Abstract
Cervical cancer is malignant neoplasm of the cervix uteri or cervical area. Human Papillomaviruses (HPVs) which are heterogeneous groups of small double stranded DNA viruses are considered as the primary cause of cervical cancer, involved in 90% of all Cervical Cancers. Two early HPV genes, E6 and E7, are known to play crucial role in tumor formation. E6 binds with p53 and prevents its translocation and thereby inhibit the ability of p53 to activate or repress target genes. E7 binds to hypophosphorylated Rb and thereby induces cells to enter into premature S-phase by disrupting Rb-E2F complexes. The strategy of the research work was to target the site of interaction of Rb1 -E7 & p53-E6. A total of 88 compounds were selected for molecular screening, based on comprehensive literature survey for natural compounds with anti-cancer activity. Molecular docking analysis was carried out with Molegro Virtual Docker, to screen the 88 chosen compounds and rank them according to their binding affinity towards the site of interaction of the viral oncoproteins and human tumor suppressor proteins. The docking result revealed that Nicandrenone a member of Withanolides family of chemical compounds as the most likely molecule that can be used as a candidate drug against HPV induced cervical cancer.
Abbreviations
HPV - Human Papiloma Virus, HTSP - Human Tumor Suppressor Proteins, VOP - Viral oncoproteins.
Keywords: Bioinformatics, Cervical cancer, Human Papiloma Virus, Structure Based Drug Design, Protein-Protein interaction, Nicandrenone, Molecular Docking
Background
Cervical cancer is malignant neoplasm of the cervix uteri or cervical area. Cervical cancers start in the cells on the surface of the cervix. Most cervical cancers originate from squamous cells. Undetected precancerous changes can develop into cervical cancer and spread to the bladder, intestines, lungs, and liver [1]. Papilloma viruses are heterogeneous groups of viruses. They are small double stranded DNA viruses that infect mucosae and cutaneous surfaces, causing warts and epithelial tumours [2]. Human Papillomaviruses (HPVs) are the primary cause of cervical cancer, involved in 90% of all Cervical Cancers. The HPVs can be divided into two groups, the ‘low risk’ such as type 6 and 11) and the ‘high risk’ (such as type 16 and 18) [3]. Persistent HPV infection with a high risk virus is essentially observed in cervical intraepithelial neoplasia (CIN) II and III stages [4]. There is sufficient morphological and epidemiological consensus for the assumption that CIN III is a dynamic disease in which some cases regress spontaneously, whereas others progress to the invasive state, invasive cervical carcinoma (ICC) [5, 6]. Two early HPV genes, E6 and E7, are known to play crucial role in tumor formation. Both in vitro and in vivo studies show that the function of E6 and E7 proteins, particularly of the ‘high risk types’, are essential for cellular transformation [7, 8]. These oncogenes from the ‘high risk’ viruses have been shown to alter pathways involved in cell cycle control, interacting with and neutralizing the regulatory functions of two important tumor suppresser proteins, p53 and Rb [9], and also in conjunction with cellular genes, like deregulating key signal transduction pathways [10].
HPV 16 E6 is a 151 amino acid protein with two zinc finger domains. E6 is one of the primary oncogenes of the virus; causes immortalization of cells and along with E7 brings about transformation. Hence, it is obvious that E6 induces several important changes in the host cell by interacting with a plethora of cellular proteins. E6 can associate with the product of the tumor suppresser gene, p53 [11]. Analysis of human tumors reveals that p53 is one of the most commonly mutated genes. P53 is a transcription factor. It stimulates the expression of genes involved in cell cycle arrest and apoptosis, for example the cyclin-dependent kinase inhibitor (p21CIP). E6 binds to p53 and leads to ubiquitin mediated degradation of the latter. E6 binding to p53 can also lead to retention of p53 in the cytoplasm, blocking its translocation to the nucleus and hence abrogating its function independently of degradation [12]. This prevention of p53 translocation inhibits the ability of p53 to activate or repress target genes [13]. HPV 16 E7, a nuclear protein of 98 amino acids, has casein kinase II phosphorylation sites at serine residues 31 and 32. It is divided into three domains, CR1, CR2 and CR3. The ability of E7 to bind to members of the retinoblastoma (Rb) protein (p107, p130) is perhaps the most characterized property of this viral oncoprotein [14]. When cells progress in mitosis from G0 through G1 to S-phase, Rb gets hyperphosphorylated by G1 cyclin–cyclin dependent kinases, releasing the transcription factor E2F, which in turn activates genes involved in DNA synthesis and cell cycle progression [15]. E7 binds to hypophosphorylated Rb and thereby induces cells to enter into premature S-phase by disrupting Rb-E2F complexes [16]. Cervical cancer treatment has remained highly variable across the globe, largely owing to variances in disease burden in developed and developing nations. Hysterectomy (whole uterus removal including part of the vagina) has been employed in Microinvasive cancer (stage IA) [17]. The most commonly used drugs in chemotherapy against cervical cancer are combination of two drugs - hycamtin and cisplatin usually employed in women with late-stage (IVB) cervical cancer [18].
Inspite of the availability of chemotherapeutic compounds, there seems to be no specific drug tailor made for cervical cancer. Hycamtin and cisplatin are generally targeted towards the viral DNA and hycamtin is also known to suppress the bone marrow production. The combination treatment has significant risk of adverse effects like neutropenia, anemia, and thrombocytopenia. In this atmosphere, identifying a lead compound that would specifically target the HPV oncoproteins or the Human tumor suppressor genes which they interact or both would be highly critical in treatment of HPV induced cervical carcinoma with high precision and efficacy.
Methodology
Lead Identification & optimization:
A comprehensive search of all eligible studies on natural compounds which possess antineoplastic property was made by searching the electronic literature (PubMed database) for relevant published reports and by manual searching of reference lists of articles on this topic. A complete survey of Dr. Duke's Phytochemical and Ethnobotanical Databases [19] was also made to enlist natural compounds which have an inhibitory effect on the HPV onco proteins.
ADMET Prediction:
Elementary physical descriptors like molecular weight, logP, number of hydrogen bond acceptors and hydrogen bond donors were calculated for all the short listed compounds using molinspiration [20]. The compounds which abide by the “Lipinski's Rule of 5” filter was chosen for further analysis.
Binding Site Identification:
The site of interaction of the HPV oncoproteins and human tumor suppressor proteins was identified with SHARP2 [21]. SHARP2 was used for prediction of predicting potential protein–protein interaction sites on protein structures. A series of surface residue patches are defined and a maximum of 6 parameters are calculated for each. The parameters such as Solvation Potential, Hydrophobicity, Accessible Surface Area, Residue Interface Propensity, Protrusion, and Planarity are calculated. Based on these parameters, the site of interaction of the proteins is calculated.
Docking studies:
The selected compounds were docked against E6 (PDB ID: 1VZN), p53 (PDB ID: 1TUP), Rb1 (PDB ID: 1AD6), E7 (PDB ID: 2B9D), using Molegro Virtual Docker (MVD) [22]. MolDock scoring system is employed by MVD and it is based on a new hybrid search algorithm, called guided differential evolution. The guided differential evolution algorithm combines the differential evolution optimization technique with a cavity prediction algorithm. Differential evolution (DE) was introduced by Storn and Price in 1995 [22] and has previously been successfully applied to molecular docking. The use of predicted cavities during the search process, allows for a fast and accurate identification of potential binding modes (poses). Moldock score scoring function has been used. The intact protein structure was loaded on to MVD platform for docking process. Potential binding sites (also referred to as cavities or active sites) has been identified using the built-in cavity detection algorithm of MVD. The search algorithm is taken as Moldock SE and numbers of runs are taken 10 and max iterations were 2000 with population size 50 with an energy threshold of 100. At each step least ‘min’ torsions/translations/rotations were tested and the one giving lowest energy was chosen. After the docking simulation was over, the poses which were generated were sorted by rerank score. The Rerank Score uses a weighted combination of the terms used by the MolDock score mixed with a few addition terms (the Rerank Score includes the Steric (by LJ12-6) terms which are Lennard-Jones approximations to the steric energy – the MolDock score uses a piecewise linear potential to approximate the steric energy) [23]. The chosen ligands were manually prepared using ligand preparation module of Molegro virtual docker. Bond order and hybridization assign wherever missing and flexible torsion and the ligands were deducted. The target protein structures were prepared after careful removal of hetero atoms and water molecules and its electrostatic surface was generated. The docking was subjected towards the amino acid residues which were found to be part of interaction between HTSPs and VOPs. The grid resolution was set at 0.3 A0.The maximum interaction was set at 1500 and maximum population size 50.
Results
Interaction site Prediction:
Role of E6 and E7 oncoprotein from Human papiloma virus in tumor formation and it's interaction with human tumor suppressor proteins p53 and Rb1 respectively is well characterized by numerous studies. It becomes quite imperative that their site of interaction be targeted in order to court a cure against cervical carcinoma. The protein-protein interaction between Rb1 -E7 & p53-E6 was analyzed and the precise site of interaction was identified by employing SHARP2.The amino acid residues of each of the aforementioned human tumor suppressor proteins and viral oncoproteins taking part in the interaction, subsequently leading to pathogenesis of the cervical carcinoma were mapped out accurately. Table 1 (see supplementary material) reveals the entire list of amino acids involved in the interaction. Table 1 (see supplementary material) contains the complete list of amino acid residues involved in the interaction between E7-Rb1, E7-p53.
Docking results:
All the 88 natural compounds listed from the thorough literature survey were docked against E6 (PDB ID: 1VZN), p53 (PDB ID: 1TUP), Rb1 (PDB ID: 1AD6), E7 (PDB ID: 2B9D) at the specific sites of interaction predicted from SHARP2 results. The docking results were tabulated for all compounds against the four receptors. For each compound, out of the many docking poses, only those which posses the highest moldock score and relatively good Hydrogen bond interaction was chosen. The best few compounds which exhibited very good affinity towards the interaction site of all the four receptors were picked out and the best ligand binding pose was picked out on the basis of aforementioned criteria. Out of the 88 compounds, Nicandrenone was found to possess best binding affinity towards all the four receptors and was found to form Hydrogen bond interaction with amino acid residues predicted to be crucial in the protein-protein interaction between human tumor suppressor proteins and viral oncoproteins. Nicandrenone was found to be capable of binding with all the four receptor under consideration. It was found to be binding with Rb1 by interacting with surrounding amino acids like ser534, ser463, and val531 by forming hydrogen bonds. It formed five hydrogen bonds with high interaction energy of -95.0387. Similarly Nicandrenone was found to bind with E7 by forming six Hydrogen bonds by interacting with amino acids residues Glu74, Asn82, Leu77, Val49, and Glu56, exhibiting interaction energy of -84.8187. Nicandrenones binding affinity towards p53-E6 interaction site was found to be even better. Very high interaction energy of -121.578 was found for its interaction towards E6, forming five hydrogen bond with the residues Lys79, Tyr99, Glu120, and Glu97. With p53 Nicandrenone, formed five hydrogen bond with the residues Thr231, Thr230, Glu221 Glu224 and Arg202 showed interaction energy of - 106.56.The complete information of Nicandrenone interaction with the 4 proteins of interest are depicted in Table 2 (see supplementary material) and illustrations of the docking pose of Nicandrenone binding with HTSPs & VOPs is shown in (Figure 1).
Figure 1.
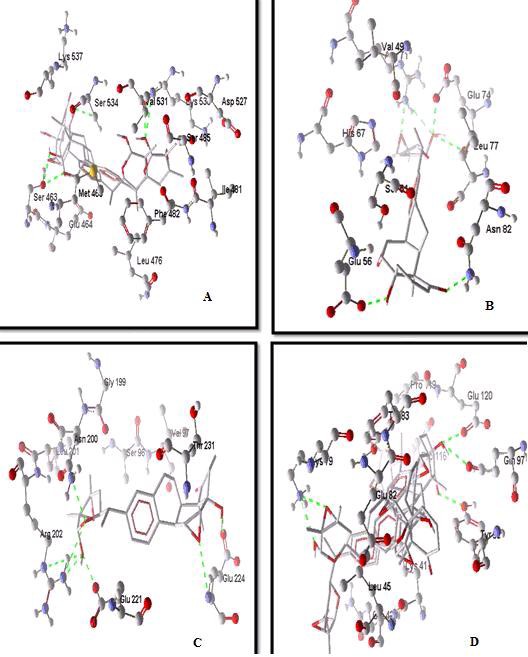
Illustrations of the docking pose of Nicandrenone binding with HTSPs & VOPs carried out with Molegro Virtual Docker 5.0. A. Nicandrenone interacting with Rb1 (1AD6) B. Nicandrenone interacting with E7oncoprotein (2B9D) C. Nicandrenone interacting with P53 (1TUP) D. Nicandrenone interacting with E6 oncoprotein (1VZN). Images depict ligand interaction with the site of interaction of the viral oncoproteins and human tumor suppressor proteins. Green dotted lines represent hydrogen bonds. The Receptor is depicted in ball and stick model and the ligand in Wireframe. CPK coloring convention has been employed.
Discussion
P53 and RB1 are highly vital tumor suppressor proteins and discrepancy in their normal function is implicated in pathogenesis of several cancers. The Inactivation of these two cellular tumor-suppressor proteins through their interaction with E6 and E7 oncoproteins of found in HPV has been widely hypothesized to play pivotal role in cervical carcinogenesis. This binding is postulated to be the functional equivalent of specific mutations in the p53 and RB genes [23]. The E7 oncoprotein from HPV is partly responsible in mediating cell transformation by binding to the human Rb1 tumor suppressor protein and E2F transcription factors, consequently dissociating Rb1 from E2F transcription factors and the premature cell progression into the S-phase of the cell cycle [24]. On the other hand, E6 oncoproteins from both high and low risk HPVs have been found to be capable of binding to p53 without inducing its degradation [25]. This interaction prevents the p53-mediated transcriptional repression of TATA-containing promoters [26], and also represses p53's transactivation of promoters containing p53- responsive elements [27]. Several studies over the past two decades have consistently suggested that strategies aimed at the therapeutic targeting of the E6-p53 interaction would be most likely to succeed if the actual physical association between E6 and p53 could be disrupted [28].
The principle goal of our study was to target the site of interaction of the E6-p53 and E7-Rb1 and thereby sabotages the physical interaction between these tumor suppressor protein and viral oncoproteins. The primary step was to enumerate the amino acid residues involved at the site of interaction. The result from SHARP2 server gave comprehensive list of all the residues which would be a part of this vital interaction. Our study results showed that amino acids residues 110 ARG - 231 THR of p53 were involved in its binding with E6 residues 22 LEU- 99 TYR. Our results were consistent with the finding of Crook et al (1991), who observed that HPV E6 can bind to the core region, (amino acids 66-326) which correlate with the induction of p53 degradation [25]. E7 amino acid residues 45PRO- 93 GLN were found to interact with 378VAL- 531VAL of Rb1. Liu, X et al (1990) established that the CR3 zinc binding domain of E7 (amino acids 44-93) is responsible in dissociation of pRb from E2F transcription factors and the premature cell progression into the S-phase of the cell cycle [29]. As the CR3 region of HPV E7 has no detectable homology to other human proteins, the structure-function studies presented here provide an avenue for developing small molecule compounds that inhibit HPV-E7-mediated cell transformation. The molecular docking studies carried out with the shortlisted compounds, targeting the site of interaction between E6-p53 & E7- Rb1.Nicandrenone was found to be most viable compound to bind to the site of interaction and disrupt the physical interaction between HTSPs & the VOPs. The nicandrenones are structurally complex steroids obtained from Nicandra physalodes and belong to group of naturally occurring chemical compounds called withanolides. Traditionally they have been known to posses with insecticidal effects and antibacterial properties. This compound as also been identified to exhibit cytotoxic effect against certain cancerous cells in Homo sapiens and Mus musculus [30]. The Docking results clearly indicate that Nicandrenone interacts with the list of amino acids as predicted by SHARP2. As the site of binding coincides with the site of interaction between viral oncoprotein and the human tumor suppressor proteins, there is a great chance that Nicandrenone would be highly capable of disrupting the physical interaction between HTPs and VOPs. The comparable affinity of nicanderone towards all the 4 receptors of interest makes it viable drug candidate for cervical carcinoma. Other compounds amongst the list which were chosen for screening- pregnane and steroidal glycoside also exhibited similar moldock score as that of nicanderanone indicating that they can also be subjected to further analysis and subsequently be treated as a candidate for drug discovery pipe line. Nicandrenone's hypothesized ability to act as a lead compound against HPV induced cerviacal carcinoma as revealed by our Insilico studies makes it an interesting case to investigate other Withanolides against the same protein target at the identical sites. Such a future studies might prove to be invaluable in finding other leads which might be structurally similar to Nicandrenone but with even better binding affinity or drug like properties.
Conclusion
The molecular interaction between the human tumor suppressor proteins p53 & Rb1 and the HPV oncoproteins E6 & E7 is crucial in the pathogenesis of cervical carcinoma. Molecular screening of natural compounds was carried out, targeting the site of interaction of E6-p53 and E7-Rb1. The docking results revealed that Nicandrenone exhibited the highest binding affinity towards the site of interactions and hence it could be an ideal molecule to disrupt the physical interaction between the human tumor suppressor proteins and HPV oncoproteins. Thus our study indicates that Nicandrenone could be a potential candidate against Human Papilloma Virus induced cervical cancer.
Supplementary material
Acknowledgments
We would like to extend our heartfelt thanks to Molegro ApS for giving us a fully functional trial version for a period of 30 days during which all the insilico docking work was carried out.
Footnotes
Citation:Shaikh et al, Bioinformation 8(13): 607-612 (2012)
References
- 1.S Khan, et al. Intl j Infect Dis. 2007;11:313. [Google Scholar]
- 2.H Zur Hausen. Prog Med Virol. 1985;32:15. [PubMed] [Google Scholar]
- 3.GY Ho, et al. J Natl Cancer Inst. 1995;87:1365. doi: 10.1093/jnci/87.18.1365. [DOI] [PubMed] [Google Scholar]
- 4.L Gustafsson, HO Adami. Br J Cancer. 1989;60:132. doi: 10.1038/bjc.1989.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.AG Ostor. Int J Gynecol Pathol. 1993;12:186. [PubMed] [Google Scholar]
- 6.MA Bedell, et al. J Virol. 1987;61:3635. doi: 10.1128/jvi.61.11.3635-3640.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.G Matlashewski, et al. EMBO J. 1987;6:1741. doi: 10.1002/j.1460-2075.1987.tb02426.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.K Vousden, et al. FASEB J. 1993;7:872. [Google Scholar]
- 9.A Rangarajan, et al. Virology. 2001;286:23. [Google Scholar]
- 10.T Kiyono, et al. Proc Natl Acad Sci U S A. 1997;94:11612. [Google Scholar]
- 11.F Mantovani, L Banks. Oncogene. 1999;18:3309. doi: 10.1038/sj.onc.1202688. [DOI] [PubMed] [Google Scholar]
- 12.JA Mietz, et al. EMBO J. 1992;11:5013. doi: 10.1002/j.1460-2075.1992.tb05608.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.N Dyson, et al. Science. 1989;243:934. [Google Scholar]
- 14.N Dyson. Genes Dev. 1998;12:2245. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- 15.PS Huang, et al. Mol Cell Biol. 1993;13:953. doi: 10.1128/mcb.13.2.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Medeiros, et al. Int Gynecol Cancer. 2009;19:1166. doi: 10.1111/IGC.0b013e3181a3d100. [DOI] [PubMed] [Google Scholar]
- 17. http://www.webmd.com/cancer/cervical-cancer/conebiopsy- conization-for-abnormal-cervical-cell-changes.
- 18. http://www.fda.gov/NewsEvents/Newsroom/PressAnn ouncements/default.htm.
- 19. http://www.ars-grin.gov/duke/
- 20. http://www.molinspiration.com/
- 21. http://www.bioinformatics.sussex.ac.uk/SHARP2.
- 22.R Thomsen, MH Christensen. J Med Chem. 2006;49:3315. doi: 10.1021/jm051197e. [DOI] [PubMed] [Google Scholar]
- 23.D Wolf, et al. Cell. 1984;38:119. [Google Scholar]
- 24.X Liu, et al. J Biol Chem. 2006;281:578. doi: 10.1074/jbc.M508455200. [DOI] [PubMed] [Google Scholar]
- 25.T Crook, et al. Oncogene. 1991;6:873. [Google Scholar]
- 26.M Lechner, et al. EMBO J. 1992;11:3045. [Google Scholar]
- 27.D Pim, et al. Oncogene. 1994;9:1869. [Google Scholar]
- 28.M Thomas, et al. Oncogene. 1999;18:7690. doi: 10.1038/sj.onc.1202953. [DOI] [PubMed] [Google Scholar]
- 29.X Liu, et al. J Biol Chem. 2006;281:578. doi: 10.1074/jbc.M508455200. [DOI] [PubMed] [Google Scholar]
- 30.G Adam, et al. Pharmazie. 1976;31:647. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


