Abstract
The present studies sought to define whether checkpoint kinase 1 (CHK1) inhibitors and poly(ADP-ribose) polymerase 1 (PARP1) inhibitors interact in vitro and in vivo to kill breast cancer cells. PARP1 and CHK1 inhibitors interacted to kill estrogen receptor (ER)+, ER+ fulvestrant-resistant, HER2+, or triple-negative mammary carcinoma cells in a manner that was not apparently affected by phosphatase and tensin homolog deleted on chromosome 10 functional status. Expression of dominant-negative CHK1 enhanced and overexpression of wild-type CHK1 suppressed the toxicity of PARP1 inhibitors in a dose-dependent fashion. Knockdown of PARP1 enhanced the lethality of CHK1 inhibitors in a dose-dependent fashion. PARP1 and CHK1 inhibitors interacted in vivo both to suppress the growth of large established tumors and to suppress the growth of smaller developing tumors; the combination enhanced animal survival. PARP1 and CHK1 inhibitors profoundly radiosensitized cells in vitro and in vivo. In conclusion, our data demonstrate that the combination of PARP1 and CHK1 inhibitors has antitumor activity in vivo against multiple mammary tumor types and that translation of this approach could prove to be a useful anticancer therapeutic approach.
Introduction
DNA damage leads to the activation of checkpoint responses that result in cell cycle arrest or apoptosis. DNA damage checkpoints are a mechanism that retards cell cycle progress and ensures that false genetic information does not pass to daughter cells before the damage is completely repaired. The checkpoint responses are orchestrated by signal transduction cascades, primarily the ataxia telangiectasia and Rad3-related -CHK1 and ataxia telangiectasia-mediated-CHK2 pathways (Lukas et al., 2003; Fernandez-Capetillo et al., 2004; Lee and Paull, 2007). Upon phosphorylation and activation by ataxia telangiectasia and Rad3-related, CHK1 phosphorylates downstream targets that regulate DNA repair and cell cycle progression, such as the protein phosphatase CDC25A. Phosphorylation of CDC25A and CDC25C by CHK1 can result in their degradation and therefore prevent them from dephosphorylating and activating the CDKs that drive cell cycle progression (Sancar et al., 2004; Eymin et al., 2006). It is noteworthy that CHK1 and CHK2 share several downstream substrates such as CDC25A/C and p53 for cell cycle control and apoptosis regulation, which potentially suggests their redundant roles in damage response (Bartek and Lukas, 2003). However, several lines of evidence also argue that CHK1 instead of CHK2 plays an essential role in regulating S and G2 checkpoints in response to double-strand DNA breaks, and CHK1 presents as a promising anticancer therapeutic target (Zhao et al., 2002; Carrassa et al., 2004; Cho et al., 2005; Morgan et al., 2006; Carlessi et al., 2007).
Multiple CHK1 inhibitors are currently being evaluated as antineoplastic agents in clinical trials, both alone and in combination with radiotherapy and chemotherapeutic agents that induce DNA damage (Mow et al., 2001; Prudhomme, 2006; Morgan et al., 2010). These agents were proposed to enhance the toxicity of chemotherapeutic drugs by inhibition of CHK1 with subsequent inappropriate cell cycle progression after DNA damage (Graves et al., 2000). Inhibition of CHK1 may directly promote activation of the protein phosphatase CDC25C and can also interfere with CDC25C elimination by blocking its binding to 14-3-3 proteins and subsequent degradation (Peng et al., 1997; Graves et al., 2000). The CHK1 inhibitor 7-hydroxystaurosporine (UCN-01) is known to have many additional intracellular kinase targets including the downstream effector of phosphatidylinositol 3-kinase, as well as “classic” protein kinase C isoforms (Komander et al., 2003).
We have noted in a variety of tumor cell types that the CHK1 inhibitor UCN-01 and, more recently, the CHK1 inhibitor 5-(3-fluoro-phenyl)-3-ureido-thiophene-2-carboxylic acid (S)-piperidin-3-ylamide hydrochloride (AZD7762) activates the ERK1/2 pathway and that pharmacologic or genetic inhibition of the ERK1/2 pathway potentiates apoptosis and suppresses tumor growth in vivo (Dai et al., 2001, 2002, 2008; Hamed et al., 2008; Mitchell et al., 2010b). We also noted that UCN-01, in addition to activating ERK1/2, promotes increased phosphorylation of histone H2AX, indicative of the fact that DNA damage was occurring because of the inhibition of CHK1 function and that inhibition of ERK1/2 further enhanced histone H2AX phosphorylation before induction of apoptosis (Dai et al., 2008). Thus, CHK1-dependent regulation of ERK1/2 may play an important role in DNA damage sensing and repair in multiple human cancer cells.
One central protein in the regulation of multiple forms of DNA repair processes is poly(ADP-ribose) polymerase (PARP), which is essential for repairing DNA damage through the base excision repair pathway (Rouleau et al., 2010). Among the PARP family, only PARP1 and PARP2 have been shown to be activated in response to DNA damage, with PARP-1 accounting for approximately 85 to 90% of the activity. PARP1 binds to damaged DNA where its catalytic activity is stimulated, which catalyzes the synthesis of branched, protein-conjugated poly(ADP-ribose) to itself and other acceptor proteins involved in base excision repair and those involved in modulating chromatin structure (Amé et al., 1999; Schreiber et al., 2002). Multiple PARP1 inhibitors have been developed including 10-(4-methyl-piperazin-1-ylmethyl)-2H-7-oxa-1,2-diaza-benzo[de]anthracen-3-one (GPI15427), 10-(4-methyl-piperazin-1-ylmethyl)-2H-7-oxa-1,2-diaza-benzo[de]anthracen-3-one; 10-(aminomethyl)-4,5,6,7-tetrahydro-1H-cyclopenta[a]pyrrolo[3,4-c]carbazole-1,3(2H)-dione (CEP6800), veliparib (ABT-888), 8-hydroxy-2-methyl-4(3H)-quinazolinone (NU1025), and olaparib (AZD2281) (Graziani and Szabó, 2005). Inhibitors that block PARP1-mediated ADP ribosylation synergize with conventional genotoxic chemotherapies, including topoisomerase I inhibitors, ionizing radiation, and DNA alkylating agents (Ben-Hur et al., 1985; Arundel-Suto et al., 1991; Boulton et al., 1995; Weltin et al., 1997; Bowman et al., 1998, 2001; Delaney et al., 2000; Tentori et al., 2002). PARP inhibitors have also shown single-agent activity against tumors deficient in homologous recombination repair, such as BRCA1/2 mutant cells (Bryant et al., 2005; Farmer et al., 2005; Martin et al., 2008; Tutt et al., 2010).
The present studies extended our analyses and determined whether this drug combination has antitumor effects in vivo. Our findings demonstrate that the combination of PARP1 and CHK1 inhibitors has antitumor activity in vivo against multiple mammary tumor types and can enhance tumor control after radiotherapy.
Materials and Methods
Materials
Human breast cancer cell lines, BT474 (PTEN WT), BT549 (PTEN mutant), HCC38 (PTEN mutant), HCC1187 (PTEN WT), and HCC1954 (PTEN WT) were purchased from American Type Culture Collection (Manassas, VA) and were not further validated. Parental and fulvestrant-resistant MCF7 cells were a kind gift from Dr. K. Nephew (University of Indiana, Bloomington, IN) (Fan et al., 2006). Fetal bovine serum was purchased from HyClone (Logan, UT). Antibiotics-antimycotics (100 units/ml penicillin, 100 μg/ml streptomycin, and 250 μg/ml amphotericin B) and trypsin-EDTA were purchased from Invitrogen (Carlsbad, CA). TUNEL kits were purchased from PerkinElmer Life and Analytical Sciences (Waltham, MA). All the primary antibodies used in the present study were purchase from Cell Signaling Technology (Danvers, MA). The validated siRNA molecules used to knock down PARP1 were from Ambion (Austin, TX): reference number S1098; S1099; Silencer negative control 1, Silencer negative control 2. siPORT NeoFX transfection agent was purchased from Ambion. Lipofectamine 2000 transfection reagent was purchased Invitrogen. The CHK1 inhibitor AZD7762 and the PARP1 inhibitors AZD2281 and ABT-888 were purchased from Axon Medchem (Groningen, The Netherlands). The CHK1 inhibitor UCN-01 was purchased from Sigma-Aldrich (St. Louis MO).
Methods
Culture and In Vitro Exposure of Cells to Drugs.
All breast cancer cells were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum and 1% antibiotic-antimycotic in a humidified incubator under an atmosphere containing 5% CO2 at 37°C. In vitro vehicle/UCN-01/AZD7762/AZD2281 treatment was from a 10 mM stock solution of each drug, and the maximal concentration of vehicle (DMSO) in media was 0.02% (v/v).
Cell Treatments, SDS-PAGE, and Western Blot Analysis.
For in vitro analyses of short-term apoptosis effects, cells were treated with vehicle/drugs or their combination for the indicated times. Cells were isolated at the times indicated in the figures by trypsinization. Cell viability, which is based on the traditional cell viability method of trypan blue exclusion, was measured with Vi-CELL Series cell viability analyzers (Beckman Coulter, Fullerton, CA). Cells for colony formation assays were plated at 250 to 4000 cells per well in sextuplicate and for in vitro assays 14 h after plating were treated with the individual drug or the drug combination(s) for the indicated time followed by drug removal. Ten to 14 days after exposure or tumor isolation, plates were washed in phosphate-buffered saline, fixed with methanol, and stained with a filtered solution of crystal violet (5% w/v). After being washed with tap water, the colonies were counted both manually (by eye) and digitally using a ColCount plate reader (Oxford Optronics, Oxford, UK). Data presented are the arithmetic means (±S.E.M.) from both counting methods from multiple studies. Colony formation was defined as a colony of 50 cells or greater.
For SDS-PAGE and immunoblotting, cells were plated at 5 × 105 cells/cm and treated with therapeutic drugs at the indicated concentrations and after the indicated time of treatment and lysed with whole-cell lysis buffer (0.5 M Tris-HCl, pH 6.8, 2% SDS, 10% glycerol, 1% β-mercaptoethanol, and 0.02% bromphenol blue) in the presence of a protease inhibitor cocktail (Sigma-Aldrich), and the samples were sonicated and boiled for 5 min. The boiled samples were loaded onto 10 to 14% SDS-PAGE and were fractionated by SDS-PAGE gels in a Protean II system (Bio-Rad, Hercules, CA). After proteins were transferred to the Immobilon-FL polyvinylidene difluoride membrane, the membrane was blocked with Odyssey Blocking buffer from LI-COR Biosciences (Lincoln, NE) for 60 min at room temperature and incubated with the primary antibody at appropriate dilutions in Odyssey Blocking buffer at 4°C overnight. After overnight incubation with appropriate primary antibodies, the membrane was washed (three times) with Tris-buffered saline-Tween 20 for a total of 15 min, probed with fluorescently labeled secondary antibody (1:5000) for 80 min at room temperature and washed (three times) with Tris-buffered saline-Tween 20 for a total of 15 min. The immunoblots were visualized by an Odyssey Infrared Imaging System (LI-COR Biosciences).
siRNA and Plasmid Transfection In Vitro.
siRNA transfection was performed with siPORT NeoFX transfection agent following the manufacturer's procedures. In brief, 10 nM prevalidated siRNA was diluted into 50 μl of serum-free media. On the basis of the manufacturer's instructions, an appropriate amount of siPORT NeoFX transfection agent was diluted into a separate vial containing serum-free media. The two solutions were incubated separately at room temperature for 15 min and mixed together by pipetting up and down several times, and the mixture was added dropwise to the target cells. Twenty-four hours after transfection, the transfection medium was replaced with complete medium, and 12 h later the cells were subjected to treatments. Procedures used for plasmid transfection were similar to those for siRNA, but instead Lipofectamine 2000 was used as the transfection reagent.
In Vivo Exposure of Mammary Carcinoma Tumors to Drugs.
Four- to 6-week-old athymic female NCr-nu/nu mice were obtained from The Jackson Laboratory (Bar Harbor, ME). Mice were maintained under pathogen-free conditions in facilities approved by the American Association for Accreditation of Laboratory Animal Care. For each mouse, a total of 5 × 106 BT474 or BT549 cells were injected subcutaneously into the fourth mammary fat pad. For animal administration, AZD7762 and AZD2281 were first dissolved in DMSO, and an equal volume of Cremophor (Sigma-Aldrich) was added. After mixing, a 1:10 dilution was made with sterile phosphate-buffered saline. Animals were injected intraperitoneally AZD7762 (50 or 12.5 mg/kg body mass), AZD2281 (50 or 12.5 mg/kg body mass), or AZD7762 plus AZD2281. Each animal in the control group was given an intraperitoneal injection of diluent alone in a volume equal to the amount given with the drug. The resulting palpable tumors were measured using a vernier caliper, and tumor volume was calculated using the formula: (width)2 × length × 0.5. Tumor growth was expressed as relative fold change in tumor volume, (Tx/T0), where T is the mean tumor volume of all tumors at a particular time in days x and T0 was the mean tumor volume at day 0. At the end of experiments, animals were euthanized using CO2. In our studies, drug treatment(s) did not have a negative impact on animal body mass.
Immunohistochemistry and Staining of Fixed Tumor Sections.
Tumors were removed using small scissors, forceps, and a disposable scalpel. The collected tumor was placed in 5 ml of Streck tissue fixative (Thermo Fisher Scientific, Waltham, MA) in a 50-ml conical tube for fixation. Fixed tumors were embedded in paraffin wax, and 10-μm slices were obtained using a microtome. Tumor sections were deparaffinized and rehydrated, and antigen retrieval was performed in a 10 mM (w/v) sodium citrate-citric acid buffer, pH 6.7. Prepared sections were then blocked and subjected to immunohistochemical analysis as per the instructions of the manufacturer for each primary antibody (Ki67, CD31, and cleaved caspase-3). The tissue sections were dehydrated, cleared, and mounted with cover slips using Permount.
Data Analysis.
The effects of various in vitro drug treatments were compared by analysis of variance using Student's t test. Differences with p < 0.05 were considered statistically significant. Experiments shown are the means of multiple individual points from multiple studies (± S.E.M.). For statistical examination of in vivo animal survival data, log-rank statistical analyses between the different treatment groups were used. Experiments shown are the means of multiple individual points from multiple experiments (±S.E.M.). Median dose-effect isobologram colony-formation analyses to determine synergism of drug interaction were performed according to the methods of Chou and Talalay (1984) using the CalcuSyn program for Windows (Biosoft, Cambridge, UK). Cells were treated with agents at an escalating fixed concentration drug dose. A combination index of <1.00 indicates synergy of interaction between the two drugs, a combination index of ∼1.00 indicates an additive interaction, and a combination index value of >1.00 indicates antagonism of action between the agents.
Results
Previously we have published results indicating that MEK1/2 inhibitors interact with CHK1 inhibitors in a synergistic manner to kill mammary tumor cells in vitro (Dai et al., 2008; Hamed et al., 2008; Mitchell et al., 2010b). In prior studies we had also determined in ER+ and in HER2+ breast cancer cells that PARP1 and CHK1 inhibitors interacted to synergistically cause tumor cell death (Mitchell et al., 2010a). Multiple novel therapeutic options for patients whose tumors are either ER+ or HER2+ have been developed in the last 25 years, whereas patients with so called “triple-negative” mammary tumors (lacking ER, progesterone receptor, and HER2) have yet to fully benefit from the more recent application of cell signaling-based therapeutic options.
Treatment of triple-negative mammary carcinoma cells with PARP1 inhibitors (AZD2281 and ABT-888) and CHK1 inhibitors (UCN-01 and AZD7762) caused a greater than additive induction of tumor cell killing (Figs. 1, A–C) (Donawho et al., 2007; Riches et al., 2008; Zabludoff et al., 2008; Fong et al., 2009; Khan et al., 2011). Of particular note was the finding that the cell death response of tumor cells lacking PTEN function (BT549 and HCC38) was not significantly different from that of cells expressing wild-type PTEN (HCC1187 and HCC1954). CHK1 and PARP1 inhibitors synergized to kill BT549 cells as judged by combination index values of less than 1.00 (Table 1). Similar data were also observed in HCC1957 cells (data not shown). In estrogen-dependent mammary tumors. one therapeutic option is to treat patients with the antiestrogen fulvestrant (Faslodex, ICI-182780); however, over time such cells become resistant to antiestrogen therapy. PARP1 and CHK1 inhibitors were competent in killing both estrogen-dependent and fulvestrant-resistant MCF7 breast cancer cells with, again, very little difference in killing between the cell types (Fig. 1D). We next determined the mechanism(s) by which PARP1 and CHK1 inhibitors killed mammary tumor cells. Expression of dominant-negative caspase 9 or BCL-XL, but not of c-FLIP-s, suppressed drug combination lethality, implying that death proceeded via the intrinsic apoptosis pathway rather than the extrinsic pathway (Fig. 1E). Treatment of cells with CHK1 and PARP1 inhibitors activated JNK1/2 (Fig. 1F). Incubation of cells with the JNK inhibitory peptide blocked JNK activation and suppressed drug combination toxicity.
Fig. 1.
PARP1 and CHK1 inhibitors interact in a greater than additive fashion in killing triple-negative and fulvestrant-resistant mammary carcinoma cells. A, BT549 and HCC1187 cells were treated with CHK1 inhibitor, UCN-01 (50 nM), PARP1 inhibitor, AZD2281 (1 μM), or UCN-01+AZD2281 for 24 or 48 h. Floating and attached cells were isolated after drug exposure, and cell viability was measured by trypan blue exclusion (±S.E.M., n = 3). B, BT549 and HCC1187 cells were treated with CHK1 inhibitor, AZD7762 (50 nM), PARP1 inhibitor, ABT888 (1.0 μM), PARP1 inhibitor, AZD2281 (1 μM), AZD7762+AZD2281, or ABT888+AZD7762 for 48 h. Floating and attached cells were isolated after drug exposure, and cell viability was measured by trypan blue exclusion (±S.E.M., n = 3). C, HCC38 and HCC1954 cells were treated with AZD7762 (50 nM), ABT888 (1.0 μM), AZD2281 (1 μM), AZD7762+AZD2281, or ABT888+AZD7762 for 48 h. Floating and attached cells were isolated after drug exposure, and cell viability was measured by trypan blue exclusion (±S.E.M., n = 3). D, MCF7 and fulvestrant-resistant MCF7 (MCF7 F) cells were treated with AZD7762 (50 nM), AZD2281 (1 μM), or AZD7762+AZD2281 for 48 h. Floating and attached cells were isolated after drug exposure, and viability was measured by trypan blue exclusion (±S.E.M., n = 3). E, BT549 cells were infected with empty vector virus (CMV) or with viruses to express dominant-negative caspase 9 (dn Casp9), BCL-XL, or c-FLIP-s. Twenty-four hours after infection, cells were treated with vehicle (DMSO) or with AZD7762 (50 nM)+AZD2281 (1 μM). Cells were fixed 48 h later, and viability was determined using TUNEL assay (±S.E.M., n = 3). Upper inset blot, BT549 cells were isolated 24 h after drug exposure, and immunoblotting was performed to detect the levels of cleaved caspase 3 (n = 2). F, MCF7 cells were pretreated for 30 min with vehicle (DMSO) or the JNK inhibitory peptide (10 μM, JNK IP). After 30 min, cells were treated with vehicle (DMSO) or with AZD7762 (50 nM)+AZD2281 (1 μM). Cells were isolated 48 h later, and viability was determined by trypan blue exclusion (±S.E.M., n = 3). Upper inset blot, cells were isolated 24 h after drug exposure, and immunoblotting was performed to detect the levels of JNK1/2 phosphorylation (n = 2). VEH, vehicle; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
TABLE 1.
PARP1 and CHK1 inhibitors synergize to kill mammary carcinoma cells
BT549 cells were plated as single cells in sextuplicate. Twelve hours after plating, cells were treated with AZD7762 (25–75 nM), UCN-01 (20–60 nM), or AZD2281 (500–1500 nM) as indicated in this table. Forty-eight hours after treatment the media was removed and replaced with drug-free media. Colonies were permitted to form for 10 days after which colonies were fixed and stained. A group of >50 cells was defined as a colony. An assessment of drug interaction was made using the CalcuSyn for Windows program. A combination index <1.00 was indicative of a synergistic drug interaction.
| Fa | CI | |
|---|---|---|
| AZD2281/AZD7762 | ||
| 500 nM/25 nM | 0.09 | 0.49 |
| 1000 nM/50 nM | 0.22 | 0.37 |
| 1500 nM/75 nM | 0.37 | 0.28 |
| AZD2281/UCN-01 | ||
| 500 nM/20 nM | 0.16 | 0.38 |
| 1000 nM/40 nM | 0.29 | 0.25 |
| 1500 nM/60 nM | 0.38 | 0.24 |
Fa, fraction affected; CI, combination index.
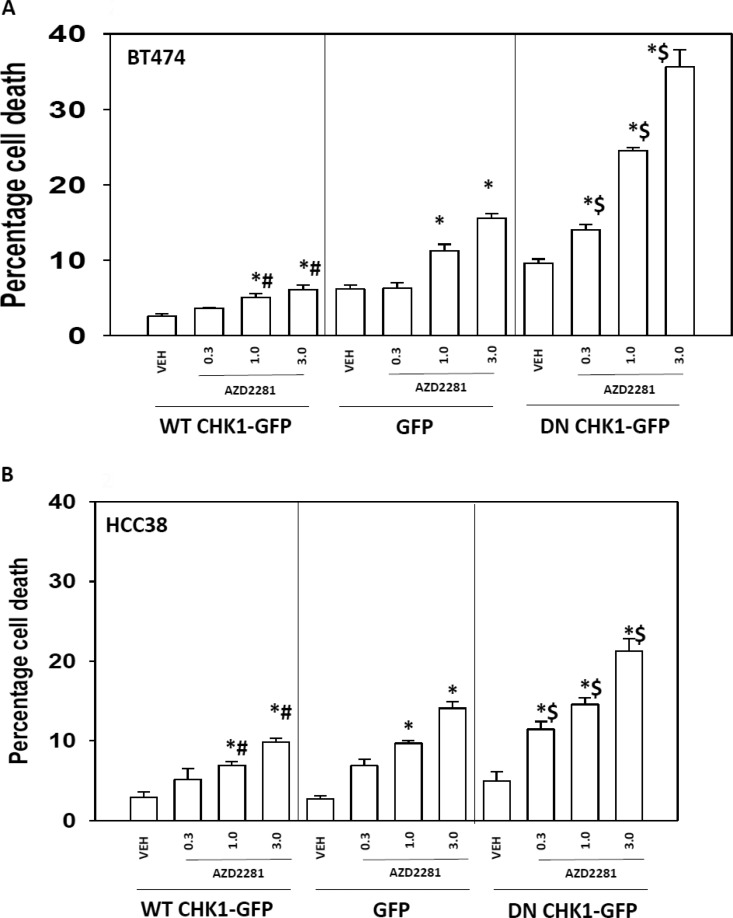
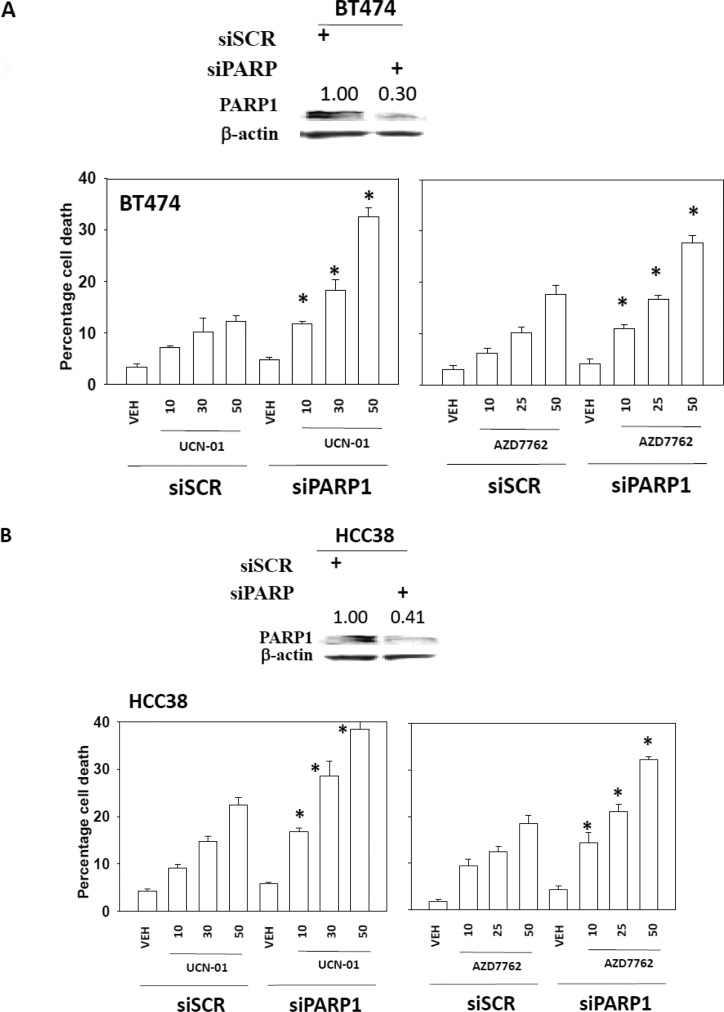
We next sought to further define the “on- and off-target effects” of the drugs in our system. BT474 (HER2+) and HCC38 (triple-negative) cells were transfected with a vector control plasmid that expresses GFP or plasmids to express dominant-negative CHK1-GFP or wild type CHK1-GFP. Treatment of vector control-transfected BT474 or HCC38 cells with increasing concentrations of the PARP1 inhibitor AZD2281 resulted in a dose-dependent increase in cell death (Figs. 2, A and B). Treatment of dominant-negative CHK1-GFP-transfected BT474 or HCC38 cells with increasing concentrations of a PARP1 inhibitor caused a significantly greater amount of cell death than observed in parallel treatments in vector control cells. Expression of wild-type CHK1-GFP suppressed PARP1 inhibitor lethality in BT474 cells and to a lesser extent in HCC38 cells. Transfection of dominant-negative GFP-CHK1 blocked radiation (6 Gy)-induced phosphorylation of CDC25C S216 (data not shown). In parallel to our CHK1 expression studies with PARP inhibitor treatment, we next assessed the impact of knocking down PARP1 expression on CHK1 inhibitor lethality. Knockdown of PARP1 enhanced CHK1 inhibitor lethality in BT474 and HCC38 cells (Fig. 3, A and B). Similar data for PARP1 knockdown were observed using a different siRNA molecule (Supplemental Fig. 1).
Fig. 2.
Dominant-negative Chk1 increased the sensitivity of the cells toward PARP inhibitor treatment. BT474 (A) or HCC38 (B) cells were transfected with WT CHK-GFP, dominant-negative (DN) CHK1-GFP, or GFP control vector. Twenty-four hours after transfection, the cells were treated with the PARP1 inhibitor AZD2281 (0.3, 1, or 3 μM) for 48 h. Floating and attached cells were isolated after drug exposure, and cell viability was measured by trypan blue exclusion (± S.E.M., n = 3). *, p < 0.05 greater than corresponding vehicle control; #, p < 0.05 less than corresponding value in GFP; $, p < 0.05 greater than corresponding value in GFP.
Fig. 3.
Knockdown of PARP1-sensitized breast cancer cells toward CHK1 inhibitors. BT474 (A) or HCC38 (B) cells were infected with siRNA control (siSCR) or a siRNA to knock down PARP1 (siPARP1). Thirty-six hours after transfection, the cells were treated with vehicle (VEH, DMSO), UCN-01 (10, 30, or 50 nM), or AZD7762 (10, 30, or 50 nM) for 48 h. Floating and attached cells were isolated after drug exposure, and cell viability was measured by trypan blue exclusion (±S.E.M., n = 3). *, p < 0.05 greater than corresponding value in siSCR cells. Top insets, knockdown of PARP1 was validated by Western blot in BT474 and HCC38 cells.
A key question remaining with respect to any new drug combination that kills tumor cells is whether the approach will translate from an in vitro cell culture setting into animal models of the disease. Multiple PARP1 and CHK1 inhibitors have been/are being developed by drug companies. We established large (∼250 mm3) BT549 tumors in the fourth mammary fat pad; this would represent the equivalent of a ∼1-kg tumor in a patient. Tumors were permitted to form for 10 days, and then animals were treated with vehicle, AZD2281, AZD7762, or the drug combination for an additional 5 days (Fig. 4A). Tumors treated with either AZD2281 or AZD7762 exhibited modest declines in tumor growth rate compared with the vehicle control value, whereas tumors exposed to both drugs showed a significant decline in their growth (Fig. 4A). This occurred with no apparent loss in animal body mass (data not shown). Based on Institutional Animal Care and Use Committee regulations, animals with tumor volumes greater than 1500 mm3 must be humanely sacrificed, and for animals treated with the vehicle control this resulted in a rapid decline in animal survival (Fig. 4B). Animals treated with AZD2281 or AZD7762 exhibited a trend showing some increase in survival compared with that of vehicle control animals; however, animals treated with both AZD2281 and AZD7762 showed a significant increase in survival compared with that of any other group, which was associated with the stabilization of tumor mass (Fig. 4, A and B). Tumor growth data similar to that in BT549 cells were obtained using smaller established (∼75 mm3) BT474 HER2+ tumors (Fig. 4C). These effects correlated with disruption of tumor cytoarchitecture and increased levels of apoptosis within the tumor as judged by elevated TUNEL-positive staining levels and reduced Ki67/4,6-diamidino-2-phenylindole staining (Fig. 4D).
Fig. 4.
PARP1 inhibitor interacted with CHK1 inhibitor in a greater than additive fashion in inhibiting tumor growth and in enhancing animal survival in vivo. A, BT549 cells (5 × 106) were injected into the fourth mammary fat pad. Tumors were permitted to form to ∼250 mm3. Initial volumes of the tumor groups were as follows: vehicle, 256 mm3; AZD7762, 241 mm3; AZD2281, 281 mm3; and AZD7762+AZD2281, 245 mm3. Animals were injected with vehicle, AZD7762 (50 mg/kg), AZD2281 (50 mg/kg), or AZD7762+AZD2281 for 5 days. Tumors were measured with a caliper to determine tumor volume as described under Materials and Methods. The mean ± S.E.M. tumor volume for all animals in each treatment condition was plotted (n = 7 animals per group, two separate studies). *, p < 0.05, less than the vehicle control value. Top panel, tumors isolated 7 days after the start of treatment were fixed and stained with TUNEL staining to examine tumor cell death. B, animals carrying BT549 tumors in A after receiving the indicated drug treatment were monitored daily and when tumor volumes were >1.5 cm3, animals were sacrificed. Survival was plotted as a percentage of animals alive on any given day. C, BT474 cells (8 × 106) were injected into the fourth mammary fat pad. Tumors were permitted to form to ∼75 mm3. Initial volumes of the tumor groups were as follows: vehicle (VEH), 81 mm3; AZD7762, 70 mm3; AZD2281, 78 mm3; and AZD7762+AZD2281, 68 mm3. Animals were injected with vehicle, AZD7762 (50 mg/kg body mass), AZD2281 (50 mg/kg body mass), or AZD7762+AZD2281 for 5 days. Tumors were measured with a caliper to determine the tumor volume as described under Materials and Methods. The mean ± S.E.M. tumor volume for all animals in each treatment condition was plotted (n = 10 animals/group). *, p < 0.05, less than vehicle control value. Results are representative of two independent studies. D, the tissues collected from BT474-derived tumors (day 15) were fixed and stained with TUNEL staining to examine tumor cell morphology and tumor cell death and with an anti-Ki67 antibody to measure the proliferative index. DAPI, 4,6-diamidino-2-phenylindole.
Radiotherapy is a standard of care treatment for primary diagnoses of breast cancer as well as palliative treatment of metastatic disease. CHK1 and PARP1 inhibitor treatment profoundly radiosensitized cells (Fig. 5, A and B). Expression of BCL-XL or dominant negative caspase 9, but not c-FLIP-s, blunted the radiosensitization effect (Fig. 5A). We then determined the impact that expression of wild-type and dominant-negative CHK1 had on tumor cell radiosensitivity. Overexpression of wild-type CHK1-GFP was radioprotective compared with the GFP control, and expression of dominant negative CHK1-GFP was radiosensitizing compared with the GFP control.
Fig. 5.
PARP and CHK inhibitors radiosensitize tumor cells. A, BT474 cells were infected with empty vector virus (CMV) or with viruses to express dominant negative caspase 9 (dn Casp9), BCL-XL, or c-FLIP-s. Twenty-four hours after infection, cells were treated with vehicle (DMSO) or with AZD7762 (50 nM)+AZD2281 (1 μM). Cells were irradiated (4 Gy) 30 min after drug treatment. Cells were fixed 24 h later, and viability was determined using TUNEL assay (±S.E.M., n = 3). B, BT474 cells were transfected with WT CHK-GFP, dominant-negative (DN) CHK1-GFP, or GFP control vector. Twelve hours after transfection, single cells were replated in sextuplicate. Twelve hours after plating, cells were treated with vehicle (VEH, DMSO), AZD2281 (1 μM), AZD7762 (50 nM), or AZD7762+AZD2281. Cells were irradiated or mock-exposed 30 min after initiation of drug treatment (2 Gy). Colonies were permitted to form over the following 10 to 14 days. #, p < 0.05 greater than corresponding value in GFP-transfected cells; *, p < 0.05 less than corresponding value in GFP-transfected cells. C, BT549 cells (5 × 106) were injected into the fourth mammary fat pad. Tumors were permitted to form to ∼75 mm3. Initial volumes of the tumor groups were as follows: vehicle, 84 mm3; AZD7762, 73 mm3; AZD2281, 66 mm3; and AZD7762+AZD2281, 76 mm3. Animals were injected with vehicle, AZD7762 (12.5 mg/kg), AZD2281 (12.5 mg/kg), or AZD7762+AZD2281 for 5 days. Tumors were irradiated on day 2 and day 4 (4 Gy). Tumors were measured with a caliper to determine tumor volume as described under Materials and Methods. The mean ± S.E.M. tumor volume for all animals in each treatment condition was plotted (n = 8 animals/group, 2 separate studies). D, for animals carrying BT549 tumors in C after receiving the indicated drug treatment, animals were monitored daily and when tumor volumes were >1.5 cm3, animals were sacrificed and survival of animals is plotted as a percentage of animals alive on any given day. P+C, PARP and CHK.
Finally, we determined in vivo whether CHK1 inhibitor plus PARP1 inhibitor treatment, using lower dose levels of the CHK1 and PARP inhibitory drugs, radiosensitized tumors. Treatment of established BT549 tumors with lower doses of CHK1 inhibitor plus PARP1 inhibitor modestly suppressed tumor growth (Fig. 5B). Exposure to radiation as a single agent also suppressed growth. The combination of CHK1 inhibitor plus PARP1 inhibitor plus radiation almost abolished tumor growth and prolonged animal survival (Fig. 5, C and D, data not shown). Collectively our findings indicate that CHK1 inhibitor plus PARP1 inhibitor treatment is effective for suppressing mammary tumor growth and for radiosensitizing mammary tumors.
Discussion
Previous studies by this group have demonstrated that mitogen-activated extracellular-regulated kinase 1/2 inhibitors as well as PARP1 inhibitors interact with CHK1 inhibitors to promote tumor cell-specific killing in a wide variety of malignancies including breast, prostate, and multiple hematological cell types (Mitchell et al., 2010a,b). The output of the protective RAS-mitogen-activated extracellular-regulated kinase 1/2-ERK1/2 pathway has previously been shown to be a critical determinant of tumor cell survival in many cell types (Riches et al., 2008). Activation of this cascade has been observed as a compensatory response of tumor cells to various environmental stresses, including cytotoxic drugs and ionizing radiation. The present studies were initiated to determine in further detail the molecular mechanisms by which PARP1 inhibitors interact with CHK1 inhibitors to promote breast cancer cell killing and to determine whether this drug combination can prove effective for controlling mammary tumor growth in vivo.
PARP1 and CHK1 inhibitors interacted in a synergistic fashion to kill mammary tumor cells. Expression of proteins that block the actions of the intrinsic apoptosis pathway (BCL-XL and dominant-negative caspase 9) inhibited drug combination lethality. In contrast, expression of the caspase 8 inhibitor c-FLIP-s did not decrease cell killing, arguing that the extrinsic pathway was not involved in drug combination lethality. Similar findings were seen in examining the ability of the combination of PARP1 and CHK1 inhibitors to radiosensitize cells; inhibition of the intrinsic pathway protected cells. Prior studies by our group noted that in fibroblasts loss of BAX and BAK also suppressed PARP and CHK inhibitor lethality. Collectively, these findings argue that our drug combination targets mitochondria for its generation of an apoptotic response.
Cells have a variety of conserved pathways to sense and overcome DNA damage and therefore preserve genomic integrity. Certain types of chemoradiotherapy lead to DNA lesions and trigger checkpoint activation and consequent cell cycle arrest that permit DNA repair or apoptosis. An additional hallmark of the cellular DNA damage response is activation of PARP1 (Rodon et al., 2009). PARP1 activation results in ADP ribosylation of multiple DNA repair complex proteins and transcription factors as well as PARP1 itself. As a result of this effect on multiple repair proteins, loss of PARP1 function promotes genomic instability and leads to hyperactivation of CHK1 with increased cell numbers in the G2 phase (Lu et al., 2006). This finding is also of interest because other groups have postulated that the chemotherapy-sensitizing effect of CHK1 inhibitors is due to abrogation of the G2 checkpoint (Prudhomme, 2006).
In our studies, two chemically distinct CHK1 inhibitors (AZD7762 and UCN-01) rapidly promoted CHK1 and ERK1/2 phosphorylation. UCN-01 has undergone phase I and II evaluation with poor pharmacokinetic/pharmacodynamic issues, preventing full exploitation of the drug, and AZD7762 was removed from clinical testing because of cardiac toxicity (Dent et al., 2011; http://www.clinicaltrials.gov/ct2/results?term=AZD7762). In prior studies, we have noted that inhibition of CHK1 inhibitor-induced H2AX and ERK1/2 phosphorylation by PARP inhibition is probably explained by the requirement of ataxia telangiectasia-mediated for PARP1 function and vice versa (Golding et al., 2007). Previously we presented evidence that inhibition of CHK1-induced ERK1/2 activation further enhanced H2AX phosphorylation, indicating that loss of ERK1/2 signaling increased the amount of DNA damage induced by the CHK1 inhibitor. This correlated with a subsequent profound induction of apoptosis.
In addition to interacting in a greater than additive fashion to kill breast cancer cells in vitro, the PARP1 inhibitor (AZD2281) and the CHK1 inhibitor (AZD7762) drug combination significantly inhibited the growth of tumors derived from either triple-negative BT549 or HER2+ BT474 cells in vivo. It is noteworthy that BT549 and HCC38 cells also lack expression of PTEN, suggesting that loss of this tumor suppressor gene does not have a significant impact on drug combination lethality. AZD2281 and AZD7762 drug combination treatment increased the survival of animals carrying BT549-derived tumors (as defined by a tumor mass of 1.5 cm3 requiring animal sacrifice). The inhibition of tumor growth in mice treated with AZD2281 and AZD7762 was in parallel associated with reduced levels of the growth marker Ki67 and with elevated apoptosis as evidenced by increased TUNEL-positive staining.
Radiotherapy is a mainstay of breast cancer therapy, and particularly of triple-negative disease. Our present studies demonstrated that concomitant irradiation after drug treatment radiosensitized tumor cells in vitro. Expression of wild-type CHK1 was protective compared with that of control vector, whereas expression of dominant-negative CHK1 caused further radiosensitization. In vivo, AZD7762 plus AZD2281 also promoted radiation toxicity using the PTEN-null BT549 cell line. These data argue that the drug combination could be combined with established breast cancer therapeutic modalities in some of the most therapeutically resistant tumor types.
In conclusion, we have demonstrated that multiple PARP1 and CHK1 inhibitors interact to kill a diverse range of breast cancer cell types in vitro and in vivo. Further studies will be required to more fully define the pathway by which PARP1 and CHK1 inhibitors interact to cause tumor cell death.
Supplementary Material
Acknowledgments
P.D. thanks Dr. N. Cruickshanks for assistance during these studies.
 The online version of this article (available at http://molpharm.aspetjournals.org) contains supplemental material.
The online version of this article (available at http://molpharm.aspetjournals.org) contains supplemental material.
This work was supported the Department of Defense [Grant W81XWH-10-1-0009]; National Institutes of Health National Cancer Institute [Grants R01-CA141703, R01-CA150214, R01-CA100866]; and Massey Cancer Center [Training Grant T32-CA085159].
Article, publication date, and citation information can be found at http://molpharm.aspetjournals.org.
- CHK
- checkpoint kinase
- UCN-01
- 7-hydroxystaurosporine
- AZD7762
- 5-(3-fluoro-phenyl)-3-ureido-thiophene-2-carboxylic acid (S)-piperidin-3-ylamide hydrochloride
- ERK
- extracellular regulated kinase
- PARP
- poly(ADP-ribose)polymerase
- GPI15427
- 10-(4-methyl-piperazin-1-ylmethyl)-2H-7-oxa-1,2-diaza-benzo[de]anthracen-3-one
- CEP6800
- 10-(aminomethyl)-4,5,6,7-tetrahydro-1H-cyclopenta[a]pyrrolo[3,4-c]carbazole-1,3(2H)-dione;ABT-888, veliparib
- NU1025
- 8-hydroxy-2-methyl-4(3H)-quinazolinone
- AZD2281
- olaparib
- WT
- wild-type
- PTEN
- phosphatase and tensin homolog deleted on chromosome 10
- TUNEL
- terminal deoxynucleotidyl transferase dUTP nick-end labeling
- si
- small interfering
- DMSO
- dimethyl sulfoxide
- PAGE
- polyacrylamide gel electrophoresis
- ER
- estrogen receptor
- JNK
- c-Jun NH2-terminal kinase
- GFP
- green fluorescent protein.
Authorship Contributions
Participated in research design: Tang, Poklepovic, Grant, and Dent.
Conducted experiments: Tang and Hamed.
Contributed new reagents or analytic tools: Grant.
Performed data analysis: Tang and Dent.
Wrote or contributed to the writing of the manuscript: Tang and Dent.
References
- Amé JC, Rolli V, Schreiber V, Niedergang C, Apiou F, Decker P, Muller S, Höger T, Ménissier-de Murcia J, de Murcia G. (1999) PARP-2, a novel mammalian DNA damage-dependent poly(ADP-ribose) polymerase. J Biol Chem 274:17860–17868 [DOI] [PubMed] [Google Scholar]
- Arundel-Suto CM, Scavone SV, Turner WR, Suto MJ, Sebolt-Leopold JS. (1991) Effect of PD 128763, a new potent inhibitor of poly(ADP-ribose) polymerase, on X-ray-induced cellular recovery processes in Chinese hamster V79 cells. Radiat Res 126:367–371 [PubMed] [Google Scholar]
- Bartek J, Lukas J. (2003) Chk1 and Chk2 kinases in checkpoint control and cancer. Cancer Cell 3:421–429 [DOI] [PubMed] [Google Scholar]
- Ben-Hur E, Chen CC, Elkind MM. (1985) Inhibitors of poly(adenosine diphosphoribose) synthetase, examination of metabolic perturbations, and enhancement of radiation response in Chinese hamster cells. Cancer Res 45:2123–2127 [PubMed] [Google Scholar]
- Boulton S, Pemberton LC, Porteous JK, Curtin NJ, Griffin RJ, Golding BT, Durkacz BW. (1995) Potentiation of temozolomide-induced cytotoxicity: a comparative study of the biological effects of poly(ADP-ribose) polymerase inhibitors. Br J Cancer 72:849–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman KJ, Newell DR, Calvert AH, Curtin NJ. (2001) Differential effects of the poly (ADP-ribose) polymerase (PARP) inhibitor NU1025 on topoisomerase I and II inhibitor cytotoxicity in L1210 cells in vitro. Br J Cancer 84:106–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman KJ, White A, Golding BT, Griffin RJ, Curtin NJ. (1998) Potentiation of anti-cancer agent cytotoxicity by the potent poly(ADP-ribose) polymerase inhibitors NU1025 and NU1064. Br J Cancer 78:1269–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ, Helleday T. (2005) Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 434:913–917 [DOI] [PubMed] [Google Scholar]
- Carlessi L, Buscemi G, Larson G, Hong Z, Wu JZ, Delia D. (2007) Biochemical and cellular characterization of VRX0466617, a novel and selective inhibitor for the checkpoint kinase Chk2. Mol Can Ther 6:935–944 [DOI] [PubMed] [Google Scholar]
- Carrassa L, Broggini M, Erba E, Damia G. (2004) Chk1, but not Chk2, is involved in the cellular response to DNA damaging agents: differential activity in cells expressing or not p53. Cell Cycle 3:1177–1181 [PubMed] [Google Scholar]
- Cho SH, Toouli CD, Fujii GH, Crain C, Parry D. (2005) Chk1 is essential for tumor cell viability following activation of the replication checkpoint. Cell Cycle 4:131–139 [DOI] [PubMed] [Google Scholar]
- Chou TC, Talalay P. (1984) Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv Enzyme Regul 22:27–55 [DOI] [PubMed] [Google Scholar]
- Dai Y, Chen S, Pei XY, Almenara JA, Kramer LB, Venditti CA, Dent P, Grant S. (2008) Interruption of the Ras/MEK/ERK signaling cascade enhances Chk1 inhibitor-induced DNA damage in vitro and in vivo in human multiple myeloma cells. Blood 112:2439–2449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Y, Landowski TH, Rosen ST, Dent P, Grant S. (2002) Combined treatment with the checkpoint abrogator UCN-01 and MEK1/2 inhibitors potently induces apoptosis in drug-sensitive and -resistant myeloma cells through an IL-6-independent mechanism. Blood 100:3333–3343 [DOI] [PubMed] [Google Scholar]
- Dai Y, Yu C, Singh V, Tang L, Wang Z, McInistry R, Dent P, Grant S. (2001) Pharmacological inhibitors of the mitogen-activated protein kinase (MAPK) kinase/MAPK cascade interact synergistically with UCN-01 to induce mitochondrial dysfunction and apoptosis in human leukemia cells. Cancer Res 61:5106–5115 [PubMed] [Google Scholar]
- Delaney CA, Wang LZ, Kyle S, White AW, Calvert AH, Curtin NJ, Durkacz BW, Hostomsky Z, Newell DR. (2000) Potentiation of temozolomide and topotecan growth inhibition and cytotoxicity by novel poly(adenosine diphosphoribose) polymerase inhibitors in a panel of human tumor cell lines. Clin Cancer Res 6:2860–2867 [PubMed] [Google Scholar]
- Dent P, Tang Y, Yacoub A, Dai Y, Fisher PB, Grant S. (2011) CHK1 inhibitors in combination chemotherapy: thinking beyond the cell cycle. Mol Interv 11:133–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donawho CK, Luo Y, Luo Y, Penning TD, Bauch JL, Bouska JJ, Bontcheva-Diaz VD, Cox BF, DeWeese TL, Dillehay LE, et al. (2007) ABT-888, an orally active poly(ADP-ribose) polymerase inhibitor that potentiates DNA-damaging agents in preclinical tumor models. Clin Cancer Res 13:2728–2737 [DOI] [PubMed] [Google Scholar]
- Eymin B, Claverie P, Salon C, Brambilla C, Brambilla E, Gazzeri S. (2006) p14ARF triggers G2 arrest through ERK-mediated Cdc25C phosphorylation, ubiquitination and proteasomal degradation. Cell Cycle 5:759–765 [DOI] [PubMed] [Google Scholar]
- Fan M, Yan PS, Hartman-Frey C, Chen L, Paik H, Oyer SL, Salisbury JD, Cheng AS, Li L, Abbosh PH, et al. (2006) Diverse gene expression and DNA methylation profiles correlate with differential adaptation of breast cancer cells to the antiestrogens tamoxifen and fulvestrant. Cancer Res 66:11954–11966 [DOI] [PubMed] [Google Scholar]
- Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, et al. (2005) Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 434:917–921 [DOI] [PubMed] [Google Scholar]
- Fernandez-Capetillo O, Lee A, Nussenzweig M, Nussenzweig A. (2004) H2AX: the histone guardian of the genome. DNA Repair 3:959–967 [DOI] [PubMed] [Google Scholar]
- Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, Mortimer P, Swaisland H, Lau A, O'Connor MJ, et al. (2009) Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med 361:123–134 [DOI] [PubMed] [Google Scholar]
- Golding SE, Rosenberg E, Neill S, Dent P, Povirk LF, Valerie K. (2007) Extracellular signal-related kinase positively regulates ataxia telangiectasia mutated, homologous recombination repair, and the DNA damage response. Cancer Res 67:1046–1053 [DOI] [PubMed] [Google Scholar]
- Graves PR, Yu L, Schwarz JK, Gales J, Sausville EA, O'Connor PM, Piwnica-Worms H. (2000) The Chk1 protein kinase and the Cdc25C regulatory pathways are targets of the anticancer agent UCN-01. J Biol Chem 275:5600–5605 [DOI] [PubMed] [Google Scholar]
- Graziani G, Szabó C. (2005) Clinical perspectives of PARP inhibitors. Pharmacol Res 52:109–118 [DOI] [PubMed] [Google Scholar]
- Hamed H, Hawkins W, Mitchell C, Gilfor D, Zhang G, Pei XY, Dai Y, Hagan MP, Roberts JD, Yacoub A, et al. (2008) Transient exposure of carcinoma cells to RAS/MEK inhibitors and UCN-01 causes cell death in vitro and in vivo. Mol Cancer Ther 7:616–629 [DOI] [PubMed] [Google Scholar]
- Khan OA, Gore M, Lorigan P, Stone J, Greystoke A, Burke W, Carmichael J, Watson AJ, McGown G, Thorncroft M, et al. (2011) A phase I study of the safety and tolerability of olaparib (AZD2281, KU0059436) and dacarbazine in patients with advanced solid tumours. Br J Cancer 104:750–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D, Kular GS, Bain J, Elliott M, Alessi DR, Van Aalten DM. (2003) Structural basis for UCN-01 (7-hydroxystaurosporine) specificity and PDK1 (3-phosphoinositide-dependent protein kinase-1) inhibition. Biochem J 375:255–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Paull TT. (2007) Activation and regulation of ATM kinase activity in response to DNA double-strand breaks. Oncogene 26:7741–7748 [DOI] [PubMed] [Google Scholar]
- Lu HR, Wang X, Wang Y. (2006) A stronger DNA damage-induced G2 checkpoint due to over-activated CHK1 in the absence of PARP-1. Cell Cycle 5:2364–2370 [DOI] [PubMed] [Google Scholar]
- Lukas C, Falck J, Bartkova J, Bartek J, Lukas J. (2003) Distinct spatiotemporal dynamics of mammalian checkpoint regulators induced by DNA damage. Nat Cell Biol 5:255–260 [DOI] [PubMed] [Google Scholar]
- Martin SA, Lord CJ, Ashworth A. (2008) DNA repair deficiency as a therapeutic target in cancer. Curr Opin Genet Dev 18:80–86 [DOI] [PubMed] [Google Scholar]
- Mitchell C, Park M, Eulitt P, Yang C, Yacoub A, Dent P. (2010a) Poly(ADP-ribose) polymerase 1 modulates the lethality of CHK1 inhibitors in carcinoma cells. Mol Pharmacol 78:909–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell C, Yacoub A, Hossein H, Martin AP, Bareford MD, Eulitt P, Yang C, Nephew KP, Dent P. (2010b) Inhibition of MCL-1 in breast cancer cells promotes cell death in vitro and in vivo. Cancer Biol Ther 10:903–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan MA, Parsels LA, Parsels JD, Lawrence TS, Maybaum J. (2006) The relationship of premature mitosis to cytotoxicity in response to checkpoint abrogation and antimetabolite treatment. Cell Cycle 5:1983–1988 [DOI] [PubMed] [Google Scholar]
- Morgan MA, Parsels LA, Zhao L, Parsels JD, Davis MA, Hassan MC, Arumugarajah S, Hylander-Gans L, Morosini D, Simeone DM, et al. (2010) Mechanism of radiosensitization by the Chk1/2 inhibitor AZD7762 involves abrogation of the G2 checkpoint and inhibition of homologous recombinational DNA repair. Cancer Res 70:4972–4981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mow BM, Blajeski AL, Chandra J, Kaufmann SH. (2001) Apoptosis and the response to anticancer therapy. Curr Opin Oncol 13:453–462 [DOI] [PubMed] [Google Scholar]
- Peng CY, Graves PR, Thoma RS, Wu Z, Shaw AS, Piwnica-Worms H. (1997) Mitotic and G2 checkpoint control: regulation of 14-3-3 protein binding by phosphorylation of Cdc25C on serine-216. Science 277:1501–1505 [DOI] [PubMed] [Google Scholar]
- Prudhomme M. (2006) Novel checkpoint 1 inhibitors. Recent Pat Anticancer Drug Discov 1:55–68 [DOI] [PubMed] [Google Scholar]
- Riches LC, Lynch AM, Gooderham NJ. (2008) Early events in the mammalian response to DNA double-strand breaks. Mutagenesis 23:331–339 [DOI] [PubMed] [Google Scholar]
- Rodon J, Iniesta MD, Papadopoulos K. (2009) Development of PARP inhibitors in oncology. Expert Opin Investig Drugs 18:31–43 [DOI] [PubMed] [Google Scholar]
- Rouleau M, Patel A, Hendzel MJ, Kaufmann SH, Poirier GG. (2010) PARP inhibition: PARP1 and beyond. Nat Rev Cancer 10:293–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar A, Lindsey-Boltz LA, Unsal-Kaçmaz K, Linn S. (2004) Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem 73:39–85 [DOI] [PubMed] [Google Scholar]
- Schreiber V, Amé JC, Dollé P, Schultz I, Rinaldi B, Fraulob V, Ménissier-de Murcia J, de Murcia G. (2002) Poly(ADP-ribose) polymerase-2 (PARP-2) is required for efficient base excision DNA repair in association with PARP-1 and XRCC1. J Biol Chem 277:23028–23036 [DOI] [PubMed] [Google Scholar]
- Tentori L, Portarena I, Graziani G. (2002) Potential clinical applications of poly(ADP-ribose) polymerase (PARP) inhibitors. Pharmacol Res 45:73–85 [DOI] [PubMed] [Google Scholar]
- Tutt A, Robson M, Garber JE, Domchek SM, Audeh MW, Weitzel JN, Friedlander M, Arun B, Loman N, Schmutzler RK, et al. (2010) Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and advanced breast cancer: a proof-of-concept trial. Lancet 376:235–244 [DOI] [PubMed] [Google Scholar]
- Weltin D, Holl V, Hyun JW, Dufour P, Marchal J, Bischoff P. (1997) Effect of 6(5H)-phenanthridinone, a poly (ADP-ribose)polymerase inhibitor, and ionizing radiation on the growth of cultured lymphoma cells. Int J Radiat Biol 72:685–692 [DOI] [PubMed] [Google Scholar]
- Zabludoff SD, Deng C, Grondine MR, Sheehy AM, Ashwell S, Caleb BL, Green S, Haye HR, Horn CL, Janetka JW, et al. (2008) AZD7762, a novel checkpoint kinase inhibitor, drives checkpoint abrogation and potentiates DNA-targeted therapies. Mol Cancer Ther 7:2955–2966 [DOI] [PubMed] [Google Scholar]
- Zhao H, Watkins JL, Piwnica-Worms H. (2002) Disruption of the checkpoint kinase 1/cell division cycle 25A pathway abrogates ionizing radiation-induced S and G2 checkpoints. Proc Natl Acad Sci USA 99:14795–14800 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.