Abstract
Mutations in retinal-specific guanylate cyclase (Gucy2d) are associated with Leber congenital amaurosis-1 (LCA1). Zebrafish offer unique advantages relative to rodents, including their excellent color vision, precocious retinal development, robust visual testing strategies, low cost, relatively easy transgenesis and shortened experimental times. In this study we will demonstrate the feasibility of using gene-targeting in the zebrafish as a model for the photoreceptor-specific GUCY2D-related LCA1, by reporting the visual phenotype and retinal histology resulting from Gucy2f knockdown. Gucy2f zebrafish LCA-orthologous cDNA was identified and isolated by PCR amplification. Its expression pattern was determined by whole-mount in-situ hybridization and its function was studied by gene knockdown using two different morpholino-modified oligos (MO), one that blocks translation of Gucy2f and one that blocks splicing of Gucy2f. Visual function was assessed with an optomotor assay on 6-days-post-fertilization larvae, and by analyzing changes in retinal histology. Gucy2f knockdown resulted in significantly lower vision as measured by the optomotor response compared with uninjected and control MO-injected zebrafish larvae. Histological changes in the Gucy2f-knockdown larvae included loss and shortening of cone and rod outer segments. A zebrafish model of Gucy2f-related LCA1 displays early visual dysfunction and photoreceptor layer dystrophy. This study serves as proof of concept for the use of zebrafish as a simple, inexpensive model with excellent vision on which further study of LCA-related genes is possible.
Keywords: leber congenital amaurosis, animal model, GUCY2D, optomotor assay
Introduction
Leber congenital amaurosis (LCA) is a family of severe, early-onset, autosomal-recessive forms of pediatric blindness. LCA has been linked to more than 16 genes, often exhibiting clinical heterogeneity.1, 2 LCA1 is caused by mutations in the gene GUCY2D, which encodes the retinal Gucy2d-1 (GC1) and accounts for ∼20% of all cases of LCA.3 LCA2 is caused by mutations in the RPE65 gene, which is expressed in the retinal pigment epithelium. Breakthrough research has demonstrated visual improvement following gene therapy trials in humans with LCA2.4, 5 Human trials relied on a decade of proof-of-concept studies in canine and rodent models.6, 7, 8 Unlike LCA2, in which gene therapy is aimed at correcting an RPE defect, searching for gene therapy for LCA1 will involve an attempt at treating the photoreceptors.
Research in inherited retinal disease relies heavily on animal models, commonly mammals such as mice and dogs. These animal models have significant advantages, including the availability of knockout technology. Two specific drawbacks slow the applicability of rodent or canine models for study of relatively rare genetic diseases such as LCA and other retinal dystrophies. The first difficulty is the high cost and length of time required to create a new knockout line; thus, it is applicable only for common or large-scale genetic studies. The second drawback when using rodent or canine models for retinal disease is their rod-dominated vision. They lack a cone-only fovea, the area of the human retina, which is essential for acute, daylight vision.
Zebrafish have surged in importance as an animal model in almost every field of medicine,9, 10, 11 owing to their publicly available genomic sequence,12 their external fertilization, rapid development, accessibility of embryos (larvae) and the evolutionary conservation of the vertebrate body plan. Their precocious development of vision and remarkable evolutionary conservation of the eye make the zebrafish retina an excellent model for human visual disorders.13, 14, 15, 16 Another advantage of the zebrafish as a model for human retinal disease is their cone-dominant retina, opening the possibility to specifically analyze cone-mediated disorders that are more difficult to study in the rod dominant nocturnal mouse. Robust visually-guided assays, including the optomotor and optokinetic tests allow quantitative measurements of visual impairment in zebrafish.17, 18, 19, 20, 21, 22 Yan, Sun and Engelhardt stressed the advantage of non-mouse animal models for accelerated gene-targeting research.23 Knockdown techniques, which are readily available in zebrafish, are particularly suited for research of early onset genetic blindness such as LCA and can be used to develop alternative LCA-models. In a landmark article, Baye et al24 showed that knockdown of zebrafish Cep290 via MOs resulted in a reduction of Kupffer's vesicle size and delays melanosome transport.23, 24 Supplementation with the N-terminal region of the CEP290 protein alone was sufficient to restore visual function suggesting that a similar method may be effective in patients with mutations in CEP290.24
Here we demonstrate that knockdown of zebrafish photoreceptor-specific Gucy2d gene (Gucy2f), a gene associated with arguably the most prevalent form of LCA,1 can serve as a readily-available, inexpensive animal model for LCA1.
Methods
Zebrafish handling
Adult zebrafish (Danio rerio; AB and TL strains) were raised in a recirculation water system under 12:12 h of light–dark cycles at 28.5 °C, adhering to the ARVO statement for the use of animals in Ophthalmic and Vision Research. To produce embryos, male and female zebrafish were paired in the evening. Spawning occurred the next morning within 1 h of light. Embryos were placed in 10-centimeter Petri dishes with egg water (60 p.p.m. sea salts) containing methylene blue (0.3 p.p.m.) and raised in a light-controlled incubator at 28.5 °C (light intensity 12 W/m2). In preparation for whole mount in situ hybridization, 1-phenyl-2-thiourea (0.2 mM) was added to the water to prevent pigmentation.
Cloning of the zebrafish GUCY2D ortholog
The zebrafish genome was searched for GUCY2D orthologous genes (the Sanger zebrafish genomic sequence project database, Zv7), using the human GUCY2D (GeneBank accession number NM_000180) as an input and the tblastn algorithm (http://www.ncbi.nlm.nih.gov). The most similar zebrafish gene is annotated as Gucy2f (ref_seq NM-131864.2, encoding NP_571939.2). An 879-bp fragment was PCR amplified using specific forward primer, 5′-AGGAGACACTGCCACCAG-3′ and reverse primer 5′-AGACCCACGACTCCAGCA-3′. The amplified fragment was cloned into pGEM T-easy (Promega, Madison, WI, USA) and the identity of the resulting clone was verified by sequencing and by comparison with the zebrafish genome. This clone was used as a template to synthesize DIG-labeled antisense riboprobe for whole mount in-situ hybridization.
Whole mount in-situ hybridization (ISH)
The expression pattern of the putative Gucy2D ortholog, the zebrafish Gucy2f, was determined by whole mount in situ hybridization of zebrafish embryos and larvae. Zebrafish embryos were fixed overnight with 4% paraformaldehyde (PFA) in phosphate-buffered saline (PBS) and stored in 100% methanol. Whole-mount ISH analysis was carried out according to established protocol.25, 26 Briefly, fixed and methanol-preserved embryos were rinsed in cold acetone for 8 min, dehydrated through a graded series of methanol concentrations, rinsed twice for 5 min with PBS and treated with Proteinase K (10 μg/ml in PBS) for 5–60 min (depending on the developmental stage of the animal). Samples were then fixed for 30 min (4% PFA in PBS), washed in PBTw (0.1% Tween-20 in PBS), and prehybridized for 3 h at 65 °C in hybridization buffer composed of 50% formamide 5 × standard sodium citrate (SSC), 5 mM EDTA, 0.1% Tween-20, 0.1% CHAPS, 50 mg/ml heparin and 1 mg/ml yeast RNA. Hybridization was carried out overnight (65°C, 1 μg/ml riboprobe in hybridization buffer), the samples were then rinsed in washing buffer 1 (2 × 30 min, 50% formamide, 2 × SSC, 0.15% CHAPS), washing buffer 2 (15 min, 2XSSC, 0.3% CHAPS), washing buffer 3 (2 × 30 min, 0.2% SSC, 0.3% CHAPS) and finally MAB buffer (0.1 M maleic acid, 150 mM NaCl, pH 7.5) at room temperature. After 1 h of pre-incubation in blocking solution (2% blocking reagent, 5% calf serum in MAB buffer), the samples were incubated with anti-DIG antibody conjugated to alkaline phosphatase (AP; 1:5000; Roche, Rotkreuz, Switzerland) in blocking solution for 3 h. The riboprobe-antibody complex was detected by the enzymatic reaction of AP with a chromogenic substrate (BM purple AP substrate, Roche) yielding a purple precipitate. Staining reaction was stopped by replacing BM purple AP substrate with PBS followed by fixation in 4% PFA. Embryos were then placed in 70% glycerol and observed under a dissecting stereoscope and photographed using a digital camera.
Gucy2f knockdown procedure
Knockdown of zebrafish Gucy2f was performed via microinjection of morpholino modified antisense oligos (MO) (Genetools, LLC, Philomath, OR, USA) to one- or two-cell-stage zebrafish zygotes as described.27, 28 Two different, non-overlapping MOs were injected on separate batches of larvae to reconfirm the observed functional effect. The first MO sequence, 5′-AGAGACCTGAAAGGGAACAGAATGT-3′ (Gucy2fI2E3MO), was designed to target the intron2-exon3 boundary of the zebrafish Gucy2f gene, to interfere with splicing. The second MO sequence was targeted to the translational start site, AUG, to block translation initiation: 5′-GAGGTGAATATGACGCATTGTCCTC-3′ (Gucy2fATGMO). BLAST (Basic local alignment search tool) analysis was performed using UCSC genome browser (http://genome.ucsc.edu/) to verify that this MO does not bind gene sequences other than its target. Standard control MO, 5′-CCTCTTACCTCAGTTACAATTTATA-3′, which has not been reported to have other targets or generate any phenotypes in any known test, was injected as a negative control.
Visual testing procedure
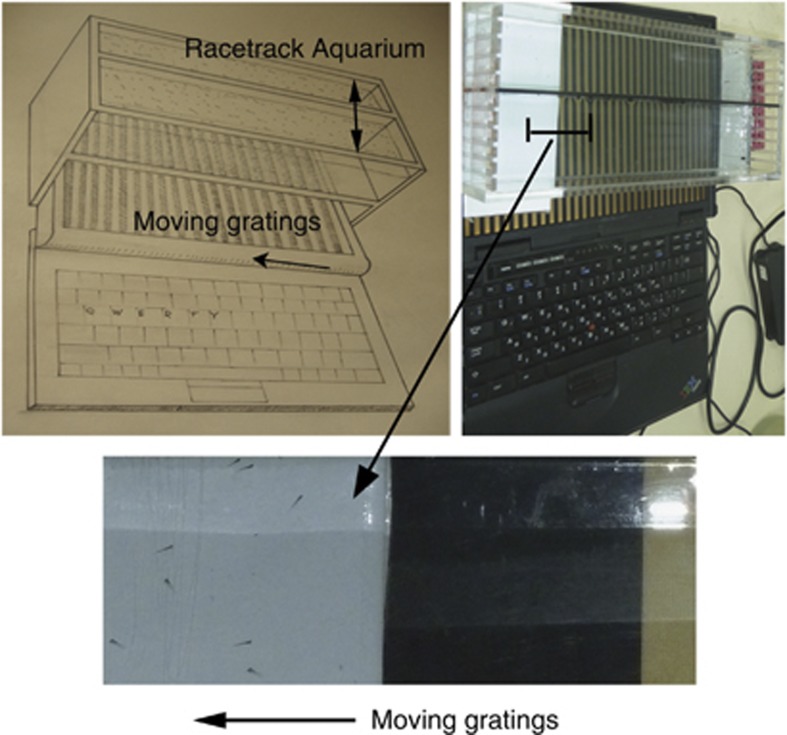
Repeated stimulation by moving grating stimuli (256 pixels per cycle, corresponding to 0.16 cycles/degree at a distance of 8 cm, speed 144 pixels/s; we make the moving stimulus program freely available at http://michaelbach.de/stim/) was presented to groups of 36-days-post-fertilization zebrafish larvae in a transparent racetrack aquarium placed on top of a laptop computer screen (Figure 1). The side walls of the racetracks were opaque, thus the fish could only view the laptop screen. Larvae were tested specifically at this age because no schooling behavior is observed at this stage of development,29 yet their vision is already well developed. Larvae were repeatedly exposed to gratings moving in two different directions; six runs of 90-sec duration for six times, three in each direction. Each set of six tests was repeated on four different days with new fish for each test, to assess inter-individual variability. Altogether, responses were recorded 24 times from four different sets of 30 zebrafish larvae. To quantify the population response to the optomotor assay, the tank was subdivided into four equal sections orthogonally to the racetracks: For each grating motion direction, the last quarter section of the computer screen and aquarium area was denoted as the ‘target area' (left or right side). After the stimulus ran for 90 s, a digital photograph of the target area of the aquarium was taken. From this documentation the proportion of the fish in the target section was counted in relation to the total number of fish. Blind mutant fish are expected to either exhibit no optomotor responses, or to respond only to coarse gratings if visually impaired.20, 30
Figure 1.
The optomotor visual assay; diagram (left), photograph (right), close-up (below). A fish aquarium containing two ‘racetracks' is placed above a computer screen. Thirty wild-type larvae are placed in each track. Moving gratings are presented on the screen to the fish larvae through the transparent aquarium. After 90 s of grating presentation, the number of fish in the last one quarter of the track, the target area, is counted and recorded as a proportion of the total number of fish.
Retinal histology
Larvae were fixed for 24 h in 4% PFA, followed by rinsing-off the PFA with zebrafish egg water twice (The Zebrafish Book, https://wiki.zfin.org/display/prot/ZFIN+Protocol+Wiki). Mirror-agarose molds were prepared according to the technique described by Tsao et al.31 The mirror-agarose gels enabled adequate alignment of the small larvae, needed for accurate microtome sectioning. The agarose blocks were immersed in 70% ethanol for 24 h, followed by dehydration until Paraffin fixation was performed. Three-micron-thick sections were prepared and stained with hematoxylin–eosin (H&E) and with immune-histochemical staining of the zebrafish double cones. For double-cone staining, the zpr1 primary antibody (Zebrafish International Resource Center, Eugene OR, USA, at a dilution of 1:20) was used followed by secondary-biotin stains, using the technique described by Larison and BreMiller.32 From each study group, 15 different larvae were randomly selected and screened. Four representative non-overlapping high-power fields ( × 100) at mid-lens sections from different animals of each study group (wild-type, Gucy2fI2E3 knockdown and Gucy2fATG knockdown) were selected. Mean double-cone counts from each group were compared using ANOVA.
Results
Identification of the conserved Gucy2f ortholog gene
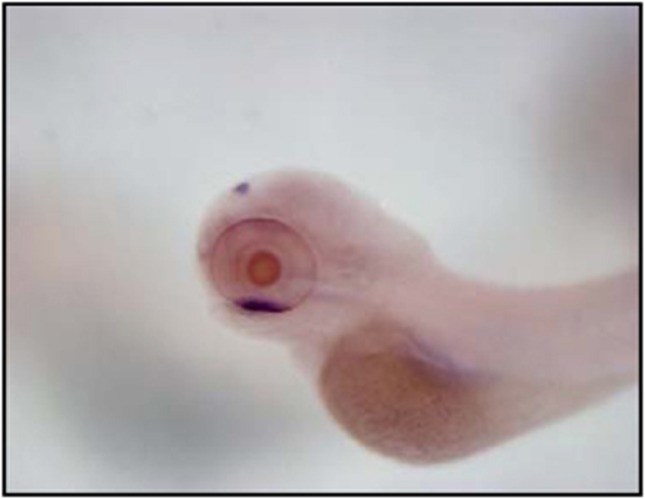
Using bioinformatic tools and the available zebrafish genome data base, we have identified a short sequence with relatively high similarity (80% identity) to the human GUCY2D mRNA. A 879 bp fragment was then PCR amplified and cloned. This sequence is part of a larger gene, located on zebrafish chromosome 15, which has been annotated by the Sanger genome project Gucy2f (refSeq NP_131864.2). Importantly, expression of this Gucy2f ortholog is highly specific for the zebrafish retina. Figure 2 displays dense uptake in the ventral patch of the DIG-labeled Gucy2f- antisense riboprobe. The ventral patch is the area in which the retina develops at 3 days post fertilization.33
Figure 2.
Early developmental expression pattern of the zebrafish Gucy2f gene by whole-mount in situ hybridization. Wild-type zebrafish larva at 3 days post fertilization is observed under a stereo dissecting microscope embryo (lateral view above, superior view below), demonstrating dense uptake of the DIG-labeled Gucy2f antisense riboprobe in the retina ventral patch.
The whole mount ISH additionally displays some uptake in the pineal gland. The sequence conservation and retinal expression together suggest that zebrafish Gucy2f is the ortholog gene of the LCA-related human GUCY2D.
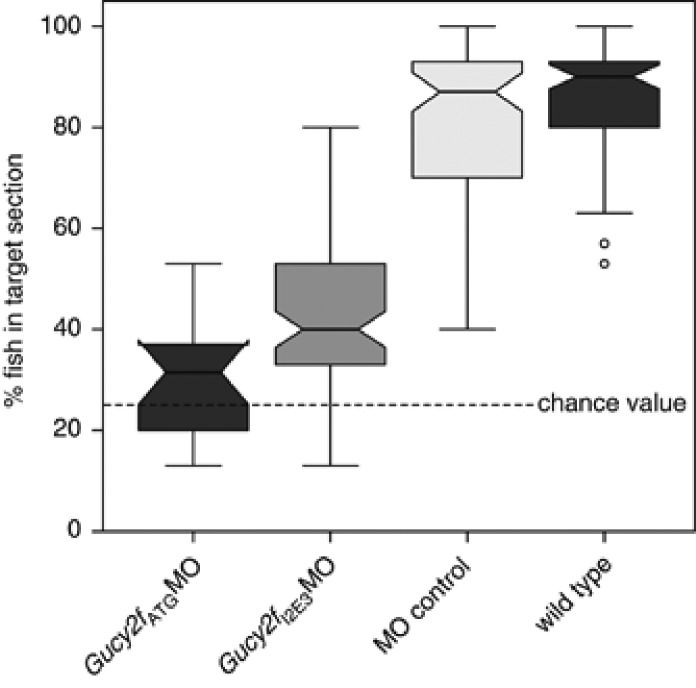
Gucy2f knockdown impairs vision
Following injection of the two non-overlapping MOs, Gucy2fI2E3MO and Gucy2fATGMO on separate batches of larvae, both batches displayed markedly impaired visual acuity in the optomotor assay when compared with wild-type and control MO-injected larvae. Figure 3 displays the percent of larvae within the target area after 90 s of grating motion. As the tank was divided into quarters, the random chance value of being in any quarter would be 25%. Of the wild-type zebrafish larvae and of the control MO-injected 6-day-old larvae a median of 90% (confidence interval (CI) 61.7–100%) and 87% (CI 50–97%), respectively, were in the target section, while the median of the Gucy2fI2E3MO batch and the Gucy2fATGMO-injected zebrafish following the 0.16 cycles/degree grating that were in the target section was 40.0% (CI 22.8–77.2%) and 31.5% (CI 14.3–48.8), respectively, and were significantly above the chance level of 25%, differing significantly from both wild type and control (P<0.0001 in both cases after Bonferroni correction for multiple testing).
Figure 3.
Visual response of the zebrafish Gucy2fI2E3MO, Gucy2fATGMO, wild-type larvae and control MO-injected zebrafish. Group results are depicted as boxplots: the thick line indicating the median, the notch the 95% CI for the median, the boxes are the ±25% percentiles, the antennas the ranges and the outliers (1.5 × the interquartile range outside the quartiles) are represented as circles.
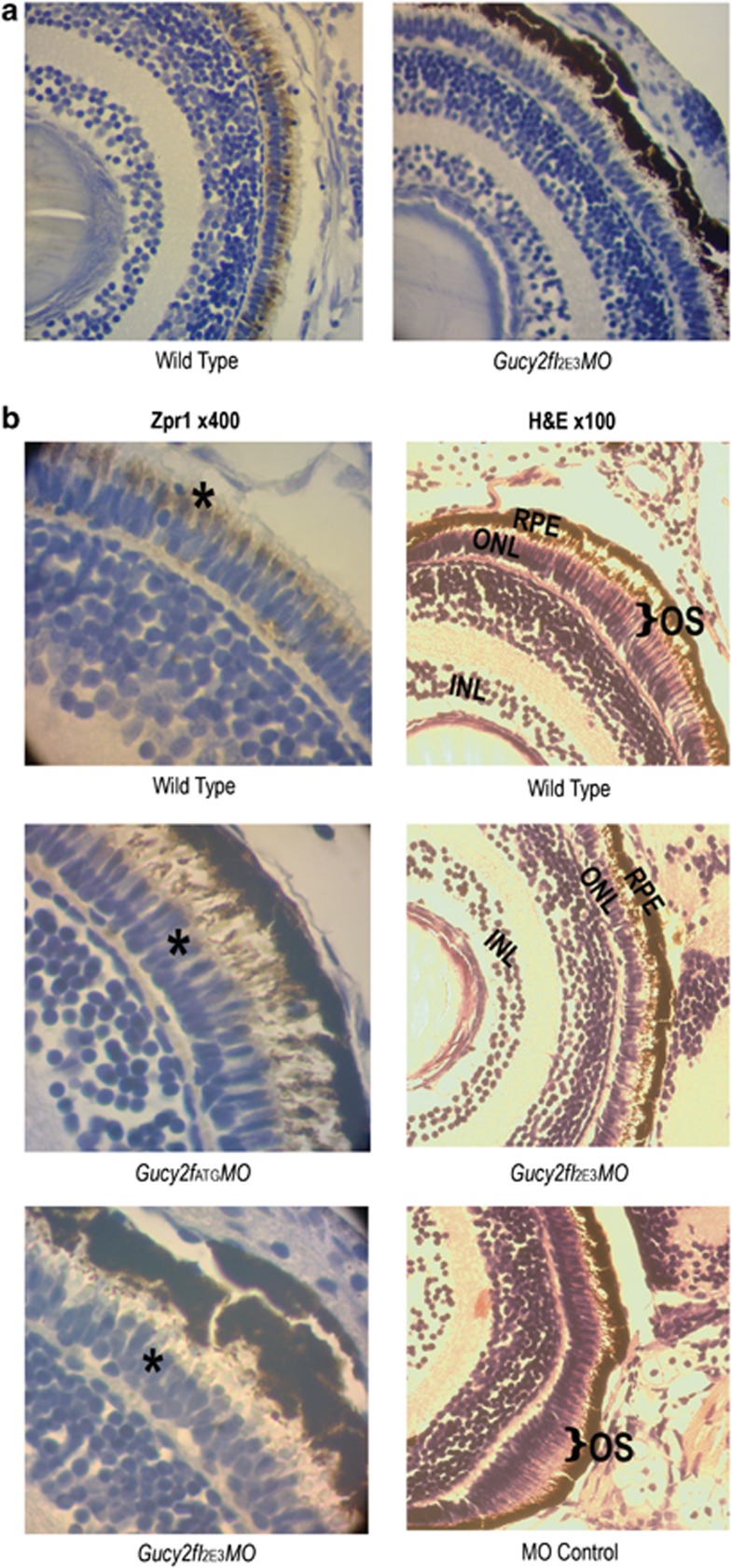
Gucy2f knockdown leads to retinal dystrophy – histology
Knockdown of the Gucy2f in zebrafish larvae led to early-onset immunohistochemical changes in the retina. Double-cone staining with the zpr1 antibody was markedly reduced in both Gucy2fI2E3MO, Gucy2fATGMO compared with wild-type larvae. Representative slides were used to count double-cone staining in the wild-type group (mean count per × 40 field =39.5, 95% CI: 33.5–45.6, standard deviation (SD)=9.0); almost no double-cone staining in the Gucy2fATGMO-injected larvae (mean=0.75, 95% CI: −5.29–6.79, SD=1.50) nor in the Gucy2fI2E3MO-injected larvae (C) (mean=1.25, 95% CI: −4.79 through 7.29, SD=1.50) was seen. The probability of this result using an ANOVA, assuming the null hypothesis, is <0.01.
Figure 4a demonstrates representative low-magnification slides from zpr1 staining. One may appreciate the diffuse zpr1 staining of the double cones in wild-type larvae, which is not present in Gucy2f knockdown larvae. Figure 4b, left column, displays higher magnifications showing details of double-cone staining patterns in the wild-type larvae as compared with the reduced staining in Gucy2fI2E3MO and Gucy2fATGMO.
Figure 4.
Histological findings reveal that Gucy2f knockdown leads to retinal dystrophy. (a) First row: overall view of the retina demonstrates abundant zpr1 staining in wild-type larvae, absent in Gucy2fI2E3MO. (b) Left column: representative sections ( × 400) following zpr-1 antibody staining of double cones reveals (brown) double-cone staining (asterix) in the wild type in contrast to reduced staining in Gucy2fATGMO and in Gucy2fI2E3MO larvae. Right column: light microscopy, hematoxylin–eosin stain (H&E, × 100. Wild-type zebrafish larvae reveal normal retinal structures and development of the photoreceptor layers. Black bracket indicates length of photoreceptor outer segments. Gucy2fI2E3MO larvae display shortening of photoreceptor outer segments (no bracket). In the MO control, note the normal outer segments (black brackets).
Figure 4b displays representative slides of H&E from wild-type larvae, Gucy2fI2E3MO larvae and fish injected with control MO. Structures were assessed by morphology, based on studies describing the zebrafish retina and photoreceptors.27, 32, 34 Prominent outer segment shortening was observed in Gucy2f-knockdown larvae, as compared with wild-type larvae and control MO-injected larvae.
Discussion
Recessive mutations in the photoreceptor-specific Gucy2d gene (retGC-1); GUCY2D are associated with LCA1, one of the most prevalent forms of this disease family.3 Dominant mutations in GC1 are also a major cause of cone-rod (including CORD6) and cone dystrophies, accounting for up to 35% of such cases.35, 36, 37, 38 LCA1 is part of a family of severe, early-onset, autosomal recessive blinding disorders characterized by extinguished electroretinogram, which precedes photoreceptor degeneration. LCA1 patients present in infancy with severely impaired vision and extinguished ERG despite a normal fundus and retained photoreceptors in both their macular and peripheral retina for decades.39, 40, 41, 42 There is relatively better maintenance of retinal structure in LCA1 patients than that seen in other forms of the disease.42 Taken together, this suggests an important role for the zebrafish as an animal model on which rapid visual screening and retinal histology may offer a unique advantage in the study of GC1 disease through both gene knockdown or future studies of dominant gene supplementation.
In this study we have found that the zebrafish ortholog for human GUCY2D has a crucial and early effect on vision and on retinal organization. At 6 days post-fertilization, larvae with Gucy2f gene knockdown exhibited markedly decreased vision, with only 32–40% of larvae responding to a stimulus normally seen by ∼90% of wild-type larvae.
The fact that vision was measured slightly higher than the 25% chance value may be explained in several ways. A visual response that is markedly reduced but higher than chance response following Gucy2f knockdown could imply the existence of residual vision. This is in accord with the slight preservation of vision documented in some human GUCY2D patients. Li et al1 found a visual acuity of at least finger counting in 3 out of 13 patients (Table S6), whereas Perrault et al43 recorded vision of 1/15 and 1/17 in one GUCY2D-LCA case. Another, noncontradictory, explanation is that the MO knockdown is not 100% efficient thus or that not all larvae were equally injected.
In our report, in situ hybridization results display expression of this Gucy2f ortholog that is highly specific for the zebrafish retina. This concurs with prior detailed expression studies in the zebrafish, using in situ hybridization and immunohistochemistry by Rätscho et al.44 These results confirmed the location of the mRNA of photoreceptor-specific Gucy2ds by in situ hybridization techniques for all Gucy2ds between 3 and 4 days post fertilization suggesting that their controlled expression coincides with the onset of visual function. Studies in mice with the GC1 null allele45 reveal that, although GC1 is also expressed in rods, it has a crucial role in cone function.
The described retinal changes observed in LCA1 patients are compatible with those found in this study of zebrafish with the Gucy2f knockdown. One study reported the post-mortem retinal findings in a child with GUCY2D mutation and another study reported the optical coherence study findings in three patients with LCA1.42, 46 Milam et al46 reported a lack of cone labeling using anti red-green cone opsin, which may resemble the lack of zpr1 staining in our report. Additional findings in zebrafish reported here include morphologically visible outer-segment loss of both rods and cones, compatible with the human finding of rods and cones without outer segments.46 The inner nuclear layer is unaffected, as seen in both this zebrafish study and in the retinal structure of a patient with LCA1.46 The changes observed in our 6-day-post-fertilization model may resemble the earliest changes observable in humans following loss of GUCY2D function. This complementary model provides an additional window into the earlier stages of disease progression in an animal model with highly developed cone vision. Our understanding of the early effects on photoreceptors would be enriched with immune-electron microscopy studies of these cells.
The costs, time frame and equipment required for zebrafish screening are far less than those involved in mammalian knockout models. Such rapid testing opportunities in the zebrafish offer an appealing method to assay whether gene knockdown or introduction of dominant, gain-of-function proteins related to human disease will result in a phenotype before embarking on large-scale genetic studies or gene therapy trials.
Three well-characterized animal models of GC1-deficiency exist, all carrying null mutations in GUCY2D: the naturally occurring Gucy1*B chicken,47 the Gucy2d-1 (GC1) knockout mouse48 and the Gucy2d-1/Gucy2d-2 double knockout (GCdko) mouse.47 Therapy results on the GCdko mouse have yet to be published,49 but talks and posters have indicated that gene replacement was a success, conferring long-term therapy to both rods and cones (Boye et al45, personal communication), demonstrating this model as a true phenotypic mammalian model of LCA1.45 Using the optomotor response assay, untreated GC1KO mouse eyes perform poorly with a visual acuity of 0.16±0.04 cycles per degree, whereas isogenic GC1+/+ control eyes responded significantly better, showing an average acuity of 0.42±0.05 cycles per degree. Gene replacement-treated eyes displayed an average acuity of 0.39±0.08 cycles per degree, a level essentially identical to control +/+ eyes and significantly better than untreated GC1KO eyes (P<0.0001).
When wishing to screen animal models for rare gene mutations, the zebrafish offers an appealing alternative to the rodent or canine models owing to their excellent cone and rod vision, the simple and inexpensive nature by which they can be genetically manipulated and the availability of open access genomic information.
In summary, we have identified the conserved Gucy2f ortholog gene in zebrafish, which is highly expressed in the retina, have shown that targeting this gene with antisense oligonucleotides leads to significantly decreased visual function in zebrafish larvae and leads to retinal photoreceptor changes. This study provides additional proof of concept that zebrafish can be used as an efficient alternative model to study the effects of LCA-related, and possibly other retinal dystrophy-associated genes. Future work will focus on evaluating the phenotype of zebrafish injected with MO in conjunction with wild-type Gucy2f plasmid with a goal of further developing the zebrafish as a viable model in which to study gene replacement therapies for retinal disease.
Acknowledgments
We thank Anat Milgrum for her involvement during her BA project.
The authors declare no conflict of interest.
References
- Li L, Xiao X, Li S, et al. Detection of variants in 15 genes in 87 unrelated Chinese patients with leber congenital amaurosis. PLoS One. 2011;6:e19458. doi: 10.1371/journal.pone.0019458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergouniotis PI, Davidson AE, Mackay DS, et al. Recessive mutations in KCNJ13, encoding an inwardly rectifying potassium channel subunit, cause leber congenital amaurosis. Am J Hum Genet. 2011;89:183–190. doi: 10.1016/j.ajhg.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrault I, Rozet JM, Gerber S, et al. Spectrum of retGC1 mutations in leber's congenital amaurosis. Eur J Hum Genet. 2000;8:578–582. doi: 10.1038/sj.ejhg.5200503. [DOI] [PubMed] [Google Scholar]
- Cideciyan AV. Leber congenital amaurosis due to RPE65 mutations and its treatment with gene therapy. Prog Retin Eye Res. 2010;29:398–427. doi: 10.1016/j.preteyeres.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguire AM, Simonelli F, Pierce EA, et al. Safety and efficacy of gene transfer for leber's congenital amaurosis. N Engl J Med. 2008;358:2240–2248. doi: 10.1056/NEJMoa0802315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acland GM, Aguirre GD, Ray J, et al. Gene therapy restores vision in a canine model of childhood blindness. Nat Genet. 2001;28:92–95. doi: 10.1038/ng0501-92. [DOI] [PubMed] [Google Scholar]
- Pang J, Cheng M, Haire SE, et al. Efficiency of lentiviral transduction during development in normal and rd mice. Mol Vis. 2006;12:756–767. [PubMed] [Google Scholar]
- Dejneka NS, Surace EM, Aleman TS, et al. In utero gene therapy rescues vision in a murine model of congenital blindness. Mol Ther. 2004;9:182–188. doi: 10.1016/j.ymthe.2003.11.013. [DOI] [PubMed] [Google Scholar]
- Sprague J, Bayraktaroglu L, Bradford Y, et al. The zebrafish information network: The zebrafish model organism database provides expanded support for genotypes and phenotypes. Nucleic Acids Res. 2008;36 (database issue:D768–D772. doi: 10.1093/nar/gkm956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldman MB, Lin S. Zebrafish as a developmental model organism for pediatric research. Pediatr Res. 2008;64:470–476. doi: 10.1203/PDR.0b013e318186e609. [DOI] [PubMed] [Google Scholar]
- Penberthy WT, Shafizadeh E, Lin S. The zebrafish as a model for human disease. Front Biosci. 2002;7:d1439–d1453. doi: 10.2741/penber. [DOI] [PubMed] [Google Scholar]
- Sprague J, Bayraktaroglu L, Clements D, et al. The zebrafish information network: the zebrafish model organism database. Nucleic Acids Res. 2006;34 (database issue:D581–D585. doi: 10.1093/nar/gkj086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gestri G, Link BA, Neuhauss SC. The visual system of zebrafish and its use to model human ocular diseases. Dev Neurobiol. 2011. [DOI] [PMC free article] [PubMed]
- Maurer CM, Huang YY, Neuhauss SC. Application of zebrafish oculomotor behavior to model human disorders. Rev Neurosci. 2011;22:5–16. doi: 10.1515/RNS.2011.003. [DOI] [PubMed] [Google Scholar]
- Fadool JM, Dowling JE. Zebrafish: a model system for the study of eye genetics. Prog Retin Eye Res. 2008;27:89–110. doi: 10.1016/j.preteyeres.2007.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockerhoff SE, Fadool JM. Genetics of photoreceptor degeneration and regeneration in zebrafish. Cell Mol Life Sci. 2011;68:651–659. doi: 10.1007/s00018-010-0563-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller KP, Neuhauss SC. Quantitative measurements of the optokinetic response in adult fish. J Neurosci Methods. 2009;186:29–34. doi: 10.1016/j.jneumeth.2009.10.020. [DOI] [PubMed] [Google Scholar]
- Huang YY, Neuhauss SC. The optokinetic response in zebrafish and its applications. Front Biosci. 2008;13:1899–1916. doi: 10.2741/2810. [DOI] [PubMed] [Google Scholar]
- Brockerhoff SE. Measuring the optokinetic response of zebrafish larvae. Nat Protoc. 2006;1:2448–2451. doi: 10.1038/nprot.2006.255. [DOI] [PubMed] [Google Scholar]
- Muto A, Orger MB, Wehman AM, et al. Forward genetic analysis of visual behavior in zebrafish. PLoS Genet. 2005;1:e66. doi: 10.1371/journal.pgen.0010066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhauss SC, Biehlmaier O, Seeliger MW, et al. Genetic disorders of vision revealed by a behavioral screen of 400 essential loci in zebrafish. J Neurosci. 1999;19:8603–8615. doi: 10.1523/JNEUROSCI.19-19-08603.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhauss SC. Behavioral genetic approaches to visual system development and function in zebrafish. J Neurobiol. 2003;54:148–160. doi: 10.1002/neu.10165. [DOI] [PubMed] [Google Scholar]
- Yan Z, Sun X, Engelhardt JF. Progress and prospects: techniques for site-directed mutagenesis in animal models. Gene Ther. 2009;16:581–588. doi: 10.1038/gt.2009.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baye LM, Patrinostro X, Swaminathan S, et al. The N-terminal region of centrosomal protein 290 (CEP290) restores vision in a zebrafish model of human blindness. Hum Mol Genet. 2011;20:1467–1477. doi: 10.1093/hmg/ddr025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abraham E, Palevitch O, Gothilf Y, et al. Targeted gonadotropin-releasing hormone-3 neuron ablation in zebrafish: Effects on neurogenesis, neuronal migration, and reproduction. Endocrinology. 2010;151:332–340. doi: 10.1210/en.2009-0548. [DOI] [PubMed] [Google Scholar]
- Palevitch O, Kight K, Abraham E, et al. Ontogeny of the GnRH systems in zebrafish brain: In situ hybridization and promoter-reporter expression analyses in intact animals. Cell Tissue Res. 2007;327:313–322. doi: 10.1007/s00441-006-0279-0. [DOI] [PubMed] [Google Scholar]
- Avanesov A, Malicki J. Approaches to study neurogenesis in the zebrafish retina. Methods Cell Biol. 2004;76:333–384. doi: 10.1016/s0091-679x(04)76016-1. [DOI] [PubMed] [Google Scholar]
- Avanesov A, Malicki J. Approaches to study neurogenesis in the zebrafish retina. Methods Cell Biol. 2004;76:333–384. doi: 10.1016/s0091-679x(04)76016-1. [DOI] [PubMed] [Google Scholar]
- Fleisch VC, Neuhauss SC. Visual behavior in zebrafish. Zebrafish. 2006;3:191–201. doi: 10.1089/zeb.2006.3.191. [DOI] [PubMed] [Google Scholar]
- Neuhauss SC, Biehlmaier O, Seeliger MW, et al. Genetic disorders of vision revealed by a behavioral screen of 400 essential loci in zebrafish. J Neurosci. 1999;19:8603–8615. doi: 10.1523/JNEUROSCI.19-19-08603.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsao-Wu GS, Weber CH, Budgeon LR, et al. Agarose-embedded tissue arrays for histologic and genetic analysis. Biotechniques. 1998;25:614–618. doi: 10.2144/98254st02. [DOI] [PubMed] [Google Scholar]
- Larison KD, Bremiller R. Early onset of phenotype and cell patterning in the embryonic zebrafish retina. Development. 1990;109:567–576. doi: 10.1242/dev.109.3.567. [DOI] [PubMed] [Google Scholar]
- Gothilf Y, Coon SL, Toyama R, et al. Zebrafish serotonin N-acetyltransferase-2: marker for development of pineal photoreceptors and circadian clock function. Endocrinology. 1999;140:4895–4903. doi: 10.1210/endo.140.10.6975. [DOI] [PubMed] [Google Scholar]
- Malicki J. Development of the retina. Methods Cell Biol. 1999;59:273–299. doi: 10.1016/s0091-679x(08)61830-0. [DOI] [PubMed] [Google Scholar]
- Kitiratschky VB, Wilke R, Renner AB, et al. Mutation analysis identifies GUCY2D as the major gene responsible for autosomal dominant progressive cone degeneration. Invest Ophthalmol Vis Sci. 2008;49:5015–5023. doi: 10.1167/iovs.08-1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne AM, Morris AG, Downes SM, et al. Clustering and frequency of mutations in the retinal guanylate cyclase (GUCY2D) gene in patients with dominant cone-rod dystrophies. J Med Genet. 2001;38:611–614. doi: 10.1136/jmg.38.9.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Nakamura M, Nuno Y, et al. Novel complex GUCY2D mutation in Japanese family with cone-rod dystrophy. Invest Ophthalmol Vis Sci. 2004;45:1480–1485. doi: 10.1167/iovs.03-0315. [DOI] [PubMed] [Google Scholar]
- Hunt DM, Buch P, Michaelides M. Guanylate cyclases and associated activator proteins in retinal disease. Mol Cell Biochem. 2010;334:157–168. doi: 10.1007/s11010-009-0331-y. [DOI] [PubMed] [Google Scholar]
- Perrault I, Rozet JM, Gerber S, et al. Leber congenital amaurosis. Mol Genet Metab. 1999;68:200–208. doi: 10.1006/mgme.1999.2906. [DOI] [PubMed] [Google Scholar]
- Chung DC, Traboulsi EI. Leber congenital amaurosis: Clinical correlations with genotypes, gene therapy trials update, and future directions. J AAPOS. 2009;13:587–592. doi: 10.1016/j.jaapos.2009.10.004. [DOI] [PubMed] [Google Scholar]
- Simonelli F, Ziviello C, Testa F, et al. Clinical and molecular genetics of leber's congenital amaurosis: a multicenter study of Italian patients. Invest Ophthalmol Vis Sci. 2007;48:4284–4290. doi: 10.1167/iovs.07-0068. [DOI] [PubMed] [Google Scholar]
- Pasadhika S, Fishman GA, Stone EM, et al. Differential macular morphology in patients with RPE65-, CEP290-, GUCY2D-, and AIPL1-related leber congenital amaurosis. Invest Ophthalmol Vis Sci. 2010;51:2608–2614. doi: 10.1167/iovs.09-3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrault I, Rozet JM, Calvas P, et al. Retinal-specific guanylate cyclase gene mutations in leber's congenital amaurosis. Nat Genet. 1996;14:461–464. doi: 10.1038/ng1296-461. [DOI] [PubMed] [Google Scholar]
- Ratscho N, Scholten A, Koch KW. Expression profiles of three novel sensory guanylate cyclases and guanylate cyclase-activating proteins in the zebrafish retina. Biochim Biophys Acta. 2009;1793:1110–1114. doi: 10.1016/j.bbamcr.2008.12.021. [DOI] [PubMed] [Google Scholar]
- Boye SL, Conlon T, Erger K, et al. Long-term preservation of cone photoreceptors and restoration of cone function by gene therapy in the guanylate cyclase-1 knockout (GC1KO) mouse. Invest Ophthalmol Vis Sci. 2011;52:7098–7108. doi: 10.1167/iovs.11-7867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milam AH, Barakat MR, Gupta N, et al. Clinicopathologic effects of mutant GUCY2D in leber congenital amaurosis. Ophthalmology. 2003;110:549–558. doi: 10.1016/S0161-6420(02)01757-8. [DOI] [PubMed] [Google Scholar]
- Karan S, Frederick JM, Baehr W. Novel functions of photoreceptor guanylate cyclases revealed by targeted deletion. Mol Cell Biochem. 2010;334:141–155. doi: 10.1007/s11010-009-0322-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semple-Rowland SL, Lee NR, Van Hooser JP, et al. A null mutation in the photoreceptor guanylate cyclase gene causes the retinal degeneration chicken phenotype. Proc Natl Acad Sci USA. 1998;95:1271–1276. doi: 10.1073/pnas.95.3.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haire SE, Pang J, Boye SL, et al. Light-driven cone arrestin translocation in cones of postnatal guanylate cyclase-1 knockout mouse retina treated with AAV-GC1. Invest Ophthalmol Vis Sci. 2006;47:3745–3753. doi: 10.1167/iovs.06-0086. [DOI] [PMC free article] [PubMed] [Google Scholar]