Abstract
Nascent-peptide modulation of translation is a common regulatory mechanism of gene expression. In this mechanism, while the nascent peptide is still in the exit tunnel of the ribosome, it induces translational pausing, thereby controlling the expression of downstream genes. One example is SecM, which inhibits peptide-bond formation in the ribosome's peptidyl transferase center (PTC) during its own translation, upregulating the expression of the protein translocase SecA. Although biochemical experiments and cryo-electron microscopy data have led to the identification of some residues involved in SecM recognition, the full pathway of interacting residues that connect SecM to the PTC through the ribosome has not yet been conclusively established. Here, using the cryo-electron microscopy data, we derived the first (to our knowledge) atomic model of the SecM-stalled ribosome via molecular-dynamics flexible fitting, complete with P- and A-site tRNAs. Subsequently, we carried out simulations of native and mutated SecM-stalled ribosomes to investigate possible interaction pathways between a critical SecM residue, R163, and the PTC. In particular, the simulations reveal the role of SecM in altering the position of the tRNAs in the ribosome, and thus demonstrate how the presence of SecM in the exit tunnel induces stalling. Finally, steered molecular-dynamics simulations in which SecM was pulled toward the tunnel exit suggest how SecA interacting with SecM from outside the ribosome relieves stalling.
Introduction
Effectively all protein synthesis in cells begins with the ribosome, a complex RNA-based molecular machine that translates genetic information into the polypeptide chains that subsequently fold into mature proteins (1). Amino acids are delivered to the ribosome by transfer RNA molecules (tRNAs), which cycle through discrete sites in the ribosome termed the A, P, and E sites. The critical catalytic step, bond formation between a nascent protein attached to the P-site tRNA and the next amino acid in the sequence at the A site, occurs deep within the ribosome at the peptidyl transferase center (PTC) (2).
After the addition of a new amino acid, the nascent chain leaves the ribosome through a 100-Å-long tunnel, the exit tunnel, which has an average diameter of 15 Å (3,4). Rather than being a passive conduit, the exit tunnel is increasingly being found to play a functional role in protein development (5,6). For example, interactions between the nascent chain and the tunnel can bias the folding of nascent polypeptides (7–12) and recruit external factors to the ribosome (13–16). The tunnel can also play a regulatory role by modulating the rate of translation (17), even to the point of stalling (18–22).
Translational stalling of the ribosome via nascent-chain-tunnel interactions serves to control the expression of downstream genes. The mechanisms of this leader-peptide-induced stalling are varied. Some require the presence of additional molecules (e.g., tryptophan in the case of TnaC (20,23,24)). The expression of specific antibiotic resistance genes can also be upregulated by the translation of a stalling peptide concomitant with binding of the corresponding antibiotic in the exit tunnel (e.g., ErmCL and erythromycin (25,26)). Whereas for all known stalling peptides translational arrest is caused by inhibition of reactions at the PTC, the sequence of interactions that signal the peptide's presence in the tunnel to the PTC is varied (27).
Stalling peptides of another class have an innate inhibition propensity and do not require the presence of any additional molecules. One example of this class is SecM, a 170-amino-acid protein that controls the expression of the protein translocase SecA (28). In Escherichia coli, the minimum sequence necessary to induce stalling is 150FXXXXWIXXXXGIRAGP166; however, a comparison across SecM homologs in different species reveals only I162, R163, and P166 to be invariant (29). In the SecM-stalled ribosome, P166 resides at the A site and is likely critical due in part to the naturally slow peptide-bond-formation rate for proline (30).
In addition to residues in SecM, elements of the ribosome involved in stalling have been identified. Specifically, the 23S rRNA bases A2058, A2062, and C2-methylated A2503 have been shown to be required for SecM-mediated stalling (E. coli numbering) (28,31,32), along with proteins L22 and L4, which form a constriction point in the exit tunnel (3). Interestingly, these bases are not universally required by stalling peptides (32).
Stalling is typically relieved through the interaction of SecA with the nascent chain and ribosome (33,34), although given enough time, peptide-bond formation (i.e., elongation at P166) can still occur spontaneously without SecA and thus overcome stalling (35). A recent cryo-electron microscopy (cryo-EM) map of a SecM-stalled ribosome revealed that the linkage between the nascent chain and the P-site tRNA is shifted by 2 Å from its position in a nonstalled ribosome, suggesting a possible mechanism of SecM-mediated stalling (22). However, no A-site tRNA was present in the 5.6-Å-resolution reconstruction used to discern the position of this linkage, although it was present in the 9.3-Å map. Thus, it remains possible that additional rearrangements within the A site also contribute to stalling.
Although biochemical and structural studies have demonstrated the involvement and organization of specific residues during SecM-induced translational arrest, the dynamics of the stalled system have not been investigated previously. Here, we used the cryo-EM map of Bhushan et al. (22) along with the molecular-dynamics flexible fitting (MDFF) method (36–38) to resolve an atomic-resolution model of SecM in the exit tunnel, complete with P- and A-site tRNAs. MD simulations of this structure and derived mutants reveal a relay of ribosomal residues connecting SecM's R163 to the PTC, and thus provide a plausible model for the cascade of interactions through which SecM arrests translation.
Methods
Modeling and fitting of the full ribosome-SecM-tRNA complex
The model of the ribosome presented here is based on the one developed by Trabuco et al. (39), which was used as a starting point. A structure for tRNAGly was generated as a homology model of tRNAGly type 3 based on tRNAPhe included in the ribosome model 2XQD (40). The model for tRNAPro was constructed as tRNAPro type 2 based on the tRNAPro included in the ribosome model 2WWL (24). The constructed models contained all modified residues, and the sequences of the fully modified tRNAs were retrieved from the MODOMICS database (http://modomics.genesilico.pl/). The initial positions of the SecM backbone were taken from previously published work (22). After completion of the side chains, the model was relaxed in the presence of the segmented map for SecM.
In the first step, a structural model was obtained for the 70S/SecM/tRNAGly complex. Because the initial structures of all components did not fit together properly, we performed manual adjustments using interactive MDFF (41) after prefitting the components into segmented densities. The adjustments included generation of proper CCA-23S rRNA interactions and reinforcements of basepairing within the tRNA model. Additionally, the N-terminal part of L27 had to be released because of clashes with the tRNA.
Fitting of the SecM-stalled ribosome model to the cryo-EM density map in Bhushan et al. (22) was carried out in multiple stages. For fitting of the full ribosome system, different protocols were tested. Optimal results were obtained by initially coupling each of the components to the full map, and additionally coupling SecM and the P-site tRNA to the corresponding segmented density. The coupling of the SecM backbone to the segmented map was five times stronger than that of the rest of the system. Subsequently, the coupling of all the components to the maps was increased by a factor of 3, and only the SecM backbone was additionally coupled to the segmented map. The rationale behind the chosen protocol was to allow the ribosomal residues and the SecM side chains to compete in the regions of the map that could not be unambiguously attributed to either of the components while ensuring the proper positioning of the SecM backbone. Due to the relatively high resolution of the map, the quality of the SecM fit could be assessed visually. By the end of the fitting procedure, the root mean-square deviation (RMSD) of the ribosome changed <0.05 Å within the last nanosecond of simulation. The total fitting time was 3.5 ns.
The study by Bushan et al. (22) contains a high-resolution (5.6 Å) map of a stalled ribosome with an empty A site and a lower-resolution (9.3 Å) map of a stalled ribosome in which the A site was occupied by a Pro-tRNAPro. To generate a structure of a SecM stalled ribosome in which also the A site is occupied, we added a Pro-tRNAPro. Initially, distance restraints based on 2WDK-2WDL (42) were used to ensure proper interactions of the CCA with the large subunit. To get maximal value from the data in Bushan et al. (22), we combined both maps reported in the following way: Whereas the 70S/SecM/tRNAGly complex was coupled to the high-resolution map, the A-site tRNA was only coupled to the corresponding segmented density obtained from the low-resolution map. To minimize possible contamination of the more highly resolved regions by the lower-resolution data, only the immediate environment of the A-site tRNA, namely, residues within 15 Å, with the exception of the PTC, was allowed to move.
Preparation and fitting of the reduced model
To obtain a more realistic picture of the structural changes induced by the presence of SecM at an acceptable computational cost, we constructed a reduced, solvated model. The reduced model includes all residues within 25 Å of the core defined by the acceptor stems and elbow regions of the tRNAs as well as SecM. Atoms >23 Å away were spatially restrained. We neutralized the obtained subsystem by adding diffusively bound Mg2+ ions (minimum ion-solute distance of 6.5 Å) using cIonize, a GPU-accelerated tool in VMD (43) that iteratively places ions at the minima of the electrostatic potential (44). Subsequently, we completed the octahedral solvation shell of each Mg2+ ion using MgSolvate (37,45), and performed the initial solvation of the system using the Solvate program (46). Finally, the system was placed in a water box with the VMD plugin Solvate, and the VMD plugin AutoIonize was used to establish a KCl concentration of 0.1 mol/L. The dimensions of the water box were chosen such that the minimum distance between ribosomal residues and their periodic images was 10 Å, resulting in a total system size of ∼360,000 atoms.
After preparation of the reduced system, fitting was continued. First, water and ions were allowed to relax for 2 ns while the rest of the system was restrained. In subsequent stages, only atoms of the ribosome at the subsystem boundary were restrained. In the second stage, fitting was continued for 15 ns in the presence of secondary structure restraints, restraints to maintain chirality and prevent trans/cis isomerization of peptide bonds (47), and restraints between nucleic acid basepairs. Restraints for ribosomal basepairs and for protein secondary structure were removed for the next 5.5 ns, followed by removal of all restraints for an additional 10 ns of fitting. Finally, the system was minimized while it was strongly coupled to the map to eliminate prior thermal fluctuations.
Simulation
MD simulations were carried out with the use of NAMD 2.8 (48) with the AMBER99SB force field (49,50), including modified nucleosides (51), and adapted to the CHARMM format for ease of use in VMD (39,43). The equations of motion were integrated every 1 fs, with nonbonded forces evaluated every other time step. Periodic boundary conditions were used, along with a cutoff of 10 Å for short-range, nonbonded interactions, modified beginning at 9 Å to decay smoothly to zero. A constant temperature of 300 K and pressure of 1 atm were maintained.
Steered MD was used in two simulations: one at constant force (CF) and one at constant velocity (CV) (52,53). In the CF simulation, a force of 1.25 nN was applied to the Cα atom of the N-terminal SecM residue (here, K131) in a direction approximately aligned with the exit tunnel, and the resulting extension of SecM peaked at ∼40 Å. In the CV simulation, the same atom was attached to a virtual spring (k = 5 kcal/mol · Å2) that moved with a velocity of 5 Å/ns.
The total time of simulations carried out for this study was ∼300 ns.
Results
Modeling the SecM-stalled ribosome
Development of an atomic-scale model of SecM in the ribosome began from a previously constructed, complete model of the empty E. coli ribosome including all modified residues (39). Additionally, structures of two tRNA species, tRNAGly in the P site and tRNAPro in the A site, were obtained by homology modeling (see Materials and Methods). The tRNAGly was added to the model first, and the full ribosome-SecM-tRNAGly was flexibly fitted via MDFF (36) to the high-resolution (5.6 Å) cryo-EM density map of Bhushan et al. (22). In the next stage of modeling, the tRNAPro was added to the A site of the model and fitted with the use of an additional, lower-resolution (9.3 Å) map that was derived from the same source but also contained density for the A-site tRNAPro. The discrepancy in resolution between the two maps is a result of the low occupancy of the A site after sample preparation, which necessitated sorting the data into two sets, one with and one without an A-site tRNA present (22). During the entire fitting procedure, additional restraints were used to preserve secondary structure and specific interactions, as well as to ensure the stereochemical integrity of the obtained model (47).
After the full ribosome-SecM model was fitted in vacuo, a subsystem consisting of SecM, the majority of the tRNAs, and surrounding residues, including all of those lining the exit tunnel, was prepared (see Materials and Methods, and Fig. S1 in the Supporting Material). This subsystem enabled subsequent modeling and simulations to focus on interactions within the PTC and exit tunnel at a greatly reduced computational cost (24,54). Without such a reduction in system size, investigations of the complete ribosome would require either significant computational resources for all-atom simulations (∼3 million atoms) (55,56) or reduced representations (e.g., coarse-grained models (57–62)). The subsystem was fully solvated and subjected to multiple stages of further refinement via MDFF with varying levels of structural restraints (see Materials and Methods for a full description of the different stages). Convergence of the fitting was monitored, in particular, by the RMSD of SecM and the peptide-bonding distance between residues G165 and P166 on the P- and A-tRNAs, respectively (see Fig. S2). The latter required a total of at least 30 ns to stabilize. It is interesting to note that no large-scale, cascade-like rearrangements in the ribosome were observed, in contrast to an earlier report (63) based on an interpretation of a 15-Å cryo-EM map.
Interactions between SecM and the ribosome in the fitted structural model are similar to those proposed on the basis of the cryo-EM map alone (22), and are shown in Fig. 1 and Fig. S3. Specifically, R163 of SecM interacts with the base of A2062 of the 23S rRNA and also forms a hydrogen bond with U2586 (donor-acceptor distance of 2.7 Å), although only the former base is known to be required for stalling (32). Near the L4/L22 constriction point of the exit tunnel, A751 of the 23S stacks with W155 of SecM (minimum separation: 3 Å). Mutation of W155 or I156 relieves stalling in E. coli, as do insertions in the region of A751 (28). Other residues of 23S rRNA that interact with SecM include U2585 near the PTC, U2609 near A159 and Q160, and A752 near Q158 of SecM (defined through polar or hydrophobic heavy atoms on each residue within 4 Å, respectively), although none are essential for stalling.
Figure 1.
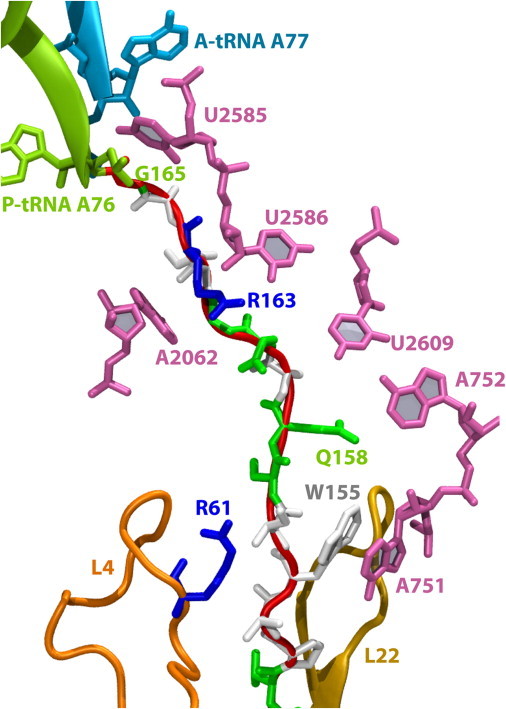
Selected interactions between SecM and the exit tunnel in the MDFF-fitted model. SecM is shown in both cartoon (red/gray) and stick (colored by residue type: blue/dark gray for basic, green/light gray for hydrophilic, white for hydrophobic) representations. Key ribosome residues are highlighted, including the A- and P-tRNAs, the L4/L22 constriction region, and those bases lining the tunnel that interact with SecM. A more complete depiction of the full-length SecM is given in Fig. S3. Note that A77 in the A-tRNAPro is also sometimes denoted A76 by convention.
By comparing the fitted structure of the SecM-stalled ribosome with that of a nonstalled ribosome, one can discern unique features that are potentially relevant to stalling. For this comparison, we aligned the Thermus thermophilus 70S ribosome (PDBs 2WDL and 2WDK), which contains both A- and P-tRNAs (42), to the fitted structure using the portion of the 23S rRNA present in the subsystem (see Fig. 2 C). The most apparent difference is an upward (away from the PTC) shift of the ends of both the P- and A-tRNAs in the SecM-stalled structures. To rule out the possibility that the observed shift by 1–2 Å of the tRNAs is an artifact of the choice of residues used for the alignment, we performed an alignment using tRNAs only, which resulted in essentially the same displacement. A shift of the P-tRNA was also noted on the basis of the map alone, and was proposed to be the inactivation mechanism utilized by SecM (22). Although this shift is below the nominal resolution of the cryo-EM map, because of the known stereochemical limits that are imposed by the structure of SecM and built into the MDFF simulations, such a shift can be reliably identified from the fitted models. Distinct positions for individual side chains, on the other hand, cannot typically be determined from the map (36,64). The movement of the tRNAs increases the distance between the peptide-bonding atoms on their respective amino acids from 3.3 Å in 2WDK to 7.1 Å in the SecM-stalled ribosome, which is sufficient to prevent catalysis and thus induce translational arrest.
Figure 2.
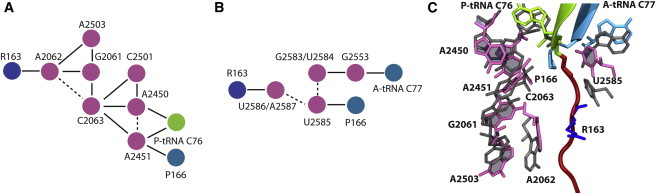
Proposed relays connecting SecM to the PTC. Residues are colored as in Fig. 1. (A and B) Relay between R163 of SecM and (A) P-tRNA and (B) A-tRNA based on interactions listed in Table 1. Solid lines represent side-chain interactions, and dotted lines represent connections through the backbone. (C) Positions of key relay residues after fitting compared with canonical positions in 3WDK/3WDL. Residues are shown as sticks colored as in Fig. 1 (fitted structure) or in dark gray (2WDK/2WDL).
Because no A-tRNA was present in the high-resolution map used for comparison in Bhusan et al. (22), its altered position was not investigated previously. To examine the validity of the shift of the A-tRNA, we repeated the MDFF of the entire SecM-tRNA-exit-tunnel subsystem using only the high-resolution map that did not contain the A-tRNA. Although no density existed for the A-tRNA in this fitting, the A-tRNA still moved from its initial position. This motion was driven by interactions with the rest of the ribosome, particularly between the CCA (the three residues at the 3′ end of A-tRNA) and U2585, which intercalates between SecM and the A-tRNA. Although the shift of the P-tRNA alone may be sufficient to induce stalling, it is possible that repositioning of the A-tRNA could also contribute by restraining its ability to move closer to the P-tRNA, which would compensate for the latter's movement.
Identification of potential relays connecting SecM to the PTC
Although the fitted structure of SecM in the ribosomal exit tunnel displays unique contacts that may be relevant for stalling translation, it is not clear that these contacts constitute a communication pathway between SecM and the PTC. Dynamics information, however, can provide evidence that these contacts are sufficiently stable and overlapping to induce stalling (65,66). Therefore, the fitted structure of the SecM-stalled ribosome was equilibrated for 20 ns in the absence of all restraints, including those from the map used during fitting. The persistence of interactions between residues near SecM and the PTC during equilibration, here defined as those that are characteristically hydrophobic, hydrophilic, or hydrogen-bonding (67), was quantified (the interactions are listed in Table 1 and Table S1).
Table 1.
Interactions that relay SecM-mediated stalling of the ribosome
| Residue 1 | Residue 2 | w/SecM | Q158A | ΔSecM | Pull (CF) | Pull (CV) | R163S | A2062U | A2503G |
|---|---|---|---|---|---|---|---|---|---|
| SecM R162 | 23S A2062 | 0.00 | 0.00 | - | 0.00 | 0.20 | 0.00 | 0.00 | 0.00 |
| SecM R163 | 23S A2062 | 0.97 | 0.99 | - | 0.60 | 0.57 | 0.47 | 0.04 | 0.97 |
| SecM R163 | 23S U2441 | 0.99 | 0.80 | - | 1.00 | 0.99 | 0.00 | 0.96 | 1.00 |
| SecM R163 | 23S U2586 | 1.00 | 1.00 | - | 0.40 | 0.72 | 0.42 | 0.94 | 1.00 |
| SecM R163 | 23S A2587 | 0.99 | 0.97 | - | 0.99 | 0.75 | 0.00 | 1.00 | 1.00 |
| 23S G2061 | 23S C2063 | 0.34 | 0.21 | 0.20 | 0.85 | 0.98 | 0.33 | 0.99 | 0.00 |
| 23S G2061 | 23S A2503 | 0.99 | 0.75 | 0.96 | 0.90 | 0.86 | 0.89 | 1.00 | 1.00 |
| 23S A2062 | 23S A2503 | 0.71 | 0.91 | 0.00 | 0.97 | 0.80 | 0.43 | 0.00 | 0.00 |
| 23S A2062 | 23S G2061 | 0.99 | 1.00 | 0.57 | 0.16 | 1.00 | 0.99 | 1.00 | 0.99 |
| 23S C2063 | 23S A2450 | 0.54 | 0.68 | 0.52 | 0.79 | 0.21 | 0.92 | 0.38 | 0.68 |
| 23S C2063 | 23S A2451 | 0.88 | 0.93 | 0.96 | 0.95 | 0.97 | 1.00 | 0.59 | 1.00 |
| 23S C2063 | 23S C2501 | 1.00 | 1.00 | 0.79 | 1.00 | 1.00 | 0.93 | 0.97 | 1.00 |
| 23S C2501 | 23S A2450 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 23S G2583 | 23S G2553 | 0.84 | 0.94 | 0.89 | 0.88 | 0.96 | 0.95 | 0.84 | 0.92 |
| 23S A2450 | P-tRNA A76 | 1.00 | 1.00 | 1.00 | 1.00 | 0.87 | 1.00 | 1.00 | 1.00 |
| 23S A2451 | P-tRNA A76 | 0.92 | 0.91 | 0.77 | 0.73 | 0.90 | 0.77 | 0.83 | 0.77 |
| 23S G2553 | A-tRNA A77 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 23S A2451 | SecM P166 | 0.89 | 0.83 | 0.98 | 0.77 | 0.68 | 0.98 | 0.89 | 1.00 |
| 23S U2585 | SecM P166 | 0.54 | 0.29 | 0.50 | 0.76 | 0.85 | 0.46 | 0.96 | 0.56 |
Interactions are either hydrophobic or hydrophilic in nature, including hydrogen bonds, with a minimum distance of <4 Å. The percentages correspond to the fraction of time interactions are present and are calculated over the last 10 ns of a 20-ns simulation; those highlighted in bold exhibit a decrease of >25% compared with the wild-type simulation with SecM. See also Table S1.
The patterns of interactions between SecM and the ribosome, as well as within the ribosome, constitute putative relays connecting SecM to the PTC, including both A- and P-tRNAs. Two pathways, one leading from R163 of SecM to the P-tRNA and one to the A-tRNA, are shown schematically in Fig. 2, A and B. On one side, R163 forms a very stable hydrophobic interaction (frequency of 97%) with A2062 of the ribosome, which also interacts with G2061 and A2503. A2062 and A2503 are both required for SecM-mediated stalling, whereas G2061 is involved in the PTC (32). G2061 interacts with C2063 through their bases (frequency of 34%) and by shifting of the rRNA backbone. From there, C2063 contacts A2450 and A2451, which also interact with A76 of the P-tRNA as well as with the amino acid, P166, on the A-tRNA. On the other side of SecM, R163 interacts with U2586 and A2587. However, when the simulation of the SecM-stalled ribosome is repeated, the interaction between R163 and U2586/A2587 is reduced (see Table S1). U2586 has a compensating interaction with I162 in this simulation (frequency of 26%), suggesting that the interaction between U2586 and SecM may be nonspecific. Similarly, favorable interactions between U2585 and P166 on the A-tRNA observed in the first simulation are eliminated in the second, although the two remain in close contact (average center-of-mass separation of 8.1 Å in the first wild-type simulation and 7.8 Å in the second). Therefore, whereas the relay from SecM to the P-tRNA involves residues known to be critical for stalling, the relay to the A-tRNA is much more speculative.
During equilibration of the SecM-ribosome complex, the increased separation between the A- and P-tRNAs compared with other ribosome x-ray structures is maintained (see Fig. 3 A). SecM appears to directly control the positioning of the residue A2062, which can be communicated through the putative interaction relays shown in Fig. 2, A and B, to the tRNAs. To investigate the role of SecM in positioning ribosomal residues, we compared the fluctuations of A2062 observed in two wild-type SecM simulations; an earlier simulation of TnaC (24), which does not require A2062 for stalling (32); and a simulation of the stalled ribosome in which the SecM peptide was removed. In the latter two simulations, the flexibility of A2062 increased dramatically, as shown in Fig. 4; for example, the root mean-square fluctuations for N1 of A2062 were 0.9–1.0 Å for SecM, 2.1 Å for TnaC, and 1.5 Å for the empty tunnel. Furthermore, the increased flexibility of A2062 without SecM present completely abolished the interactions between it and A2503, severing the putative interaction relay. These findings support A2062 as a sensor of the exit tunnel's occupancy that communicates its position to the PTC when it is restrained (32). On the other side of the exit tunnel, the position and flexibility of U2585 are not significantly affected by deletion of SecM (data not shown).
Figure 3.
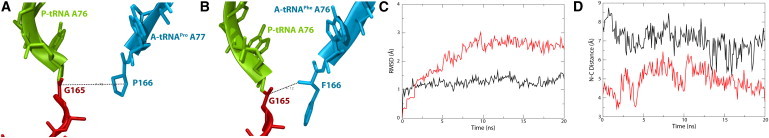
Comparison of tRNAPro and tRNAPhe in the A site. (A and B) PTC of the SecM-stalled ribosome with tRNAPro (A) and tRNAPhe (B) in the A site, colored as in Fig. 1. The distances between reacting atoms, namely, the carbonyl carbon of G165 and the nitrogen of P166 or F166 are indicated. (C) RMSD for the A-tRNA backbone during equilibration. tRNAPro is shown in black and tRNAPhe in red/gray. (D) Peptide-bonding distance over time for tRNAPro (black) and tRNAPhe(red/gray).
Figure 4.
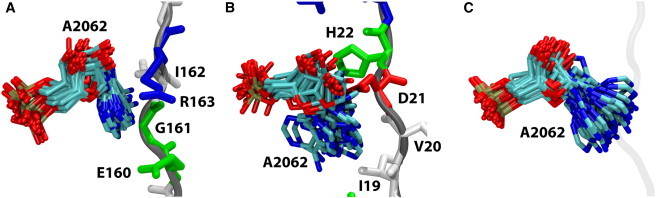
Flexibility of A2062. Shown are conformations sampled by A2062, taken every 1 ns over the course of 20 ns of unconstrained simulations. SecM/TnaC residues are shown in stick representation, colored by residue type. (A) Case of SecM bound in the exit tunnel. (B) Case of TnaC bound in the exit tunnel. (C) Empty tunnel. The original position of SecM before its deletion is indicated by a transparent gray line.
In addition to SecM influencing the positions of ribosomal residues, specific components in the ribosome also control the structure and position of SecM in the exit tunnel. Because R163 appears to be a key residue in the proposed interaction relay connecting SecM to the PTC, its proper positioning in the tunnel should be critical. Stabilizing the extension of the side chain of R163 in our model is a hydrogen bond between R163 and the backbone of U2441 of 23S; however, this hydrogen bond was not consistently observed (see Table 1 and Table S1). Base stacking of W155 of SecM with A751 is maintained during equilibration, as are other interactions identified in the fitted structure. These interactions cause a compaction in the upper region of SecM, namely, between residues W155 and R163. The Cα-Cα distance between these residues in the fitted structure is 24 Å, whereas in a completely extended peptide it would be ∼31 Å (assuming an average extended length of one amino acid of 3.4 Å). The compaction serves to localize R163 to the vicinity of A2062 and, furthermore, to stabilize it there. Therefore, as predicted from mutagenesis and bioinformatics studies (29), only R163 is directly involved in communicating with the PTC, and other residues interact with the ribosome to correctly position R163.
Testing the role of specific residues through in silico mutation
A number of point mutations to residues in both the ribosome and SecM that relieve stalling have been discovered (28,29,31,32). Many of these residues are involved in the putative interaction relay connecting SecM to the PTC as described above. To further validate their roles in the proposed relay, we generated three mutations (R163S in SecM, and A2062U and A2503G in 23S) in silico and compared the resulting dynamics over 20-ns equilibrations against the wild type. We also examined a mutation in SecM that does not alleviate stalling, Q158A, as well as the role of A-tRNAPro.
All three mutations that abolish SecM-mediated translational arrest had a detrimental effect on the interaction relay. In the case of R163S, interactions between residue 163 and the ribosome are significantly reduced or eliminated (see Table 1). Similarly, in the A2062U mutant, interactions that include residue 2062 are diminished as U2062 flips up, away from its position between R163 and A2503, as shown in Fig. S5 B. Alternatively, in the A2503G mutant, G2061 flips down, between A2062 and G2503. Although the mechanisms are distinct, in all three mutants it is apparent that the primary interaction relay between SecM and the PTC is disrupted. Repeated simulations of each of the three mutants displayed moderately different interaction patterns (see Table S1), although the effect of disrupting the putative relays was always the same.
In contrast to the three detrimental mutations, the Q158A mutation only slightly disturbs the proposed relay. In particular, interactions involving G2061 are reduced in frequency from 34% to 21% with C2063, and from 99% to 75% with A2503. The interaction relay is otherwise maintained, as expected due to the mutant's retained ability to stall the ribosome (29).
In an attempt to determine the most relevant features of R163 for stalling, we simulated two novel mutants: R163K and R163Q. In the case of R163K, numerous interactions along the proposed relay were broken, precipitated by a shift in the position of A2062 akin to that observed in the A2062U mutant (see Table S1). In the case of R163Q, however, only the interaction between residue 163 of SecM and A2062 was significantly disturbed, with the rest of the relay remaining intact. Closer examination revealed that the hydrophobic interactions between A2062 and R163 were replaced by interactions with I162, a residue that is also conserved across all species (29). Whether this new interaction with I162 would be sufficient to maintain the relay over longer timescales is uncertain. Nonetheless, the hydrophobic character of the residues immediately adjacent to A2062 appears to be more important for recognition of SecM by the ribosome than the charge of residue 163.
One of the requirements for SecM stalling is a Pro-tRNAPro in the A site. The fitted structure displays a shifted position of the A-tRNA, and simulations of the native SecM-stalled ribosome suggest an interaction relay connecting SecM to the A-tRNA. However, it is not evident whether the observed shift of A-tRNA is caused by SecM or is intrinsic to tRNAPro. To distinguish between the two possibilities, the A-tRNA in the fitted structure was replaced by tRNAPhe and equilibrated for 20 ns. After alignment based on the position of tRNAPro, the tRNAPhe in the A site displayed a slight downward shift, toward the PTC (see Fig. 3, A and B). During simulation, further movement of tRNAPhe is prevented by basepairing interactions between the CCA of tRNAPhe and the ribosome, as well as by U2585, although simulation without SecM suggests that U2585's position is not uniquely established by SecM's presence (see Table 1). Additionally, the RMSD of the two A-tRNAs (shown in Fig. 3 C) indicates that tRNAPhe is less stable, and therefore larger rearrangements over longer timescales may occur.
A comparison of the distances between the reacting atoms, namely, the carbonyl carbon of G165 on the P-tRNA and the nitrogen of the A-tRNA amino acid (either P166 or F166), reveals the most significant distinction between the two A-tRNAs. In the case of the native tRNAPro, the distance is 7 Å on average, whereas for the tRNAPhe it is <5 Å. Of note, the separation between the two distances is greater than the fluctuations in each (see Fig. 3 D). Although the latter distance is still greater than that observed in other ribosome structures (e.g., 3.3 Å in 2WDK (42)), the reduction to 5 Å could potentially enhance the rate of peptide-bond formation sufficiently to overcome SecM-induced stalling.
SecA-mediated relief of stalling
Translational pausing by SecM upregulates the expression of the secA gene, downstream of secM (28). The newly synthesized SecA proteins then interact with the stalled ribosome and SecM to alleviate stalling, resulting in a negative feedback loop (34). It is unknown how SecA interacts with the ribosome and SecM, although a pulling action of SecA and the translocon, SecYEG, was inferred from experiments in which a stop-transfer sequence was inserted into SecM (33). To test this potential action, we applied a force directly to the N-terminal end of SecM, exterior to the ribosome, in two separate simulations. In the first simulation, the force was constant (denoted CF) at 1.25 nN, which is much larger than that applied by SecA but sufficient to generate a conformational change in SecM on the nanosecond simulation timescale. In the second simulation, the force was varied such that a constant velocity, i.e., rate of extension (5 Å/ns, denoted CV), was maintained. The need to apply forces of nonphysiological strength is common in simulations due to the much shorter timescale covered compared with that required for many biological processes. Nonetheless, the stronger force only accelerates conformational changes and does not alter their character, such that the conformational changes observed in these simulations can still be interpreted as typical for cellular processes (68).
As noted above, compaction of SecM at various places in the exit tunnel is important for the proper localization of R163 of SecM near A2062 of the ribosome. In both the CF and CV simulations, the applied force induced straightening of SecM in the exit tunnel, including unfolding, as shown in Fig. 5, A and B, respectively. The initial fast rise in extension, plotted in Fig. 5 C, occurs due to unfolding of the N-terminus of SecM (residues 131–150). After this rise, the extension plateaus until the interaction between W155 of SecM and A751 of the ribosome, supported also by a hydrogen bond formed between Q158 and G748, begins to give way at 2.5 ns, demonstrating its role in maintaining the compact structure of SecM in the exit tunnel (see also Fig. S6). A maximal extension of ∼40 Å was finally reached after ∼5–6 ns. At this extension, the native interactions that maintain the stalled state, i.e., those between R163 and A2062 along with R163 and U2586 (see Table 1), are significantly reduced.
Figure 5.
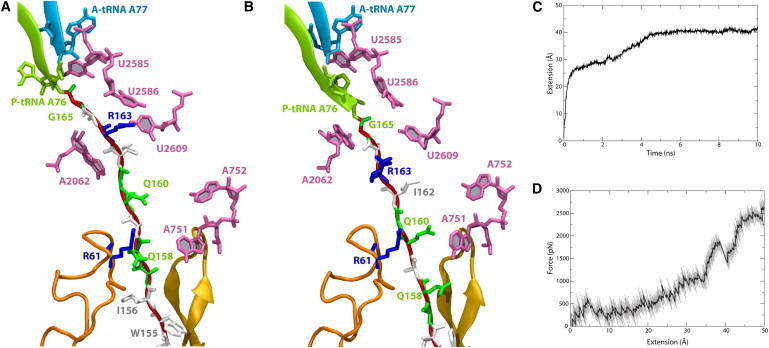
Result of applied force on SecM. (A and B) SecM in the exit tunnel along with key residues highlighted, colored as in Fig. 1, at the end of the (A) CF and (B) CV simulations. (C) Extension of SecM as a function of time for the CF simulation. (D) Force as a function of extension for the CV simulation. Note that due to the large forces encountered during pulling, particularly in the CV simulation shown in D, the final magnitude of the extension may be unphysiological.
In the CV simulation, relatively little force is required to extend SecM by 25 Å, i.e., the same unfolding of the N-terminus arises as observed in the CF simulation until ∼2.5 ns. The force required to maintain the constant extension velocity rises to nearly 2 nN for extensions up to 40 Å, at which point the interaction between W155 and A751 is broken and the applied force momentarily drops. However, in contrast to the CF simulation, the extension continues but requires significantly larger forces. As SecM is covalently bonded to the P-tRNA, continued pulling displaces the CCA of the P-tRNA into the exit tunnel (compare Fig. 5, A and B), a nonphysiological result. Beyond displacing R163, extension of SecM further disturbs the proposed interaction relay, with interactions between G2061 and A2062 being lost in the CF simulation, and those between C2063 and A2450 being lost in the CV simulation. Although to ensure the fast rate of pulling, the simulated forces are two to three orders of magnitude greater than those applied by molecular motors in vivo, the observed change in interactions is expected to be independent of pulling speed due to the limited conformational space for SecM available in the exit tunnel, which confines its possible motions drastically.
Discussion
Numerous structural and mutagenesis experiments have uncovered many of the critical elements involved in SecM-mediated stalling of ribosomes. Residues that are essential for stalling have been identified (28,31,32), and a shifted position of the ester linkage in the P-tRNA that could impede peptide-bond formation was proposed on the basis of a cryo-EM density map (22). A relay of residues through the ribosome connecting SecM to the PTC was also previously suggested as the means of communicating SecM's presence (22). In the work presented here, we derived the first (to our knowledge) atomic model of a SecM-stalled ribosome applying MDFF (36) to the cryo-EM map of Bhushan et al. (22). Subsequent simulations of the structure demonstrated stable interactions between residues along the proposed relay, thus directly connecting SecM in the exit tunnel to perturbations in the PTC through the dynamics of residues in the ribosome.
Although it appears to be quite rigid, the exit tunnel of the ribosome is actually capable of responding to the nascent protein contained within it (69). However, it should be emphasized that the responsiveness of the tunnel is at the level of individual side chains, and does not involve global alterations in its structure, in agreement with normal-mode analyses (70). As an example of local changes in the tunnel, the flexibility of A2062, the nascent-chain sensor (32), is notably reduced when SecM is present (see Fig. 4). Stalling induced by the nascent peptide ErmCL combined with erythromycin was previously proposed to occur in the same fashion, i.e., by restricting the movement of A2062 and pressing it up against the tunnel wall (25). The global rigidity of the exit tunnel may in fact enhance communication through the ribosome by allowing small perturbations to cascade through stable relays. In the case of SecM, two separate putative relays end at the PTC, shifting the ends of both the P- and A-tRNAs away from their canonical positions; however, the relevance of the latter relay to stalling is uncertain. Although the increased separation is small (∼2 Å), it persists in simulation of the wild-type SecM-stalled ribosome. It can be overcome, however, by replacing the tRNAPro in the A-site with a tRNAPhe, whose side chain was observed to possess greater flexibility and extension into the PTC in the simulation of a hypothetical A-site tRNAPhe model.
Although a number of interactions between SecM and the ribosome were observed during simulation, only R163 was found to be involved in potential interaction relays between SecM and the PTC (see Fig. 2, A and B). In silico mutation of R163 to serine or lysine, as well as deletion of SecM entirely, affected the positioning of successive residues, abolishing the interaction relay. Other conserved residues in SecM do not contribute directly to stalling, but rather serve to stably position R163 to interact with A2062, primarily through hydrophobic contacts, as well as with U2586 and A2587, although the latter interactions may be incidental. Therefore, communication between SecM and the ribosome is two-way (71), with SecM altering the positions of key ribosomal residues and vice versa. External relief of stalling by SecM comes from SecA pulling on the nascent peptide (33). Simulated pulling of SecM's N-terminus was found to abrogate critical interactions in the putative interaction relay between SecM and the PTC due to SecM's extension. Because of the necessarily short simulation timescale, the applied forces had to be chosen to be much stronger than typically seen in vivo. However, SecA has been demonstrated to be capable of unfolding proteins (72), suggesting that the enforced extension of SecM is reasonable. It is expected that over a longer timescale, breaking the relay leads to a subsequent shift of the tRNAs toward their canonical positions, thereby permitting peptide-bond formation. However, because the precise interactions between SecM and SecA, along with other translocase components (e.g., SecY (55)), including the magnitude and timing of applied forces, are still unknown, a more complex mechanism cannot be ruled out.
Despite the disparity in timescales between our simulations and stalling in vivo, the results obtained should be relevant for multiple reasons. First, the initial atomic structure of the SecM-ribosome system we used for our simulations is based on experimental cryo-EM data and thus provides a guide for future experimental studies on stalling. Additionally, the simulations were focused on alterations of individual residue positions and interactions within or near the ribosome's polypeptide exit tunnel, which equilibrate quickly (i.e., within the short simulation time), rather than on global ribosome motion or the propagation of signals over large distances, which require more time to complete (70). Finally, our computationally derived observations correlate well with mutagenesis results, yielding a molecular mechanism for the underlying events. In particular, the agreement between residues found in the simulations to be most relevant to the simulated stalling process and those identified in biochemical experiments lends support to the broader conclusions drawn.
Acknowledgments
J.G., E.S., and K.S. thank Leonardo Trabuco for helpful discussions and comments.
This work was supported by grants from the National Institutes of Health (R01 GM067887 and P41 RR005969) and the National Science Foundation (PHY0822613). This work used the Extreme Science and Engineering Discovery Environment, which is supported by National Science Foundation grant number OCI-1053575. J.G. is supported by a Director's Postdoctoral Fellowship from Argonne National Laboratory.
Footnotes
Eduard Schreiner's present address is Materials Modeling Department, BASF SE, Ludwigshafen, Germany.
Supporting Material
References
- 1.Schmeing T.M., Ramakrishnan V. What recent ribosome structures have revealed about the mechanism of translation. Nature. 2009;461:1234–1242. doi: 10.1038/nature08403. [DOI] [PubMed] [Google Scholar]
- 2.Simonović M., Steitz T.A. A structural view on the mechanism of the ribosome-catalyzed peptide bond formation. Biochim. Biophys. Acta. 2009;1789:612–623. doi: 10.1016/j.bbagrm.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nissen P., Hansen J., Steitz T.A. The structural basis of ribosome activity in peptide bond synthesis. Science. 2000;289:920–930. doi: 10.1126/science.289.5481.920. [DOI] [PubMed] [Google Scholar]
- 4.Voss N.R., Gerstein M., Moore P.B. The geometry of the ribosomal polypeptide exit tunnel. J. Mol. Biol. 2006;360:893–906. doi: 10.1016/j.jmb.2006.05.023. [DOI] [PubMed] [Google Scholar]
- 5.Kramer G., Boehringer D., Bukau B. The ribosome as a platform for co-translational processing, folding and targeting of newly synthesized proteins. Nat. Struct. Mol. Biol. 2009;16:589–597. doi: 10.1038/nsmb.1614. [DOI] [PubMed] [Google Scholar]
- 6.Wilson D.N., Beckmann R. The ribosomal tunnel as a functional environment for nascent polypeptide folding and translational stalling. Curr. Opin. Struct. Biol. 2011;21:274–282. doi: 10.1016/j.sbi.2011.01.007. [DOI] [PubMed] [Google Scholar]
- 7.Woolhead C.A., McCormick P.J., Johnson A.E. Nascent membrane and secretory proteins differ in FRET-detected folding far inside the ribosome and in their exposure to ribosomal proteins. Cell. 2004;116:725–736. doi: 10.1016/s0092-8674(04)00169-2. [DOI] [PubMed] [Google Scholar]
- 8.Lu J., Deutsch C. Folding zones inside the ribosomal exit tunnel. Nat. Struct. Mol. Biol. 2005;12:1123–1129. doi: 10.1038/nsmb1021. [DOI] [PubMed] [Google Scholar]
- 9.Lu J., Deutsch C. Secondary structure formation of a transmembrane segment in Kv channels. Biochemistry. 2005;44:8230–8243. doi: 10.1021/bi050372q. [DOI] [PubMed] [Google Scholar]
- 10.Tu L.W., Deutsch C. A folding zone in the ribosomal exit tunnel for Kv1.3 helix formation. J. Mol. Biol. 2010;396:1346–1360. doi: 10.1016/j.jmb.2009.12.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bhushan S., Gartmann M., Beckmann R. α-Helical nascent polypeptide chains visualized within distinct regions of the ribosomal exit tunnel. Nat. Struct. Mol. Biol. 2010;17:313–317. doi: 10.1038/nsmb.1756. [DOI] [PubMed] [Google Scholar]
- 12.Bhushan S., Meyer H., Beckmann R. Structural basis for translational stalling by human cytomegalovirus and fungal arginine attenuator peptide. Mol. Cell. 2010;40:138–146. doi: 10.1016/j.molcel.2010.09.009. [DOI] [PubMed] [Google Scholar]
- 13.Bornemann T., Jöckel J., Wintermeyer W. Signal sequence-independent membrane targeting of ribosomes containing short nascent peptides within the exit tunnel. Nat. Struct. Mol. Biol. 2008;15:494–499. doi: 10.1038/nsmb.1402. [DOI] [PubMed] [Google Scholar]
- 14.Berndt U., Oellerer S., Rospert S. A signal-anchor sequence stimulates signal recognition particle binding to ribosomes from inside the exit tunnel. Proc. Natl. Acad. Sci. USA. 2009;106:1398–1403. doi: 10.1073/pnas.0808584106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pool M.R. A trans-membrane segment inside the ribosome exit tunnel triggers RAMP4 recruitment to the Sec61p translocase. J. Cell Biol. 2009;185:889–902. doi: 10.1083/jcb.200807066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peterson J.H., Woolhead C.A., Bernstein H.D. The conformation of a nascent polypeptide inside the ribosome tunnel affects protein targeting and protein folding. Mol. Microbiol. 2010;78:203–217. doi: 10.1111/j.1365-2958.2010.07325.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu J., Deutsch C. Electrostatics in the ribosomal tunnel modulate chain elongation rates. J. Mol. Biol. 2008;384:73–86. doi: 10.1016/j.jmb.2008.08.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lovett P.S., Rogers E.J. Ribosome regulation by the nascent peptide. Microbiol. Rev. 1996;60:366–385. doi: 10.1128/mr.60.2.366-385.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tenson T., Ehrenberg M. Regulatory nascent peptides in the ribosomal tunnel. Cell. 2002;108:591–594. doi: 10.1016/s0092-8674(02)00669-4. [DOI] [PubMed] [Google Scholar]
- 20.Seidelt B., Innis C.A., Beckmann R. Structural insight into nascent polypeptide chain-mediated translational stalling. Science. 2009;326:1412–1415. doi: 10.1126/science.1177662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ito K., Chiba S., Pogliano K. Divergent stalling sequences sense and control cellular physiology. Biochem. Biophys. Res. Commun. 2010;393:1–5. doi: 10.1016/j.bbrc.2010.01.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bhushan S., Hoffmann T., Beckmann R. SecM-stalled ribosomes adopt an altered geometry at the peptidyl transferase center. PLoS Biol. 2011;9:e1000581. doi: 10.1371/journal.pbio.1000581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gong F., Yanofsky C. Instruction of translating ribosome by nascent peptide. Science. 2002;297:1864–1867. doi: 10.1126/science.1073997. [DOI] [PubMed] [Google Scholar]
- 24.Trabuco L.G., Harrison C.B., Schulten K. Recognition of the regulatory nascent chain TnaC by the ribosome. Structure. 2010;18:627–637. doi: 10.1016/j.str.2010.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vazquez-Laslop N., Thum C., Mankin A.S. Molecular mechanism of drug-dependent ribosome stalling. Mol. Cell. 2008;30:190–202. doi: 10.1016/j.molcel.2008.02.026. [DOI] [PubMed] [Google Scholar]
- 26.Ramu H., Mankin A., Vazquez-Laslop N. Programmed drug-dependent ribosome stalling. Mol. Microbiol. 2009;71:811–824. doi: 10.1111/j.1365-2958.2008.06576.x. [DOI] [PubMed] [Google Scholar]
- 27.Wilson D.N. Peptides in the ribosomal tunnel talk back. Mol. Cell. 2011;41:247–248. doi: 10.1016/j.molcel.2011.01.017. [DOI] [PubMed] [Google Scholar]
- 28.Nakatogawa H., Ito K. The ribosomal exit tunnel functions as a discriminating gate. Cell. 2002;108:629–636. doi: 10.1016/s0092-8674(02)00649-9. [DOI] [PubMed] [Google Scholar]
- 29.Yap M.N., Bernstein H.D. The plasticity of a translation arrest motif yields insights into nascent polypeptide recognition inside the ribosome tunnel. Mol. Cell. 2009;34:201–211. doi: 10.1016/j.molcel.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pavlov M.Y., Watts R.E., Forster A.C. Slow peptide bond formation by proline and other N-alkylamino acids in translation. Proc. Natl. Acad. Sci. USA. 2009;106:50–54. doi: 10.1073/pnas.0809211106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lawrence M.G., Lindahl L., Zengel J.M. Effects on translation pausing of alterations in protein and RNA components of the ribosome exit tunnel. J. Bacteriol. 2008;190:5862–5869. doi: 10.1128/JB.00632-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vázquez-Laslop N., Ramu H., Mankin A.S. The key function of a conserved and modified rRNA residue in the ribosomal response to the nascent peptide. EMBO J. 2010;29:3108–3117. doi: 10.1038/emboj.2010.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Butkus M.E., Prundeanu L.B., Oliver D.B. Translocon “pulling” of nascent SecM controls the duration of its translational pause and secretion-responsive secA regulation. J. Bacteriol. 2003;185:6719–6722. doi: 10.1128/JB.185.22.6719-6722.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huber D., Rajagopalan N., Bukau B. SecA interacts with ribosomes in order to facilitate posttranslational translocation in bacteria. Mol. Cell. 2011;41:343–353. doi: 10.1016/j.molcel.2010.12.028. [DOI] [PubMed] [Google Scholar]
- 35.Muto H., Nakatogawa H., Ito K. Genetically encoded but nonpolypeptide prolyl-tRNA functions in the A site for SecM-mediated ribosomal stall. Mol. Cell. 2006;22:545–552. doi: 10.1016/j.molcel.2006.03.033. [DOI] [PubMed] [Google Scholar]
- 36.Trabuco L.G., Villa E., Schulten K. Flexible fitting of atomic structures into electron microscopy maps using molecular dynamics. Structure. 2008;16:673–683. doi: 10.1016/j.str.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trabuco L.G., Villa E., Schulten K. Molecular dynamics flexible fitting: a practical guide to combine cryo-electron microscopy and X-ray crystallography. Methods. 2009;49:174–180. doi: 10.1016/j.ymeth.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trabuco L.G., Schreiner E., Schulten K. Applications of the molecular dynamics flexible fitting method. J. Struct. Biol. 2011;173:420–427. doi: 10.1016/j.jsb.2010.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trabuco L.G., Schreiner E., Schulten K. The role of L1 stalk-tRNA interaction in the ribosome elongation cycle. J. Mol. Biol. 2010;402:741–760. doi: 10.1016/j.jmb.2010.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Voorhees R.M., Schmeing T.M., Ramakrishnan V. The mechanism for activation of GTP hydrolysis on the ribosome. Science. 2010;330:835–838. doi: 10.1126/science.1194460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Armache J.-P., Jarasch A., Beckmann R. Localization of eukaryote-specific ribosomal proteins in a 5.5-Å cryo-EM map of the 80S eukaryotic ribosome. Proc. Natl. Acad. Sci. USA. 2010;107:19754–19759. doi: 10.1073/pnas.1010005107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Voorhees R.M., Weixlbaumer A., Ramakrishnan V. Insights into substrate stabilization from snapshots of the peptidyl transferase center of the intact 70S ribosome. Nat. Struct. Mol. Biol. 2009;16:528–533. doi: 10.1038/nsmb.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. 27–28. [DOI] [PubMed] [Google Scholar]
- 44.Stone J.E., Phillips J.C., Schulten K. Accelerating molecular modeling applications with graphics processors. J. Comput. Chem. 2007;28:2618–2640. doi: 10.1002/jcc.20829. [DOI] [PubMed] [Google Scholar]
- 45.Eargle J., Black A.A., Luthey-Schulten Z. Dynamics of Recognition between tRNA and elongation factor Tu. J. Mol. Biol. 2008;377:1382–1405. doi: 10.1016/j.jmb.2008.01.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grubmüller H., Heymann B., Tavan P. Ligand binding: molecular mechanics calculation of the streptavidin-biotin rupture force. Science. 1996;271:997–999. doi: 10.1126/science.271.5251.997. [DOI] [PubMed] [Google Scholar]
- 47.Schreiner E., Trabuco L.G., Schulten K. Stereochemical errors and their implications for molecular dynamics simulations. BMC Bioinformatics. 2011;12:190. doi: 10.1186/1471-2105-12-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Phillips J.C., Braun R., Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cornell W.D., Cieplak P., Kollman P.A. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J. Am. Chem. Soc. 1995;117:5179–5197. [Google Scholar]
- 50.Hornak V., Abel R., Simmerling C. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aduri R., Psciuk B.T., SantaLucia J. AMBER force field parameters for the naturally occurring modified nucleosides in RNA. J. Chem. Theory Comput. 2007;3:1464–1475. doi: 10.1021/ct600329w. [DOI] [PubMed] [Google Scholar]
- 52.Izrailev S., Stepaniants S., Schulten K. Molecular dynamics study of unbinding of the avidin-biotin complex. Biophys. J. 1997;72:1568–1581. doi: 10.1016/S0006-3495(97)78804-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sotomayor M., Schulten K. Single-molecule experiments in vitro and in silico. Science. 2007;316:1144–1148. doi: 10.1126/science.1137591. [DOI] [PubMed] [Google Scholar]
- 54.Petrone P.M., Snow C.D., Pande V.S. Side-chain recognition and gating in the ribosome exit tunnel. Proc. Natl. Acad. Sci. USA. 2008;105:16549–16554. doi: 10.1073/pnas.0801795105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gumbart J., Trabuco L.G., Schulten K. Regulation of the protein-conducting channel by a bound ribosome. Structure. 2009;17:1453–1464. doi: 10.1016/j.str.2009.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Whitford P.C., Onuchic J.N., Sanbonmatsu K.Y. Connecting energy landscapes with experimental rates for aminoacyl-tRNA accommodation in the ribosome. J. Am. Chem. Soc. 2010;132:13170–13171. doi: 10.1021/ja1061399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang Y., Rader A.J., Jernigan R.L. Global ribosome motions revealed with elastic network model. J. Struct. Biol. 2004;147:302–314. doi: 10.1016/j.jsb.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 58.Trylska J., Tozzini V., McCammon J.A. Exploring global motions and correlations in the ribosome. Biophys. J. 2005;89:1455–1463. doi: 10.1529/biophysj.104.058495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Elcock A.H. Molecular simulations of cotranslational protein folding: fragment stabilities, folding cooperativity, and trapping in the ribosome. PLOS Comput. Biol. 2006;2:e98. doi: 10.1371/journal.pcbi.0020098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kurkcuoglu O., Doruker P., Jernigan R.L. The ribosome structure controls and directs mRNA entry, translocation and exit dynamics. Phys. Biol. 2008;5:046005. doi: 10.1088/1478-3975/5/4/046005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Whitford P.C., Geggier P., Sanbonmatsu K.Y. Accommodation of aminoacyl-tRNA into the ribosome involves reversible excursions along multiple pathways. RNA. 2010;16:1196–1204. doi: 10.1261/rna.2035410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sanbonmatsu K.Y. Computational studies of molecular machines: the ribosome. Curr. Opin. Struct. Biol. 2012;22:168–174. doi: 10.1016/j.sbi.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mitra K., Schaffitzel C., Frank J. Elongation arrest by SecM via a cascade of ribosomal RNA rearrangements. Mol. Cell. 2006;22:533–543. doi: 10.1016/j.molcel.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 64.Frank J. Single-particle reconstruction of biological macromolecules in electron microscopy—30 years. Q. Rev. Biophys. 2009;42:139–158. doi: 10.1017/S0033583509990059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ghosh A., Vishveshwara S. A study of communication pathways in methionyl- tRNA synthetase by molecular dynamics simulations and structure network analysis. Proc. Natl. Acad. Sci. USA. 2007;104:15711–15716. doi: 10.1073/pnas.0704459104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sethi A., Eargle J., Luthey-Schulten Z. Dynamical networks in tRNA:protein complexes. Proc. Natl. Acad. Sci. USA. 2009;106:6620–6625. doi: 10.1073/pnas.0810961106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Frauenfeld J., Gumbart J., Beckmann R. Cryo-EM structure of the ribosome-SecYE complex in the membrane environment. Nat. Struct. Mol. Biol. 2011;18:614–621. doi: 10.1038/nsmb.2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lee E.H., Hsin J., Schulten K. Discovery through the computational microscope. Structure. 2009;17:1295–1306. doi: 10.1016/j.str.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lu J., Hua Z., Deutsch C. Nascent peptide side chains induce rearrangements in distinct locations of the ribosomal tunnel. J. Mol. Biol. 2011;411:499–510. doi: 10.1016/j.jmb.2011.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kurkcuoglu O., Kurkcuoglu Z., Jernigan R.L. Collective dynamics of the ribosomal tunnel revealed by elastic network modeling. Proteins. 2009;75:837–845. doi: 10.1002/prot.22292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Woolhead C.A., Johnson A.E., Bernstein H.D. Translation arrest requires two-way communication between a nascent polypeptide and the ribosome. Mol. Cell. 2006;22:587–598. doi: 10.1016/j.molcel.2006.05.021. [DOI] [PubMed] [Google Scholar]
- 72.Nouwen N., Berrelkamp G., Driessen A.J. Bacterial sec-translocase unfolds and translocates a class of folded protein domains. J. Mol. Biol. 2007;372:422–433. doi: 10.1016/j.jmb.2007.07.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


