Abstract
Long-distance migrants are, by definition, highly mobile but it is poorly understood if this leads to high rates of gene flow and an essentially panmictic global population structure. Genetic divergence in migratory species could be promoted, for example, by fidelity to distinct migratory pathways. In this study, we investigate the population genetic structure of tufted duck (Aythya fuligula), a long-distance migrant with a largely continuous breeding distribution across Eurasia. Distinct, longitudinally oriented flyways have been postulated based on geographically disjunct wintering areas and are supported by evidence from ringing data. We generated sequences of the mitochondrial control region and multi-locus microsatellite genotypes for several hundreds of samples from the European and Asian breeding and wintering grounds including some individuals infected with highly pathogenic avian influenza virus H5N1. Significant differentiation between breeding sites was observed for both marker types, but FST values were approximately 10 times higher for maternally inherited mitochondrial DNA than for biparentally transmitted nuclear markers. The genetic differentiation between the postulated European and Asian flyways was similar to that observed within continents and, in general, genetic divergence was not associated with geographic distance. Neither marker type showed evidence of genetic substructure among aggregations on the European wintering grounds. Our results suggest some breeding site fidelity, especially in females, but extensive population admixture on the wintering grounds. Several scenarios may explain the observed lack of genetic divergence between Europe and Asia including non-equilibrium conditions following a recent range expansion or contemporary gene flow across the postulated migratory divides.
Keywords: bird migration, population structure, flyway, microsatellites, mitochondrial DNA, avian flu
Introduction
Migration refers to the species' movements that are predictable both in time and space and very often involve seasonal shifts between different feeding and breeding grounds, which may be separated by thousands of kilometres (Berthold, 2001). Some of the most spectacular examples of long-distance migration come from birds, for example, the Arctic tern (Sterna paradisaea), which completes round trips between Arctic breeding grounds and Antarctic wintering areas each year (Berthold, 2001). Dispersal, on the other hand, is defined as the movement between natal and breeding areas, or two consecutive breeding sites, and may or may not result in actual gene flow between populations (Clobert et al., 2001; Schweizer et al., 2007). The high mobility of long-distance migrants could have profound implications for their population structure as it may allow dispersal and gene flow at macrogeographic scales. In the most extreme case, an entire species could then be composed of a single panmictic population.
The recognition of population substructure or the absence thereof is relevant both for fundamental and applied research. Depending on the rate and spatial patterns of gene flow, a species may be composed of subpopulations that are genetically distinct and may have unique adaptations to local environments. The rate of dispersal will also determine the extent to which demographic patterns of growth or decline are coupled between populations, which may have significant implications for population persistence (Clobert et al., 2001). Finally, migrating animals can carry and potentially spread parasites and pathogens (Altizer et al., 2011), which, in the case of zoonotic pathogens, may have direct consequences also for human health. A debated issue in this context has been the role of migratory waterfowl in the global spread of highly pathogenic avian influenza virus H5N1 (Gilbert et al., 2006). Infected individuals were repeatedly found among various species of wild birds, especially ducks including the tufted duck (Aythya fuligula; World Organization for Animal Health, online resources), and it seems likely that wild birds may have contributed to the spatial spread of H5N1, probably together with human activities like poultry trade (Gilbert et al., 2006; Kilpatrick et al., 2006).
Population subdivision in spite of high mobility may exist if individuals are philopatric and consistently return to the same breeding grounds. In fact, evidence of different subpopulations characterized by some genetic divergence is available from several long-distance migrants, like the dunlin Calidris alpina (Wennerberg, 2001) or Swainson's thrush Catharus ustulatus (Ruegg and Smith, 2002). Three main factors are typically thought to favour philopatry (Lawson Handley and Perrin, 2007): high direct costs of dispersal, advantages of familiarity with feeding or breeding grounds and, finally, benefits of kin cooperation. Clearly, the latter two explanations would seem more relevant for long-distance migrants where direct costs of dispersal are probably comparatively low. Site fidelity may also be observed outside the breeding season and, again, could be favoured, for example, if familiarity with feeding sites is advantageous. Individual fidelity to non-breeding grounds may also have direct consequences for genetic population structure if pairs are formed there (Robertson and Cooke, 1999). In fact, in many migratory waterfowl, including the tufted duck, pairs are formed on the wintering grounds (Robertson and Cooke, 1999) and, as a result, non-random use of different wintering grounds could lead to genetic divergence between different populations.
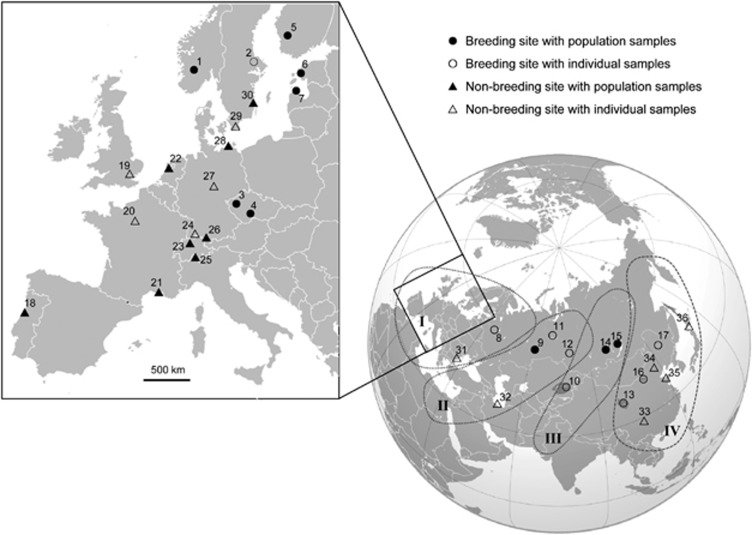
The tufted duck, A. fuligula, is the most numerous and widespread diving duck in the world (population size estimates 2.6–2.9 million individuals; Birdlife International, 2011), with most populations being migratory. There are no morphologically recognized subspecies, and the breeding range is mostly continuous covering the temperate and northern regions of the entire Palaearctic (Cramp and Simmons, 1977; Scott and Rose, 1996). Blums and Baumanis (1990) distinguished four geographical subpopulations of tufted ducks based on their spatially disjunct wintering grounds in Europe, the Caspian Sea/Nile area, India and Southeast Asia, respectively. Within Europe, a further subdivision has been proposed into a north-western population wintering around the North Sea and a south-eastern population wintering in central Europe and around the Mediterranean and Black Seas (Scott and Rose, 1996). This split is in line with the observation that birds ringed on Swiss wintering grounds are reencountered only in continental Europe in the same or subsequent winters but not in Britain (Fränzi Korner-Nievergelt, personal communication). However, the wintering areas of the two European subpopulations overlap to some extent along the North Sea coast, and they share breeding grounds in Europe and in Siberia east to the river Ob (Hofer et al., 2005). Consequently, we would not predict significant genetic divergence between these two European wintering populations. However, genetic isolation seems more likely between the European and Asian flyways. Limited information from ringing data suggests considerable fidelity to large-scale wintering regions (Hofer et al., 2005) and quite clear longitudinal separation between the breeding grounds of the birds wintering in Europe and India, respectively (Figure 14 of Hofer et al., 2005). Based on these observations, we may postulate several genetically differentiated subpopulations within Eurasian tufted ducks associated with migratory divides (see Figure 1).
Figure 1.
Sampling localities of the tufted duck (A. fuligula) across its distribution range in Europe and Asia. Circles and triangles represent samples from the breeding and the wintering grounds, respectively. Filled symbols indicate sites used for population genetic analyses with sample size n⩾8 and open symbols indicate smaller samples used for phylogeographic analyses only. Coordinates and sample sizes for each site are displayed in Supplementary Table S1. The approximate positions of four hypothetical flyways are indicated. Note that the shown flyway boundaries indicate the grouping used in our analyses (Figures 2 and 3, Table 2) but probably overemphasize the separation between flyways on the Siberian breeding grounds. (I) European, (II) Western Siberian, (III) Eastern Siberian and (IV) East Asian flyway.
The primary aim of this study was to quantify the extent of such genetic differentiation and to delineate potential subpopulations of Eurasian tufted ducks. We combined markers with maternal and biparental modes of inheritance to investigate sex-specific differences in the rates of gene flow and provide a detailed picture of the range-wide genetic population structure of this long-distance migrant. Our samples included some highly pathogenic avian influenza H5N1-positive individuals from the Swiss and German wintering grounds, and an additional goal was to identify the potential breeding origin of these ducks using genetic assignment methods.
Materials and methods
Sample collection and DNA extraction
Samples of tufted ducks were collected at different localities throughout the species' Palaearctic distribution range during our field expeditions to the Baltic States, Russia and China in 2008 and 2009, and through ringing and research schemes and hunting associations during 2006–2009. The majority of samples were collected on breeding (June–August) or wintering grounds (November–February; Supplementary Table S1). We collected mostly feathers in a noninvasive manner, but also some muscle, blood or egg-membrane samples. Tissue samples were also obtained from museum collections. In addition, DNA samples from three and eleven individuals identified as H5N1-positive by the national reference laboratories were available from Switzerland and Germany, respectively. In total, 366 samples were available from 36 localities in 19 countries (Figure 1, Supplementary Table S1). Localities with n⩾8 were defined as population samples and included in all population genetic analyses, whereas samples from other sites were incorporated in phylogeographic analyses only. Tissue samples were stored at −25 °C or in absolute ethanol. Genomic DNA was extracted using a standard phenol-chloroform extraction protocol.
Mitochondrial DNA (mtDNA) sequencing
The complete mitochondrial control region (ctr, approximately 1300, bp) was amplified using a primer pair, L1 (5′-ACAAAATAAGTCATTATTCCTGCTC-3′) and H1 (5′-TTTGCACTCATTGCTTAATGTT-3′), newly designed based on a sequence from mallard (Anas platyrhynchos; EMBL accession no.: L22477). The amplification was performed in a GeneAmp PCR System 9700 (Applied Biosystems, Carlsbad, CA, USA) in a reaction volume of 25 μl containing 12.5 μl H2O, 2 μl template DNA (20–100 ng μl−1), 4.8 μl dNTPs (2.5 mℳ), 2.5 mℳ MgCl2, 1 μl of each primer (10 pmol μl−1), 0.2 μl Taq polymerase (5 units μl−1; Qiagen, Hilden, Germany) and the associated buffer at 1x concentration. The PCR cycling protocol was the following: 2 min at 93 °C, followed by 35 cycles of 30 s at 93 °C, 30 s at 54 °C, 1 min at 72 °C and a final extension step of 7 min at 72 °C. PCR products were purified with the GenElute PCR clean-up kit (Sigma-Aldrich, Dorset, UK) according to the manufacturer's protocol.
Two additional internal primers L78 (Sorenson and Fleischer, 1996) and L736 (Sorenson et al., 1999) were used to sequence two 650-bp fragments, which, together, yielded a 1162-bp concatenated sequence covering both hypervariable regions of ctr. The sequencing reaction was carried out in a volume of 10 μl using Terminator Ready Reaction Mix ‘Big Dye' (v.3.1; Applied Biosystems) according to the guidelines of the manufacturer. The temperature profiles were as follows: 50 s at 96 °C, followed by 35 cycles of 10 s at 96 °C, 10 s at 50 °C and 4.5 min at 60 °C. The products were purified by ethanol precipitation and detected on an ABI Prism 3100 Genetic Analyser (Applied Biosystems).
Sorenson and Fleischer (1996) reported the presence of nuclear copies (Numts) of the mitochondrial control region in the tribe Aythyini, including the genus Aythya. We took the following steps in order to minimize the potential amplification of Numts: First, as Numts are often short, our PCR primers were chosen to amplify a long fragment of approximately 1300, bp. Second, if multiple bands still occurred on the agarose gel, the target band was cut and reamplified prior to sequencing. Finally, we carefully examined all the sequences by comparing them with published sequences of Aythya ctr and Numts available on GenBank. Because of these precautionary measures, we are confident that the sequences we included in further analyses are of mitochondrial origin.
Microsatellite genotyping
A total of 366 individuals were screened at 14 autosomal microsatellite loci initially developed for other related species (multiplex set 1: Caud13, MM07, Sfiμ3, Smo11, Sfiμ4, MM05 and CmAAT28; multiplex set 2: Apl12, CmAAT38, Smo4, Aph13, MM03, Apl36 and Sfiμ2; see Table A2 in Liu et al., 2011; Stovicek et al., 2011). The loci were combined into two multiplex PCRs and the 5′-end of each reverse primer was modified with a pig-tail extension to facilitate genotyping. Amplification was carried out in a 10-μl reaction volume containing 5 μl of PCR mix (Qiagen Multiplex Kit), 3 μl H2O, 1 μl of a primer mix and 1 μl of template DNA. All the details regarding the multiplex set up and exact primer concentrations are given in Table S2 of Liu et al., (2011) The temperature profiles were as follows: 15 min at 96 °C, followed by 30 cycles of 30 s at 94 °C, 1.5 min at 57 °C, 60 s at 72 °C and a final step for 15 min at 60 °C. Fragments were separated on an ABI Prism 3100 Genetic Analyser (Applied Biosystems) and their length was determined using GeneMapper software v.3.7 (Applied Biosystems) against an internal size standard (GeneScan-500LIZ; Applied Biosystems). Approximately 25% of the samples were reamplified and genotyped independently to ensure genotyping consistency.
Genetic diversity indices
The mtDNA sequences were aligned using the CLUSTAL W algorithm implemented in BioEdit v.7.0 (Hall, 1999), and the alignment was checked by eye and manually revised if necessary. The average number of pairwise nucleotide differences (K), the number of haplotypes (NH), haplotype diversity (H) and nucleotide diversity (π) were calculated for all population samples using DnaSP v.5.0 (Librado and Rozas, 2009).
For microsatellites, mean allelic richness (AR), number of alleles at each locus (NA), observed (HO) and expected heterozygosities (HE) were estimated for all population samples using FSTAT v.2.9.3.2 (Goudet, 2002). Deviations from Hardy–Weinberg equilibrium and genotypic linkage equilibrium within populations were assessed using Arlequin v.3.5 (Excoffier and Lischer, 2010) and FSTAT, respectively. Significance levels were adjusted using the Bonferroni procedure within each population. Further, FIS was calculated for each population sample and its significance assessed based on 10 000 permutations in Arlequin.
Phylogeographic analysis
Based on the ctr sequences, we constructed separate unrooted haplotype networks for breeding and wintering birds using the median-joining algorithm (Bandelt et al., 1999) implemented in Network v.4.516 (http://www.fluxus-engineering.com). Additionally, phylogenetic trees were constructed using Bayesian inference in MrBayes v.3.1.2 (Huelsenbeck and Ronquist, 2001). The Hasegawa-Kishino-Yano model allowing for among site substitution rate variation and invariable sites was selected as the best-fitting nucleotide substitution model using the Akaike Information Criterion in jModelTest v.0.1.1 (Posada, 2008). Four independent runs with default heating temperatures were run for five million steps and sampled every 1000th step. The first 25% of samples were discarded as burn-in. Convergence of the Markov chain Monte Carlo chains was assessed using Tracer v.1.4 (http://tree.bio.ed.ac.uk/software/tracer/). Phylogenetic trees were rooted using a homologous sequence from redhead (Aythya americana, GenBank accession no.: NC_000877). The phylogeographic structure in the nuclear microsatellite markers was investigated with principal coordinates analyses based on pairwise Euclidian distances between individual genotypes in GENALEX v.6.2 (Peakall and Smouse, 2006), again separately for breeding and wintering birds.
Population genetic analyses
All population genetic analyses were carried out separately for the nine breeding sites and the eight samples from wintering grounds. Analyses of molecular variance implemented in Arlequin were performed for both marker sets to assess the proportion of genetic variance explained by the hypothesized migratory divides among breeding or wintering grounds (breeding grounds: Europe (1 and 3–7; Figure 1), Western Siberia (9) and Eastern Siberia (14 and 15); wintering grounds: Northwest Europe (18, 22, 28 and 30) and Southeast Europe (21, 23, 25 and 26). For mtDNA, we calculated pairwise ΦST with the Tamura-Nei model, and for the microsatellite data, pairwise FST using the Weir and Cockerham estimator in Arlequin. Significance was assessed based on 10 000 permutations, with significance levels adjusted for multiple testing using the sequential Bonferroni procedure. Pairwise genetic differentiation was also assessed for these population samples where enough samples from females only were available (that is, n⩾8; CzSb, CzDi and LaEn). Similar analyses were not possible for males only because of the relatively limited number of samples from individuals with known sex. For breeding populations we tested for an association between FST/(1−FST) and the logarithm of geographical distance in kilometres using a Mantel test implemented in GENALEX based on 999 permutations.
For the microsatellite data, we further used the programme STRUCTURE v.2.3.1 (Falush et al., 2003) to test for genetic substructure among breeding and wintering populations, respectively. An admixture model with correlated allele frequencies was used and each run consisted of a burn-in of 100 000 steps and 500 000 Markov chain Monte Carlo steps. We conducted 10 independent runs for each K-value (K=1–9 for breeding and K=1–8 for wintering ducks, respectively) and determined the most likely number of clusters following the appraoch described by Evanno et al., (2005), We further used discriminant analysis of principal components (PCs), a model-free multivariate method, to identify genetic clusters when prior grouping information is lacking (Jombart et al., 2010). We performed discriminant analysis of PCs and graphically displayed our results using the package ‘adegenet' (Jombart, 2008) in R v.2.12.0 (R Development Core Team, 2008). In all the analyses, 33 PCs were retained in the data transformation step, which accounted for >80% of the total genetic variability. The inference of the most likely number of clusters was based on the Bayesian information criterion.
The power of our microsatellite markers to detect given levels of population differentiation was assessed in POWSIM v.4.0 (Ryman and Palm, 2006). We generated 1000 data sets each for six pre-defined levels of population differentiation (FST=0.001, 0.0025, 0.005, 0.01, 0.02 and 0.025) with samples sizes, numbers of markers and allele frequencies corresponding to the empirical data. Statistical power was defined as the proportion of times the null hypothesis of equal allele frequencies across populations was rejected using a Chi-square test or a Fisher's exact test.
Results
Genetic diversity
The diversity indices for mtDNA and microsatellites showed high levels of genetic variation in tufted ducks (Table 1). Overall, 1162, bp of the control region sequence from 300 individuals from 36 locations revealed 66 polymorphic sites (5.68%) defining 94 haplotypes (GenBank accession numbers JQ422386–JQ422479). Of these, 34 haplotypes were shared by 2–79 individuals each and the remaining 60 were singletons. The average number of pairwise nucleotide differences (K) was similar in samples from breeding and wintering grounds (2.75 versus 2.73; Mann–Whitney U-test, P=0.61).
Table 1. Estimates of genetic variability in tufted ducks inferred from 1162, bp of the mitochondrial control region and 11 microsatellite loci.
| Map ref. | Location labels |
Mitochondrial DNA |
Microsatellites |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Nmt | K | NH | H±s.d. | π±s.d. (%) | Nnuc | AR | HOa | HEa | FISa | ||
| 1 | NOSo | 9 | 3.44 | 5 | 0.883±0.098 | 0.298±0.053 | 9 | 5.01 | 0.49 | 0.55 | 0.14 |
| 3 | CZSb | 19 | 0.98 | 5 | 0.579±0.114 | 0.085±0.028 | 23 | 4.85 | 0.53 | 0.56 | 0.04 |
| 4 | CZDi | 22 | 2.86 | 14 | 0.952±0.026 | 0.247±0.027 | 16 | 5.16 | 0.50 | 0.58 | 0.14 |
| 5 | FISo | 12 | 2.56 | 7 | 0.894±0.063 | 0.223±0.036 | 12 | 4.70 | 0.59 | 0.60 | 0.03 |
| 6 | ESMa | 12 | 3.55 | 6 | 0.864±0.072 | 0.305±0.052 | 16 | 4.75 | 0.61 | 0.57 | −0.07 |
| 7 | LAEn | 28 | 1.86 | 13 | 0.788±0.078 | 0.163±0.030 | 39 | 4.95 | 0.60 | 0.63 | 0.03 |
| 9 | RUYe | 8 | 2.89 | 7 | 0.964±0.077 | 0.249±0.046 | 10 | 4.54 | 0.64 | 0.67 | 0.08 |
| 14 | RUTv | 20 | 2.54 | 7 | 0.853±0.043 | 0.274±0.023 | 20 | 4.77 | 0.55 | 0.57 | 0.05 |
| 15 | RUBl | 17 | 4.10 | 13 | 0.963±0.033 | 0.354±0.041 | 21 | 5.46 | 0.64 | 0.69 | 0.07 |
| 18 | PTAv | 10 | 2.53 | 7 | 0.933±0.062 | 0.221±0.039 | 12 | 5.33 | 0.63 | 0.68 | 0.03 |
| 21 | FRNo | 11 | 3.35 | 9 | 0.964±0.051 | 0.288±0.046 | 11 | 5.16 | 0.56 | 0.60 | 0.03 |
| 22 | NLMa | 8 | 2.77 | 6 | 0.929±0.084 | 0.166±0.036 | 10 | 4.85 | 0.51 | 0.58 | 0.14 |
| 23 | CHOb | 54 | 2.71 | 26 | 0.863±0.045 | 0.234±0.030 | 90 | 5.24 | 0.59 | 0.59 | 0.01 |
| 25 | ITVa | 8 | 2.18 | 7 | 0.964±0.077 | 0.187±0.035 | 8 | 4.95 | 0.67 | 0.67 | 0.01 |
| 26 | DERd | 14 | 2.65 | 8 | 0.858±0.063 | 0.232±0.035 | 17 | 5.45 | 0.61 | 0.63 | 0.03 |
| 28 | DEOs | 8 | 2.33 | 6 | 0.833±0.127 | 0.201±0.048 | 12 | 4.53 | 0.56 | 0.62 | 0.08 |
| 30 | SWBi | 9 | 3.28 | 8 | 0.972±0.064 | 0.282±0.059 | 9 | 4.79 | 0.67 | 0.68 | 0.03 |
Map ref. corresponds to Figure 1 with bold numbers indicating samples from breeding grounds. The numbers of individuals analysed for mtDNA (Nmt) and microsatellites (Nnuc) are given. For mtDNA, the average number of nucleotide differences (K), the number of haplotypes (NH), haplotype diversity (H±s.d.) and nucleotide diversity (π±s.d., in percent) are indicated. For microsatellites, means are given for allelic richness (AR), observed heterozygosity (HO), and expected heterozygosity (HE). Multi-locus inbreeding coefficients (FIS) were calculated for each sample. There were no significant deviations from null-expectations.
Calculated without locus MM03, which showed high frequency null alleles (see text for details).
A total of 366 tufted ducks from 35 localities were genotyped at 14 microsatellite loci. Three loci (Sfiμ3, MM07 and CmAAT 38) were monomorphic and not included in further statistical analysis. The overall number of alleles at the eleven polymorphic loci ranged from 4 (Sfiμ02) to 28 (Smo4) and the average allelic richness was similar in samples from breeding (4.91) and wintering grounds (5.04; Mann–Whitney U-test, P=0.39). A total of 12 pairs of loci showed significant deviations from linkage equilibrium in population-specific tests after Bonferroni correction, however, this did not consistently involve the same pairs. Significant heterozygote deficits were observed in 24 out of 187 locus-specific tests with locus MM03 being affected 15 times (Supplementary Table S2). Without this locus, FIS ranged from −0.07 to 0.14, and no values were significantly different from zero (Table 1). Exclusion of locus MM03 from further population genetic analyses had no significant quantitative or qualitative effect (results not shown).
Absence of phylogeographic pattern
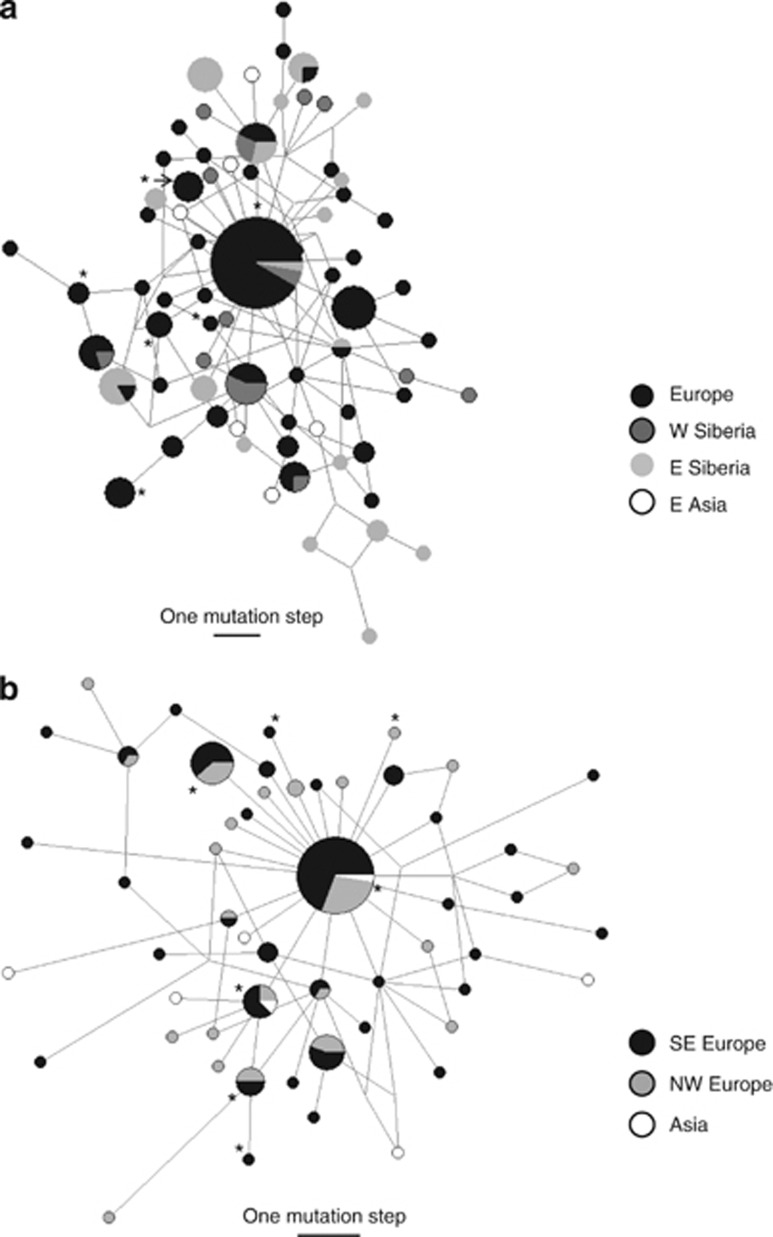
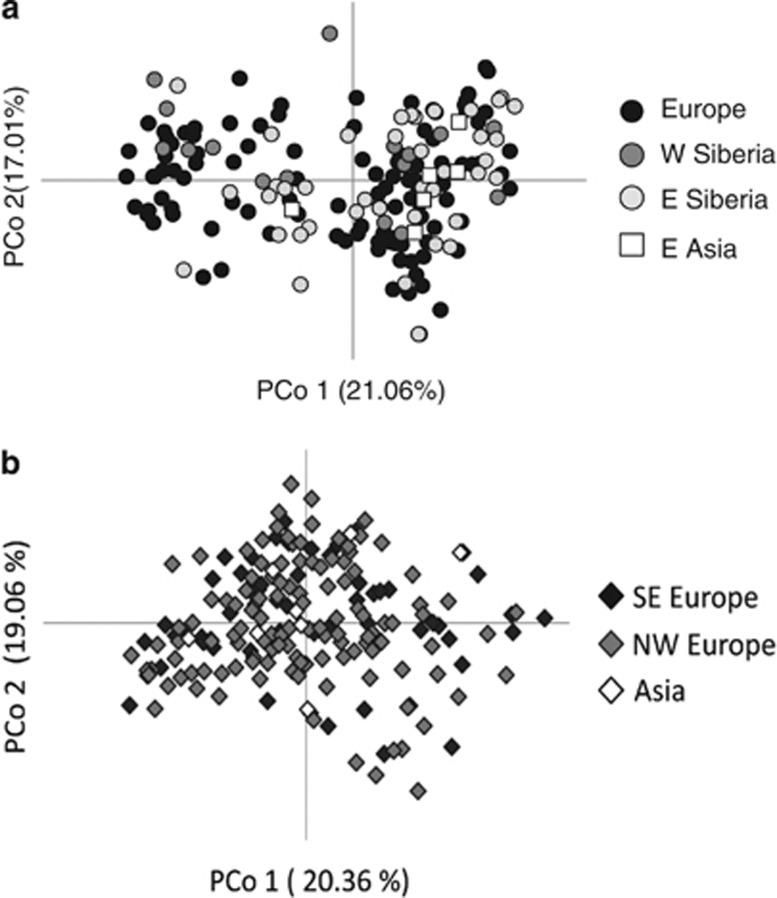
The haplotype networks based on mtDNA sequences from breeding and wintering birds, respectively, showed a star-like topology with no clear geographical patterns (Figure 2). This result was consistent with the Bayesian trees that were marked by shallow sequence divergence and contained no well-supported basal clades (Supplementary Figures S2 and S3). Again, the individuals did not cluster based on their geographic origin and, in particular, we observed no basal split between Europe and Asia. The 14 H5N1-positive individuals found on Swiss or German wintering grounds showed seven different mtDNA haplotypes (Figure 2b). Six infected ducks from three different localities in Germany and Switzerland (DEOs, DERd and CHBu; Supplementary Table S1) had the most common haplotype found in all wintering regions (and also across all breeding grounds; see Figure 2a). Three haplotypes were singletons and the other three were shared by ducks from different wintering grounds. The principal coordinates analyses based on the microsatellite data revealed no clear geographical structure among individuals from breeding (Figure 3a) or wintering grounds (Figure 3b).
Figure 2.
Unrooted median-joining networks based on 1162, bp sequences of the mitochondrial control region for (a) breeding (n=162) and (b) wintering (n=138) tufted ducks. Samples from the breeding season were grouped as Europe, Western Siberia (west of river Ob), Eastern Siberia and Eastern Asia. Wintering ducks were partitioned into hypothetical subpopulations in south-eastern and north-western Europe and in Asia (Iran, China and Japan). The size of the circles is proportional to the number of individuals with a particular haplotype. Asterisks mark the haplotypes observed in H5N1-positive tufted ducks sampled on European wintering grounds (see text). Note that the corresponding haplotypes are also indicated in (a) to illustrate their spatial distribution on the breeding grounds.
Figure 3.
Plots of the first two coordinates from a principal coordinates analysis based on Euclidian distances between multi-locus microsatellite genotypes for (a) breeding individuals (n=180) and (b) wintering individuals (n=186) of tufted duck. Different symbols represent different geographic regions (see legend of Figure 2 for details).
Population genetic analyses
Significant differentiation between breeding sites
The postulated major migratory divides in tufted ducks across Eurasia were not supported by the population-based analyses. The three geographical groups (Europe, Western Siberia and Eastern Siberia) did not explain a significant proportion of the genetic variation for mtDNA or microsatellites (Table 2). This between-group component remained nonsignificant for both mtDNA (FCT=0.007, P=0.387) and microsatellites (FCT=0.0006, P=0.435) when the small Western Siberian sample was excluded (site 9, Figure 1, Table 2) and the analysis of molecular variance was based only on two groups (Europe versus Eastern Siberia)
Table 2. Hierarchical analysis of molecular variance (AMOVA) based on mitochondrial DNA (mtDNA) and microsatellite data from tufted duck.
| Markers | Grouping | Variation among groups (%) | Variation among populations within groups (%) | Variation within populations (%) |
|---|---|---|---|---|
| mtDNA | Europe (1, 3, 4, 5, 6, 7), | −0.77 | 17.00** | 83.76 |
| Western Siberia (9) and | ||||
| Eastern Siberia (14, 15) | ||||
| microsatellites | 0.05 | 1.72** | 98.23 | |
| mtDNA- | 0.79 | 1.34 | 97.87 | |
| North-western Europe (18, 22, 28, 30) and | ||||
| South-eastern Europe (21, 23, 25, 26) | ||||
| microsatellites | −0.29 | 0.53 | 99.76 |
Samples from breeding grounds (top) were grouped by three major breeding areas (Europe, Western Siberia and Eastern Siberia). Wintering ducks (bottom) were partitioned according to two major European wintering regions. Numbers in brackets represent the map reference numbers (shown in Figure 1) of the sample within each group.
** P<0.001.
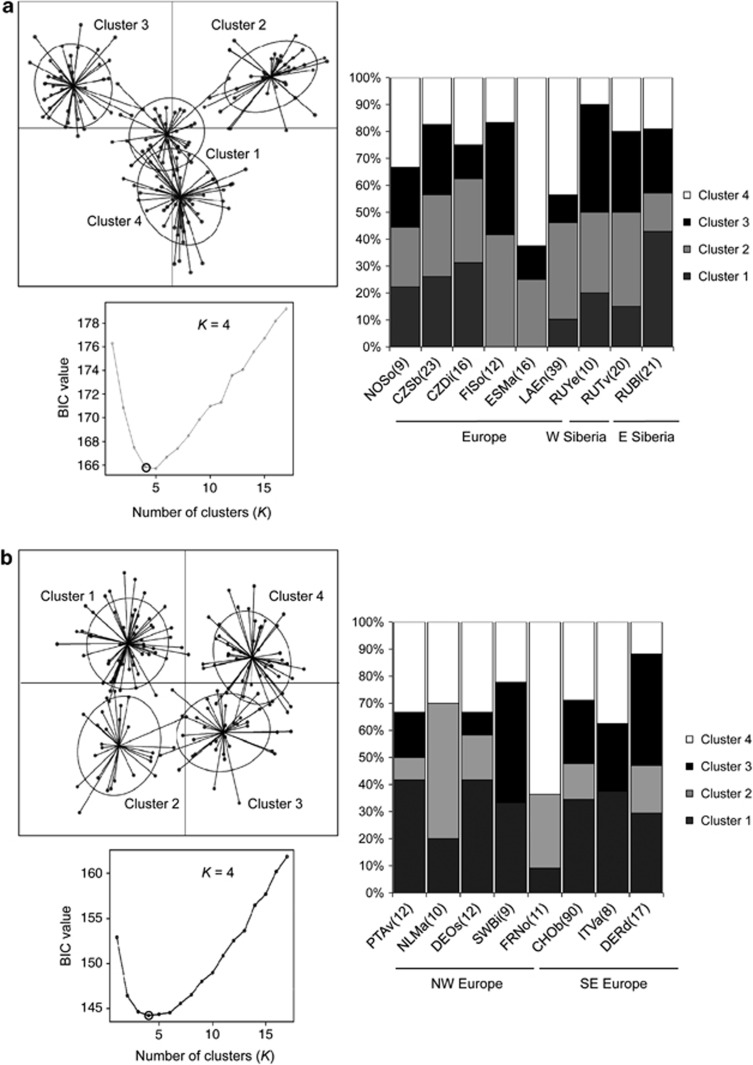
The molecular variation attributed to differences among populations within geographical groups, however, was significant for both marker types but larger for mtDNA (mtDNA: 17.0%, P<0.0001; microsatellites: 1.72%, P<0.0001). For mtDNA, 20 out of 36 ΦST values between pairs of breeding sites were significant after Bonferroni correction (Supplementary Table S3; global ΦST=0.166, P<0.0001). Based on the microsatellite markers, we obtained a global FST estimate of 0.017 (P<0.0001). Only 2 out of 36 pairwise comparisons exhibited significant FST values and these significant results involved one sample from Eastern Siberia (RUBl) and samples from Estonia and Latvia, respectively (Supplementary Table S3). FST values calculated for population samples with females only were very similar to estimates based on all individuals. Neither of the marker types showed a significant association between geographical and genetic distance between breeding sites (Mantel tests; mtDNA: R2=0.003, P=0.35; microsatellites: R2=0.014, P=0.26). The STRUCTURE analysis of microsatellites did not provide clear support for a particular number of clusters. Posterior probabilities were very similar for different values of K, including K=1 (Supplementary Figure S4a). Discriminant analysis of PCs inferred the optimal number of genetic clusters as four, but each sample contained components of at least three of these clusters (Figure 4a), and their relative frequencies did not show a geographical pattern.
Figure 4.
Inference of genetic clusters using discriminant analysis of PCs (DAPCs) based on microsatellite data for tufted duck samples from breeding (a) and wintering (b) grounds. In each representation, the bottom-left plot shows the Bayesian information criterion for different numbers of clusters (K), with the optimal K indicated by a red circle. The scatter plots display the first two PCs of the DAPC for the optimal K. The inferred genetic clusters are displayed by inertia ellipses with dots representing individuals. The barplots illustrate the proportion of individuals from each sample assigned to each of these genetic clusters. Sample sizes are given in parenthesis after the sample names.
No genetic structure among European wintering grounds
The proportion of genetic variation associated with the a priori defined wintering regions was not significantly different from zero for mtDNA or microsatellites (Table 2). There was also no evidence of significant substructure within wintering regions for mtDNA (global ΦST=0.018, P=0.092) or microsatellites (FST=−0.001; P=0.765), and none of the pairwise comparisons were statistically significant (Supplementary Table S3). No evidence of genetic substructure was detected by STRUCTURE (Supplementary Figure S4b). Again, discriminant analysis of PCs inferred the optimal number of genetic clusters as four but in each sample, individuals were assigned to at least three different clusters (Figure 4b). The relative frequency of the inferred clusters was not associated with geography (Figure 4b).
The power analysis showed that with our specific markers and sample sizes, we had high statistical power (>0.9) to detect significant population structure for FST⩾0.005 (Supplementary Figure S1).
Discussion
Genetic differentiation and migratory divides
Genetic divergence between A. fuligula sampled across Eurasia is apparently not associated with the hypothesized migratory flyways. This was unexpected because ring recovery data suggested high fidelity of individuals to large-scale wintering regions. For example, no bird ringed on Indian wintering grounds was ever recovered in Europe (Hofer et al., 2005). Ring recoveries also suggested quite clear longitudinal separation on the breeding grounds of birds wintering in Europe (European flyway) and India (Central Asian flyway), respectively. Our samples from Eastern Siberia should represent the proposed Central Asian flyway (Blums and Baumanis, 1990; Hofer et al., 2005), but their genetic distinctness from the populations in the European flyway was not supported by any of our phylogeographic or population genetic analyses. It is possible that the degree of overlap between the breeding grounds of the postulated subpopulations of A. fuligula was underestimated due to the limited number of ring recoveries available from Eastern Siberia.
Many migratory bird species with a wide geographic distribution are composed of different subpopulations following distinct migratory routes. In most cases, these flyways have been delimited based on mark-recapture data but, in many species, they are also supported by the molecular data (mostly mtDNA), that is, in passerines (Ruegg and Smith, 2002; Clegg et al., 2003; Pérez-Tris et al., 2004), waders (Wennerberg, 2001) and several species of waterfowl (Scribner et al., 2001; Gay et al., 2004). Similarly, Friesen et al., (2007) found that a majority of seabirds with discontinuous distributions on the breeding or wintering grounds showed some genetic substructure. Sometimes, genetic divergence across migratory divides is only observed for one marker type, typically mtDNA in species with male-biased dispersal (Scribner et al., 2001; Gay et al., 2004), but we are aware of only two other studies where it was absent altogether (Pearce et al., 2004; Liu et al. 2011).
A lack of genetic divergence between migratory flyways may suggest that the migratory behaviour is generally flexible or that the migration route has recently changed in some populations. The presence of distinct flyways is often interpreted as a legacy of the ice ages. Different subpopulations would have been confined to distinct refugia, which they retained as wintering grounds following a post-glacial range expansion (Ruegg and Smith, 2002). Such periods in allopatry would lead to some genetic divergence between subpopulations, which may then be gradually eroded through gene flow. In species like tufted duck, where mating pairs are formed on the wintering grounds, gene flow would require that individuals occasionally switch between wintering regions, which, in turn, may suggest some behavioural plasticity. Alternatively, distinct migration routes can result if novel migratory strategies are adopted by some populations. Such changes may sometimes occur rapidly especially if migratory behaviour is culturally transmitted (Sutherland 1998). These alternative scenarios, that is, recent migratory change versus current gene flow, could be distinguished, for example, through a detailed characterization of the demographic history of populations and the migratory behaviour of individuals. Overall, the lack of genetic divergence between migratory flyways detected in A. fuligula appears to be the exception rather than the rule among migratory birds.
Genetic divergence and spatial distance
The extent of genetic divergence between breeding populations of A. fuligula sampled across Eurasia seems to be largely independent of geographical distance. Although many of the pairwise comparisons between Asian and European breeding sites are significant for mtDNA (and two also for microsatellites; Supplementary Table S3), the extent of divergence is similar to values observed within continents resulting in no detectable isolation by distance. We see two possible explanations for this result: first, the populations may not be at migration-drift equilibrium and, consequently, the observed divergence may poorly reflect the current rates of gene flow. For example, a recent range expansion could lead to the absence of isolation by distance even if gene flow is spatially restricted (Slatkin, 1993). The western breeding range of tufted duck has expanded in the last century (Cramp and Simmons, 1977), but no information is available about potential population size changes in other parts of the range or in the more distant past. With relatively large population sizes, it may take many thousands of generations for FST to reach its equilibrium value (Whitlock and McCauley, 1999). Hence, it cannot be excluded that the absence of isolation by distance across Eurasia in A. fuligula could still be a consequence of a range expansion, for example, after the last glacial maximum.
Alternatively, the absence of isolation by distance may be influenced by non-neutral processes affecting gene flow. For example, adaptation to particular local conditions unrelated to spatial distance might result in selection against immigrants and consequently against cross-breeding between some populations (Marshall et al., 2010). Of course, the extent of local adaptation in tufted ducks is unknown and, a priori, we may not expect it to be very pronounced in a species with high active dispersal abilities, which may counteract the effects of divergent selection (Lenormand, 2002).
Female breeding site fidelity
The extent of genetic divergence between breeding sites was 10 times higher for mtDNA than for nuclear markers. Such a pattern of elevated divergence for maternally inherited markers may suggest that gene flow is mainly mediated by males. However, several other biological and methodological factors can potentially contribute to such differences. First, the estimates of genetic differentiation may be influenced by differences in genetic variability arising from different mutation rates (Meirmans and Hedrick, 2011). Second, the haploidy and uniparental mode of inheritance of the mitochondrial genome leads to a roughly four-fold elevation of the rates of random genetic drift compared with the nuclear genome. Finally, as a nonrecombining molecule, mtDNA may also be more strongly affected by selective sweeps and background selection (Ballard and Whitlock, 2004), which could further reduce genetic variation. However, a strong effect of selective sweeps on mitochondrial sequence divergence between tufted duck populations is at odds with the generally high levels of genetic diversity and the presence of common and spatially widespread haplotypes. Consequently, it seems likely that sex-specific differences in the rates of gene flow at least contribute to the observed 10-fold difference in genetic differentiation between mitochondrial and nuclear markers.
High female philopatry to breeding sites is consistent with observations from Latvia where 97% of females returned to breed at the same lake where they had been ringed (Blums et al., 2002). Unfortunately, similar estimates are not available for males. Studies on related species have also found considerably higher genetic differentiation at mitochondrial than nuclear markers, for example, in spectacled eider (Scribner et al., 2001), red-crested pochard (Gay et al., 2004), common eider (Tiedemann et al., 2004) and common pochard (Liu et al., 2011), and female philopatry and male-biased dispersal is probably a general pattern among Anatidae (Rohwer and Anderson, 1988). This corresponds to the situation in many mammals while female-mediated gene flow is typically more common in other birds (Lawson Handley and Perrin, 2007). The relative costs of dispersal for the two sexes are thought to be strongly influenced by the mating system, and familiarity with a site may be advantageous in birds where males are territorial (Lawson Handley and Perrin, 2007). In tufted ducks, as in other Anatidae, offspring care is mostly provided by females (Cramp and Simmons, 1977) and advantages of local experience in the choice of feeding and breeding sites may favour female philopatry.
Population admixture on European wintering grounds
For the European wintering grounds, some subdivision between the ducks wintering in the Northwest and those wintering in Central and Southern Europe has been proposed based on information from ring recoveries (Scott and Rose, 1996). Although individual ducks probably show high fidelity to one of these large geographical regions (Fränzi Korner-Nievergelt, personal communication), the two postulated flyways overlap extensively along the North Sea coast (Hofer et al., 2005), which is expected to promote gene flow. In general, ringing data suggest that tufted ducks are highly mobile also outside the migration season and, within a winter, may move hundreds of kilometres between different feeding sites (Korner-Nievergelt et al., 2009). Against this background, it is not surprising that we found no evidence of genetic differentiation among European wintering grounds. Although our southern sampling sites should be representative of the Southeast wintering population, the sites grouped as Northwest (Table 2) may be located in the zone where the proposed wintering populations overlap along the North Sea coast. Still, these samples do not exhibit any obvious genetic signals expected in admixed populations like elevated genetic diversity or systematic deviations from Hardy–Weinberg or linkage equilibrium.
The absence of spatially separate subpopulations on the wintering grounds may suggest that pathogens, like avian influenza viruses, could be spread widely within Europe through within–winter movements of ducks. It is clear, however, that the likelihood of disease spread and transmission will depend heavily on the consequences of infection, which are very poorly understood especially in the wild. A laboratory study found that among the eight tufted ducks experimentally infected with highly pathogenic avian influenza H5N1, the individuals showing high levels of virus excretion were also the ones exhibiting severe disease symptoms (Keawcharoen et al., 2008). Given that the laboratory environments are usually relatively benign, the course of infection might be even more severe in the wild. Consequently, the role of tufted ducks as vectors of H5N1 may be limited, but this remains to be tested in specific studies. It is clear, however, that the lack of a clear association between genetic variation and geography strongly limits the inferences possible with respect to the likely origin of infected individuals. In particular, the extent of nuclear genetic differentiation (global FST=0.017) may be insufficient to use individual-based assignment methods to allocate individuals to particular breeding populations or regions based on multi-locus microsatellite genotypes (Riffaut et al., 2005; Latch et al., 2006; Gomez-Diaz and Gonzalez-Solis, 2007). Genomic-scale marker numbers could potentially provide enough resolution for such endeavours in the future, and current progress in molecular biology will likely put these within reach soon.
Data archiving
Sequence data have been deposited at GenBank: accession numbers JQ422386–JQ422479. Genotype data have been deposited at Dryad: http://dx.doi.org/10.5061/dryad.73n69m01
Acknowledgments
We acknowledge the following institutions and people for kindly providing samples: the Swiss Ornithological Institute, Sempach; Natural History Museum at the University of Oslo, Norway; Swedish Museum of Natural History; Max Planck Institute for Ornithology, Radolfzell, Germany; Museum of Natural Science, Louisiana State University, USA; Burke Museum of Natural History and Culture, University of Washington, USA; A Boto, M Boos, N Bulatova, A Caizergues, I Fefelov, M Ilieva, M Janaus, K Kaisel, R Hearn, J Kreisinger, R Lezalova, K Rabii, S van Rijn, M Ritz, D Rodrigues, A Sauter, Y Shi, T Shimada, J Viksne, V-M Väänänen, X.M Wang, S.P Zhang and ZW Zhang. We are grateful to the Swiss and the German National Reference Laboratory for Avian Influenza (Institute of Veterinary Bacteriology, University of Zürich and Friedrich-Löffler Institute) for providing samples of H5N1-positive tufted ducks. We gratefully acknowledge V Polyakov and S Pyzhjanov, who assisted his field work in Russia. We thank F Korner-Nievergelt for sharing her unpublished manuscripts. We thank S Tellenbach, E Kindler, T Jenkins, M Fischer and M Beysard for technical assistance, discussion and comments on an early version of the manuscript. This project was supported by the Bern University Research Foundation and a grant from the Swiss Veterinary Office (BVET) to G Heckel and I Keller.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Altizer S, Bartel RA, Han BA. Animal migration and infectious disease risk. Science. 2011;331:296–302. doi: 10.1126/science.1194694. [DOI] [PubMed] [Google Scholar]
- Ballard JWO, Whitlock MC. The incomplete natural history of mitochondria. Mol Ecol. 2004;13:729–744. doi: 10.1046/j.1365-294x.2003.02063.x. [DOI] [PubMed] [Google Scholar]
- Bandelt HJ, Forster P, Röhl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- Berthold P. Bird Migration: A General Survey. Oxford University Press USA; 2001. [Google Scholar]
- BirdLife International 2011Species Factsheet: Aythya fuligulaDownloaded fromhttp://www.birdlife.orgon, 7 April 2011.
- Blums P, Baumanis J. Migration and geographical distribution of pochard and tufted duck populations in the USSR. Baltic Birds. 1990;5:49–57. [Google Scholar]
- Blums P, Nichols J, Hines J, Mednis A. Sources of variation in survival and breeding site fidelity in three species of European ducks. J Animal Ecol. 2002;71:438–450. [Google Scholar]
- Clegg SM, Kelly JF, Kimura M, Smith TB. Combining genetic markers and stable isotopes to reveal population connectivity and migration patterns in a Neotropical migrant, Wilson's warbler (Wilsonia pusilla) Mol Ecol. 2003;12:819–830. doi: 10.1046/j.1365-294x.2003.01757.x. [DOI] [PubMed] [Google Scholar]
- Clobert J, Danchin E, Dhondt AA, Nichols JD. Dispersal. Oxford University Press New York; 2001. [Google Scholar]
- Cramp S, Simmons K. Handbook of the Birds of Europe, the Middle East and North Africa: the Birds of the Western Palearctic. Vol.1. Oxford University Press Oxford; 1977. [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Lischer HE. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Res. 2010;3:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesen VL, Burg TM, McCoy KD. Mechanisms of population differentiation in seabirds. Mol Ecol. 2007;16:1765–1785. doi: 10.1111/j.1365-294X.2006.03197.x. [DOI] [PubMed] [Google Scholar]
- Gay L, Defos Du Rau P, Mondain-Monval J, Crochet P. Phylogeography of a game species: the red-crested pochard (Netta rufina) and consequences for its management. Mol Ecol. 2004;13:1035–1045. doi: 10.1111/j.1365-294X.2004.02117.x. [DOI] [PubMed] [Google Scholar]
- Gilbert M, Xiao X, Domenech J, Lubroth J, Martin V, Slingenbergh J, et al. Anatidae migration in the western Palearctic and spread of highly pathogenic avian influenza H5N1 virus. Emerg Infect Dis. 2006;12:1650–1656. doi: 10.3201/eid1211.060223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Diaz E, Gonzalez-Solis J. Geographic assignment of seabirds to their origin: combining morphologic, genetic, and biogeochemical analyses. Ecol Appl. 2007;17:1484–1498. doi: 10.1890/06-1232.1. [DOI] [PubMed] [Google Scholar]
- Goudet J.2002FSTAT, A program to estimate and test gene diversities and fixation indices (version 2.9.3.2). Lausanne, SwitzerlandAvailable athttp://www2.unil.ch/popgen/softwares/fstat.htmUpdated from Goudet (1995).
- Hall T. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Hofer J, Korner-Nievergelt F, Korner-Nievergelt P, Kestenholz M, Jenni L. Herkunft und Zugverhalten von in der Schweiz überwinternden Reiherenten Aythya fuligula: eine Ringfundanalyse. Ornithol Beobacht. 2005;102:181–204. [Google Scholar]
- Huelsenbeck JP, Ronquist F. MrBayes: a program for the Bayesian inference of phylogeny. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Jombart T. adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics. 2008;24:1403–1405. doi: 10.1093/bioinformatics/btn129. [DOI] [PubMed] [Google Scholar]
- Jombart T, Devillard S, Balloux F. Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet. 2010;11:94. doi: 10.1186/1471-2156-11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keawcharoen J, van Riel D, van Amerongen G, Bestebroer T, Beyer WE, van Lavieren R., et al. Wild ducks as long-distance vectors of highly pathogenic avian influenza virus (H5N1) Emerg Infect Dis. 2008;14:600–607. doi: 10.3201/eid1404.071016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilpatrick AM, Chmura AA, Gibbons DW, Fleischer RC, Marra PP, Daszak P, et al. Predicting the global spread of H5N1 avian influenza. Proc Nat Acad Sci USA. 2006;103:19368–19373. doi: 10.1073/pnas.0609227103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korner-Nievergelt F, Hofer J, Sauter A, Jenni L. Measuring within-winter movement rates of tufted duck Aythya fuligula and common pochard A. ferina based on ring re-encounter data. Wildfowl. 2009;S2:24–41. [Google Scholar]
- Latch E, Dharmarajan G, Glaubitz J, Rhodes O. Relative performance of Bayesian clustering software for inferring population substructure and individual assignment at low levels of population differentiation. Cons Genet. 2006;7:295–302. [Google Scholar]
- Lawson Handley LJ, Perrin N. Advances in our understanding of mammalian sex-biased dispersal. Mol Ecol. 2007;16:1559–1578. doi: 10.1111/j.1365-294X.2006.03152.x. [DOI] [PubMed] [Google Scholar]
- Lenormand T. Gene flow and the limits to natural selection. Trends Ecol Evol. 2002;17:183–189. [Google Scholar]
- Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Liu Y, Keller I, Heckel G. Range-wide genetic population structure in common pochard (Aythya ferina) a potentially important vector of highly pathogenic avian influenza viruses. Ecol Evol. 2011;1:529–545. doi: 10.1002/ece3.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall DJ, Monro K, Bode M, Keough MJ, Swearer S. Phenotype–environment mismatches reduce connectivity in the sea. Ecol Lett. 2010;13:128–140. doi: 10.1111/j.1461-0248.2009.01408.x. [DOI] [PubMed] [Google Scholar]
- Meirmans PG, Hedrick PW. Assessing population structure: Fst and related measures. Mol Ecol Res. 2011;11:5–18. doi: 10.1111/j.1755-0998.2010.02927.x. [DOI] [PubMed] [Google Scholar]
- Peakall ROD, Smouse PE. GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes. 2006;6:288–295. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce J, Talbot S, Pierson B, Scribner K, Dickson L, et al. Lack of spatial genetic structure among nesting and wintering king eiders. The Condor. 2004;106:229–240. [Google Scholar]
- Pérez-Tris J, Bensch S, Carbonell R, Helbig A, Tellería J. Historical diversification of migration patterns in a passerine bird. Evolution. 2004;58:1819–1832. doi: 10.1554/03-731. [DOI] [PubMed] [Google Scholar]
- Posada D. jModelTest: phylogenetic model averaging. Mol Biol Evol. 2008;25:1253–1258. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- R Development Core Team 2008R: A language and environment for statistical computing. ISBN 3-900051-07-0 . www.R-project.orgR Foundation for Statistical Computing Vienna, Austria [Google Scholar]
- Riffaut L, McCoy K.D, Tirard C, Friesen V.L, Boulinier T. Population genetics of the common guillemot Uria aalge in the North Atlantic: geographic impact of oil spills. Marine Ecol Prog Ser. 2005;291:263–273. [Google Scholar]
- Robertson G, Cooke F. Winter philopatry in migratory waterfowl. The Auk. 1999;116:20–34. [Google Scholar]
- Rohwer F, Anderson M. Female-biased philopatry, monogamy, and the timing of pair formation in migratory waterfowl. Current Ornithol. 1988;5:187–221. [Google Scholar]
- Ruegg KC, Smith TB. Not as the crow flies: a historical explanation for circuitous migration in Swainson's thrush (Catharus ustulatus) Proc R Soc B. 2002;269:1375–1381. doi: 10.1098/rspb.2002.2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryman N, Palm S. POWSIM: a computer program for assessing statistical power when testing for genetic differentiation. Mol Ecol Notes. 2006;6:600–602. doi: 10.1046/j.0962-1083.2001.01345.x. [DOI] [PubMed] [Google Scholar]
- Schweizer M, Excoffier L, Heckel G. Fine-scale genetic structure and dispersal patterns in the common vole Microtus arvalis. Mol Ecol. 2007;16:2463–2473. doi: 10.1111/j.1365-294X.2007.03284.x. [DOI] [PubMed] [Google Scholar]
- Scott D, Rose P. Atlas of Anatidae Populations in Africa and Western Eurasia. Wetlands International Wageningen; 1996. [Google Scholar]
- Scribner KT, Petersen MR, Fields RL, Talbot SL, Pearce JM, Chesser RK, et al. Sex-biased gene flow in spectacled eiders (Anatidae): inferences from molecular markers with contrasting modes of inheritance. Evolution. 2001;55:2105–2115. doi: 10.1111/j.0014-3820.2001.tb01325.x. [DOI] [PubMed] [Google Scholar]
- Slatkin M. Isolation by distance in equilibrium and non-equilibrium populations. Evolution. 1993;47:264–279. doi: 10.1111/j.1558-5646.1993.tb01215.x. [DOI] [PubMed] [Google Scholar]
- Sorenson MD, Cooper A, Paxinos EE, Quinn TW, James HF, Olson SL, et al. Relationships of the extinct moa-nalos, flightless Hawaiian waterfowl, based on ancient DNA. Proc R Soc B. 1999;266:2187–2193. doi: 10.1098/rspb.1999.0907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorenson MD, Fleischer RC. Multiple independent transpositions of mitochondrial DNA control region sequences to the nucleus. Proc Nat Acad Sci USA. 1996;93:15239–15243. doi: 10.1073/pnas.93.26.15239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stovicek O, Cizkova D, Liu Y, Albrecht T, Heckel G, Vyskocilova M, et al. Development of microsatellite markers for a diving duck, the common pochard (Aythya ferina) Cons Genet Res. 2011;3:573–576. [Google Scholar]
- Sutherland WJ. Evidence for flexibility and constraint in migration systems. J Avian Biol. 1998;29:441–446. [Google Scholar]
- Tiedemann R, Paulus KB, Scheer M, von Kistowski KG, Skírnisson K, Bloch D, et al. Mitochondrial DNA and microsatellite variation in the eider duck (Somateria mollissima) indicate stepwise postglacial colonization of Europe and limited current long-distance dispersal. Mol Ecol. 2004;13:1481–1494. doi: 10.1111/j.1365-294X.2004.02168.x. [DOI] [PubMed] [Google Scholar]
- Wennerberg L. Breeding origin and migration pattern of dunlin (Calidris alpina) revealed by mitochondrial DNA analysis. Mol Ecol. 2001;10:1111–1120. doi: 10.1046/j.1365-294x.2001.01256.x. [DOI] [PubMed] [Google Scholar]
- Whitlock MC, McCauley DE. Indirect measures of gene flow and migration: FST≠1/(4Nm+1) Heredity. 1999;82:117–125. doi: 10.1038/sj.hdy.6884960. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.