Abstract
Patrilineal heritable surnames are widely used to select autochthonous participants for studies on small-scale population genetic patterns owing to the unique link between the surname and a genetic marker, the Y-chromosome (Y-chr). Today, the question arises as to whether the surname origin will be informative on top of in-depth genealogical pedigrees. Admixture events that happened in the period after giving heritable surnames but before the start of genealogical records may be informative about the additional value of the surname origin. In this context, an interesting historical event is the demic migration from French-speaking regions in Northern France to the depopulated and Dutch-speaking region Flanders at the end of the sixteenth century. Y-chr subhaplogroups of individuals with a French/Roman surname that could be associated with this migration event were compared with those of a group with autochthonous Flemish surnames. Although these groups could not be differentiated based on in-depth genealogical data, they were significantly genetically different from each other. Moreover, the observed genetic divergence was related to the differences in the distributions of main Y-subhaplogroups between contemporary populations from Northern France and Flanders. Therefore, these results indicate that the surname origin can be an important feature on top of in-depth genealogical results to select autochthonous participants for a regional population genetic study based on Y-chromosomes.
Keywords: admixture, genetic genealogy, historical gene flow, human population structure, Y-chromosome
Introduction
Gene flow is an essential evolutionary force, which homogenizes genetic differentiation that is generated between populations by selection, mutation and genetic drift (including founder effects; Jobling et al., 2004). The effect of historical gene flow on the population genetic structure of organisms can be studied based on the present genetic variation within a species by means of phylogeographic analyses (Larmuseau et al., 2009). Nevertheless, Western-European human populations have been strongly influenced by recent migration and expansion events, especially since the Industrial Revolution, which blur the picture of fine-scaled historical population structure and admixture events (Bowden et al., 2008). Thanks to the unique link between a heritable cultural element, the patrilineal surname and a genetic marker, the Y-chromosome (Y-chr); it is common to select autochthonous DNA donors based on their surnames authentic to the location of residence (Manni et al., 2005; Winney et al., 2012). Using this specific surname-based approach, several genetic studies observed historical population genetic structure or past admixture events, which were otherwise invisible (Hill et al., 2000; Bowden et al., 2008).
With the exponential increase of genetic genealogical data combining qualitative genealogical pedigrees with certain Y-chr variants, population genetic studies will make more and more use of these highly informative data sets instead of surname-sampling approaches to study the impact of historical migration events on current population structure. Using concrete genealogical information, one will directly analyze pre-industrial population structure and will quantify the effect of gene flow during the last centuries on population genetic structure (King and Jobling 2009b; Larmuseau et al., 2011). A recent study on the West-European region Brabant found significant temporal differentiation along a north–south gradient based on in-depth genetic–genealogical data (Larmuseau et al., 2012). Generally speaking, a substantial series of genealogical documents are present in Western-European history ever since the beginning of the seventeenth century. The Council of Trent, closed in 1563, had required the registration of baptisms and weddings; yet as a result of wars from the sixteenth through to the twentieth century, the oldest parish registrations were mostly lost (van Uytven et al., 2011). Therefore, most of the Western-European (amateur) genealogists have a reliable oldest reported paternal ancestor (ORPA) born between 1600 and 1750 (Rasmuson, 2008; Larmuseau et al., 2012). Yet, the use of paternally inherited surnames started within the thirteenth–fourteenth century in Western Europe (Winkler and Twilhaar, 2006), and the question arises if the anthroponymy, which is the study of the names of human beings, can provide more insight into admixture events since the time of surname establishment complementing in-depth genealogical trees (King and Jobling, 2009b). The heritable surname often gives information about the residence of the patrilineal ancestor to whom the surname was applied. For example, the language, dialect and/or the appearance of a toponym within the surname may indicate the original geographic location of the ancestor (Winkler and Twilhaar, 2006). To assess this added value of anthroponymy to study fine-scaled population structure and migration, we use it here to detect the genetic signature of an admixture event that happened in the period after giving heritable surnames but before the start of genealogical records.
At the end of the sixteenth century, a demic migration event happened from Northern France to the Dutch-speaking Flanders (here defined as the contemporary Flanders and not as the sixteenth century County of Flanders). In the last quarter of the sixteenth century, Flanders was seriously devastated and depopulated because of flu and plague epidemics, war and emigration to contemporary Germany, The Netherlands and England for economical and mainly religious reasons (van Roosbroeck, 1968; Briels, 1985). The census population decreased by 30 to 50% between 1570 and 1585, of course with strong local differences (Scholliers and Vandenbroeke, 1980; Daelemans, 1988). In several regions of Flanders, the decrease is even estimated to be two-third (Parker, 1979). After 1585, the census population size quickly increased again, partly because of the aforementioned immigration of French-speaking individuals from regions of Northern France, namely Picardy, the region of Lille and the County of Artois. At the beginning of the seventeenth century, almost 10% of the Flemish families had their origin in Northern France (Van Acker, 1986). After some generations, the only remainder of their origin was their French/Roman surname (FRS). Many of these foreign French surnames were distorted over time to a Flemish pronunciation but are today still clearly recognized as originating from the French language. This is the reason for the huge number of FRSs of Flemish families although their ORPA lived in (Dutch-speaking) Flanders before 1750 based on their genealogical records (Debrabandere, 2003). In this study, we contrasted the frequency distribution of Y-chr subhaplogroups in Flanders between individuals with French and authentic Flemish surnames (AFSs), and checked whether the migratory history from Northern France to Flanders at the end of the sixteenth century had indeed left a genetic signal.
Materials and methods
Ethics statement
Samples were selected via genealogical societies from the Benelux (Belgium, The Netherlands and Grand Duchy of Luxembourg) for the open genealogical project ‘DNA Brabant/Belgium'. All samples have been collected with written consent from the donors who gave permission for the DNA analyses, storage of the samples and scientific publication of their genealogical data and their DNA results. Genealogical and genetic data of all donors of this open genealogical project are available in Van den Cloot (2010), Van den Cloot (2011).
As well as a DNA sample, the requirement for participation was the availability of patrilineal genealogical data with the ORPA born before 1750. After receiving all genealogical data, only participants that had an ORPA that lived within the contemporary borders of Flanders were retained. To have an unbiased sample, this requirement and the aim of the study was not communicated to the participants.
A buccal swab sample from each selected participant was collected for DNA extraction by using the Maxwell 16 System (Promega, Madison, WI, USA) followed by real-time PCR quantification (Quantifiler Human DNA kit, Applied Biosystems, Foster City, CA, USA). In total, 38 Y-chr short tandem repeat (Y-STR) loci were genotyped as described in previous studies (Jacobs et al., 2009). All haplotypes were submitted to Whit Athey's Haplogroup Predictor (Athey, 2006) to obtain probabilities for the inferred haplogroups. Based on these results, the samples were assigned to specific Y-chr single-nucleotide polymorphisms (Y-SNPs) assays to confirm the haplogroup and to assign the subhaplogroup to the lowest possible level of the latest Y-chr tree reported by Karafet et al. (2008) and according to the update on the Y Chromosome Consortium web page (http://ycc.biosci.arizona.edu/ nomenclature_system/index.html), with the exception of the substructuring within A, R1b1b2a1 (R-U106) and R1b1b2a2g (R-U152). Also, a set of recently characterized Y-SNPs that improved resolution of the haplogroup G phylogeny were included (Sims et al., 2009). A total of 16 multiplex systems with 110 Y-SNPs were developed using SNaPshot mini-sequencing assays (Applied Biosystems) and analyzed on an ABI3130XL Genetic Analyzer (Applied Biosystems) according to previously published protocols (Caratti et al., 2009). A fraction of the participants for this study were already partly genotyped for the Y-chr in a previous study (Larmuseau et al., 2011). All primer sequences and concentrations for the analysis of the 110 Y-SNPs are available from the authors upon request.
The genealogical data from each participant underwent a high-quality control through the demonstration of their research with official documents. Pairs with a common official ancestor in paternal lineage but with a different Y-chr subhaplogroup or with Y-STR-haplotypes with more than six mutations out of the 38 Y-STRs were excluded from the data set. Based on the calculated mean mutation rate for the 38 genotyped Y-STRs using the individual mutation rates measured in Ballantyne et al. (2010), more than 6 mutations out of the 38 Y-STRs are not likely to occur between recent (<600 years) genealogical relationships (Walsh 2001). Different DNA donors with a recent (<600 years) ‘most recent common ancestor' (MRCA) based on all genotyped Y-STRs loci were excluded as well to avoid a family bias. Also known descendants of foundlings, adoptions and sons of unknown fathers based on their genealogical data were excluded owing to the known lack of a relationship between the origin of the surname and the Y-chr variant. The language (inclusive dialect) and the meaning of all surnames, and the earliest found appearance of each surname in Belgium and Northern France were examined by Debrabandere (2003), the major general reference for individual surname origins in Belgium and northern France, and based on the databank of the State Archives of Belgium (www.arch.be). Based on surname origin, two groups were defined, namely the AFS sample with all individuals with an authentic surname for Flanders and with an ORPA born in Flanders before 1750, and the FRS sample with all individuals with a FRS, which is not observed before 1600 in Belgian archives, and with an ORPA born in Flanders before 1750. Finally, the frequencies of the main subhaplogroups in two northern French regions, namely Île-de-France and Nord-Pas-de-Calais, were reconstructed based on data published in Ramos-Luis et al. (2009) and Busby et al. (2012).
The genetic relationships between the two groups AFS and FRS, between AFS, FRS, Île-de-France and Nord-Pas-de-Calais, and between the five Flemish provinces (based on the birth place of all ORPAs) were assessed by means of FST, without taking into account evolutionary distance between individual subhaplogroups on the assumption that the different Y-chr subhaplogroups were distributed independently from each other in Western Europe based on their wide and diverged distributions and their high mutual tMRCA values (more than 5000 years ago) (Karafet et al., 2008; Chiaroni et al., 2009; Cruciani et al., 2011). All FST values were estimated using ARLEQUIN v.3.1 (Excoffier et al., 2005). Significance of population subdivision was tested using a permutation test. Because of the large difference in sample size between both groups (549 individuals for AFS and 50 individuals for FRS), we used the weighted average of the haplotype diversity HS as test statistic in the permutation test (Hudson et al., 1992):
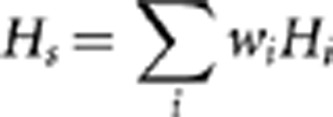 |
where
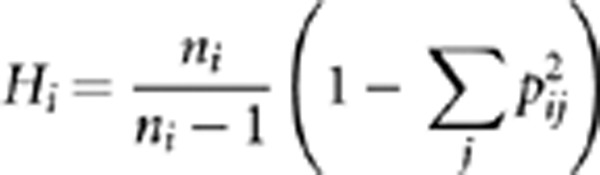 |
with pij the relative frequency of the j-th allele in the i-th subpopulation, ni the sample size for the i-th subpopulation and wi the weighting factor for the i-th subpopulation, calculated as wi=(ni−2)/(Σn−4). With unequal sample sizes, this test statistic has a higher power than other test statistics (Hudson et al., 1992). The test was implemented in R (The R Foundation for Statistical Computing, 2011) (see script in Supplementary Material). In the case of pairwise tests, the Bonferroni correction was applied to all P-values (Rice, 1989). No further tests to observe population differentiation based on the Y-STRs were done because of the insufficient power of the used set of the Y-STRs to detect population structure, as a consequence of the high homoplasy associated with these markers (Larmuseau et al., 2011).
Results
Almost 900 individuals sent their genealogical data. In total, 685 individuals could present reliable genealogical records going back before 1750 and with an ORPA born within the contemporary borders of Flanders. After selection, 622 males with a Flemish ORPA before 1750 were used for the analysis. Among these 622 males, 50 individuals have a surname with a Roman/French origin. All these individuals have Dutch as their native language. From the group of 50 individuals, 23 individuals have an ORPA who lived in West Flanders, 10 in East Flanders, 8 in Flemish Brabant, 5 in Limburg and 4 in the province of Antwerp. Before 1600, all these surnames, including related names and spelling variants, are not observed in official archive documents from Belgium (Debrabandere, 2003). Next to the 50 FRSs, 23 others refer, with a toponym, to a place outside Flanders (namely Germany, England or The Netherlands; for example, Abrath, Van Engeland, Van Gils, and so on.) or they are German derivatives (for example, Waldack, Waltenier, Wambacq, and so on). Therefore, we had a data set with 549 ‘Dutch surname samples' (for example, Van den Cloot, Van den Beemt, De Bie, and so on) and 50 ‘French surname samples' (for example, Larmuseau, Ghesquire, Seynaeve, and so on).
All individuals were correctly assigned to the main haplogroup using the Whit Athey's Haplogroup Predictor. The single exception was a Y-chr belonging to haplogroup A, which is not included in the predictor. However, according to a recent study on the root of the human Y-chromosomal phylogenetic tree by Cruciani et al. (2011), haplogroup A is not monophyletic and therefore this existing Y-chr is further referred to as belonging to paragroup Y*(xBT). In total, 41 different subhaplogroups were observed (inclusively the Y-chr assigned to Y*(xBT)). The subhaplogroup counts and relative frequencies in the two groups AFS and FRS are given in Table 1. Overall, the four most frequent subhaplogroups were: R1b1b2a1 (R-U106) with 26.78% in AFS and 12.00% in FRS; R1b1b2a2* (R-P312*) with 19.13% in AFS and 32.00% in FRS; I1* (I-M253*) with 12.02% in AFS and 10.00% in FRS; and R1b1b2a2g (R-U152) with 10.56% in AFS and 16% in FRS. The AFS and FRS groups were significantly genetically differentiated with an FST value of 0.01641 (P-value 0.0135±0.00094 s.e.; 10 000 replicates). To compare between regions within Flanders, DNA donors were attributed to contemporary Flemish provinces based on the birth place of their ORPA. No significant differences were found between regions within Flanders, including pairwise comparisons between the regions most different in proportion of FRS (West Flanders and Antwerp; P-value=0.9067±0.00317 s.e.). The observed distributions and frequencies of the different Y-chr subhaplogroups in all Flemish provinces are given in Supplementary Table S1.
Table 1. Distribution and frequency of the Y-chr subhaplogroups within the groups of the ‘Authentic Flemish surnames' and ‘French/Roman surnames'.
| Y-chr subhaplogroups |
Authentic Flemish surnames |
French/Roman surnames |
||
|---|---|---|---|---|
| Distribution | Frequency | Distribution | Frequency | |
| Y*(xBT) | 1 | 0.18% | 0 | 0% |
| E1b1b1* (E-M35*) | 1 | 0.18% | 0 | 0% |
| E1b1b1a1 (E-V12) | 1 | 0.18% | 0 | 0% |
| E1b1b1a2 (E-V13) | 16 | 2.91% | 1 | 2% |
| E1b1b1a3 (E-V22) | 2 | 0.36% | 1 | 2% |
| E1b1b1b (E-M81) | 1 | 0.18% | 0 | 0% |
| E1b1b1c* (E-M123*) | 1 | 0.18% | 0 | 0% |
| E1b1b1c1 (E-M34) | 8 | 1.46% | 0 | 0% |
| G2a* (G-P15*) | 1 | 0.18% | 0 | 0% |
| G2a3* (G-U8*) | 17 | 3.10% | 0 | 0% |
| G2a3a (G-U16) | 1 | 0.18% | 0 | 0% |
| G2a3b1 (G-U13) | 1 | 0.18% | 0 | 0% |
| I1* (I-M253*) | 66 | 12.02% | 5 | 10% |
| I1c (I-P109) | 6 | 1.09% | 0 | 0% |
| I2* (I-P215*) | 9 | 1.64% | 0 | 0% |
| I2a* (I-P37.2*) | 9 | 1.64% | 0 | 0% |
| I2b* (I-M223*) | 17 | 3.10% | 4 | 8% |
| I2b1 (I-M284) | 2 | 0.36% | 0 | 0% |
| I2b3 (I-P78) | 1 | 0.18% | 1 | 2% |
| I2b4 (I-P95) | 1 | 0.18% | 1 | 2% |
| J1* (J-M267*) | 2 | 0.36% | 0 | 0% |
| J1e* (J-P58*) | 5 | 0.91% | 1 | 2% |
| J2a* (J-M410*) | 12 | 2.19% | 1 | 2% |
| J2a2* (J-M67*) | 2 | 0.36% | 0 | 0% |
| J2a2a* (J-M92*) | 1 | 0.18% | 0 | 0% |
| J2a8 (J-M319) | 2 | 0.36% | 0 | 0% |
| J2b2* (J-M241*) | 4 | 0.73% | 0 | 0% |
| L1 (L-M27) | 1 | 0.18% | 0 | 0% |
| L2 (L-M317) | 1 | 0.18% | 0 | 0% |
| Q1* (Q-P36.1*) | 3 | 0.55% | 0 | 0% |
| R1a* (R-SRY10831.2*) | 1 | 0.18% | 0 | 0% |
| R1a1* (R-M17*) | 24 | 4.37% | 2 | 4% |
| R1b1* (R-P25*) | 1 | 0.18% | 0 | 0% |
| R1b1b* (R-P297*) | 1 | 0.18% | 0 | 0% |
| R1b1b2* (R-M269*) | 9 | 1.64% | 2 | 4% |
| R1b1b2a* (R-P310*) | 5 | 0.91% | 0 | 0% |
| R1b1b2a1 (R-U106) | 147 | 26.78% | 6 | 12% |
| R1b1b2a2* (R-P312*) | 105 | 19.13% | 16 | 32% |
| R1b1b2a2d (R-SRY2627) | 1 | 0.18% | 0 | 0% |
| R1b1b2a2g (R-U152) | 58 | 10.56% | 8 | 16% |
| T* (T-M70*) | 2 | 0.36% | 1 | 2% |
| Total | 549 | 50 | ||
The subhaplogroups with a frequency ⩾10% are given in bold.
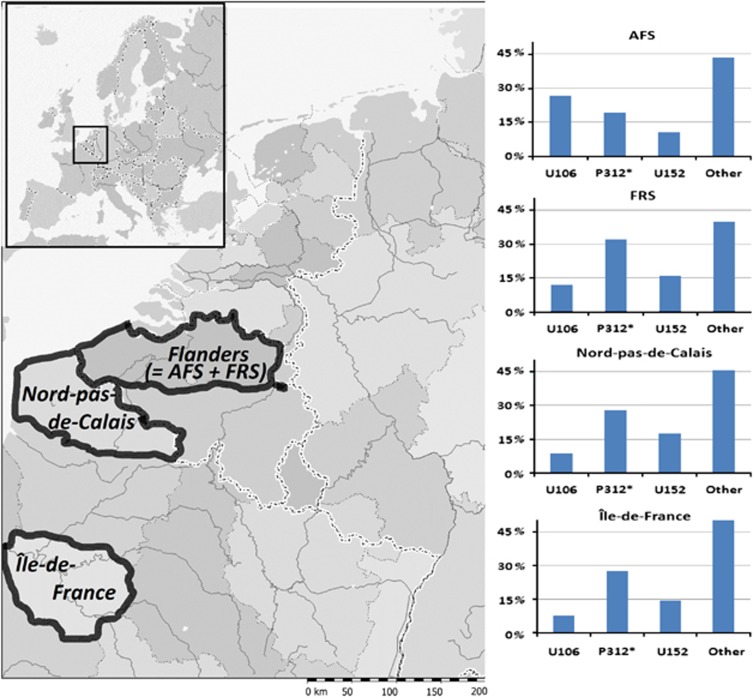
The distributions and frequencies of three main Y-chr subhaplogroups, namely R1b1b2a1 (R-U106), R1b1b2a2* (R-P312*) and R1b1b2a2g (R-U152), could be reconstructed for two northern French regions, Île-de-France and Nord-Pas-de-Calais (Table 2, Figure 1). Some SNPs within R1b1b2a2 (R-P312), namely R-M37, R-M65, R-M153 and R-P66, were not genotyped for the northern France samples by Ramos-Luis et al. (2009) and Busby et al. (2012). Nevertheless, all these SNPs are virtually absent in Western Europe (Myres et al., 2007; Karafet et al., 2008; Larmuseau et al., 2011) and therefore they will not influence the frequency of R1b1b2a2* for both northern French regions. Based on the frequencies of the three reconstructed subhaplogroups and all other ones grouped together, AFS was significantly genetically different from Île-de-France (P-value=0.0028±0.00051 s.e.) and Nord-Pas-de-Calais (P-value=0.0104±0.00061 s.e.). In contrast, FRS and both northern French regions were not differentiated from each other (P-value=0.5146±0.0047 s.e. between FRS and Île-de-France; P-value=0.8334±0.00303 s.e. between FRS and Nord-Pas-de-Calais) as also Île-de-France vs Nord-Pas-de-Calais were not significantly different (P-value=0.8618±0.00255 s.e.), all after Bonferroni correction.
Table 2. Reconstructed distribution and frequency of Y-chr subhaplogroups within the groups of the ‘Authentic Flemish surnames', ‘French/Roman surnames', ‘Île-de-France' and ‘Nord-Pas-de-Calais'.
| Y-chr subhaplogroups | Authentic Flemish surnames | French/Roman surnames | Île de France | Nord-Pas-de-Calais | ||||
|---|---|---|---|---|---|---|---|---|
| |
Distribution |
Frequency |
Distribution |
Frequency |
Distribution |
Frequency |
Distribution |
Frequency |
| R1b1b2a1 (R-U106) | 147 | 26.78% | 6 | 12.00% | 7 | 7.69% | 6 | 8.82% |
| R1b1b2a2* (R-P312*) | 105 | 19.13% | 16 | 32% | 25 | 27.47% | 19 | 27.94% |
| R1b1b2a2g (R-U152) | 58 | 10.56% | 8 | 16.00% | 13 | 14.29% | 12 | 17.65% |
| Other subhaplogroups | 239 | 43.53% | 20 | 40.00% | 46 | 50.55% | 31 | 45.59% |
| Total | 549 | 50 | 91 | 68 | ||||
Figure 1.
Map of Western Europe with the frequencies (in percentage) of the three main Y-chromosome subhaplogroups R-U106, R-P312* and R*U153, and of all other subhaplogroups in the datasets of Nord-pas-de-Calais, Île-de-France and Flanders with the AFS and FRS.
Discussion
Since the observation of a clear North–South gradient of abundant Y-chr subhaplogroups (Myres et al., 2007; Cruciani et al., 2011) and an unambiguous East–West gradient of Y-haplotypes in Europe (Roewer et al., 2005), significant genetic differences are detected between regions on a scale of <100 km in Western Europe (Larmuseau et al., 2011; Larmuseau et al., 2012). The presence of such fine-scaled geographic differences in genetic structure may also allow for the investigation of historical gene flow events and their effects on population genetic structure and admixture on a comparable regional scale (Roewer et al., 2005). Our results show evidence for a past admixture event within the Flemish population from a French-speaking population, which happened between the start of the heritable surname (fourteenth–fifteenth century) and the genealogical registration (seventeenth century) (Winkler and Twilhaar, 2006). Significant genetic differentiation was found for the Y-chr frequencies between the group of French/Roman vs autochthonous Flemish surnames for individuals that showed an ORPA born in Flanders before 1750. This suggests that the grouping based on surnames is strongly informative and that it is capable to reveal the hybrid nature of the population, which was undetectable with only in-depth genealogical data.
Nonpaternity events, including adoption and foundlings, will break the link between Y-chr and surname, and they will lead to the loss of anthroponymic information. Owing to the fact that the overall estimate for nonpaternity is between 1–5% per generation (Jobling et al., 2004; King and Jobling, 2009a), there will be a substantial confounding effect of nonpaternity events when looking back before 1600. Therefore, the genetic differentiation between two populations has to be high enough to detect a signal of historic admixture based on surnames of DNA donors, which cannot be distinguished from each other based on genealogical records. Our results thus indicate that the signal of admixture is much higher than the expected noise due to breaks of the link between the surname and the Y-chr by nonpaternity events. Moreover, because Y-SNP mutation rates are so low within the time depth of some centuries, Y-SNPs are indeed not expected to have undergone mutations since the origin of the surname (Jobling et al., 2004). This all suggests that Y-chr subhaplogroups linked to French surnames in families with an ORPA born in Flanders before 1750 are significantly differentiated from Y-chr subhaplogroups linked to AFSs.
French male Y-lineages are still poorly characterized on phylogenetic depth, especially on a regional scale (Ramos-Luis et al., 2009). Accurate frequencies of all subhaplogroups within France, inclusively Northern France, are expected soon (Ramos-Luis E, personal communication). Yet, the frequencies of three main subhaplogroups, namely R1b1b2a1 (R-U106), R1b1b2a2* (R-P312*) and R1b1b2a2g (R-U152), could be reconstructed for two northern French regions Nord-Pas-de-Calais and Île-de-France from data already available (Ramos-Luis et al., 2009; Busby et al., 2012). It has to be noted that these reconstructed frequencies are based on participants currently living in Northern France without notion about the places of residence of their paternal ancestors at the beginning of the archival records (Ramos-Luis et al., 2009) as it is the case for the AFS and FRS data sets. Nevertheless, based on these reconstructed frequencies, it was possible to associate the variation in the French surname group to the Y-chr distribution in Northern France. First, it was clear that the French surname group was not significantly differentiated from both northern French regions. This is in contrast to the AFS group, which was significantly differentiated from the French regions as well as from the French surname group. Second, the most frequent subhaplogroup observed in the two northern French regions, R-P312*, is also the most frequent in the French surname group in Flanders but not in the autochthonous Flemish surname group. The latter group, R-U106, was the most frequent subhaplogroup (Table 1). Finally, as mentioned, a clear North–South gradient in frequencies has been observed within Western Europe for R-U106 and R-U152 (Cruciani et al., 2011), which is visible even on a micro-geographical scale (<100 km) (Myres et al., 2007; Larmuseau et al., 2011). As Northern France is further south than Flanders, it is not surprising to detect that the frequencies of R-U106 and R-U152 are indeed, respectively, lower and higher in Northern France than in the AFS group (Table 2, Figure 1). In the French surname group, however, frequencies of R-U106 and R-U152 correspond with those observed in Northern France and not with those in the Flemish surname group (R-U106 with 26.78% in AFS and 12.00% in FRS, R-U152 with 10.56% in AFS and 16.00% in FRS; Tables 1 and 2). These results also show that to observe an accurate signal for admixture it is insufficient to genotype only the Y-SNPs of the main haplogroups and/or Y-STRs. Because of homoplasy which does not vary substantially enough among the used Y-STR loci, haplotypes based on 38 Y-STRs (or less) are not distinctive enough to differentiate subhaplogroups within R1b1b2 (R-M269) (Larmuseau et al., 2011). Therefore, these (DYS) Y-STR loci are useful for recent ancestry and paternity testing but not useful for population genetic studies. A new set of Y-STRs with a low mutation rate may be interesting to observe a more subtle population genetic structure in the future.
Our results are further corroborated by analysis of the occurrence of the observed French surnames to verify if these surnames are associated with the emigration from North France to Flanders at the end of the sixteenth century. First, all observed surnames of the FRS group were subjected to archival research. As mentioned, these surnames do not occur in Flemish archives before 1550, but only since the end of the sixteenth century (Debrabandere, 2003). Second, the frequency of these French surnames in our data set of individuals with an ORPA living in Flanders is 8%, and is comparable to the expected 10% of the Flemish families with an origin in Northern France at the end of the sixteenth century (Van Acker, 1986). Finally, the distribution of the ORPAs of our FRS group within Flanders is, according to the historical spreading direction from south-western to north-eastern, with the highest frequency of French surnames in the province West-Flanders (19.3%) and the lowest in the province Antwerp (2.2%) with a distance of 120 km between the two provinces. The high percentage in West Flanders reflects the higher admixture in this region, which is bordering the North French region, in comparison with the north-eastern part of Flanders. Nevertheless, when we grouped all the samples of AFS and FRS, no significant population differentiation was found within Flanders based on our present Y-chr data set.
Conclusions
The vicissitudes of war in the low countries (1568–1648) caused a substantial migration from contemporary Northern France to the ravaged and depopulated areas of Flanders in the second half of the sixteenth century, most probably after 1585. Although this migration from Northern France to Flanders happened more than 400 years ago and could not be traced based on genealogical pedigrees, it is still possible to find a signal of Y-chr differentiation between a group with AFSs and a group with FRSs associated with this northwards migration at the end of the sixteenth century. This small-scale migration led to an admixture event, which influenced the genetic variation within Flanders. The fact that such an admixture event could be traced based on anthroponymy of surnames in contrast to in-depth genealogy strongly suggests for the first time that the analysis of the surname origin can be used as an important feature to select DNA donors for population genetic studies on regional scale on top of in-depth genealogical data.
Data Archiving
All Y-chr data (Y-STRs and Y-SNPs) genotyped for this study have been submitted to the open access Y-STR Haplotype Reference Database (YHRD, www.yhrd.org): accession number YA003651–YA003652–YA003653–YA003738–YA003739–YA003740–YA003741–YA003742. The R-script of the analytical method to test the statistical significance of measurements of population differentiation based on subhaplogroups is given as Supplementary Material.
Acknowledgments
We thank all the volunteers who donated DNA samples and provided genealogical data used in this study. We acknowledge the Flemish society for genealogical research ‘Familiekunde Vlaanderen', Marc Van Den Cloot and Marc Gabriëls, who were involved in the collection of the samples and genealogical data. We want to thank Luc De Meester, Hanne Jansma, Jean-Jacques Cassiman, Peter de Knijff, Lutz Rüwer, Inge Neyens, Joost Raeymaekers and Lucrece Lernout for useful assistance and discussions, and Antonette Anandarajah and three anonymous referees for their useful corrections on an earlier version of this paper. Maarten HD Larmuseau and Joost Vanoverbeke are postdoctoral fellows of the FWO-Vlaanderen (Research Foundation—Flanders); Gert Gielis benefits from a PhD fellowship also of the FWO-Vlaanderen. This study was funded by the Flemish Society for Genealogical Research ‘Familiekunde Vlaanderen' (Antwerp), the Flanders Ministry of Culture and the K.U.Leuven BOF-Center of Excellence Financing on ‘Eco- and socio-evolutionary dynamics' (Project number PF/2010/07).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Athey WT. Haplogroup prediction from Y-STR values using a Bayesian-allele-frequency approach. JoGG. 2006;2:34–39. [Google Scholar]
- Ballantyne KN, Goedbloed M, Fang RX, Schaap O, Lao O, Wollstein A, et al. Mutability of Y-chromosomal microsatellites: rates, characteristics, molecular bases, and forensic implications. Am J Hum Genet. 2010;87:341–353. doi: 10.1016/j.ajhg.2010.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowden GR, Balaresque P, King TE, Hansen Z, Lee AC, Pergl-Wilson G, et al. Excavating past population structures by surname-based sampling: the genetic legacy of the Vikings in northwest England. Mol Biol Evol. 2008;25:301–309. doi: 10.1093/molbev/msm255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briels J. Zuid-Nederlanders in de republiek 1572–1630. Een demografische en cultuurhistorische studie. Danthee Sint-Niklaas; 1985. [Google Scholar]
- Busby GBJ, Brisighelli F, Sánchez-Diz P, Ramos-Luis E, Martinez-Cadenas C, Thomas MG, et al. The peopling of Europe and the cautionary tale of Y chromosome lineage R-M269. Proc Biol Sci. 2012;279:884–892. doi: 10.1098/rspb.2011.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caratti S, Gino S, Torre C, Robino C. Subtyping of Y-chromosomal haplogroup E-M78 (E1b1b1a) by SNP assay and its forensic application. Int J Legal Med. 2009;123:357–360. doi: 10.1007/s00414-009-0350-y. [DOI] [PubMed] [Google Scholar]
- Chiaroni J, Underhill PA, Cavalli-Sforza LL. Y chromosome diversity, human expansion, drift, and cultural evolution. Proc Natl Acad Sci USA. 2009;106:20174–20179. doi: 10.1073/pnas.0910803106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruciani F, Trombetta B, Antonelli C, Pascone R, Valesini G, Scalzi V, et al. Strong intra- and inter-continental differentiation revealed by Y chromosome SNPs M269, U106 and U152. Forensic Sci Int Genet. 2011;5:E49–E52. doi: 10.1016/j.fsigen.2010.07.006. [DOI] [PubMed] [Google Scholar]
- Daelemans F.1988Boeren in oorlogstijd. De sociale, economische en demografische gevolgen van oorlogen op het platteland in Brabant (15e-18e eeuw)In: Vanhemelryck F (ed.)Mensen in oorlogstijdUFSAL. Centrum voor Brabantse Geschiedenis:Brussel; 57–104. [Google Scholar]
- Debrabandere F. Woordenboek van de familienamen in België en Noord-Frankrijk. L.J. Veen/Het Taalfonds Amsterdam/Antwerpen; 2003. [Google Scholar]
- Excoffier L, Laval G, Schneider S. ARLEQUIN ver.3.0: an integrated software package for population genetics data analysis. Evol Bioinform Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Hill EW, Jobling MA, Bradley DG. Y-chromosome variation and Irish origins. Nature. 2000;404:351–352. doi: 10.1038/35006158. [DOI] [PubMed] [Google Scholar]
- Hudson RR, Boos DD, Kaplan NL. A statistical test for detecting geographic subdivision. Mol Biol Evol. 1992;9:138–151. doi: 10.1093/oxfordjournals.molbev.a040703. [DOI] [PubMed] [Google Scholar]
- Jacobs M, Janssen L, Vanderheyden N, Bekaert B, Van de Voorde W, Decorte R.2009Development and evaluation of multiplex Y-STR assays for application in molecular genealogy Forensic Sci Int GenetSupplement Series257–59. [Google Scholar]
- Jobling MA, Hurles ME, Tyler-Smith C. Human Evolutionary Genetics: Origins, Peoples and Disease. Garland Science Publishing London/New York; 2004. [Google Scholar]
- Karafet TM, Mendez FL, Meilerman MB, Underhill PA, Zegura SL, Hammer MF. New binary polymorphisms reshape and increase resolution of the human Y chromosomal haplogroup tree. Genome Res. 2008;18:830–838. doi: 10.1101/gr.7172008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King TE, Jobling MA. Founders, drift, and infidelity: the relationship between Y chromosome diversity and patrilineal surnames. Mol Biol Evol. 2009a;26:1093–1102. doi: 10.1093/molbev/msp022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King TE, Jobling MA. What's in a name? Y chromosomes, surnames and the genetic genealogy revolution. Trends Genet. 2009b;25:351–360. doi: 10.1016/j.tig.2009.06.003. [DOI] [PubMed] [Google Scholar]
- Larmuseau MHD, Ottoni C, Raeymaekers JAM, Vanderheyden N, Larmuseau HFM, Decorte R. Temporal differentiation across a West-European Y-chromosomal cline - genealogy as a tool in human population genetics. Eur J Hum Genet. 2012;20:434–440. doi: 10.1038/ejhg.2011.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larmuseau MHD, Van Houdt JKJ, Guelinckx J, Hellemans B, Volckaert FAM. Distributional and demographic consequences of Pleistocene climate fluctuations for a marine demersal fish in the north-eastern Atlantic. J Biogeogr. 2009;36:1138–1151. [Google Scholar]
- Larmuseau MHD, Vanderheyden N, Jacobs M, Coomans M, Larno L, Decorte R. Micro-geographic distribution of Y-chromosomal variation in the central-western European region Brabant. Forensic Sci Int Genet. 2011;5:95–99. doi: 10.1016/j.fsigen.2010.08.020. [DOI] [PubMed] [Google Scholar]
- Manni F, Toupance B, Sabbagh A, Heyer E. New method for surname studies of ancient patrilineal population structures, and possible application to improvement of Y-chromosome sampling. Am J Phys Anthropol. 2005;126:214–228. doi: 10.1002/ajpa.10429. [DOI] [PubMed] [Google Scholar]
- Myres NM, Ekins JE, Lin AA, Cavalli-Sforza LL, Woodward SR, Underhill PA. Y-chromosome short tandem repeat DYS458.2 non-consensus alleles occur independently in both binary haplogroups J1-M267 and R1b3-M405. Croat Med J. 2007;48:450–459. [PMC free article] [PubMed] [Google Scholar]
- Parker G.1979War and economic change: the economic costs of the Dutch revoltIn: Parker G (ed)Spain and the Netherlands, 1559–1659 Ten studies Short Hills; 178–203. [Google Scholar]
- Ramos-Luis E, Blanco-Verea A, Brión M, Van Huffel V, Carracedo A, Sánchez-Diz P.2009Phylogeography of French male lineages Forensic Sci Int GenetSupplement Series2439–441. [DOI] [PubMed] [Google Scholar]
- Rasmuson M. Genealogy and gene trees. Hereditas. 2008;145:20–27. doi: 10.1111/j.0018-0661.2008.2041.x. [DOI] [PubMed] [Google Scholar]
- Rice WR. Analyzing tables of statistical tests. Evolution. 1989;43:223–225. doi: 10.1111/j.1558-5646.1989.tb04220.x. [DOI] [PubMed] [Google Scholar]
- Roewer L, Croucher PJP, Willuweit S, Lu TT, Kayser M, Lessig R, et al. Signature of recent historical events in the European Y-chromosomal STR haplotype distribution. Hum Genet. 2005;116:279–291. doi: 10.1007/s00439-004-1201-z. [DOI] [PubMed] [Google Scholar]
- Scholliers E, Vandenbroeke C. Structuren en conjuncturen in de Zuidelijke Nederlanden 1480–1800. Algemene Geschiedenis Der Nederlanden. 1980;5:252–310. [Google Scholar]
- Sims LM, Garvey D, Ballantyne J. Improved resolution haplogroup G phylogeny in the Y chromosome, revealed by a set of newly characterized SNPs. PLoS One. 2009;4:e5792. doi: 10.1371/journal.pone.0005792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The R Foundation for Statistical Computing 2011R version 2.13.0.
- Van Acker L. Immigratie van Franstaligen in Westelijk Vlaanderen na 1585. Biekorf. 1986;86:113–130. [Google Scholar]
- Van den Cloot M. DNA Brabant - DNA-project 2009 oud-hertogdom Brabant. Familiekunde Vlaanderen Antwerpen; 2010. [Google Scholar]
- Van den Cloot M. DNA België - DNA-project 2010 België, exclusief oud-hertogdom Brabant. Familiekunde Vlaanderen Antwerpen; 2011. [Google Scholar]
- van Roosbroeck R. Emigranten - Nederlandse vluchtelingen in Duitsland (1550–1600) Davidsfonds Leuven; 1968. [Google Scholar]
- van Uytven R, Bruneel C, Koldeweij AM, van de Sande AWFM, van Oudheusden JAFM. Geschiedenis van Brabant van het hertogdom tot heden. Uitgeverij Waanders Zwolle; 2011. [Google Scholar]
- Walsh B. Estimating the time to the most recent common ancestor for the Y chromosome or mitochondrial DNA for a pair of individuals. Genetics. 2001;158:897–912. doi: 10.1093/genetics/158.2.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler J, Twilhaar JN. Achternamen in Nederland & Vlaanderen: oorsprong, geschiedenis en betekenis. sdu Uitgevers Den Haag; 2006. [Google Scholar]
- Winney B, Boumertit A, Day T, Davison D, Echeta C, Evseeva I, et al. People of the British Isles: preliminary analysis of genotypes and surnames in a UK-control population. Eur J Hum Genet. 2012;20:203–210. doi: 10.1038/ejhg.2011.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.