Abstract
Fifty dairy herds in Alberta were tested for the presence of Mycobacterium paratuberculosis by fecal culture and serum enzyme linked immunosorbent assay (ELISA). Individual sera (1500) were tested for antibodies to M. paratuberculosis by ELISA. Fecal samples were combined in pools of 3 (10 pools/herd) for a total of 500 pools that were cultured for M. paratuberculosis. Thirty cultures, including all 10 pools from 1 herd, were not readable due to fungal contamination. The remaining 470 cultures, representing 49 herds, yielded 16 positive pools (3.4% ± 2.1%) from 10 herds (20.4% ± 11.3%). The ELISA of each of the 1500 sera detected 105 (7.0% ± 2.4%) positive sera and 20 (40.0% ± 13.6%) positive herds, based on 2 or more individual positive sera in the herd. The true herd-level prevalence, as determined by ELISA, was 26.8% ± 9.6%. The true herd-level prevalence, as determined by M. paratuberculosis fecal culture, ranged from 27.6% ± 6.5% to 57.1% ± 8.3%, depending on whether 1, 2, or all 3 individual fecal samples in the positive fecal pool were culture positive.
Introduction
Mycobacterium paratuberculosis causes Johne's disease, a progressive, debilitating disease of adult ruminant animals. The infection is usually contracted in the first few months of life (1,2), but the first signs of disease may not be apparent for 6 mo to 15 y (3,4). Only 1 of 20 to 25 infected animals will show clinical signs of Johne's disease before being removed from the herd (5). The National Animal Health Monitoring System (NAHMS) Dairy 1996 study (6) in the United States found that about 40.6% ± 2.0% of US dairy operations had at least 1 cow that was seropositive for M. paratuberculosis, while 16.8% ± 1.5% had 2 or more seropositive cows. VanLeeuwen et al (7) found that 43.3% ± 10.4% of dairy herds in Maritime Canada had at least 1 cow that was seropositive for M. paratuberculosis, while 16.75% ± 7.9% had 2 or more seropositive cows.
There are strong incentives for dairy farmers to control M. paratuberculosis in their herds. Dairy herds with clinical Johne's disease in ≥ 10% of cull cattle have been estimated to lose USD$ 227 to 245/cow/y (6,8) for each cow in the infected herd. In addition, there is controversy and concern regarding the potential association between M. paratuberculosis and Crohn's disease in humans (9). Strong scientific arguments have been advanced both for (10) and against (11) a link between M. paratuberculosis and Crohn's disease. Mycobacterium paratuberculosis will remain under suspicion until definitive proof of the cause of Crohn's disease is presented. If a link between Crohn's disease and M. paratuberculosis is established, there may be dramatic consequences for the dairy industry (12).
There is no effective treatment for Johne's disease; therefore, identification and culling of infected cows in conjunction with on-farm biosecurity are vital to developing and maintaining a M. paratuberculosis-free herd (6,13). Mycobacterium paratuberculosis can be controlled with commitment and persistence (14), but farmers are unlikely to implement the measures needed to reduce the spread of the infection, unless this condition is identified in their herd. The long delay between infection and the appearance of Johne's disease (3,4) may result in cows being culled from the herd before clinical signs develop, leaving the farmer unaware of the presence of M. paratuberculosis in the herd.
Control programs for Johne's disease have been established in a number of countries. One intent of these programs is to prevent the spread of the disease through the sale of infected replacement stock from infected herds. The United States has initiated a voluntary 4-stage Johne's Disease Herd Status Program for Cattle in an effort to identify herds with a reduced risk of being M. paratuberculosis infected. Australia has the Market Assurance Program to identify M. paratuberculosis-free herds and to control the movement of cattle from infected to disease-free areas. Without knowing the prevalence of M. paratuberculosis in dairy herds in Alberta, the appropriateness of a similar herd status program for Alberta could not be established. This study was initiated to establish a baseline prevalence of M. paratuberculosis infection in dairy herds in Alberta and it provided incentive for the establishment of the Alberta Johne's Herd Status Program.
Materials and methods
Two hundred dairy farms were chosen randomly from the Alberta Dairy Herd Improvement (DHI) registry and invited, by mail, to participate in the project. The only criterion for a farm's participation was that at least 45 cows were being milked at the time of initial contact. On each participating farm, a nonrandom convenience selection of 44 to 45 cows currently being milked was made available by the herd owner and sampled by project personnel. All samples were coded to maintain the anonymity of the study participants. No age or production data was acquired for the cows selected for inclusion in the study.
Blood and feces were collected from each selected cow. Blood was collected into vacutainers from the median coccygeal vein. A new disposable examination glove was used to collect feces by rectal evacuation from each cow. Approximately 60 mL of feces was placed in a sterile vial for transport to the laboratory. Blood vacutainer tubes and fecal sample vials were kept cool and delivered to the Agri-Food Laboratories Branch (AFLB) within 24 h of collection.
Clotted blood samples were centrifuged and serum was removed and stored at −20°C until assayed. Individual sera were analyzed at the AFLB, using the current enzyme-linked immunosorbent assay (ELISA) (HerdChek Mycobacterium paratuberculosis Antibody Test Kit; IDEXX Laboratories, Inc., Westbrook, Maine USA), according to the manufacturer's instructions. The AFLB is accredited in the use of serum ELISA for M. paratuberculosis by the United States Department of Agriculture (USDA). The sample to positive control (S/P) ratio was used in interpretating test results, with ratios ≥ 0.25 being considered positive.
All collected sera were analyzed by ELISA. In order to reduce the selection bias resulting from the nonrandom sample cow population, the results from all 44 or 45 sera from each herd were randomized and the first 30 were chosen for subsequent statistical analysis. A herd was considered to be infected with M. paratuberculosis if 2 or more positive sera were detected among the 30 randomly chosen cows.
Fecal culture for M. paratuberculosis was carried out by the AFLB, which is accredited for M. paratuberculosis fecal culture by the USDA. Culture was not carried out on individual fecal samples. Individual 60-mL fecal samples were mixed with a sterile spatula and approximately 2 g from each of 3 samples (a total of approximately 6 g) were pooled and mixed. Samples were pooled in the order of sampling. Fifteen pooled fecal samples were prepared from each of the 50 herds in the survey for a total of 750 pools. Only 44 samples were collected from 2 herds; therefore, 1 of the fecal pools from each of these herds represented only 2 cows. All pooled fecal samples were stored at −85°C until processed.
Pooled fecal samples were thawed at room temperature and thoroughly mixed again with a sterile spatula. A 2-g aliquot was mixed with 35 mL of sterile distilled water and shaken for 30 min. Large debris was allowed to settle for 45 min at room temperature. The aqueous layer was removed and centrifuged at 1800 g for 30 min. The resulting pellet was processed through a 2-step decontamination procedure (15). First, the pellet was resuspended in 0.9% hexadecylpyridinium chloride (HPC; Sigma-Aldrich Canada, Oakville, Ontario) in half strength (0.5 ×) brain heart infusion broth (BHI; Becton Dickinson, Mississauga, Ontario), incubated overnight at 36°C, and then centrifuged at 1800 g for 30 min to form a pellet. The pellet was resuspended in 1.2 mL 0.5 × BHI containing nalidixic acid (100 μg/mL), vancomycin (100 μg/mL), and amphotericin B (50 μg/mL) (all from Sigma-Aldrich Canada) and incubated overnight at 36°C. Each of 4 Herrold's egg yolk medium (HEYM) agar slants containing antibiotics was inoculated with 0.2 mL of this decontaminated inoculum. The HEYM was prepared in-house and tested for growth with M. paratuberculosis ATCC strains 19698 and 43544, M. intracellulare ATCC 43950, and a known M. paratuberculosis-positive fecal sample. The 4 HEYM agar slants were chosen from 4 different medium lots and contained nalidixic acid (50 μg/mL), vancomycin (50 μg/mL), amphotericin B (50 μg/mL), and mycobactin J (2 μg/mL) (Allied Monitor Labs, Fayette, Missouri USA) (15). A 5th HEYM agar slant with antibiotics, but without mycobactin J, was also inoculated with 0.2 mL of inoculum. All cultures were incubated at 36°C for up to 20 wk, with periodic visual assessment.
Identification of M. paratuberculosis was based on cultural, microscopic, and molecular biological criteria (16). In culture, M. paratuberculosis is dependent on the presence of mycobactin J for growth. Typical bacterial morphology was determined by examination of Ziehl-Neelsen-stained smears. Confirmation of M. paratuberculosis isolates was carried out by polymerase chain reaction (PCR) using primers specific for the IS900 element, as described by Hermon-Taylor et al (10). All fecal culture and confirmation steps were carried out by the AFLB. Technicians carrying out fecal culture analysis were not aware of the ELISA results.
In order to reduce the selection bias resulting from the nonrandomized cow population, the results from all 15 fecal pools from each herd were randomized and the first 10 were chosen for subsequent statistical analysis. A herd was considered to be infected, if M. paratuberculosis was isolated from 1 or more of the 10 chosen fecal pools.
Descriptive statistics were calculated to determine the apparent prevalence of M. paratuberculosis at the cow and herd levels. Confidence intervals (95%) of the means of clustered samples were calculated, as outlined by Thrusfield (17). True cow-level prevalence was estimated, as described by Cameron (18), using published test sensitivity and specificity of 42% and 100%, respectively, for culture (19), and 50% and 96.8%, respectively, for ELISA (20) where fecal culture is the gold standard. The herd-level sensitivity and specificity were estimated, as described by Jordan (21), by using the calculated true cow-level prevalence for culture and ELISA. The herd-level sensitivity and specificity estimates were then entered into a software package (Survey Toolbox for Livestock Diseases, Version 1.03; Autralian Centre for International Agricultural Research, Canberra, Australia) to estimate the true herd-level prevalence.
Results
Fifty-three farmers who met the inclusion criteria agreed initially to participate in the project. During the course of the study, however, 1 farm withdrew, sample collection could not be scheduled at 1 farm, and an incomplete sample set was collected at 1 farm. Full sample sets were collected from the remaining 50 herds and were included in the analysis for this report. In total, 2248 samples were collected from the 50 herds in the study. The mean size of the dairy herds in this study was 84.0 cows (Alberta dairy herds average 94.4 cows (22)), with a range of from 44 to 182 cows. The majority of cooperating farms were located in central Alberta, but operations in the far southern and far northern regions also participated.
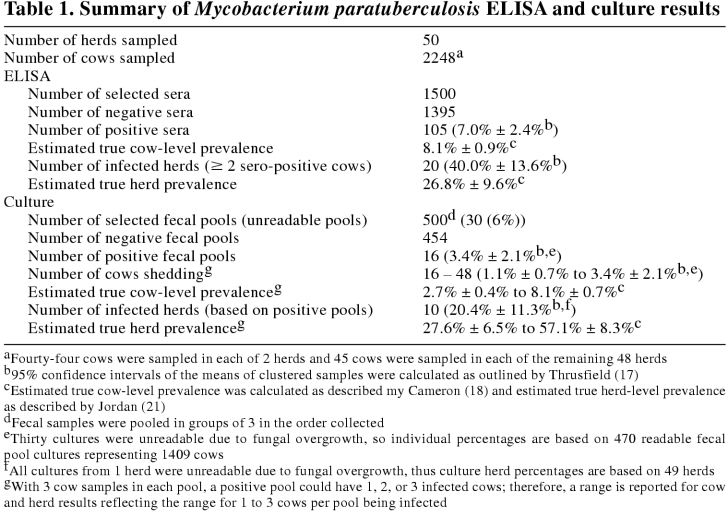
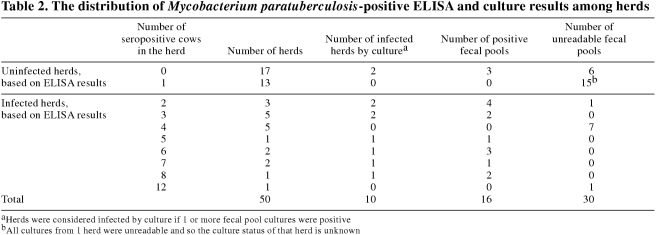
One hundred and five (7.0% ± 2.4%) of the 1500 selected sera tested positive for M. paratuberculosis (Table 1). Thus, the estimated ELISA true cow-level prevalence was calculated to be 8.1% ± 0.9%. No seropositive cows were detected in 17 herds and 13 herds had only 1 seropositive cow (Table 2). These 30 herds were not considered to be infected with M. paratuberculosis, based on the ELISA results. The remaining 20 herds (40.0% ± 13.6%) had 2 or more positive sera and were considered to be infected. The estimated ELISA herd-level sensitivity and specificity were 70.3% and 71.1%, respectively, leading to a calculated true herd-level prevalence of 26.8% ± 9.6%.
Table 1.
Table 2.
The herds deemed uninfected, based on ELISA results, averaged 73.2, standard deviation (s) = 27.4 cows per herd, with 5 herds having ≥ 100 cows. Infected herds were significantly larger (P ≤ 0.01), averaging 99.9, s = 41.1 cows per herd, with 9 herds having ≥ 100 cows. The number of seropositive cows in these infected herds ranged from 2 to 12 of the 30 cows in the random sample. Five or more seropositive cows were detected in 7 infected herds.
Thirty (6.0%) of the 500 selected fecal pools (3 cows/pool, 10 pools/herd) became overgrown with fungus (Table 1). No determination could be made with these cultures and they were removed from the subsequent analysis. All cultures from 1 herd were overgrown with fungus, resulting in the removal of that herd from the culture portion of the study; therefore, the culture analysis included only 49 herds. Twelve other farms had 1 to 4 cultures that were not readable due to fungal overgrowth.
Colonies of M. paratuberculosis were confirmed by the morphology of their acid-fast cells, by their dependence on mycobactin J for growth, and by the presence of the IS900 DNA sequence. Sixteen of the 470 readable cultures (3.4% ± 2.1%) showed growth of M. paratuberculosis (Tables 1 and 2). Thus, between 16 (1.1% ± 0.7%) and 48 (3.4% ± 2.1%) of the tested cows (from 1 to 3 cows per pool) were shedding detectable levels of M. paratuberculosis at the time of sampling. The estimated fecal culture true cow-level prevalence was from 2.7% ± 0.4% to 8.1 ± 0.7% (1 infected cow/positive fecal pool to 3 infected cows/positive fecal pool). The positive cultures were from samples distributed among 10 (20.4% ± 11.3%) of the 49 herds (Tables 1 and 2). The estimated fecal culture herd-level sensitivity and specificity were calculated to be 35.7% and 100%, respectively, for 1 infected cow per positive pool, and 74.0% and 100%, respectively, for 3 infected cows per positive pool. Thus the calculated true herd-level prevalence was calculated to be 27.6% ± 6.5% to 57.1% ± 8.3%, depending on the number of infected cows per positive fecal pool.
Herds shown, by culture, to be infected averaged 102.8, s = 40.9 cows per herd, and were not significantly larger compared with an average of 79.6, s = 34.2 cows per herd, in uninfected herds (P = 0.1).
Two of the herds deemed infected, based on culture results, were free of seropositive cows, while 12 seropositive herds (≥ 2 positive sera) could not be confirmed by culture (Table 2).
Discussion
Mycobacterium paratuberculosis has been detected in dairy herds throughout North America (6,7,23,24,25). This study has confirmed that M. paratuberculosis is also present in dairy herds in Alberta, however, the herd prevalence of M. paratuberculosis infection reported here may be biased toward an overestimate of the true prevalence. Contacted dairies were chosen at random, but farmers with herds known, or suspected, to have M. paratuberculosis may have been more willing to participate in this study. Furthermore, although all the milking cows were sampled in some herds, cows in other herds were sampled at the request or convenience of the farmer. In contrast, VanLeeuwen et al (7), in a similar study, selected both farms and cows randomly, and testing was carried out for M. paratuberculosis, bovine leukemia virus (BLV), and bovine viral diarrhea virus (BVDV). Thus, farms with any of these 3 diseases would have had an incentive to be included in the study, reducing some of the bias toward any single disease in the survey. Indeed, very few producers declined to participate in that study, whereas the participation rate in this study was only 25%.
In an attempt to reduce the bias inherent in the experimental protocol of this study, this report is based on the analysis of 30 sera, randomly selected from the 44 to 45 sera collected, and 10 fecal pools from each heard, randomly selected from the 15 pools prepared from each herd. The sera and the fecal pools tested were chosen independently, so they were not necessarily from the same cows: fecal samples were pooled in the order that the cows were sampled; thus, the 3 cows represented by a single fecal pool were not random and were potentially biased. The 30 random sera chosen for inclusion in the analysis were independent of the fecal pools to remove this possible bias. Our decision to select 30 sera and the equivalent number of fecal pools for analysis was based on the following considerations: the random selection of 30 sera allowed for the removal of 14 or 15 of the potentially biased sera collected, while retaining a significant set of sera for analysis; analysis of 30 sera has been shown to provide acceptable herd sensitivity and specificity (21) and it allowed a direct comparison to be made with the most recent Canadian survey of M. paratuberculosis in dairy cattle (7).
Fecal culture has been shown to have a sensitivity of 42% and to be most effective in detecting cattle in the later stages of Johne's disease (19). The clinical status of cows sampled for this study is not known. The estimated true cow-level prevalence by culture was between 2.7% ± 0.4% and 8.1% ± 0.7%, and 20.4% ± 11.3% of the dairy herds tested had at least 1 cow shedding M. paratuberculosis. Thus, the range in estimated true herd-level prevalence was 27.6% ± 6.5% to 57.1% ± 8.3%, depending on the number of infected cows per positive fecal pool. Culture was carried out on pooled samples only, because the intent of the study was to identify infected herds, not infected cows. Although the samples were pooled in the order collected, we believe that this did not significantly reduce the herd-level sensitivity of fecal culture. Indeed, Kalis et al (26) showed that fecal pooling by age did not decrease the herd-level sensitivity.
By ELISA, the apparent cow-level prevalence for Alberta was 7.0% ± 2.4%. Similar levels of infection have been reported in Wisconsin (7.3%) (25) and Michigan (6.9%) (24). These levels of infection are higher than those reported by NAHMS Dairy 1996 for herds of equivalent size (2.3%) (6) and that reported for Maritime Canada (1.3% to 3.3%, depending on the province) (7). The true cow-level prevalence of M. paratuberculosis infection in dairy cattle in Alberta was estimated to be 8.1% ± 0.9%.
Jordan (21) has shown that analysis of 30 sera by ELISA, with a cut-point of 2, gives an acceptable herd-level sensitivity, while maintaining a high herd-level specificity, for M. paratuberculosis. Therefore, nearly all truly uninfected herds will be correctly classed as test-negative by this process, although some infected herds may be falsely classified as uninfected. This study and that by VanLeeuwen et al (7) tested the sera from 30 cows per herd. Other surveys have used other sampling strategies. The NAHMS Dairy 1996 survey (6) tested between 25 to 40 cows per herd by ELISA, depending on herd size, while other studies have sampled all or statistically defined portions of the subject herd (23,24,25). For herds of equivalent size to those in this study, NAHMS Dairy 1996 (6) sampled 30 cows and found that 17.2% ± 2.3% of these herds had 2 or more seropositive cows based on ELISA results. The data reported here show that 40.0% ± 13.6% of the dairy herds in Alberta that were tested had 2 or more seropositive cows, as compared with 16.7% ± 7.9% of dairy herds in Maritime Canada (7). The apparent herd-level seroprevalence reported here (40%) is similar to that reported for Michigan (55%) (24).
The discrepancy between the apparent prevalence estimates reported here for Alberta and those reported for Maritime Canada (7) may have resulted from the differences in the herd and cow sampling protocols, as discussed above. Other factors, such as herd size and geography, may also impact the comparison of M. paratuberculosis prevalence between Alberta and Maritime Canada. Alberta tends to have larger dairy herds than does Maritime Canada, an average of 94.4 cows as compared with 50.9 to 60.1 cows (22), in the year of this study. Indeed, the average herd size of both seropositive and culture positive herds in this study was larger than that of negative herds (99.9 versus 73.2 (P ≤ 0.01) and 102.8 versus 79.6 (P = 0.10), respectively). The NAHMS Dairy 1996 Study showed that there was a small increase in M. paratuberculosis prevalence with increased herd size (6). The NAHMS Dairy 1996 study also showed a statistically significant difference in the prevalence of M. paratuberculosis between the northeastern and northwestern states that were tested (13.3% versus 18.6%, respectively). It is not known whether this is a reflection of climatic conditions or environmental factors, such as soil pH, which has been shown to influence M. paratuberculosis infection (24). Thus, a combination of herd characteristics, and climatic and environmental factors may influence the differences in prevalence of M. paratuberculosis observed in Alberta and Maritime Canada.
Although the apparent herd-level prevalence reported here and that reported for Maritime Canada (7) differ considerably, 40% and 16.7%, respectively, there was little difference between the estimated ELISA true herd-level prevalence reported here (26.8% ± 9.3%) and that reported for Maritime Canada (30% ± 10%) (7). These estimates, however, were calculated using different values for the sensitivity and specificity of the current ELISA test used in this study. Test sensitivity and specificity have a profound effect on the calculated true herd-level prevalence. Estimates of the sensitivity of the adsorbed ELISA have varied widely from an optimistic 58% (19) to a conservative 25% (27), while estimates of the specificity have varied from 96.8% (20) to 99% (28). VanLeeuwen et al (7) used test sensitivity and specificity of 43% and 99%, respectively, as reported by Sockett et al (28). These values were reported for the ELISA (Commonwealth Serum Laboratories (CSL)), in use in 1992, and it is not clear that they are appropriate for the current ELISA. Our estimates of the true ELISA cow-level prevalence, herd sensitivity, and herd specificity were based on the test sensitivity and specificity recently reported by Dargatz et al (20), 50% and 96.8%, respectively, for the current ELISA used in this study. Thus, it is not clear whether or not the true herd-level prevalence of M. paratuberculosis differs between Alberta and Maritime Canada. If the Dargatz et al (20) sensitivity and specificity values were to be applied to the VanLeeuwen et al (7) data, the estimated true herd-level prevalence would be much less than the reported 30% ± 10%. Certainly, were the Sockett et al (28) sensitivity and specificity values applied to the data reported here, the estimated true herd-level prevalence would be considerably higher than that reported for Maritime Canada. However, with the 25% response rate of participating producers in Alberta, and the possible bias introduced by this self-selected population, a real difference may not exist between Alberta and Maritime Canada.
Both ELISA and culture are poor methods for detecting cattle that are subclinically infected with M. paratuberculosis (19,20), which contributes to the reported low sensitivity of the tests. These tests also detect different indicators of infection, either an immune response or the shedding of the mycobacteria, and so they may not be classifying the same cows, herds, or both as infected. It is interesting to note, however, that the 95% confidence intervals of the overall true herd-level prevalence estimates reported in this study by the 2 testing methods did overlap.
This study has shown that M. paratuberculosis infection is not rare at the herd level in Alberta and has reinforced the need to identify source herds free of M. paratuberculosis infection. Because there is no effective treatment, obtaining M. paratuberculosis-free replacement stock and adhering to biosecurity and management recommendations are important for disease control. Current testing methods cannot offer the dairy farmer certainty of M. paratuberculosis-free status of individual replacement cattle. Nonetheless, today's tests can be used to lessen the risk of purchasing M. paratuberculosis-infected replacement stock by identifying herds with a reduced risk of infection. Toward this end, a voluntary Alberta Johne's Herd Status Program was initiated in Alberta in 2001 (29) to identify Johne's disease test negative herds.
Footnotes
Acknowledgments
The authors acknowledge Clarissa Snyder, Annette Visser, Catherine Taylor, Laurie Reay, and Ludovic Silasi for collecting the samples; Vicki Maitland for sample processing; Arlene Otto for culture identification; Pat Layton and Robin King for PCR confirmation of M. paratuberculosis; Suzanne Gibson for media preparation; and Eva Chow for the ELISA. The authors thank David Renter for his assistance in the application and interpretation of the epidemiological analysis. We also wish to thank Western Canadian Dairy Herd Improvement Services and the Alberta Milk Producers for their kind assistance in our initial contacts with participating farmers. CVJ
Funding for this project was provided by Alberta Milk Producers, the Alberta Agricultural Research Institute, Alberta Agriculture, Food and Rural Development and Government of Canada, Western Economic Diversification through a Western Economic Partnership Agreement.
Address all correspondence to Dr. O. Sorensen; e-mail: ole.sorensen@gov.ab.ca
No reprints will be available from the authors.
References
- 1.McIntyre WIM, Selman IE. Johne's disease paratuberculosis. Curr Top Vet Med Anim Sci 1981;6:287–296.
- 2.Stabel JR. Johne's disease: a hidden threat. J Dairy Sci 1998;81:283–288. [DOI] [PubMed]
- 3.Chiodini RJ, Van Kruiningen HJ, Merkal RS. Ruminant paratuberculosis (Johne's disease): the current status and future prospects. Cornell Vet 1984;74:218–262. [PubMed]
- 4.Manning EJ, Collins MT. Mycobacterium avium subsp. paratuberculosis: pathogen, pathogenesis and diagnosis. Rev Sci Tech Off Int Epiz 2001;20:133–150. [DOI] [PubMed]
- 5.Riemann HP, Abbas B. Diagnosis and control of bovine paratuberculosis (Johne's disease). Adv Vet Sci Comp Med 1983;27:481–506. [PubMed]
- 6.National Animal Health Monitoring System. Johne's disease on U.S. dairy operations. USDA:APHIS:VS, CEAH, National Animal Health Monitoring System 1997; Fort Collins, Colorado.
- 7.VanLeeuwen JA, Keefe GP, Tremblay R, Power C, Wichtel JJ. Seroprevalence of infection with Mycobacterium avium subspecies paratuberculosis, bovine leukemia virus, and bovine viral diarrhea virus in maritime Canada dairy cattle. Can Vet J 2001;42:193–198. [PMC free article] [PubMed]
- 8.Ott SL, Wells SJ, Wagner BA. Herd-level economic losses associated with Johne's disease on US dairy operations. Prev Vet Med 1999;40:179–192. [DOI] [PubMed]
- 9.Collins MT. Paratuberculosis and Crohn's disease: A relationship. Proc US Animal Health Assoc 1994;98:293–298.
- 10.Hermon-Taylor J, Bull TJ, Sheridan JM, Cheng J, Stellakis ML, Sumar N. Causation of Crohn's disease by Mycobacterium avium subspecies paratuberculosis. Can J Gastroenterol 2000;14:521–539. [DOI] [PubMed]
- 11.Van Kruiningen HJ. Lack of support for a common etiology in Johne's disease of animals and Crohn's disease in humans. Inflamm Bowel Dis 1999;5:183–191. [DOI] [PubMed]
- 12.Hammer P, Knappstein K, Hahn G. Significance of Mycobacterium paratuberculosis in milk. Bull Int Dairy Fed 1998;330:12–16.
- 13.Wells SJ, Wagner BA. Herd-level risk factors for infection with Mycobacterium paratuberculosis in US dairies and association between familiarity of the herd manager with the disease or prior diagnosis of the disease in that herd and use of preventive measures. J Am Vet Med Assoc 2000;216:1450–1457. [DOI] [PubMed]
- 14.Collins MT, Manning EJ. Control: Overview. Johne's Information Center, University of Wisconsin School of Veterinary Medicine 2002.
- 15.Stabel JR. An improved method for cultivation of Mycobacterium paratuberculosis from bovine fecal samples and comparison to three other methods. J Vet Diagn Invest 1997;9:375–380. [DOI] [PubMed]
- 16.Secott TE, Ohme M, Barton KS, Wu CC, Rommel FA. Mycobacterium paratuberculosis detection in bovine feces is improved by coupling agar culture enrichment to an IS900-specific polymerase chain reaction assay. J Vet Diagn Invest 1999;11:441–447. [DOI] [PubMed]
- 17.Thrusfield M. Veterinary Epidemiology. 2nd ed. Oxford: Blackwell Science, 1995.
- 18.Cameron A. Survey Toolbox for Livestock Diseases: A Practical Manual and Software Package for Active Surveillance in Developing Countries, Version 1.03. Vol. ACIAR Monograph No. 54. Australian Centre for International Agriculture Research, 1999.
- 19.Whitlock RH, Wells SJ, Sweeney RW, Van Tiem J. ELISA and fecal culture for paratuberculosis (Johne's disease): sensitivity and specificity of each method. Vet Microbiol 2000;77:387–398. [DOI] [PubMed]
- 20.Dargatz DA, Byrum BA, Barber LK, et al. Evaluation of a commercial ELISA for diagnosis of paratuberculosis in cattle. J Am Vet Med Assoc 2001;218:1163–1166. [DOI] [PubMed]
- 21.Jordan D. Aggregate testing for the evaluation of Johne's disease herd status. Aust Vet J 1996;73:16–19. [DOI] [PubMed]
- 22.Agriculture and Agri-Food Canada. Statistics — Market Info. Canadian Dairy Information Centre: Agriculture and Agri-Food Canada, 2002. http://www.dairyinfo.agr.ca
- 23.McNab WB, Meek AH, Duncan JR, Martin SW, Van Dreumel A. An epidemiological study of paratuberculosis in dairy cattle in Ontario: Study design and prevalence estimates. Can J Vet Res 1991;55:246–251. [PMC free article] [PubMed]
- 24.Johnson-Ifearulundu Y, Kaneene JB. Distribution and environmental risk factors for paratuberculosis in dairy cattle herds in Michigan. Am J Vet Res 1999;60:589–596. [PubMed]
- 25.Collins MT, Sockett DC, Goodger WJ, Conrad TA, Thomas CB, Carr DJ. Herd prevalence and geographic distribution of, and risk factors for, bovine paratuberculosis in Wisconsin. J Am Vet Med Assoc 1994;204:636–641. [PubMed]
- 26.Kalis CH, Hesselink JW, Barkema HW, Collins MT. Culture of strategically pooled bovine fecal samples as a method to screen herds for paratuberculosis. J Vet Diagn Invest 2000;12:547–551. [DOI] [PubMed]
- 27.Rossiter C, Hansen D. Johne's Disease — Information for Veterinarians. Johne's disease diagnostic tests — the ELISA. Am Assoc Bovine Pract. 2001. http://www.aabp.org
- 28.Sockett DC, Conrad TA, Thomas CB, Collins MT. Evaluation of four serological tests for bovine paratuberculosis. J Clin Microbiol 1992;30:1134–1139. [DOI] [PMC free article] [PubMed]
- 29.Scott HM, Sorensen O, Manninen K, Ollis G. An Integrated Johne's Disease Control Program for Cattle in Alberta. Anim Health Forum 2001;6:7–8.