Abstract
Summary: RNA silencing is a complex, highly conserved mechanism mediated by small RNAs (sRNAs), such as microRNAs (miRNAs), that is known to be involved in a diverse set of biological functions including development, pathogen control, genome maintenance and response to environmental change. Advances in next generation sequencing technologies are producing increasingly large numbers of sRNA reads per sample at a fraction of the cost of previous methods. However, many bioinformatics tools do not scale accordingly, are cumbersome, or require extensive support from bioinformatics experts. Therefore, researchers need user-friendly, robust tools, capable of not only processing large sRNA datasets in a reasonable time frame but also presenting the results in an intuitive fashion and visualizing sRNA genomic features. Herein, we present the UEA sRNA workbench, a suite of tools that is a successor to the web-based UEA sRNA Toolkit, but in downloadable format and with several enhanced and additional features.
Availability: The program and help pages are available at http://srna-workbench.cmp.uea.ac.uk.
Contact: vincent.moulton@cmp.uea.ac.uk
1 INTRODUCTION
RNA silencing is a complex, highly conserved mechanism mediated by small RNAs (sRNAs) (Chapman and Carrington, 2007). sRNAs, such as micro RNAs (miRNAs), are typically 20–24 nt in length and act as guide molecules to regulate gene expression. Next generation sequencing (NGS) technologies have become the de facto way to generate sRNA datasets. These datasets often come from several samples, each consisting of millions of distinct reads. Therefore, there is an increasing need for computational tools for exploring and analysing such data.
Several tools have been developed for NGS sRNA analysis (see Li et al. (2012) for a recent overview of tools for miRNA analysis). These are typically command-line driven, focused on a specific application (e.g. miRNA detection) and often require installation of supporting software packages. In addition, many of them lack tools for visualizing results.
Herein, we present the UEA sRNA workbench, a suite of interactive Java-based sRNA analysis tools, which provides an easy-to-use, well-documented platform to create workflows for processing sRNA NGS data. The workbench is a successor to the web-based UEA sRNA Toolkit (Moxon et al., 2008), which, since its establishment in 2009, has received an average of 12 000 page views per month and processed over 5400 sRNA datasets. As well as being downloadable, the new workbench provides many improved features compared with the toolkit, with new tools for visualizing data and processing multiple datasets. In addition, the workbench is limited only by local memory whereas the toolkit had strict datasize limits due to the nature of its web-based interfaces. The workbench can also be command-line driven to allow users to easily plug it into existing pipelines (Studholme, 2011).
2 DESCRIPTION OF THE WORKBENCH
2.1 Helper tools
Adaptor remover removes adaptor fragments from raw short read sequence data and outputs data to FASTA format.
Filter produces a filtered version of an sRNA dataset, controlled by several user-defined criteria, including sequence length, abundance, complexity, transfer and ribosomal RNA removal.
Sequence alignment provides a graphical front-end for the PatMaN (Prüfer et al., 2008) sequence alignment tool.
2.2 Computational analysis tools
miRCat (miRNA categorization) predicts mature miRNAs and their precursors from an sRNA dataset and a genome. The algorithm uses the technique in Moxon et al. (2008) for predicting miRNA precursor hairpins. The algorithm has been enhanced to handle animal datasets and to use multi-threading hardware, improving run-time performance. Users are presented with a table containing details of predicted miRNA candidates and precursors. miRNA candidates are reported as they are discovered allowing users to immediately begin down-stream analysis. miRNAs in miRBase (Griffiths-Jones, 2010) are also reported as such.
SiLoCo (short interfering RNA locus comparison) compares sRNA expression levels by grouping sRNAs into loci based on genomic location (Moxon et al., 2008). SiLoCo has been extended to allow comparison of multiple samples. The output is displayed in a table containing normalized abundance of the sequences and a weighted count on the number of occurrences in the genome. In addition, the mean abundance score for the locus and the maximum fold change between normalized abundances over each sample are reported.
ta-siRNA (trans-acting short interfering RNA) prediction is an implementation of an algorithm proposed in Chen et al. (2007) for prediction of phased ta-siRNAs in plant sRNA datasets. Users are presented with the coordinates of the TAS loci and the phased sRNAs (Fig. 1c).
miRProf (miRNA profiler) determines normalized expression levels of sRNAs matching known miRNAs in miRBase. This tool has been enhanced for comparison of miRNA expression levels across multiple samples. Users are presented with a table containing raw, normalized and weighted counts of miRNAs. Users can also combine results through several criteria including miRNAs containing mismatches, miRNAs found in different organisms and miRNA variants.
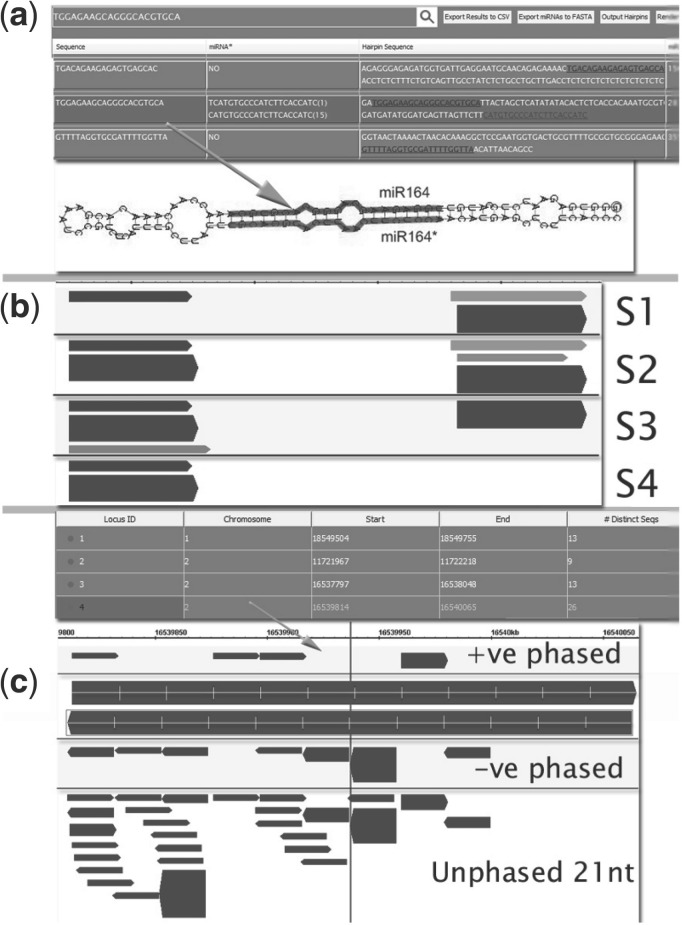
Fig. 1.
Workflow examples from the UEA sRNA workbench (data from Rajagopalan et al. (2006)). (a) After miRCat has classified miR164, a secondary structure plot is generated through the Hairpin Annotation tool. (b) After SiLoCo predicts the miR164 locus in four sRNA samples (including the sample used in (a)), output is sent to VisSR for visualization. Reads of lengths 19, 20–21 and 22–23 are coloured pink, red and green, respectively. (c) ta-siRNA prediction of a TAS gene. The locus is visualized using VisSR
2.3 Visualization tools
Hairpin annotation generates a secondary structure from an RNA sequence and highlights regions of interest using RNAplot (Hofacker et al., 1994). This tool can be activated from miRCat to display predicted miRNA precursors with both the miRNA and the miRNA* highlighted (Fig. 1a). Alternatively, a user can input long and up to 14 short sequences to generate a secondary structure. For each displayed miRNA precursor, the minimum free energy of the structure is also reported.
VisSR (visualization of sRNAs) Visualization of sRNA features can be used to gain insight into their likely biogenesis or function. To do this, VisSR uses components from GenoViz (Helt et al., 2009) to generate a visual representation of sRNAs and user-imported genomic features. Arrows are used to depict the strand to which a short read maps to, colour is used to represent the size class of the short read and thickness illustrates abundance. VisSR can be activated from various tools to display predicted sRNA loci with each sample displayed in separate tiers (Fig. 1b), phased sRNAs for both strands in separate tiers and all unphased 21nt sRNAs in a further tier (Fig. 1c), and predicted miRNAs with miRNA* if present with the precursor as it appears on the genome. Output is generated in an image format, suitable for publication.
3 DISCUSSION
The relative low cost of NGS has generated an abundance of sRNA data necessitating a demand for dedicated bioinformatics support. The sRNA workbench enables biologists to gain an understanding of the sRNAome in plants and animals through the efficient processing of large NGS sRNA datasets, complemented with easily accessible visualization tools. It is hoped that these tools will enable biologists to shed light on the key molecular pathways involved in RNA silencing.
Funding: Biotechnology and Biological Sciences Research Council (grant BBSRC BB/I00016X/1).
Conflict of Interest: none declared.
Footnotes
†The authors wish it to be known that, in their opinion, the first two authors should be regarded as joint First Authors.
REFERENCES
- Chapman E.J., Carrington J.C. Specialization and evolution of endogenous small RNA pathways. Nat. Rev. Genet. 2007;8:884–896. doi: 10.1038/nrg2179. [DOI] [PubMed] [Google Scholar]
- Chen H.-M., et al. Bioinformatic prediction and experimental validation of a microRNA-directed tandem trans-acting siRNA cascade in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2007;104:3318–3323. doi: 10.1073/pnas.0611119104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S. miRBase: microRNA sequences and annotation. Curr. Protoc. Bioinformatics. 2010 doi: 10.1002/0471250953.bi1209s29. Chapter 12Unit 12.9.1–Unit 12.910. [DOI] [PubMed] [Google Scholar]
- Helt G.A., et al. Genoviz software development kit: Java tool kit for building genomics visualization applications. BMC Bioinformatics. 2009;10:266. doi: 10.1186/1471-2105-10-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofacker I.L., et al. Fast folding and comparison of RNA secondary structures. Monatshefte für Chemie/Chemi. Mon. 1994;125:167–188. [Google Scholar]
- Li Y., et al. Performance comparison and evaluation of software tools for microRNA deep-sequencing data analysis. Nucleic Acids Res. 2012;40:4298–4305. doi: 10.1093/nar/gks043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moxon S., et al. A toolkit for analysing large-scale plant small RNA datasets. Bioinformatics. 2008;24:2252–2253. doi: 10.1093/bioinformatics/btn428. [DOI] [PubMed] [Google Scholar]
- Prüfer K., et al. PatMaN: rapid alignment of short sequences to large databases. Bioinformatics. 2008;24:1530–1531. doi: 10.1093/bioinformatics/btn223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan R., et al. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 2006;20:3407–3425. doi: 10.1101/gad.1476406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studholme D.J. Deep sequencing of small RNAs in plants: applied bioinformatics. Brief Funct. Genom. 2012;11:71–85. doi: 10.1093/bfgp/elr039. [DOI] [PubMed] [Google Scholar]