Abstract
Streptococcus agalactiae remains an important pathogen of dairy herds in Québec, but data about antigenic characteristics of this microorganism are sparse. This study was conducted to determine the variety of S. agalactiae serotypes in dairy herds in Québec. Two hundred and ninety-five isolates cultured from the milk of individual cows from 7 regions of Québec were serotyped. Sixty-two percent of the isolates were untypeable. Among the 38% of typeable isolates, serotype III was found most frequently.
In conclusion, the heterogeneity found among antigenic determinants of isolates from bovine milk suggests that an immunological method for the detection of S. agalactiae performed directly on bovine milk would not be a practical approach.
Introduction
Streptococcus agalactiae is a highly contagious, obligate bacterium of the bovine mammary gland. Its presence is frequently associated with high somatic cell counts in milk and decreased milk yield. Unidentified carrier cows are responsible for spreading the pathogen to herdmates (1). In 1991, the herd prevalence of S. agalactiae infection among Québec dairies was estimated to be 47% (2). Data from 7 regional Veterinary Diagnostic Laboratories of the Ministère de l'Agriculture, des Pêcheries et de l'Alimentation du Québec (MAPAQ) show that, from 1988 to 2000, S. agalactiae was isolated from 8% to 14% of all bovine milk samples (individual quarter, composite, and bulk tank milk) submitted for bacteriological analysis (Nadeau M, personal communication, 2001).
The routine method for detection of S. agalactiae in milk samples is bacteriological culture. Other methods, such as latex agglutination, slide coagglutination, enzyme-linked immunoadsorbent assay (ELISA), and immunofluorescent assay (IFA), have been developed, but isolation of the bacterium is often essential for their execution (1). Detection of S. agalactiae by polymerase chain reaction (PCR) is also possible and may be particularly useful with milk tank specimens, if eradication of the infection from dairy herds is the objective (3). The sensitivity of the culture method for bulk tank milk samples is low to moderate, depending on the protocol used and the type and quality of samples (1). Therefore, any procedure that would be more sensitive, rapid, available at a reasonable price, and carried out directly on bulk tank milk samples would be a valuable tool for managing udder health. When considering the development of other techniques, such as immunological methods, for the detection of S. agalactiae directly from milk samples, accurate information regarding the antigenic determinants of the bacterium in the population under study is needed, because such detection techniques require a certain uniformity among antigenic determinants. If numerous serotypes of S. agalactiae are present among dairy herds, then the technique is less effective. Serotyping of S. agalactiae is based on the capsular carbohydrate antigens (I to VII) and surface exposed proteins (R and X) (4). Studies conducted in different countries have shown that there are considerable variations in the distribution of the different serotypes and that numerous combinations of antigens are possible. In Kenya, the most common serotypes among both human and bovine isolates were Ia, Ib, and III (5). A German study demonstrated the presence of serotypes III, IV, IV/X, and untypeable/X (NT/X) strains in bovine milk (6). A similar study, conducted in Scotland, showed a predominance of serotypes X (48%), II/X (35%), II (4%), and NT (10%) (7). In Denmark, 27 different antigenic combinations were found, but type II was predominant and the protein X was widespread among the isolates (8). A study in New York State indicated the presence of serotypes I, II, III, and X in bovine milk samples, with prevalences of 70%, 17%, 10%, and 3%, respectively (9).
The present study was conducted by the bacteriology laboratory of the Faculté de médecine vétérinaire (FMV), Université de Montréal, with the primary objective of describing the variety of S. agalactiae serotypes in dairy herds in the province of Québec.
Materials and methods
Streptococcus agalactiae isolates, collection and sampling protocol
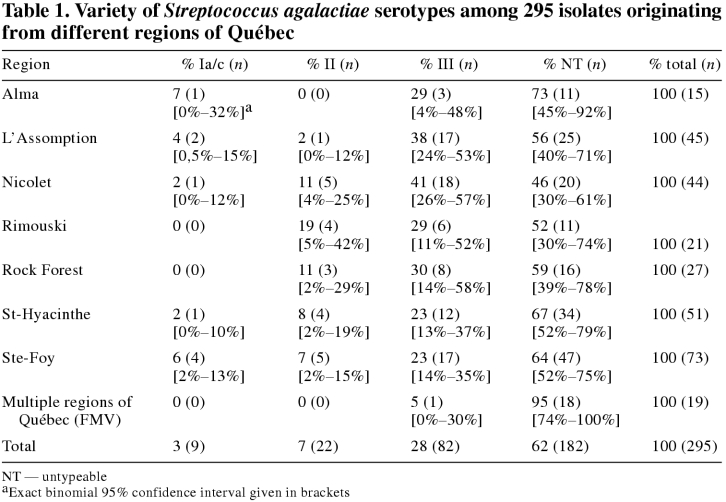
Isolates of S. agalactiae used in the study were cultured from milk samples submitted to the provincial veterinary diagnostic laboratories of the MAPAQ and to the bacteriology laboratory of the FMV from October 1996 to May 1997 by bovine practitioners. Because bulk tank milk samples were submitted for bacterial analysis less frequently, only S. agalactiae cultured from individual quarters or composite milk samples were selected for the study. The number of isolates required from each region was proportional to its relative number of farms (Table 1).
Table 1.
It was hypothesized that serotypes identified from milk samples collected from the same farm were more likely to be similar than were those originating from different farms. Therefore, since the primary objective was to describe the variety of S. agalactiae serotypes across farms and regions, only 1 isolate per farm was included in the study. A total of 295 isolates of S. agalactiae were collected.
Isolation and identification procedures
Collaborating laboratories received milk samples from practitioners as part of their routine diagnostic service. The protocol used by these laboratories for the identification of S. agalactiae in milk samples was according to the procedures of the National Mastitis Council (NMC) (10,11).
On blood agar, S. agalactiae strains show small colonies that are moist, convex, and translucent. Most are beta hemolytic; however, some can be nonhemolytic and a few are alpha hemolytic (10).
Standard bacterial procedures used by collaborating laboratories for identifying colonies isolated from milk, included Gram staining and catalase testing. All catalase negative, gram-positive cocci producing alpha hemolysis or no hemolysis were submitted to the Christie; Atkins; Munch; Peterson (CAMP) reaction, esculin hydrolysis, raffinose fermentation, and growth in 6.5% NaCl.
Tests used for catalase negative, gram-positive cocci producing beta hemolytic colonies were the CAMP reaction, esculin hydrolysis, and Lancefield serogrouping. Isolates of S. agalactiae identified by collaborating laboratories were forwarded on blood or brain heart infusion agar by express courier services to the bacteriology laboratory of the FMV.
Validation of the bacterial identification
Upon arrival, isolates were grown on trypticase soy agar (TSA), supplemented with 5% sheep blood. The plates were incubated in a 5% CO2 atmosphere at 37°C for 18 to 24 h. Identification of the organisms was verified, prior to the serotyping procedure, by the CAMP reaction (10) in a blood-esculin plate with a streak of Staphylococcus aureus (American Type Culture Collection (ATCC) 25923a), incubated at 37°C for 18 to 24 h. Additionally, serogrouping was conducted by using the latex agglutination method (Patho DX; Diagnostic Products, Los Angeles, California, USA) according to the manufacturer's instructions. An isolate was identified as S. agalactiae and selected for the study when a positive CAMP reaction was observed, esculin was not hydrolyzed, and a positive agglutination test for Lancefield group B was recorded.
In order to estimate the proportion of S. agalactiae isolates able to grow in the presence of NaCl, all isolates were inoculated in 5 mL of broth containing 6.5% NaCl and incubated at 37°C for 18 to 24 h (12).
Serotyping procedure
Antisera — Commercial antisera (Oxoid, Basingstoke, England) were obtained for serotypes II and III. For serotypes Ia/c, IV, V, VI, and VII, monospecific rabbit antisera were supplied by the National Centre for Streptococcus (NCS) in Edmonton, Alberta.
Antigens — Antigens for serological typing were prepared by the acid-heat procedure (13). Only the detection of carbohydrate capsular antigens was performed in the present study. Briefly, a loopful of S. agalactiae culture was inoculated into 5 mL of Todd-Hewitt broth (THB) and then incubated at 37°C with agitation for 6 h. Subsequently, 2, 20-mL tubes of THB were inoculated with 1 mL of the first broth and incubated at 37°C with agitation for 18 h. After 2 consecutive centrifugations, the supernatant fluid was discarded and the bacterial sediment was mixed with 0.4 mL of 0.2 M HCl. Following this procedure, 1 tube was heated at 50°C for 2 h and the other was boiled at 100°C for 10 min. After cooling to room temperature, a drop of phenol red indicator was added and the solutions were rapidly neutralized with 0.2 M NaOH. Then, the mixtures were centrifuged for 10 min. The supernatants were harvested and centrifuged for a further 10 min. Finally, a drop of thimerosal 1% was added as a preservative to the antigen extracts.
Immunodiffusion — In the preparation for the immunodiffusion procedure, 3 mL of 1% agarose was poured onto a microscope slide. When the gel was set, 7 wells were made with a 3.5-mm gel punch. A center well was surrounded by 6 outer wells numbered 1 to 6, counterclockwise, with number 1 at the top position. Wells 1 and 4 were positive controls, while well 2 served as a negative control. Wells 3, 5, and 6 were filled with antigens extracted from the streptococcal cultures. The center well was filled with the antiserum specific for each type (I to VII). The slides were kept at room temperature for 24 h in a moist environment. Slides showing no reactions in wells 3, 5, or 6 were incubated under the same conditions for an additional 24 h. Positive and negative controls were the Laboratory Centre for Disease Control, Health Canada (LCDC 7914: Ia; LCDC 7915: Ib; LCDC 7916: Ia/c; LCDC 7917: II; LCDC 7919: III; LCDC 6519-B: IV; LCDC 9841: V; LCDC 9842: VI; LCDC 6519-A: VII).
Validation of the serotyping technique
To validate the technique performed at the FMV, all 9 ATCC reference strains used in the study were serotyped. Additionally, 25 isolates were randomly selected and sent to the NCS for serotyping.
Statistical analyses
Confidence limits were calculated for the proportion of the different serotypes in each region with the exact binomial calculations. Results for the different tests and seroytyping for each isolate were recorded in a spread sheet (LOTUS 1-2-3; Lotus Development Corporation, Cambridge, Massachusetts, USA) and transfered for further analysis with the statistical analysis system (Statistical Analysis System, version 6.10; SAS Institute, Cary, North Carolina, USA and Power Analysis and Sample Size 2000; Number Cruncher Statistical System, Kaysville, Utah, USA). Fisher's exact test was used to determine if there was a difference in the proportion of isolates that are able to grow in NaCl among serotypes Ia/c, II, III, and untypeable isolates. The Fisher's exact test was applied instead of the chi-square test because of expected low counts in many cells. The null hypothesis was rejected for P values below or equal to 0.05.
Results
A total of 295 S. agalactiae isolates were selected for the study. All bacterial isolates were confirmed at the FMV laboratory to have a positive CAMP reaction, not to hydrolyse esculin, and to react with Lancefield group B antiserum. Ninety-five percent (280/295) of the isolates produced a beta-hemolysis and 5% (15/295) were nonhemolytic.
The serotyping results from the 25 isolates sent to the NCS for validation were in complete agreement with those obtained at the FMV. Furthermore, all the ATCC reference strains were correctly serotyped by the FMV.
Table 1 outlines the classification of serotypes by region. Three different, recognized serotypes were found in the province of Québec, along with numerous untypeable strains. Serotype III representing 28% (82/295) of all isolates was found most frequently Serotypes II and serotype Ia/c had a distribution of 7% (22/295) and 3% (9/295), respectively. Sixty-two percent (182/295) of the isolates were untypeable.
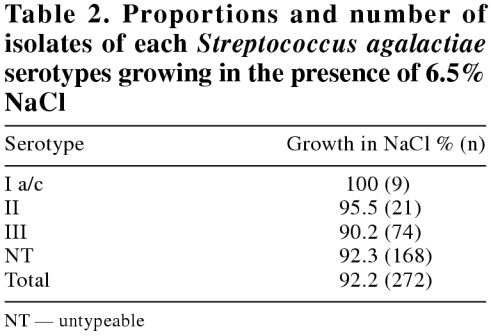
Ninety-two percent (272/295) of S. agalactiae isolates were able to grow in the presence of 6.5% NaCl (Table 2). There was no statistical correlation between the ability of the isolates to grow in NaCl and their respective serotype (P = 0.89).
Table 2.
Discussion
Stretococcus agalactiae serotypes
This study demonstrated that at least 3 serotypes are present among the isolates of S. agalactiae found in bovine milk samples in the province of Québec. An explanation for the high proportion of untypeable strains found in the study could be that the isolates were unencapsulated; it has been reported that cultivation of encapsulated isolates of S. agalactiae, in vitro, caused a shift to the unencapsulated form (16). Similar shifts between unencapsulated and encapsulated variants might occur in the bovine udder. Finally, there is a slight possibility that new serotypes, different from those presently known, could be present in this part of Canada.
An interesting observation was that most of the isolates were able to grow in a broth containing 6.5% NaCl. Capability to grow in 6.5% NaCl is routinely used by several laboratories to differentiate Streptococcus spp. from Enterococcus spp., which are able to grow at that concentration of NaCl (10,11). Numerous reference manuals indicate that S. agalactiae is either unable to grow in this medium (10,11,14) or has a variable capacity to do so (15). Prior to the study, observations were made by technical staff at collaborating laboratories that, in addition to enterococci, numerous nonhemolytic S. agalactiae isolates were able to grow in 6.5% NaCl. However, this test was not performed routinely by the laboratories in the standard procedure for the identification of beta hemolytic streptococci. Therefore, to establish the proportion of S. agalactiae isolates that were able to grow in 6.5% NaCl, all isolates selected for the study, regardless of the type of hemolysis produced, were submitted to the test. Subsequent to the initial testing of the 295 samples, the ingredients included in the NaCl broth and their proportions were verified and found to be consistent with those found in the literature (12). For further validation, 72 S. agalactiae isolates were randomly selected from the pool of 295 and tested for their ability to grow in both agar and broth containing 6.5% NaCl. All of the 72 isolates showed identical results in both agar and broth media. Finally, 30 of the 295 isolates were randomly selected and sent to the bacteriology laboratory of the Hôpital Maisonneuve-Rosemont of Montréal, Québec, to assess their ability to grow or not in 6.5% NaCl broth. Ninety-seven percent (29/30) of the isolates were able to do so.
Our results suggest that a considerable proportion of S. agalactiae strains have the ability to grow in 6.5% NaCl. Further research should be conducted to determine if other streptococci isolated from milk can grow as frequently in NaCl. If that is the case, routine utilization of this test in the identification procedure should not be recommended, because isolates could be incorrectly identified as enterococci. Further studies should be undertaken to evaluate and revise the testing system used for the identification of pathogenic streptococci and enterococci found in milk.
In conclusion, the heterogeneity found among antigenic determinants of isolates from bovine milk suggests that an immunological method for the detection of S. agalactiae performed directly on bovine milk would not be a practical approach.
Footnotes
Acknowledgments
The authors thank Marguerite Lovgren, Betty Ann Forwick (NCS), and Sonia Lacouture (FMV) for their technical assistance, and the technical staff of the MAPAQ and the clinical bacteriology laboratory of the FMV for collecting the isolates. They also thank Dre Jocelyn Delorme and his staff from Hôpital Maisonneuve-Rosemont (Montréal, Quebec) for performing NaCl tests on 30 isolates, and Dre Lucie Dutil (FMV) for her help with the statistical analyses. CVJ
Funding for this study was provided by Le Fonds du Centenaire de la Faculté de Médecine Vétérinaire de l'Université de Montréal and by Le Fonds de Recherche en Santé Animale du Québec.
Antisera for serotypes I, IV, V, VI, and VII were kindly supplied by the National Centre for Streptococcus (NCS) in Edmonton, Alberta, operated in conjunction with the Laboratory for Disease Control, Health Canada.
Address all correspondence and reprint requests to Dr. Danielle Daignault; e-mail: danielle_daignault@hc-sc.gc.ca
References
- 1.Keefe GP. Streptococcus agalactiae mastitis: A review. Can Vet J 1997;38:429–437. [PMC free article] [PubMed]
- 2.Guillemette JM. Mastitis control practices and herd prevalence of Streptococcus agalactiae and coagulase positive staphylococci in Québec dairy herds. (M. Sc. dissertation). Saint-Hyacinthe, Québec: Université de Montréal; 1995. 136p.
- 3.Martinez G, Harel J, Gottschalk M. Specific detection by PCR of Streptococcus agalactiae in milk. Can J Vet Res 2000;65:68–72. [PMC free article] [PubMed]
- 4.Lancefield RC. A serological differentiation of specific types of bovine hemolytic streptococci (group B). J Exp Med 1934;59: 441–458. [DOI] [PMC free article] [PubMed]
- 5.Mosabi JM, Arimi SM, Kang'ethe EK. Isolation and characterization of group B streptococci from human and bovine sources within and around Nairobi. Epidemiol Infect 1997;118:215–220. [DOI] [PMC free article] [PubMed]
- 6.Bopp V, Lammler C. Comparative studies on group B streptococci isolated from bovine milk samples in Thuringia and Hesse. Zentralbl Veterin armed (B) 1995;42:427–433. [DOI] [PubMed]
- 7.Morrisson JRA, Wright CL. Streptococcus agalactiae serotypes in the south west of Scotland. Vet Rec 1984;115:499. [DOI] [PubMed]
- 8.Jensen NE. Distribution of serotypes of group B streptococci in herds and cows within an area of Denmark. Acta Vet Scand 1980;21:354–366. [DOI] [PMC free article] [PubMed]
- 9.Norcross NL, Oliver N. The distribution and characterization of group B streptococci in New York State. Cornell Vet 1976; 66:240–248. [PubMed]
- 10.National Mastitis Council Inc., Laboratory Handbook on Bovine Mastitis. Revised edition, Madison, Wisconsin: National Mastitis Council Inc., 1999.
- 11.National Mastitis Council Inc, Microbiological Procedures for the Diagnosis of Bovine Udder Infection. 3rd ed., Madison, Wisconsin: National Mastitis Council Inc., 1990:7–8.
- 12.MacFaddin JF. Media for Cultivation-Identification-Maintenance of Medical Bacteria. Vol 1. Baltimore: Williams and Wilkins, 1985:365–367.
- 13.National Centre for Streptococcus. Procedures Manual, Provincial Laboratory of Public Health for Northern Alberta, Edmonton, Alberta: University of Alberta Hospitals: 1995.
- 14.Quinn PJ, Carter ME, Markey B, Carter GR. Clinical Veterinary Microbiology, London: Mosby, 1994:134.
- 15.Holt JG, Krieg NR, Sneath PHA, Staley JT, William ST. Bergey's Manual of Determinative Bacteriology, 9th ed. Baltimore: Williams and Wilkins, 1994:552.
- 16.Salasia SI, Wibawan IW, Lämmer C, Sellin M. Phase variation in streptococci of serological group B. Characteristic properties of isolates from human and bovine infection. Acta Pathol Microbiol Immunol Scand 1994;102:925–930. [PubMed]