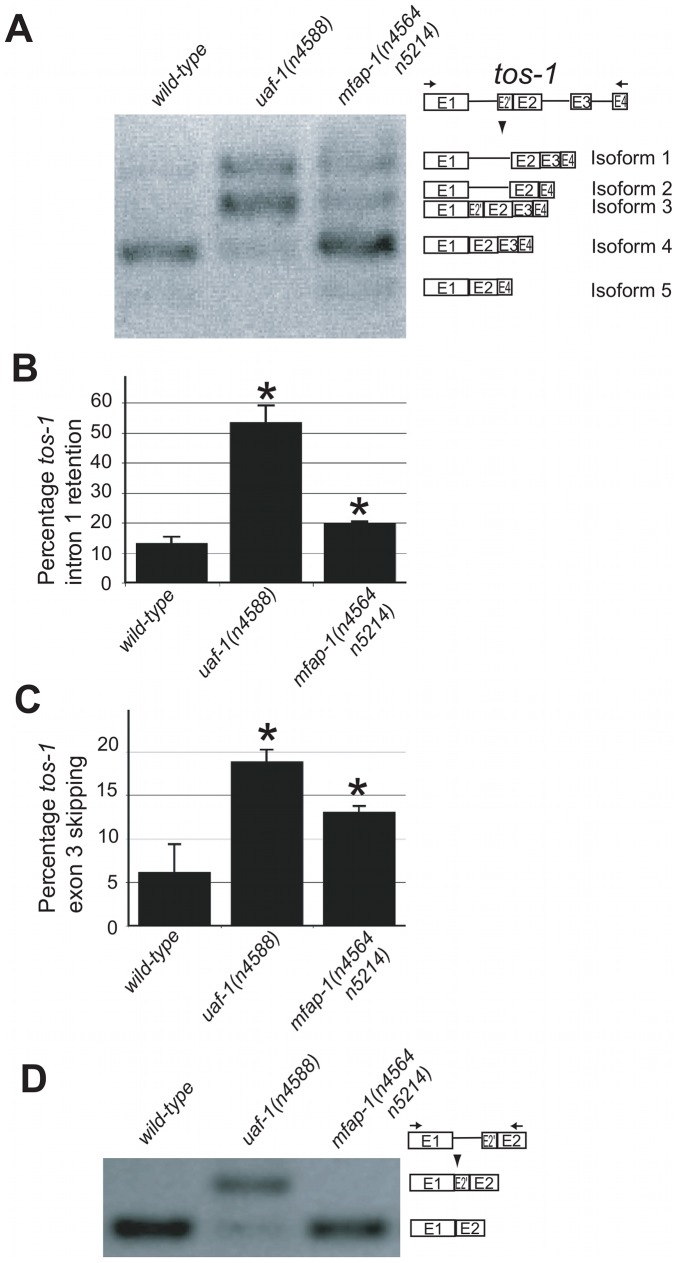
To analyze whether mfap-1 can affect alternative splicing, we examined the splicing of tos-1 in mfap-1(n4564 n5214) animals. We previously described tos-1 as a sensitive endogenous reporter for alternative splicing in C. elegans [30]. As shown in Figure 3A–3C, mfap-1(n4564 n5214) increased tos-1 intron 1 retention and exon 3 skipping, suggesting that the recognition of the 3′ splice sites in intron 1 or before exon 3 was reduced in mfap-1(n4564 n5214) animals. We note that the tos-1 splice isoforms we observed in mfap-1(n4564 n5214) and mfap-1(RNAi) animals can also be seen in wild-type animals (Figure 3A), indicating that alterations in mfap-1 affect tos-1 alternative splicing events that occur in the wild type. We also examined whether mfap-1(n4564 n5214) could cause recognition of the cryptic 3′ splice site in tos-1 intron 1, which was recognized in uaf-1(n4588) animals [30]. We did not detect obvious recognition of the cryptic 3′ splice sites in mfap-1(n4564 n5214) animals (Figure 3D), suggesting that mfap-1(n4564 n5214) did not alter the specificity of 3′ splice-site recognition. We similarly examined mfap-1(n5214) animals and did not observe obvious effects on the splicing of tos-1 or the recognition of the intron 1 cryptic 3′ splice site (Figure S2B), suggesting that mfap-1(n5214) does not alter or has a very weak effect on tos-1 splicing. This result is consistent with the notion that n5214 is a weaker mfap-1 allele than n4564 n5214.
Figure 3. mfap-1(n4564 n5214) affects the splicing of tos-1.
(A) RT-PCR experiments showing the effect of mfap-1(n4564 n5214) on tos-1 alternative splicing. tos-1 splice isoforms are illustrated on the right. (B) The molar ratios of all tos-1 splice isoforms with intron 1 retention (isoforms 1 and 2), presented as a percentage of all isoforms combined (Isoforms 1, 2, 3, 4 and 5). Error bars: standard deviations. *p<0.05. (C) The molar ratios of all tos-1 splice isoforms with exon 3 skipping (isoforms 2 and 5), presented as a percentage of all isoforms combined (isoforms 1, 2, 3, 4 and 5). Error bars: standard deviations. *p<0.05. (D) RT-PCR experiments showing the effect of the mfap-1(n4564 n5214) mutation on the recognition of the cryptic 3′ splice site of tos-1 intron 1. For all analyses, isoform intensities were obtained by analyzing biological duplicates or triplicates using NIH ImageJ software.
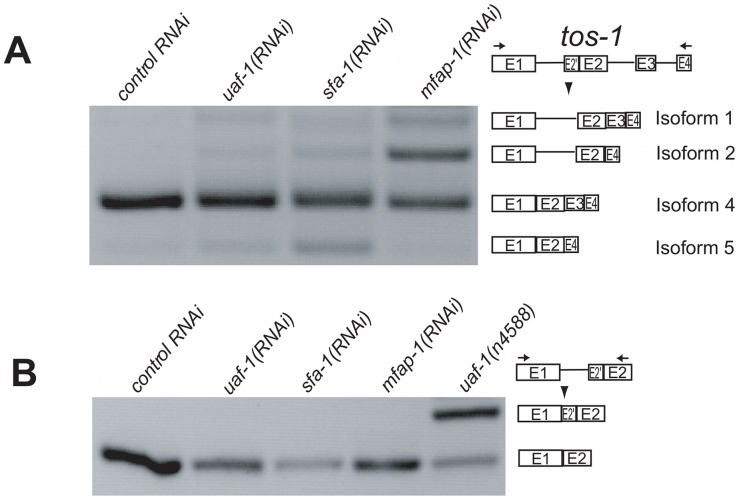
We next examined the splicing of tos-1 in animals fed dsRNA targeting mfap-1. We observed an apparent increase of tos-1 isoforms 1 and 2 (Figure 4A). Increased expression of isoforms 1 and 2 is caused by increased intron 1 retention and exon 3 skipping in tos-1 splicing, as was seen for mfap-1(n4564 n5214) animals (Figure 3). The similarity of mfap-1(n4564 n5214) and mfap-1(RNAi) in affecting tos-1 splicing further suggests that n4564 n5214 causes a reduction or loss of mfap-1 function. Reducing mfap-1 expression by RNAi feeding did not cause the recognition of the cryptic 3′ splice site in intron 1 (Figure 4B). Similarly, that cryptic splice site was not recognized in mfap-1(n4564 n5214) animals (Figure 3D).
Figure 4. Reducing mfap-1 expression by RNA interference alters the splicing of tos-1.
(A) RT-PCR experiments showing the effect of reducing mfap-1 expression by RNAi feeding on tos-1 alternative splicing. tos-1 splice isoforms are illustrated on right. (B) RT-PCR experiments showing the effect of reducing mfap-1 expression by RNAi feeding on the recognition of the cryptic 3′ splice site of tos-1 intron 1.
To determine whether uaf-1 mutations and mfap-1 mutations could interact in affecting RNA splicing, we examined the splicing of tos-1 and the recognition of the cryptic 3′ splice site of tos-1 intron 1 in mfap-1; uaf-1 mutant animals. For each mfap-1; uaf-1 mutant, the splicing of tos-1 appeared to be similar to that in the corresponding uaf-1 single mutant or mfap-1 single mutant (Figure S2A, S2B, top panels). We did not observe additive or synergistic effects of uaf-1 and mfap-1 mutations on intron 1 retention (Figure S3A, e.g., compare uaf-1(n4588 n5127) with mfap-1(n4564 n5214); uaf-1(n4588 n5127)) or exon 3 skipping (Figure S3B, e.g., compare mfap-1(n4564 n5214) with mfap-1(n4564 n5214); uaf-1(n4588 n5127)). Interestingly, the recognition of the cryptic 3′ splice site was reduced by about 50% in mfap-1(n4564 n5214); uaf-1(n4588 n5127) and mfap-1(n5214); uaf-1(n4588 n5127) mutant animals compared to that in uaf-1(n4588 n5127) mutant animals (Figure S2A, S2B, bottom panels and Figure S3C), suggesting that wild-type mfap-1 function might be required for the recognition of the cryptic 3′ splice site in uaf-1(n4588 n5127) animals.
MFAP-1 interacts physically with known splicing factors in a yeast two-hybrid experiment
To further understand the function of MFAP-1, we sought to identify interacting proteins of MFAP-1 using a yeast two-hybrid screen (see Materials and Methods). We isolated and retested nine genes encoding proteins that might potentially interact with MFAP-1 (Table S1). Two of the genes, K04G7.11 and D1054.14, encode the C. elegans orthologs of the presumptive splicing factors SYF2 [34] and Prp38 [35]. Drosophila MFAP-1 (dMFAP1) was found to be in a protein complex with dPrp38 in co-immunoprecipitation experiments and to interact with dPrp38 physically [33]. SYF2 mutations were found to cause synthetic lethality with mutations in splicing factors clf1Delta2 and prp17/cdc40Delta in S. cerevisiae [34], and mammalian orthologs of SYF2 were identified in various spliceosomal complexes by proteomic approaches [3], suggesting that SYF2 acts as a splicing factor. We also identified MFAP-1 as an interactor in the two-hybrid screen, indicating that MFAP-1 might form homodimers. The other potential interactors are two proteins involved in rRNA processing (C05C8.2 and T22H9.1), a ribosomal protein (RPS-6), a Ras-associated PH-domain containing protein (MIG-10), a Rab11 family-interacting protein 2 (Y39F10B.1) and a protein of unknown function (F43C11.9). Although the interactions between MFAP-1 and the proteins from the yeast two-hybrid screen remain to be verified by other approaches, the observation that two presumptive splicing factors (Prp38 and SYF2) are among the candidates is consistent with our findings and also those of Andersen and Tapon [33] indicating that MFAP-1 might function as a splicing factor.
We next examined the splicing of tos-1 in animals fed dsRNAs targeting mfap-1, D1054.14 and K07G7.11 (Figure S4). We found that D1054.14(RNAi) caused an increase of tos-1 splice isoform 2 similar to that seen in mfap-1(RNAi) animals (Figure S4). No apparent alteration in tos-1 splicing was detected in K07G7.11(RNAi) animals. This result is consistent with previous studies indicating that homologs of D1054.14 can function as splicing factors [33], [35] and identified tos-1 as a target of D1054.14 in C. elegans.
Discussion
MFAP-1 is essential for animal development
MFAP1 was initially identified as a putative extracellular matrix protein based on a screen of an embryonic chicken cDNA expression library using antibodies against elastic fiber microfibrils-enriched bovine ocular zonule proteins [32]. Our analysis of C. elegans mfap-1 and the study of the Drosophila ortholog dMFAP1 [33] indicate that MFAP-1 orthologs in these two species are nuclear proteins that affect RNA splicing. In addition, the human ortholog of MFAP-1 was found to be associated with spliceosomes by mass spectrometry analysis [36]–[38]. Since the human ortholog of mfap-1 can rescue the activity of C. elegans mfap-1(n4564 n5214) for suppressing the Unc phenotype of unc-93(e1500) animals, the function of MFAP-1 is likely conserved from nematodes to humans.
Drosophila dMFAP1 acts with several other splicing factors in G2/M progression during mitosis and affects the ratio of pre-mRNA to mature mRNA of the γ-tubulin gene and the mRNA level of the stg/cdc25 gene [33]. It is not known if dMFAP1 affects alternative splicing [33]. C. elegans mfap-1(n4564 n5214) mutants exhibit a temperature-sensitive phenotype with normal growth at 15°C and embryonic lethality at 25°C; mfap-1(tm3456Δ) homozygous animals arrest at L1 to L2 larval stages at 20°C, and mfap-1 RNAi-treated animals arrest at variable larval stages at 20°C (L. Ma and H. R. Horvitz, unpublished observations). These observations indicate that C. elegans mfap-1 is essential for animal development. Whether the developmental defect of mfap-1-deficient animals is caused by mitotic defects remains to be determined.
MFAP-1 is probably required for the recognition of weak 3′ splice sites
We isolated mfap-1(n4564 n5214) from the same genetic screen in which we identified mutations affecting uaf-1 and sfa-1 [29]. mfap-1(n4564 n5214) suppressed the phenotypes of different rubberband Unc mutants in patterns similar to those of uaf-1(n4588) and sfa-1(n4562) mutations. The findings led us to hypothesize that MFAP-1 might function as a splicing factor. Our analysis of tos-1 splicing in mfap-1 mutant animals and in animals treated with mfap-1 RNAi provided molecular evidence that mfap-1 can affect alternative splicing. mfap-1(n4564 n5214) and mfap-1(RNAi) both increased tos-1 intron 1 retention (with a weak 3′ splice site) and tos-1 exon 3 skipping (with a different weak 3′ splice site), and neither affected the splicing of intron 3 (with a strong consensus 3′ splice site TTTTCAG). These observations suggest that MFAP-1 might be required for the recognition of weak 3′ splice sites and not for the recognition of strong 3′ splice sites. We propose that MFAP-1 can act much like UAF-1 and SFA-1, mutations in which cause reduced recognition of the weak 3′ splice sites in introns 1 and 2 and do not affect the recognition of the consensus 3′ splice site in intron 3 of tos-1 [30]. Similar to sfa-1(n4562), mfap-1(n4564 n5214) or mfap-1(RNAi) did not cause recognition of the cryptic 3′ splice site in tos-1 intron 1, suggesting that MFAP-1 probably is not essential for determining the specificity of 3′ splice sites.
The effects of mfap-1(n4564 n5214) and mfap-1(RNAi) on tos-1 splicing are qualitatively similar but quantitatively different. We also observed differences in the effects on tos-1 splicing by genetic mutations and RNAi treatments in our previous studies of uaf-1 and sfa-1, in which we found that uaf-1(n4588) or sfa-1(n4562) caused more dramatic alterations of tos-1 splicing than did either uaf-1(RNAi) or sfa-1(RNAi). This difference was at least partially caused by an altered function of UAF-1 in uaf-1(n4588) animals [30]. Thus, differing effects on target gene splicing by RNAi-treatment and genetic mutation of a splicing factor gene could be caused by several factors, including the differing level of the splicing factor in RNAi-treated and mutant animals and the effects of mutations on the function of the splicing factor.
MFAP-1 might interact with multiple partners to affect RNA splicing
From a yeast two-hybrid screen, we identified nine candidate MFAP-1-interacting proteins. One, D1054.14, is homologous to the S. cerevisiae splicing factor Prp38 [35]. Another, K04G7.11, is orthologous to the S. cerevisiae candidate splicing factor SYF2 [34]. Drosophila splicing factors dPrp38 and dSYF1 were found in a protein complex with dMFAP1, and dPrp38 interacts with dMFAP1 in GST-pulldown experiments [33]. We did not identify orthologs of SYF1 or several other splicing factors found in the Drosophila dMFAP1 complex, possibly because the co-immunoprecipitation experiments could have identified Drosophila proteins that did not directly interact with dMFAP1, whereas our yeast two-hybrid screen could identify only proteins that directly interact with MFAP-1. Both studies suggest that MFAP-1 interacts with multiple splicing factors directly or indirectly and hence that MFAP-1 might play multiple roles in affecting RNA splicing.
We found that reducing the expression of D1054.14 by RNAi caused alterations in tos-1 splicing similar to those caused by mfap-1(RNAi) (Figure S4), suggesting that MFAP-1 and D1054.14/PRP38 might interact to regulate the splicing of the same set of genes. We did not identify alterations in tos-1 splicing in animals fed dsRNA targeting K04G7.11, indicating that K04G7.11 might not be required for the splicing of tos-1.
mfap-1 and uaf-1 might interact to affect RNA splicing
We previously concluded that the suppression of the rubberband Unc phenotype of unc-93(e1500) animals by uaf-1(n4588) and sfa-1(n4562) is probably caused by altered splicing of an unknown gene [29]. The splicing of unc-93(e1500) appears to be normal in mfap-1(n4564 n5214) animals, indicating that altered splicing of unc-93(e1500) transcripts similarly is not the cause of the suppression of unc-93(e1500) Unc phenotype by mfap-1(n4564 n5214).
The uaf-1(n4588) mutation likely causes the recognition of the cryptic 3′ splice site of tos-1 intron 1 by altering rather than decreasing the function of the UAF-1 protein [30]. uaf-1(n4588 n5127) is a weaker mutation than uaf-1(n4588), and the recognition of the cryptic 3′ splice site in uaf-1(n4588 n5127) animals but not in uaf-1(n4588) animals was reduced by the presence of the mfap-1(n4564 n5214) and mfap-1(n5214) mutations (Figure S3C). These results suggest that the recognition of the cryptic 3′ splice site of tos-1 requires MFAP-1 only if that recognition is already weak as in uaf-1(n4588 n5127) mutants. These results further support our conclusion that MFAP-1 affects alternative splicing.
In short, we propose that MFAP-1 can act as a splicing factor. Future studies should reveal how MFAP-1 interacts with other splicing factors to affect C. elegans development by regulating splicing of its target genes.
Materials and Methods
Strains
C. elegans strains were grown at 20°C as described unless otherwise indicated [39]. N2 (Bristol) was the reference wild-type strain [39]. Other strains used in the study were:
LGI: mfap-1(n4564 n5214, n5214, tm3456Δ) (this study).
LGII: sup-9(n1550) [20].
LGIII: uaf-1(n4588, n5123, n4588 n5125, n4588 n5127) [29], unc-93(e1500, n200) [23], sup-18(n1014) [21].
LGIV: sfa-1(n4562) [29].
LGX: sup-10(n983) [21].
CB4856 (Hawaiian) [40]
We used the genetic translocation hT2[bli-4(e937) let-?(q782) qIs48] LG I; LG III [41], which carries an integrated pharyngeal GFP element [42], to balance the mfap-1(tm3456Δ) mutation.
Cloning of mfap-1
We used single-nucleotide polymorphism (SNP) mapping [40] to localize n4564 n5214 to the right of nucleotide 7110 (SNP T01H8: 7110 S = AT) on cosmid T01H8 (T01H8 sequences refer to nucleotides of GenBank accession no. Z80219) and to the left of nucleotide 3334 (SNP F36A2: 3334 S = CT) on cosmid F36A2 (F36A2 sequences refer to nucleotides of GenBank accession no. Z81077) (Figure 1A). We isolated 10 Dpy recombinants after crossing dpy-5(e61) n4564 n5214 hermaphrodites with males of the Hawaiian strain CB4856, and six Unc recombinants after crossing n4564 n5214 unc-75(e950) hermaphrodites with CB4856 males. Cosmid F43G9 rescued three phenotypic characteristics of the n4564 n5214 strain: suppression of unc-93(e1500), partial sterility and temperature-sensitive lethality at 25°C (Figure 1B). We determined the coding sequences of F43G9.10 and identified two missense mutations (GAT-to-GTT (D426V) and ACA-to-GCA (T427A)) in the fourth exon of F43G9.10 (Figure 1C). The predicted proteins encoded by the F43G9.10 orthologs in C. elegans, Drosophila, chicken and human are highly conserved, and the Asp-Thr dipeptide altered in the mfap-1(n4564 n5214) mutant is completely conserved (Figure 1D). A genomic fragment containing a 2.5 kb promoter region, the entire 1.7 kb coding region and a 1.5 kb 3′ downstream region exhibited rescuing activity similar to that of the F43G9 cosmid (Figure 1B). Transgenes expressing F43G9.10 and or the human ortholog of F43G9.10 (Hsmfap-1) in body-wall muscles each rescued the suppression of unc-93(e1500) by mfap-1(n4564 n5214) (Figure 1B). A transgene carrying the mfap-1(n4564 n5214) mutation (mut) failed to rescue the suppression of unc-93(e1500) by mfap-1(n4564 n5214) (Figure 1B), suggesting that this mutation caused the suppressor activity in mfap-1(n4564 n5214) animals.
Screen for suppressors of mfap-1(n4564 n5214)
Synchronized L4 mfap-1(n4564 n5214) animals (P0) grown at 15°C were mutagenized with ethyl methanesulfonate (EMS) as described [39] and grown to gravid adults at 15°C. The P0 adults were bleached, and their F1 progeny were grown to young adults at 15°C and moved to 25°C. After three days at 25°C, the plates were examined for surviving fertile animals. From about 50,000 haploid genomes (F2) screened, two independent suppressed strains were isolated. As animals with a mfap-1(n4564 n5214) phenotype (partial sterility at 20°C and temperature-sensitive lethality at 25°C) were not identified from about 1800 progeny of mfap-1(n4564 n5214) sup/++animals, we concluded that the two suppressors were either intragenic or extragenic and very closely linked to mfap-1. DNA sequence determination identified in both suppressors the same nucleotide change (GCA-to-ACA at codon 427), which converted the mutated amino acid alanine in the mfap-1(n4564 n5214) mutant to the wild-type threonine (A427T) while retaining the D426V mutation. One of the two isolates was used for subsequent studies, and the single mutation it carried was designated mfap-1(n5214).
Yeast two-hybrid screen
We used a yeast two-hybrid screen [43], [44] to identify proteins that might interact with MFAP-1. A pACT2.2 C. elegans yeast two-hybrid library (Addgene plasmid 11523, provided by Dr. Guy Caldwell) was used to screen for MFAP-1 interactors. For the bait, a full-length mfap-1 cDNA was subcloned into the pGBK T7 plasmid (Clontech) and then transfected into the S. cerevisiae strain PJ69-4A. pGBK T7-mfap-1-positive yeast cells were transfected with 2 µg of DNA from the pACT2.2 yeast two-hybrid library, and cells were grown on a SD-Leu-Trp-His medium with 5 mM 3-AT. Positive clones were picked and grown in separate cultures using the same SD medium. Plasmids were obtained from each clone by the smash-and-grab method [45]. Purified plasmids were transfected into the bacterial strain DH5α and grown on LB-ampicillin plates. Plasmids were purified from the bacterial cultures, and sequences of the inserts were determined. Using yeast two-hybrid assays, we confirmed that these clones did not cause survival of yeast cells transfected with the pGBK T7 empty vector on SD-Leu-Trp-His medium, suggesting that proteins encoded by these clones potentially interact with MFAP-1 but not with the GAL4 DBD domain expressed by the pGBK T7 vector.
RNA interference
Young adult animals were fed HT115 (DE3) bacteria containing plasmids directing the expression of dsRNAs targeting uaf-1, sfa-1, mfap-1, D1054.14 or K04G7.11 on NGM plates with 1 mM IPTG and 0.1 mg/ml Ampicillin [46]. F1 progeny of these animals, which arrested at various developmental stages, were washed from plates, rinsed with H2O and resuspended in Trizol (Invitrogen) for preparation of total RNA. The DNA construct that expresses dsRNA targeting uaf-1 and the bacterial strain that expresses dsRNA targeting sfa-1 were described previously [29]. We obtained a bacterial strain expressing dsRNA targeting mfap-1 from a whole-genome RNAi library [47] and bacterial strains expressing dsRNAs targeting mfap-1, D1054.14 or K04G7.11 from an ORFeome-based RNAi library [48]. The sequences of plasmids from single colonies were determined to confirm the presence of coding sequences for each gene.
Body-bend assay
L4 animals were picked 16–24 hrs before being tested. One day later, young adults were individually picked to plates with OP50 bacteria, and body-bends were counted for 30 sec using a dissecting microscope as described [49].
Molecular biology
Total RNA was purified with Trizol (Invitrogen), and cDNA was generated following the protocol provided with the Superscript II kit (Invitrogen). PCR was performed with Eppendorf Cyclers, and DNA products were resolved using agarose gels. NIH ImageJ software was used to quantify tos-1 splice isoforms [30]. The percentages of tos-1 intron 1 retention, exon 3 skipping and the recognition of the intron 1 cryptic 3′ splice were analyzed as described [30]. We performed RT-PCR experiments with animals at different developmental stages and found no indication that tos-1 splicing is regulated developmentally (Figure S5). DNA sequence analysis was performed using an ABI Prism 3100 Genetic Analyzer and an ABI 3730XL DNA Analyzer.
Plasmids
mfap-1 cDNAs were amplified by RT-PCR from wild-type, mfap-1(n4564 n5214) or mfap-1(n5214) animals and subcloned to the pPD93.97 vector in-frame with GFP using BamHI blunt/AgeI restriction sites. For constructs expressing the mfap-1(n4564) transgene, the n4564 mutation was introduced into the pPD93.97::mfap-1cDNA(wt) construct using a QuickChange Site-Directed Mutagenesis Kit (Stratagene) with primers containing the n4564 mutation. For the plasmid used as bait in the yeast two-hybrid screen, full-length mfap-1 cDNA was released from the pGEM-T Easy vector (Promega) with NcoI blunt/PstI and subcloned in-frame into the pGBK T7 vector (Trp1, Kanr) (Clontech) using BamHI blunt/PstI restriction sites. To construct a Pmfap-1::GFP transcriptional fusion transgene, an mfap-1 promoter fragment of about 2.5 kb was amplified using PCR and subcloned into the pPD95.79 vector using PstI/SmaI restriction sites.
Transgene experiments
Microinjection of DNA into the syncytial gonad and the generation of animals with germline transmission of transgenes were performed as described [50] with mfap-1(n4564 n5214) animals grown at 15°C. DNA injection mixtures generally contained 20 µg/ml 1 kb DNA ladder, 20 µg/ml Arabidopsis genomic DNA and 20 µg/ml of the transgene of interest. When the transgene did not express a GFP fusion protein, 20 µg/ml pPD95.86-GFP plasmid (which expresses GFP in body-wall muscles) was added to the injection mixture as a visible fluorescence marker.
Supporting Information
mfap-1(n4564 n5214) does not obviously affect the altered splicing of unc-93(e1500) exon 9. Real-time RT-PCR was performed as described [29] to quantify the recognition of the cryptic 3′ splice site of unc-93(e1500) exon 9. No apparent difference between unc-93(e1500) and mfap-1(n4564 n5214); unc-93(e1500) animals was detected. *: results reported previously by Ma and Horvitz (2009) [29].
(TIF)
mfap-1 and uaf-1 mutations do not exhibit additive or synergistic effects on the altered splicing of tos-1. (A) RT-PCR experiments showing the splicing of tos-1 (top panel) or the recognition of the cryptic 3′ splice site of tos-1 intron 1 (bottom panel) in mfap-1(n4564 n5214) single or mfap-1(n4564 n5214); uaf-1 mutant animals. Genotypes are indicated at the top, and tos-1 splice isoforms on the right. (B) RT-PCR experiments showing the splicing of tos-1 (top panel) or the recognition of the cryptic 3′ splice site of tos-1 intron 1 (bottom panel) in mfap-1(n5214) single or mfap-1(n5214); uaf-1 double mutant animals. Genotypes are indicated at the top, and tos-1 splice isoforms on the right.
(TIF)
Quantification of tos-1 intron 1 retention, exon 3 skipping and the recognition of the intron 1 cryptic 3′ splice site in mfap-1; uaf-1 multiple mutant animals. (A) The molar ratios of all tos-1 splice isoforms with intron 1 retention, presented as a percentage of all isoforms combined. Error bars: standard deviations. (B) The molar ratios of all tos-1 splice isoforms with exon 3 skipping, presented as a percentage of all isoforms combined. Error bars: standard deviations. (C) Percentages of tos-1 isoforms spliced at the cryptic 3′ splice site of tos-1 intron 1 compared to all isoforms spliced at either the endogenous 3′ splice site or the cryptic 3′ splice site. For all analyses, isoform intensities were obtained by analyzing biological duplicates or triplicates using NIH ImageJ software. N.S., no significant difference.
(TIF)
Reducing the expression of mfap-1 or D1054.14 by RNA interference altered the splicing of tos-1. RT-PCR experiments showing the effects of reducing the expression of mfap-1, D1054.14 or K04G7.11 by RNAi feeding on tos-1 alternative splicing. mfap-1(RNAi) and D1054.14(RNAi) caused similar alterations in tos-1 splicing, while K04G7.11(RNAi) did not obviously affect the splicing of tos-1. RNAi bacterial strains were obtained from an ORFeome-based RNAi library [48].
(TIF)
The splicing of tos-1 is not regulated developmentally. RT-PCR experiments examining the splicing of tos-1 in animals at different synchronized developmental stages. tos-1 splicing was similar in all developmental stages examined. YA: young adult animals 24 hours after the L4 larval stage.
(TIF)
Candidate MFAP-1 interactors isolated from a yeast two-hybrid screen.
(DOC)
Acknowledgments
We thank members of the Horvitz laboratory and the Ma laboratory, especially Anna Corrionero, Nicolas Paquin, Takashi Hirose, Shuo Luo, Dengke Ma, Zhaofa Xu, and Ben Wu, for suggestions. We thank Nicolas Paquin for help with the yeast two-hybrid screen. We thank Na An and Yaoyu Zhong for strain maintenance and Rita Droste and Fei Wang for technical support. We thank Andy Fire for providing the pPD plasmids we used for constructing transgene constructs. We thank Shohei Mitani of Tokyo Women's Medical University for providing the tm3456Δ mutant. Some genomic sequences and polymorphisms were identified by the WUSTL Genome Center. Some C. elegans strains were provided by the Caenorhabditis Genetics Center. H. R. Horvitz is the David H. Koch Professor of Biology at MIT and an Investigator of the Howard Hughes Medical Institute.
Footnotes
The authors have declared that no competing interests exist.
This study was supported by a NSFC grant (30971639, www.nsfc.gov.cn) and a National Basic Research Program of China grant to LM (2011CB510005, www.973.gov.cn) and by NIH grant GM24663 (www.nih.gov) to HRH. Some C. elegans strains were provided by the Caenorhabditis Genetics Center, which was funded by the NIH National Center for Research Resources. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463:457–463. doi: 10.1038/nature08909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maniatis T, Tasic B. Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature. 2002;418:236–243. doi: 10.1038/418236a. [DOI] [PubMed] [Google Scholar]
- 3.Jurica MS, Moore MJ. Pre-mRNA splicing: awash in a sea of proteins. Mol Cell. 2003;12:5–14. doi: 10.1016/s1097-2765(03)00270-3. [DOI] [PubMed] [Google Scholar]
- 4.Abovich N, Rosbash M. Cross-intron bridging interactions in the yeast commitment complex are conserved in mammals. Cell. 1997;89:403–412. doi: 10.1016/s0092-8674(00)80221-4. [DOI] [PubMed] [Google Scholar]
- 5.Arning S, Gruter P, Bilbe G, Kramer A. Mammalian splicing factor SF1 is encoded by variant cDNAs and binds to RNA. RNA. 1996;2:794–810. [PMC free article] [PubMed] [Google Scholar]
- 6.Krainer AR, Maniatis T. Multiple factors including the small nuclear ribonucleoproteins U1 and U2 are necessary for pre-mRNA splicing in vitro. Cell. 1985;42:725–736. doi: 10.1016/0092-8674(85)90269-7. [DOI] [PubMed] [Google Scholar]
- 7.Madhani HD, Guthrie C. Dynamic RNA-RNA interactions in the spliceosome. Annu Rev Genet. 1994;28:1–26. doi: 10.1146/annurev.ge.28.120194.000245. [DOI] [PubMed] [Google Scholar]
- 8.Merendino L, Guth S, Bilbao D, Martinez C, Valcarcel J. Inhibition of msl-2 splicing by Sex-lethal reveals interaction between U2AF35 and the 3′ splice site AG. Nature. 1999;402:838–841. doi: 10.1038/45602. [DOI] [PubMed] [Google Scholar]
- 9.Wu S, Romfo CM, Nilsen TW, Green MR. Functional recognition of the 3′ splice site AG by the splicing factor U2AF35. Nature. 1999;402:832–835. doi: 10.1038/45590. [DOI] [PubMed] [Google Scholar]
- 10.Zamore PD, Green MR. Biochemical characterization of U2 snRNP auxiliary factor: an essential pre-mRNA splicing factor with a novel intranuclear distribution. EMBO J. 1991;10:207–214. doi: 10.1002/j.1460-2075.1991.tb07937.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zorio DA, Blumenthal T. Both subunits of U2AF recognize the 3′ splice site in Caenorhabditis elegans. Nature. 1999;402:835–838. doi: 10.1038/45597. [DOI] [PubMed] [Google Scholar]
- 12.Reed R. Mechanisms of fidelity in pre-mRNA splicing. Curr Opin Cell Biol. 2000;12:340–345. doi: 10.1016/s0955-0674(00)00097-1. [DOI] [PubMed] [Google Scholar]
- 13.Cooper TA, Wan L, Dreyfuss G. RNA and disease. Cell. 2009;136:777–793. doi: 10.1016/j.cell.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang GS, Cooper TA. Splicing in disease: disruption of the splicing code and the decoding machinery. Nat Rev Genet. 2007;8:749–761. doi: 10.1038/nrg2164. [DOI] [PubMed] [Google Scholar]
- 15.Guth S, Valcarcel J. Kinetic role for mammalian SF1/BBP in spliceosome assembly and function after polypyrimidine tract recognition by U2AF. J Biol Chem. 2000;275:38059–38066. doi: 10.1074/jbc.M001483200. [DOI] [PubMed] [Google Scholar]
- 16.Rutz B, Seraphin B. Transient interaction of BBP/ScSF1 and Mud2 with the splicing machinery affects the kinetics of spliceosome assembly. RNA. 1999;5:819–831. doi: 10.1017/s1355838299982286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tanackovic G, Kramer A. Human splicing factor SF3a, but not SF1, is essential for pre-mRNA splicing in vivo. Mol Biol Cell. 2005;16:1366–1377. doi: 10.1091/mbc.E04-11-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pleiss JA, Whitworth GB, Bergkessel M, Guthrie C. Transcript specificity in yeast pre-mRNA splicing revealed by mutations in core spliceosomal components. PLoS Biol. 2007;5:e90. doi: 10.1371/journal.pbio.0050090. doi: 10.1371/journal.pbio.0050090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blanchette M, Labourier E, Green RE, Brenner SE, Rio DC. Genome-wide analysis reveals an unexpected function for the Drosophila splicing factor U2AF50 in the nuclear export of intronless mRNAs. Mol Cell. 2004;14:775–786. doi: 10.1016/j.molcel.2004.06.012. [DOI] [PubMed] [Google Scholar]
- 20.de la Cruz IP, Levin JZ, Cummins C, Anderson P, Horvitz HR. sup-9, sup-10, and unc-93 may encode components of a two-pore K+ channel that coordinates muscle contraction in Caenorhabditis elegans. J Neurosci. 2003;23:9133–9145. doi: 10.1523/JNEUROSCI.23-27-09133.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Greenwald I, Horvitz HR. A visible allele of the muscle gene sup-10 X of C. elegans. Genetics. 1986;113:63–72. doi: 10.1093/genetics/113.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Greenwald IS, Horvitz HR. unc-93(e1500): A behavioral mutant of Caenorhabditis elegans that defines a gene with a wild-type null phenotype. Genetics. 1980;96:147–164. doi: 10.1093/genetics/96.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levin JZ, Horvitz HR. The Caenorhabditis elegans unc-93 gene encodes a putative transmembrane protein that regulates muscle contraction. J Cell Biol. 1992;117:143–155. doi: 10.1083/jcb.117.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brinkmann MM, Spooner E, Hoebe K, Beutler B, Ploegh HL, et al. The interaction between the ER membrane protein UNC93B and TLR3, 7, and 9 is crucial for TLR signaling. J Cell Biol. 2007;177:265–275. doi: 10.1083/jcb.200612056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Casrouge A, Zhang SY, Eidenschenk C, Jouanguy E, Puel A, et al. Herpes simplex virus encephalitis in human UNC-93B deficiency. Science. 2006;314:308–312. doi: 10.1126/science.1128346. [DOI] [PubMed] [Google Scholar]
- 26.Kim YM, Brinkmann MM, Paquet ME, Ploegh HL. UNC93B1 delivers nucleotide-sensing toll-like receptors to endolysosomes. Nature. 2008;452:234–238. doi: 10.1038/nature06726. [DOI] [PubMed] [Google Scholar]
- 27.Koehn J, Huesken D, Jaritz M, Rot A, Zurini M, et al. Assessing the function of human UNC-93B in Toll-like receptor signaling and major histocompatibility complex II response. Hum Immunol. 2007;68:871–878. doi: 10.1016/j.humimm.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 28.Tabeta K, Hoebe K, Janssen EM, Du X, Georgel P, et al. The Unc93b1 mutation 3d disrupts exogenous antigen presentation and signaling via Toll-like receptors 3, 7 and 9. Nat Immunol. 2006;7:156–164. doi: 10.1038/ni1297. [DOI] [PubMed] [Google Scholar]
- 29.Ma L, Horvitz HR. Mutations in the Caenorhabditis elegans U2AF large subunit UAF-1 alter the choice of a 3′ splice site in vivo. PLoS Genet. 2009;5:e1000708. doi: 10.1371/journal.pgen.1000708. doi: 10.1371/journal.pgen.1000708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma L, Tan Z, Teng Y, Hoersch S, Horvitz HR. In vivo effects on intron retention and exon skipping by the U2AF large subunit and SF1/BBP in the nematode Caenorhabditis elegans. . RNA. 2011;17:2201–2211. doi: 10.1261/rna.027458.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Levin JZ, Horvitz HR. Three new classes of mutations in the Caenorhabditis elegans muscle gene sup-9. Genetics. 1993;135:53–70. doi: 10.1093/genetics/135.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Horrigan SK, Rich CB, Streeten BW, Li ZY, Foster JA. Characterization of an associated microfibril protein through recombinant DNA techniques. J Biol Chem. 1992;267:10087–10095. [PubMed] [Google Scholar]
- 33.Andersen DS, Tapon N. Drosophila MFAP1 is required for pre-mRNA processing and G2/M progression. J Biol Chem. 2008;283:31256–31267. doi: 10.1074/jbc.M803512200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vincent K, Wang Q, Jay S, Hobbs K, Rymond BC. Genetic interactions with CLF1 identify additional pre-mRNA splicing factors and a link between activators of yeast vesicular transport and splicing. Genetics. 2003;164:895–907. doi: 10.1093/genetics/164.3.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blanton S, Srinivasan A, Rymond BC. PRP38 encodes a yeast protein required for pre-mRNA splicing and maintenance of stable U6 small nuclear RNA levels. Mol Cell Biol. 1992;12:3939–3947. doi: 10.1128/mcb.12.9.3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Makarov EM, Makarova OV, Urlaub H, Gentzel M, Will CL, et al. Small nuclear ribonucleoprotein remodeling during catalytic activation of the spliceosome. Science. 2002;298:2205–2208. doi: 10.1126/science.1077783. [DOI] [PubMed] [Google Scholar]
- 37.Rappsilber J, Ryder U, Lamond AI, Mann M. Large-scale proteomic analysis of the human spliceosome. Genome Res. 2002;12:1231–1245. doi: 10.1101/gr.473902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou Z, Licklider LJ, Gygi SP, Reed R. Comprehensive proteomic analysis of the human spliceosome. Nature. 2002;419:182–185. doi: 10.1038/nature01031. [DOI] [PubMed] [Google Scholar]
- 39.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wicks SR, Yeh RT, Gish WR, Waterston RH, Plasterk RH. Rapid gene mapping in Caenorhabditis elegans using a high density polymorphism map. Nat Genet. 2001;28:160–164. doi: 10.1038/88878. [DOI] [PubMed] [Google Scholar]
- 41.McKim KS, Peters K, Rose AM. Two types of sites required for meiotic chromosome pairing in Caenorhabditis elegans. . Genetics. 1993;134:749–768. doi: 10.1093/genetics/134.3.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Edgley ML, Baillie DL, Riddle DL, Rose AM. Genetic balancers. WormBook. 2006 doi: 10.1895/wormbook.1.89.1. doi/10.1895/wormbook.1891.1897.1891, http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chien CT, Bartel PL, Sternglanz R, Fields S. The two-hybrid system: a method to identify and clone genes for proteins that interact with a protein of interest. Proc Natl Acad Sci U S A. 1991;88:9578–9582. doi: 10.1073/pnas.88.21.9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 45.Hoffman CS. Preparation of yeast DNA. Curr Protoc Mol Biol. 2011;13:11. doi: 10.1002/0471142727.mb1311s39. [DOI] [PubMed] [Google Scholar]
- 46.Timmons L, Court DL, Fire A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 2001;263:103–112. doi: 10.1016/s0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]
- 47.Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- 48.Rual JF, Ceron J, Koreth J, Hao T, Nicot AS, et al. Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome Res. 2004;14:2162–2168. doi: 10.1101/gr.2505604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sawin ER, Ranganathan R, Horvitz HR. C. elegans locomotory rate is modulated by the environment through a dopaminergic pathway and by experience through a serotonergic pathway. Neuron. 2000;26:619–631. doi: 10.1016/s0896-6273(00)81199-x. [DOI] [PubMed] [Google Scholar]
- 50.Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
mfap-1(n4564 n5214) does not obviously affect the altered splicing of unc-93(e1500) exon 9. Real-time RT-PCR was performed as described [29] to quantify the recognition of the cryptic 3′ splice site of unc-93(e1500) exon 9. No apparent difference between unc-93(e1500) and mfap-1(n4564 n5214); unc-93(e1500) animals was detected. *: results reported previously by Ma and Horvitz (2009) [29].
(TIF)
mfap-1 and uaf-1 mutations do not exhibit additive or synergistic effects on the altered splicing of tos-1. (A) RT-PCR experiments showing the splicing of tos-1 (top panel) or the recognition of the cryptic 3′ splice site of tos-1 intron 1 (bottom panel) in mfap-1(n4564 n5214) single or mfap-1(n4564 n5214); uaf-1 mutant animals. Genotypes are indicated at the top, and tos-1 splice isoforms on the right. (B) RT-PCR experiments showing the splicing of tos-1 (top panel) or the recognition of the cryptic 3′ splice site of tos-1 intron 1 (bottom panel) in mfap-1(n5214) single or mfap-1(n5214); uaf-1 double mutant animals. Genotypes are indicated at the top, and tos-1 splice isoforms on the right.
(TIF)
Quantification of tos-1 intron 1 retention, exon 3 skipping and the recognition of the intron 1 cryptic 3′ splice site in mfap-1; uaf-1 multiple mutant animals. (A) The molar ratios of all tos-1 splice isoforms with intron 1 retention, presented as a percentage of all isoforms combined. Error bars: standard deviations. (B) The molar ratios of all tos-1 splice isoforms with exon 3 skipping, presented as a percentage of all isoforms combined. Error bars: standard deviations. (C) Percentages of tos-1 isoforms spliced at the cryptic 3′ splice site of tos-1 intron 1 compared to all isoforms spliced at either the endogenous 3′ splice site or the cryptic 3′ splice site. For all analyses, isoform intensities were obtained by analyzing biological duplicates or triplicates using NIH ImageJ software. N.S., no significant difference.
(TIF)
Reducing the expression of mfap-1 or D1054.14 by RNA interference altered the splicing of tos-1. RT-PCR experiments showing the effects of reducing the expression of mfap-1, D1054.14 or K04G7.11 by RNAi feeding on tos-1 alternative splicing. mfap-1(RNAi) and D1054.14(RNAi) caused similar alterations in tos-1 splicing, while K04G7.11(RNAi) did not obviously affect the splicing of tos-1. RNAi bacterial strains were obtained from an ORFeome-based RNAi library [48].
(TIF)
The splicing of tos-1 is not regulated developmentally. RT-PCR experiments examining the splicing of tos-1 in animals at different synchronized developmental stages. tos-1 splicing was similar in all developmental stages examined. YA: young adult animals 24 hours after the L4 larval stage.
(TIF)
Candidate MFAP-1 interactors isolated from a yeast two-hybrid screen.
(DOC)