Abstract
Therapeutic agents with a goal to eradicate cancer needs to capable of inhibiting the growth and kill, any preformed tumor and should also inhibit oncogenic transformation of normal cells to cancer cells. Bacteriocins are bacterial proteins produced to prevent the growth of competing microorganisms in a particular biological niche and have been proved to possess antineoplastic activity. The entire genome of Lactobacillus salavarius was scanned for putative bacteriocins and subsequently these bacteriocins were characterized by subjecting them as functional annotation algorithms. Azurin is a well characterized bacteriocins with proven cytostatic and apoptotic effect against human cancer cell and was taken as control. Functional characterization revealed that the three bacteriocins Lsl_003, Lsl_0510, Lsl_0554 possessed functional properties very similar to that of Azurin. Molecular screening of these bacteriocins against the common cancer targets p53, Rb1 and AR revealed that Lsl_0510 possessed highest binding affinity towards the all the three receptors making it to ideal candidate for future cancer therapeutics.
Abbreviations
P53 - Protein 53, Rb1 - Retinoblastoma 1, AR - Androgen Receptor, Lsl - Lactobacillus salavarius.
Keywords: Cancer, Bioinformatics, Computer Aided Drug Design (CADD), Insilico Protein characterization, Bacteriocins, Docking
Background
Recent research has revealed that cancer is a disease involving complex changes in the human genome. As a result of the innate complexities of the disease, no single or even combination of drugs, works for all cancers and drugs are normally targeted to specific cancers based on certain characteristics called validated targets that are often upregulated in cancer cells to allow their unchecked growth [1]. Majority of research in the afore mentioned areas is focused on cancer-specific mechanisms and the corresponding molecular targets, but the search for improved cytotoxic agents which act on ubiquitous targets such as DNA still represent an important part of new anticancer drug innovation [2]. Innovation of anticancer drug is always an attractive challenge and is classified in different ways according to their nature, sources, mechanisms of action etc, which may be of microbial products, synthetic chemicals, and plant or animal extracts [3]. Rapid advances in peptide therapeutics, has enabled the scientific community to explore wider horizons to discover novel therapeutic agents like Bacteriocins. These are small peptides (30 –60) amino acid with antimicrobial properties against bacteria usually of the same or closely related species (narrow spectrum), and sometimes against a wide spectrum of species [4, 5]. Bacteriocins promises to be effective therapeutic agent and its biochemical properties have been studied; their antineoplastic capability has also identified after its discovery in the late 1970s by using crude bacteriocin preparation toxic to mammalian cells [6]. Common bacteriocins like Pyocin, colicin, ped-iocin, and microcin have been shown to possess inhibitory properties against different neoplastic line cells [7], Azurin, is an important bacteriocins, a member of the cupredoxin family of redox proteins, secreted by the bacterium Pseudomonas aeruginosa, which can preferentially penetrate human cancer cells and exerts cytostatic and apoptotic effects with no apparent activity on normal cells. A 28 amino acid long peptide derived from Azurin, termed as p28 (consisting of amino acid residues 50 to 77 of the 128 amino acids long Azurin) possessed entry specificity in cancer cells but not the healthy ones [8]. The discovery of the unique capabilities of azurin and p28 in interfering in cancer cell growth and promote their cell death, but also its ability to prevent tumor emergence, brings about a wonderful prospect of looking out other bacteriocins, which might have similar properties or even better antineoplastic effect.
A comprehensive expedition to identify and subsequently characterize such potentially beneficial bacteriocins from common bacterial species which are usually an integral part of a human microflora can lead us to identify the novel anticancer molecules. The focus of the study was to enumerate, identify bacteriocins from commensal bacteria - Lactobacillus salivarius which is a gram-positive bacillus, inhabiting human oral cavities, intestines, and vagina. An attempt has been made to comprehensively characterize these identified bacteriocins and subsequently check its antineoplastic capacity by carrying out molecular docking studies.
Methodology
Literature Mining:
A comprehensive search of all eligible studies on bacteriocins , Lactobacillus salavarius & their antineoplastic properties (as on April 2012) was made by searching the electronic literature (PubMed database) for relevant published reports and by manual searching of reference lists of articles on this topic. Only studies in the English language were included in the analysis.
Identification of bacteriocins from Lactobacillus salavarius genome:
The entire genome of Lactobacillus salavarius was scanned in order to recognize putative bacteriocin ORFs. BAGEL (http://bioinformatics.biol.rug.nl/websoftware/bagel.) a web server that identifies putative bacteriocin ORFs in a DNA sequence using novel, knowledge-based bacteriocin databases and motif databases was employed. Bagel identified 28 putative bacteriocins, out of these 22 bacteriocins which were hypothetical were taken up for further analysis and characterization. The protein sequences of these 22 hypothetical bacteriocins were retrieved from NCBI Reference Sequences database (http://www.ncbi.nlm.nih.gov/RefSeq/).
Functional Annotations of Hypothetical bacteriocin:
All the chosen bacteriocin protein sequences were subjected to the protein function prediction server tools - Prot fun2.2 (http://www.cbs.dtu.dk/services/ProtFun/) and Kihara Bioinformatics (http://kiharalab.org/pfp/). These servers perform automated function prediction that provides the most probable annotations for a query sequence in each of the three branches of the Gene Ontology: biological process, molecular function, and cellular component. Rather than utilizing precise pattern matching to identify functional motifs in the sequences and structures of these proteins, these servers are we designed to increase the coverage of function annotation by lowering resolution of predictions when a detailed function is not predictable [9].
Sub- Cellular Localization Prediction:
Sub-cellular localization tools such as Balanced Sub-cellular Localization Predictor. BaCelLo (http://gpcr.biocomp.unibo.it/bacello/pred.htm)was utilized. BaCelLo is useful in identifying the putative location (secretory way, cytoplasm, nucleus, mitochondrion and chloroplast) of the bacteriocins under investigation; it is based on different SVMs organized in a decision tree. After thorough functional characterization, the properties of the analyzed bacteriocins were compared to that of Azurin an only those hypothetical bacteriocins which seems to posses properties very similar to azurin were chosen and subjected to further studies.
Protein Structure Prediction and Structure Assessment:
The three hypothetical bacteriocins, namely Lsl_003, Lsl_0554, and Lsl_0510 were chosen after functional annotation and sub – cellular localization analysis and after comparison against Azurin were modeled using ab-initio method employing Quark algorithm (http://zhanglab.ccmb.med.umich.edu/QUARK/). QUARK models are built from small fragments (1-20 residues long) by replica-exchange Monte Carlo simulation under the guide of an atomic-level knowledge-based force field. Protein models obtained after ab intio modeling were verified for their accuracy by using Ramachandran plot. The molprobility server (http://molprobity.biochem.duke.edu/) was employed for this purpose.
Docking Studies of Bacteriocin against the Cancer Target:
Molecular docking analysis were carried out to study the binding affinity of our characterized bacteriocins towards common cancer targets such as p53 (with mutant core domain), Rb1 and AR in order to understand the probable efficiency of these bacteriocins to act as antineoplastic agents. The protein structures 2J1X - p53 core domain mutant M133L-V203AY220C- N239Y-N268D, 1AD6 – RB1 and 1Z95 – AR were downloaded from the Protein Database (http://www.rcsb.org). Hex 4.5 was employed for performing rigid docking between the chosen bacteriocins and selected cancer targets by SP Fourier Transform, FFT steric scan, FFT final search and MM refinement [10]. The clustering histogram with the scoring function was generated to analyze the binding energy of each selected conformations. Based on the docking results, preferably in terms of ETotal, minimum energy value, the effectiveness of all cancer receptor and bacteriocins (drug molecules) to interact with each other were observed and the relative affinity of binding was studied. The detail workflow of the study has been illustrated in (Figure 1).
Figure 1.
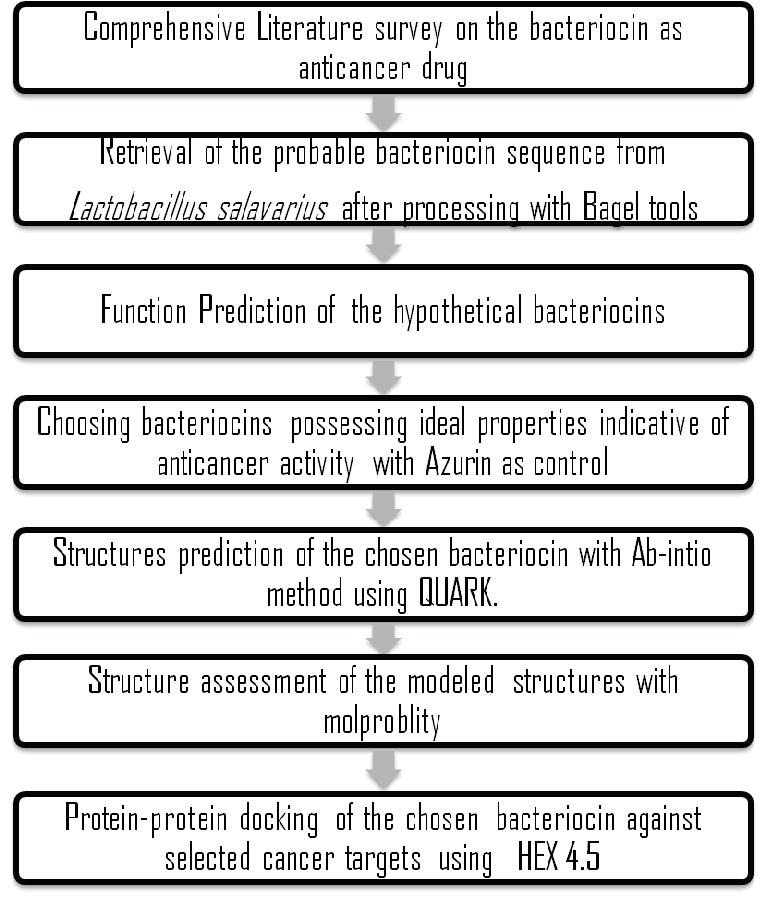
Flowchart illustrating the process of identification and subsequent characterization of hypothetical bacteriocins from Lactobacillus salavarius.
Discussion
Any therapeutic agents with a goal to eradicate cancer needs to capable of inhibiting the growth and kill, any preformed tumor and should also inhibit oncogenic transformation of normal cells to cancer cells. The biologists have a recognized a new class of anticancer agents possessing both these properties- Bacteriocins. These are bacterial proteins, which have evolved and designed efficiently by force of natural selection over eons. Goal of our study was to identify bacteriocins from Lactobacillus salavarius and characterize them as potential anticancer drug, by employing suitable Bioinformatics techniques.
Functional Annotation:
A Total of 28 bacteriocins were retrieved after comprehensive scanning of Lactobacillus salavarius genome using the Bagel tool [Anne de jong, Nucleac acid research, 2006]. Out of these 22 bacteriocins are hypothetical. To characterize them, they were subjected to function prediction servers and out of these, 3 bacteriocins were chosen which possessed the ideal properties indicative of anticancer activity comparable to that of Azurin- a member of the cupredoxin family of copper containing redox proteins capable of preferentially penetrating human cancer cells and exert cytostatic and cytotoxic (apoptotic) effects with no apparent activity on normal cells. Azurin's mode of action involves inhibition of cell signaling, inhibition of angiogenesis and stabilization of p53. The ability of Azurin to preferentially & rapidly penetrate cancer cells and promote significant anticancer activity without membrane disruption suggests this unique molecule may be effective against different types of solid tumours with fewer side effects than current anticancer therapeutics [8]. Owing to these characteristics, Azurin was chosen as the control for the study. The properties of the bacteriocins from Lactobacillus salavarius under study were compared against Azurin and inferences were accordingly drawn. Functional characterization can reveal unknown mechanisms by which the bacteriocins under study might exercise anticancer effect. The results from function prediction servers Protfun2.2, Khara bioinformatics, Interproscan, The no: of Protein phosphorylation, sites were initially predicted. Predicting the number of Protein phosphorylation, sites is crucial in any research pursuit attempting to identifying proteins which can be potential anti neoplastic agents. This is partly because of its early links to metabolic regulation and cancer [11] has been the most widely studied PTM. There is also a requirement of not only the modified residue and surrounding sequence, but also its regulation by treatments and ligands, associated biological processes, upstream and downstream interactions and location relative to protein domains and other functional regions of the protein.
Protfun results predicted that the odds of the three bacteriocins being a part of the cellular were very similar to that of azurin. The other selected functional characterization was to predict the propensity of the bacteriocins to have enzymatic activity. Our study revealed that our bacteriocins and Azurin, both possess very low proclivity to exhibit catalytic activity. This is indicative that the bacteriocins of our interest have little chance of interacting with any other substrates or alter the normal cellular kinetics. The probability of eliciting an immune response was much lower in bacteriocins of our choice compared to that of azurin. While azurin had a ~50.1% chance of eliciting immune response, Lsl_003, Lsl_0554 and Lsl_0510 showed ~15%, ~21% and ~13% respectively. This indicates that the bacteriocins that we had chosen for study have very low chances of Druginduced adverse reactions of type B which comprise idiosyncratic and immune-mediated side effects [12]. For characterizing the localization within the cell, all bacteriocins it subjected to the sub cellular localization tool – BaCelLo. The detail information on the exact subcellular location of each bacteriocin is elucidated in Table 1 (see supplementary material). These results signify that the three chosen bacteriocins Lsl_003, Lsl_0554 and Lsl_0510 have great chance of possessing antineoplastic activity similar to that of azurin. The essential information on function annotation of the bacteriocins is depicted in (Figure 2).
Figure 2.
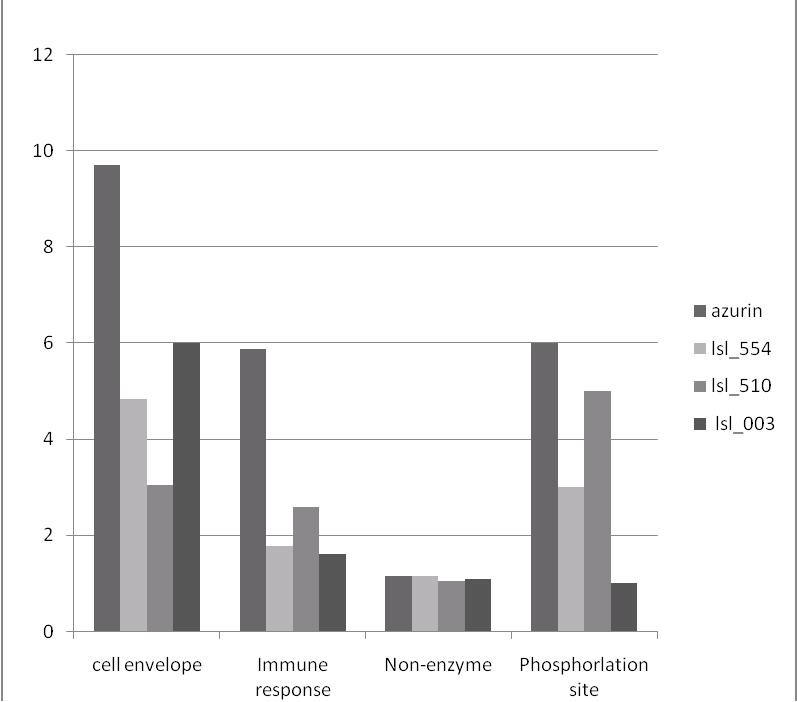
Graph depicting key functional characteristics of the three chosen hypothetical bacteriocins from Lactobacillus salavarius in comparison with Azurin. The propensity to be part of cellular envelope, eliciting immune response and being nonenzymatic in nature are represented in odds and the numbers of phosphorylation sites are denoted.
Ab-Initio Modeling Of the Bacteriocins & Structure Assessment:
Prediction of the three-dimensional structure of a protein from its amino acid sequence is imperative, as knowledge of the 3 d structure of the bacteriocins is essential to study its binding affinity towards the protein receptors, which are to be targeted to yield an anti cancer effect. Owing to the short sequence length of the bacteriocins, homology modeling was not possible, as any sequence similarity might arise due to near coincidence and might not be indicative of any evolutionary relationship. The structure of the 3 bacteriocins was predicted by ab-initio method using the quark server. De novo protein structure prediction methods attempt to predict tertiary structures from sequences based on general principles that govern protein folding energetics and statistical tendencies of conformational features that native structures acquire, without the employing explicit templates [13]. The assessment of the modeled structures was done by observing the Ramachandran plot using molprobility. Over 90% of the amino acid residues in the modeled bacteriocin structures reside in the most favorable regions, clearly indicating that the modeled structures are good. In Lsl_003 90.1% (64/71) of all residues were in favored regions, in case of Lsl_0554 -96.4% (54/56) of all residues were found in favored regions and for Lsl_0510, about 89.2% (83/93) of all residues were in favored regions.
Docking Of the Bacteriocins against Selected Cancer Targets:
The protein-protein docking analysis is highly useful in judging the strength of binding and also to analyze a spatial configuration adopted by the molecules in their bounds state. The modeled bacteriocin structures were subjected to docking procedure using HEX4.5 against three cancer targets namely p53 core domain mutant M133L-V203A-Y220C-N239Y-N268D and Rb1 protein the androgen receptor (AR). P53 is inactivated directly by mutation in _50% of human cancers. Nearly all of the oncogenic mutations occur in its core or DNA-binding domain, contained within the sequence of residues 94-292 [14, 15]. Destabilized mutants of p53 can be stabilized by the binding of other molecules, as shown by the binding of a specific double-stranded DNA [16]. Theoretically destabilized mutants of p53 can be stabilized by the binding of other molecules. We made an attempt to check for the binding affinity of the listed bacteriocins towards the destabilized core region of p53 by carrying out molecular docking studies. Rb1 is responsible in preventing excessive cell growth by inhibiting cell cycle progression until a cell is ready to divide. pRb belongs to the pocket protein family, whose members have a pocket for the functional binding of other proteins. Binding of oncogenic protein, such as those produced by cells infected by high-risk types of human papillomaviruses, can inactivate Rb1, and consequently lead to cancer [17]. Our focus was to check whether the Lactobacilus bacteriocins are capable of binding to Rb1 binding sites and thereby obstruct the physical interaction of Rb1 with viral ooncoproteins. Androgen receptor is the most common receptor targeted by most drugs which are administered against prostate cancer. Drugs like Flutamide are a nonsteroidal antiandrogen that blocks the action of both endogenous and exogenous testosterone by binding to the androgen receptor [18]. Docking analysis carried against Androgen Receptor yielded very promising results, with all three bacteriocins show very high binding affinity. Against mutant p53 and Rb1 - Lsl_0510 shows a good binding strength with an E-Total value of -46.93 and -57.85 respectively. This indicates that Lsl_0510 is a promising candidate to stabilize the destabilized core region of mutated p53. The E-total values obtained after docking against the cancer target is illustrated in Table 1 (see supplementary material). The docking poses of the three bacteriocins against the chosen cancer targets - p53, Rb1, AR is shown in (Figure 3).
Figure 3.

The docking results of the three bacteriocins Lsl_003, Lsl_0554, and Lsl_0510 against cancer targets - p53, Rb1 and AR. The cancer targets are depicted in the cartoon form and the bacteriocins depicted in surface model.
Conclusion
Our study identified 3 bacteriocins from Lactobacillus salavarius namely Lsl_003, Lsl_0554, and Lsl_0510 which were found to possess characteristics which highly similar to that of Azurin and can serve as potential leads in the quest for new cancer therapeutic drugs. These bacteriocins may be further tested by in vitro experimentation for their antioncogenic activity.
Supplementary material
Footnotes
Citation:Shaikh et al, Bioinformation 8(13): 589-594 (2012)
References
- 1. http://www.commercialbiotechnology.com.
- 2. http://www.insightsociety.org/ojaseit/index.php/ijaseit/ article/view/24/2.
- 3.PS Foster, et al. Pharmacol Ther. 2002;94:253. doi: 10.1016/s0163-7258(02)00220-6. [DOI] [PubMed] [Google Scholar]
- 4.Z Weizman, A Alsheikh. J Am Coll Nutr. 2006;25:415. doi: 10.1080/07315724.2006.10719554. [DOI] [PubMed] [Google Scholar]
- 5.TR Klaenhammer, et al. Biochimie. 1988;70:337. doi: 10.1016/0300-9084(88)90206-4. [DOI] [PubMed] [Google Scholar]
- 6.G Cornut, et al. Am J Clin Oncol. 2008;31:399. doi: 10.1097/COC.0b013e31815e456d. [DOI] [PubMed] [Google Scholar]
- 7. ttps://tuhat.halvi.helsinki.fi/portal/en/publications/nisin (7927a82d-e8b4-4ee0-8536-40837fd012bf).html.
- 8. http://www.chemistry-today.com/
- 9.T Hawkins, et al. Protein Sci. 2006;5:1550. [Google Scholar]
- 10.DW Ritchie, et al. Proteins. 2003;52:98. [Google Scholar]
- 11.H Varmus, et al. Princess Takamatsu Symp. 1989;20:63. [PubMed] [Google Scholar]
- 12.DJ Naisbitt, et al. Drug Saf. 2000;23:483. doi: 10.2165/00002018-200023060-00002. [DOI] [PubMed] [Google Scholar]
- 13. http://zhanglab.ccmb.med.umich.edu/QUARK/
- 14.M Olivier, et al. Hum Mutat. 2002;19:607. doi: 10.1002/humu.10081. [DOI] [PubMed] [Google Scholar]
- 15.AC Joerger, et al. Oncogene. 2007;26:2226. doi: 10.1038/sj.onc.1210291. [DOI] [PubMed] [Google Scholar]
- 16.AN Bullock, et al. Proc Natl Acad Sci U S A. 1997;94:14338. [Google Scholar]
- 17.AL Murphree, WF Benedict. Science. 1984;223:1028. doi: 10.1126/science.6320372. [DOI] [PubMed] [Google Scholar]
- 18.AK Roy, et al. Vitam Horm. 1999;55:309. doi: 10.1016/s0083-6729(08)60938-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


